Abstract
We have developed a rapid molecular mapping technology—Direct Linear Analysis (DLA)—on the basis of the analysis of individual DNA molecules bound with sequence-specific fluorescent tags. The apparatus includes a microfluidic device for stretching DNA molecules in elongational flow that is coupled to a multicolor detection system capable of single-fluorophore sensitivity. Double-stranded DNA molecules were tagged at sequence-specific motif sites with fluorescent bisPNA (Peptide Nucleic Acid) tags. The DNA molecules were then stretched in the microfluidic device and driven in a flow stream past confocal fluorescence detectors. DLA provided the spatial locations of multiple specific sequence motifs along individual DNA molecules, and thousands of individual molecules could be analyzed per minute. We validated this technology using the 48.5 kb λ phage genome with different 8-base and 7-base sequence motif tags. The distance between the sequence motifs was determined with an accuracy of ±0.8 kb, and these tags could be localized on the DNA with an accuracy of ±2 kb. Thus, DLA is a rapid mapping technology, suitable for analysis of long DNA molecules.
Traditionally, DNA mapping has been an important strategy to study structures and organizations of genomes. Recent advances in DNA sequencing technologies, however, have served to diminish the relative importance of traditional mapping. Nonetheless, growing interest in comparative genomics has created a need for technologies that can rapidly and efficiently characterize a genome, particularly larger genomes. Furthermore, in many cases, single-base resolution is unnecessary, as genomic differences among species (e.g., microorganisms) or among individuals within a given species (e.g., humans) can be discerned using lower-resolution mapping approaches (Olive and Bean 1999). Thus, there is a need for a practical, rapid, and highly efficient DNA mapping technology.
Currently, restriction mapping is the most practicable mapping approach that combines high resolution with high density (Brown 1999). Gel electrophoresis-based restriction enzyme mapping using just a single enzyme has been a workhorse for the human genome project and other large-scale efforts to provide a fingerprint identification of BAC clones (Soderlund et al. 2000). Traditional restriction mapping with multiple enzymes has allowed characterization and manipulation of genomic regions of interest (Brown 1999). To study human variation, restriction fragment length polymorphism (RFLP) analysis has allowed investigators to identify SNPs that correlate with disease loci (Shi 2002). Nonetheless, restriction mapping has fundamental drawbacks that limit its utility for comparative genomics. Digestion of the DNA removes information regarding the ordering of the fragments, requiring the use of multiple enzymes to construct the correct map. Furthermore, as RFLP analysis involves a mixture of molecules, haplotype information is inaccessible. For large DNA, pulsed-field gel electrophoresis is slow, requiring 10–50 h per run, depending on the size of analyzed DNA (Birren and Lai 1993). Further, traditional restriction mapping is cumbersome, difficult to automate, and requires significant amounts of DNA for analysis.
A promising approach for enhanced genomic mapping involves optical interrogation of individual DNA molecules demarcated at sequence-specific sites. Single-molecule analysis serves to reduce reagent costs and preserve physical haplotype information. Elegant studies by Schwartz and colleagues (Aston et al. 1999a; Lim et al. 2001) showed that very long strands of DNA can be physically mapped by imaging individual stretched DNA molecules attached to a surface and digested with a restriction enzyme. Thus, without DNA amplification, this group was able to reconstruct restriction maps of a 5.5-Mb microbial genome (Lim et al. 2001). Preservation of physical ordering of the fragments obviated the need for complex reassembly of map information, and the use of long DNA molecules greatly reduced the complexity of the sample. Thus, this work has revealed the power of single-molecule mapping technology for rapid genome characterization.
We have developed a new technology, termed Direct Linear Analysis (DLA), to map large DNA molecules. Herein, we report a proof of principle application of DLA for mapping the 48.5-kb λ phage genome using several sequence-motif tags. DLA is based on microfluidic unwinding and stretching of individual double-stranded DNA molecules that flow in a linear fashion through a laser-illuminated detection zone. To reveal map information, site-specific fluorescent tags are bound to the DNA. In the present study, we used fluorescent peptide nucleic acids (PNAs) targeting 7–8 bp sites. The detection sensitivity of the system allowed us to analyze single fluorophore tags on individual molecules at throughputs of thousands of molecules per minute. Several λ phage DNA maps were successfully measured with different tag designs.
RESULTS
Direct Linear Analysis Technology
Once isolated and purified, double-stranded DNA in aqueous solution assumes a random-coil conformation. Prior to linear measurement, it must be unwound and stretched. This is performed in our system by hydrodynamic forces generated in laminar flow by a tapered microfluidic channel (Fig. 1A). After injection into the loading port, the sample solution is pressure driven into the chip, and tagged DNA molecules travel with the flow in their random-coil forms. Downstream, DNA uncoils as it interacts with the post fields. These prestretched molecules stretch to completion under hydrodynamic influences in the tapered region. The short taper length produces flow acceleration over a distance comparable to the size of the DNA. The resulting differences in flow velocities around different parts of the molecule produce the force that stretches it (Perkins et al. 1995). The flow acceleration must be sufficient to provide forces on the order of 10 pN to ensure stretching DNA to its contour length (Bustamante et al. 2000). The degree of DNA stretching depends upon the flow strain rate and relaxation time of the molecule conformation (Marko and Siggia 1995; Perkins et al. 1997). These parameters are mostly set by taper design, flow velocity, and polymer contour length. A systematic study of the performance of different microfluidic geometries with a range of DNA size standards will be published elsewhere (J.W. Larson, G.R. Yantz, R. Charnas, C.M. D'Antoni, M.V. Gallo, K.A. Gillis, L.A. Neely, K.M. Phillips, G.G. Wong, S.R. Gullans, et al., in prep.). In the present study, we used a published analysis (Perkins et al. 1997) to design a microchip configuration and operating conditions that are adequate for stretching 50-kb long DNA. The chip included a post field, a funnel with a 10:1 taper reduction ratio, a taper shape providing W(x)∼1/x2 profile (W is the channel width, and x is the coordinate along the flow direction), and a 5 μm-wide interrogation channel. The interrogation channel had uniform cross-section to ensure constant solution velocity, which was 10–15 μm/msec.
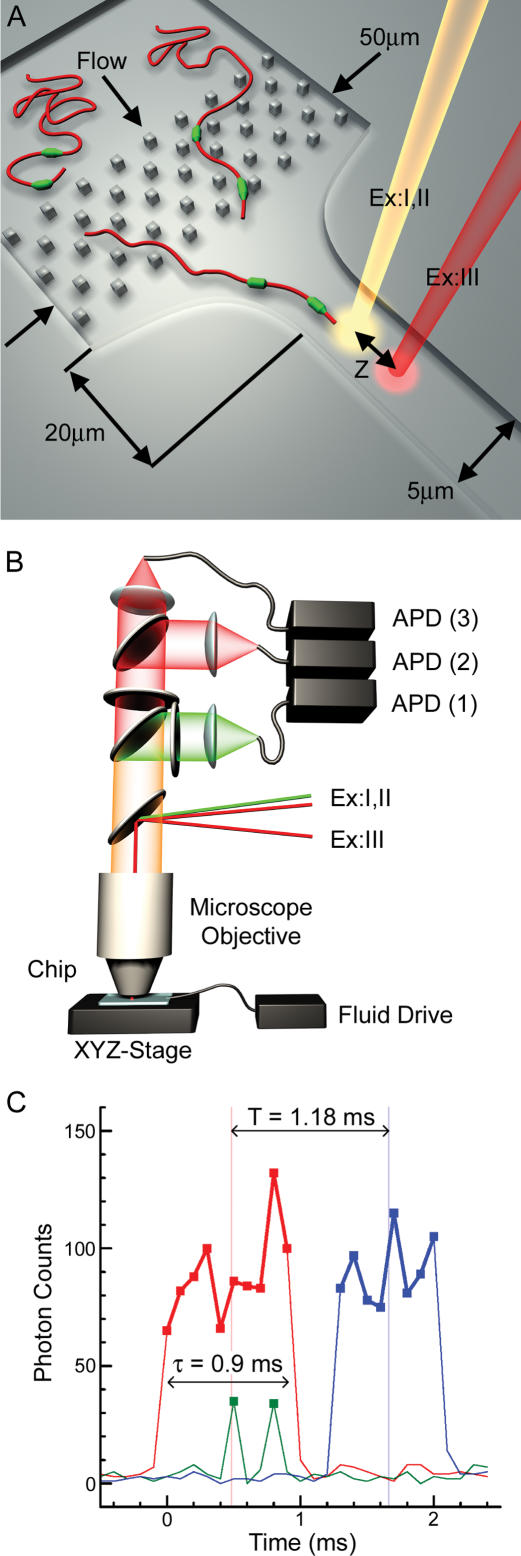
Figure 1.
Schematics of the Direct Linear Analysis (DLA) technology. (A) A cross-section of the microfluidic DNA stretching microchip (top). A typical design, one of a few, is presented. See text for the exact parameters of the chip used in this study. (B) Optical scheme (side view). The excitation and detection are arranged within a confocal fluorescence microscope. The excitation laser beams are directed into the microscope objective with a dichroic mirror that reflects the light with 532 nm (beam ExI) and 633 nm (beams ExII and ExIII) wavelengths, but is transparent to the fluorescence emission excited by these beams. The emission is further split by another dichroic mirror and bandpass filters. Fluorescence excited by the green laser is delivered by optical fiber to the photon-counting avalanche photodiode (APD) for signal detection in data channel 1. Fluorescence excited by red beams ExII and ExIII is directed to the APDs of data channels 2 and 3, respectively. (C) Typical raw data traces from data channels 1–3 for a single tagged DNA molecule. The red and blue traces arise from fluorescence of the intercalating dye when the DNA backbone travels through the excitation spots ExII and ExIII, respectively. The green spikes are detected when the DNA-bound PNA tags pass through the excitation spot ExI and emit bursts of photons.
Once inside the channel, stretched and tagged DNA molecules traveled through spots of focused laser light that excites fluorescence. Epi-illumination of the sample and confocal detection were arranged within a fluorescence microscope (Fig. 1B). This optical scheme is typical for single molecule detection (for example, see Rigler et al. 1993). The excitation spots of beams ExI and ExII were overlapped, whereas spot ExIII was displaced by a distance Z down the channel (Fig. 1A). Typical emission signals detected in data channels 1–3 are presented in Figure 1C. The DNA backbone was stained with the intercalating dye, TOTO-3, which was excited by red light (633 nm) and emitted fluorescence when DNA traveled through spots ExII and ExIII. As a result, the DNA backbone profile was detected initially in data channel 2 (red profile in Fig. 1C) and, after some delay (T), in data channel 3 (blue profile). We define a DNA event as a contiguous set of photon counts (red and blue squares connected by thick lines in Fig. 1C) above a threshold value that is determined from the average and the standard deviation of the background noise. The excitation spots were positioned sufficiently far downstream in the interrogation channel so that flow velocities were uniform around the molecules as they passed between spots to ensure unaccelerated movement of the molecules. The average velocity, V, of each molecule was calculated independently using the known distance, Z = 20 microns, between spots ExII and ExIII, and the measured transit time, T. For example, the velocity of the molecule in Figure 1C is V = Z/T = (20 microns)/(1.18 msec) = 16.9 mm/sec. Using the calculated velocity and the DNA residence time in spot ExII, τ = 0.9 msec, the DNA length, X = V*τ = 15.3 μm, was calculated for this individual event. For simplicity, we call this measurement a length, although technically, it is the DNA projection in the direction of flow, and it only relates to the DNA contour length—the length per nucleotide times the number of nucleotides—in that the two values are equal for 100% stretching.
The apparent center of the molecule (CM) was used as the main geometrical reference point along detected DNAs. For example, the time delay, T, was determined as the transit time of the CM between ExII and ExIII in Figure 1C. The CM position was determined using the backbone emission profile, under the assumption that the intensity was proportional to the amount of DNA within the excitation volume. This assumption is valid when DNA is stained close to saturation (Larsson et al. 1994; Carlsson et al. 1995). In the stretched state, DNA molecules with free termini are not stretched homogeneously, but rather adopt a stem-and-flowers conformation (Perkins et al. 1995; Manneville et al. 1996). As a result, when incomplete stretching occurs, it is concentrated in the terminal regions (flowers), whereas the middle of the DNA molecule remains highly stretched (stem). Therefore, the central portion is generally usable for analysis even for incompletely stretched molecules, and is the best position for a reference point such as the CM. The fully stretched stem portion relaxes slowly (∼60 msec for λ DNA in water; Perkins et al. 1997) in comparison with the measurement time (the time the molecule travels through spot ExII, which is <2 msec), so it stays stretched after the taper for a time sufficiently long for interrogation.
Although tension varies along the DNA backbone, the goal of uniformly linearized molecules can still be accomplished, because once almost fully stretched, DNA resists further elongation quite strongly; changing force from 5 to 70 pN only increases DNA extension from 95% to 100% (Smith et al. 1996; Bustamante et al. 2000). Thus, the inextensibility of the DNA double helix is a cornerstone of our approach. However, forces exceeding 70 pN may result in DNA overstretching, which we have observed, but which can be avoided by the proper choice of channel geometry and flow velocity.
Microfluidic Stretching and Selection of Molecules for Analysis
Figure 2A presents the distribution of calculated lengths for all detected DNA molecules. The histogram has an asymmetric peak with a maximum at 16–17 μm (Fig. 2A), which corresponds to considerable extension of λ DNA molecules from their relaxed random-coil conformations (0.9 μm average end-to-end distance; Strobl 1997). This peak asymmetry, with an abrupt border at larger sizes, is expected for a population of stretched molecules, in which a large portion is completely stretched. In this case, the position of the abrupt border corresponds to the DNA contour length. As further evidence that this population includes completely stretched molecules, the peak position was independent of flow velocity above a minimum threshold value (data not shown). This observation is consistent with complete DNA stretching, because insensitivity to increased force is a hallmark of completely stretched DNA, as discussed above. Note that the position of the maximum corresponds to the 16.5 μm contour length of unstained λ DNA molecules (Bustamante et al. 1994); our DNA did not exhibit an increase in contour length when intercalated, probably due to a low proportion of dye (see Methods for details).
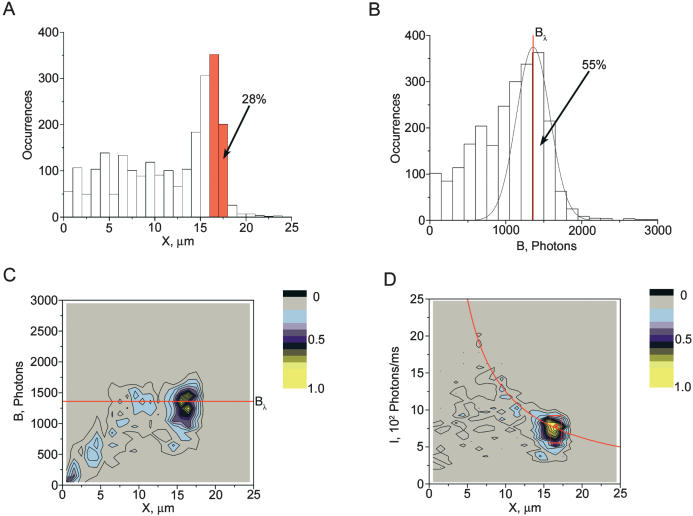
Figure 2.
Properties of detected molecules of λ phage DNA (48.5 kb) in a representative experiment. (A) The distribution of DNA lengths (sizes of DNA projections X onto the movement direction) for detected DNA molecules. (B) The distribution of burst sizes B (total number of photons per detected molecule) of the stained DNA backbone (data channel 2). The peak corresponding to full-size λ DNA is interpolated with a best fit Gaussian curve. Scatter plots of burst size B vs. DNA length X (C) and of average intensity of DNA backbone I vs. X (D). C and D include points representing single observed DNA molecules stained with the TOTO-3 intercalating dye. Because the points are concentrated in small regions, we have chosen to present their color-coded density rather than the points themselves. The solid red lines in C and D correspond to the center of the Gaussian distribution Bλ interpolating the full-size DNA molecules shown in B. A total of 2373 individual molecular traces are represented in this data file. The data points included in the red box in D were used to calculate one of the DNA maps presented in Figure 3D. This selection contains ∼400 molecules. The corresponding interval, including molecular traces with projections between 16.25 and 17.25 μm, is outlined in red in A.
Molecules that appeared shorter than 15 μm were either understretched or fragmented, and we took advantage of uniform DNA staining with intercalator to distinguish between the two possibilities. The total photon count collected per DNA event, or burst size, B (Fig. 2B), indicates molecule size (Chou et al. 1999; Foquet et al. 2002). When properly stained, DNA molecules of the same size contain comparable amounts of intercalating dye, and hence, emit similar numbers of photons, B, per DNA molecule. We corrected for molecule velocity because faster moving molecules emit fewer photons, Bi = bi*Vi/V0, where bi is the measured burst size for the i-th molecule, Vi is its velocity, and V0 = 10 μm/msec is our standard velocity. As expected, full-length λ DNA molecules, being the same size, emit a similar number of photons, observed as a peak in burst-size distribution with a maximum at 1400 photons (Fig. 2B). Gaussian interpolation of these data reveals that this sample contained 55% intact, full-length λ DNA molecules, which is smaller than the 89% ± 5% proportion independently estimated through photometry of a pulsed-field electrophoresis gel. The ∼35% discrepancy can be attributed to preferred adsorption of longer DNA molecules onto chip surfaces and/or to some DNA fragmentation during the measurement.
Only stretched and intact DNA molecules were used for mapping. These can be selected from a histogram of burst size versus length (Fig. 2C). The average burst size, Bλ, corresponding to intact λ DNA molecules (horizontal line on Fig. 2C), is defined by the center of the Gaussian fit (the red line on Fig. 2B). Only the longest molecules grouped near the Bλ level were selected for mapping. For heterogeneous DNA samples, fully stretched DNA molecules cluster, according to size, along the line of proportionality between burst size and length (data not shown).
We generally prefer a slightly different data presentation suited for heterogeneous samples, shown in Figure 2D, that lines up the fully stretched molecules horizontally in histograms of the average intensity of DNA backbone (burst size divided by the DNA residence time in spot ExII) versus length. In this approach to selecting fully stretched intact molecules, the average DNA intensity versus length revealed a hyperbolic profile (comet) for intact λ DNA (Fig. 2D). The hyperbolic relationship arises because molecules with the same contour length have the same amount of bound intercalating dye. Therefore, incompletely stretched molecules have a proportionally larger fraction of the labeled DNA backbone, and hence, a larger number of intercalating fluorophores within the excitation field. Consequently, these comet plots conveniently present DNA mixtures with each size lying along a unique hyperbola, and with the fully stretched molecules lined up horizontally at the same level determined by the dye density on the DNA. This important feature provides one extra criterion for the selection of the appropriate molecule traces for analysis. For mapping analysis, we selected only the longest molecules (16.25–17.25 μm) belonging to the comet head, and, therefore, having the same burst size. The selected data are surrounded by a red square in Figure 2D and included about 400 individual molecule traces or 28% of all detected molecules. If only intact λ DNA molecules are taken into account, which constitute 55% of all detected traces (see above), then 100%*28/55 = 51% of them were selected as being sufficiently stretched for further analysis.
Generation of a Molecular DNA Motif Map With Site-Specific Tagging
DLA technology uncovers maps by locating fluorescent tags specific for sequence motifs along individual extended DNA molecules. Motif frequency depends on length and sequence composition. For instance, short and frequent motifs, like the 7- and 8-bp recognition sequences used in this study, produce information dense maps. We chose a DNA motif-tagging chemistry based on peptide nucleic acid (PNA) probes (Nielsen and Egholm 1999) and specifically bisPNA clamps that are capable of forming stable complexes with 7- and 8-bp targets. Their detailed description, including specific designs, functional properties, methods of use, and an evaluation of tagging efficiency and specificity, is available in the Supplemental material.
Figure 3 illustrates the use of DLA to map the motif tag bisPNA H (Table 1) within the 48.5-kb λ genome. We selected this sequence motif to validate the DLA approach because it has only two target sites, and they form a simple asymmetric pattern with one site conveniently at the center (Fig. 3A). The λ genome also includes two sites with a single-end mismatch (SEMM) within. We neglected all other mismatching complexes, because SEMM sites prevail as a source of false-positives (Kuhn et al. 1999). Single- and double-tetramethylrhodamine (TMR) fluorophore versions of tag H were synthesized (H-1TMR and H-2TMR, respectively, Table 1). Tagging specificity was independently assessed for every sample using a band-shift assay and gel photometry (see Supplemental material). For bisPNA H-2TMR, 89% and 11% of target and SEMM sites, respectively, were tagged. Therefore, 7.4 targets per SEMM were bound, or roughly one SEMM site per 3–4 λ DNA molecules was tagged.
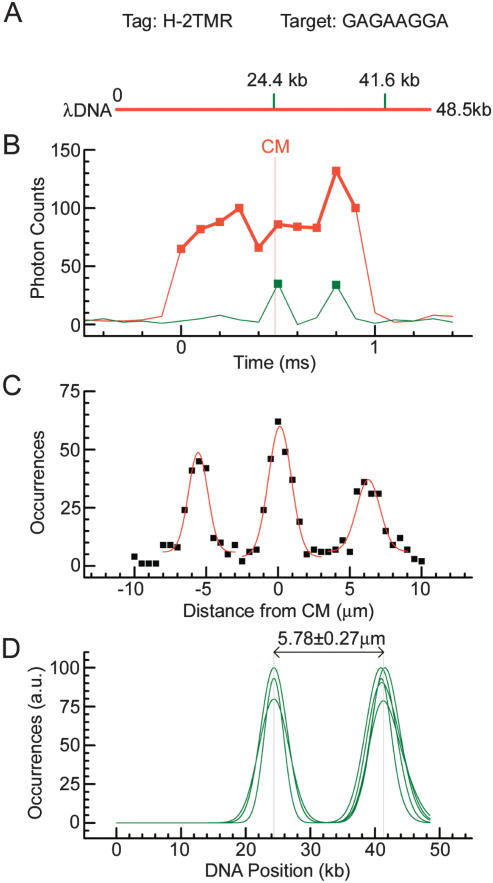
Figure 3.
Mapping of bisPNA tag H-2TMR on λ phage DNA. (A) Theoretical λ DNA (48.5 kb) map. Motif H has sequence (amino terminus)TCCTTCTC(carboxyl terminus) (Table 1), corresponding to the target (5′)GAGAAGGA(3′). Positions of motif target sites are shown with green dashes. These motifs are separated by 17.2 kb. (B) Actual fluorescence data traces from a single motif tagged λ DNA molecule. Motif tag fluorescence (green trace) and intercalated DNA fluorescence (red trace) are detected in data channels 1 and 2, respectively (see also Fig. 1C). The vertical gray line shows the position of the center of the molecule (CM). (C) A map obtained by summing of ∼500 single molecule traces. Gaussian fits to the tag position data are represented by the red line. (D) Oriented maps measured in five different experiments with different sample preparations to illustrate reproducibility. Only Gaussian fits are presented. The maps are aligned in the middle of the interpolated central peak. The maps are normalized to the higher of the two peaks. The average measured geometrical distance between the motif targets is presented.
Table 1.
Chemical Structures of bisPNA Fluorescent Tags
| Tag name | Chemical Structure |
|---|---|
| H-1TMR | TMR-OO-Lys-Lys-TCC TTC TC-OOO-JTJ TTJ JT-Lys-OO-Lys-O |
| H-2TMR | TMR-OO-Lys-Lys-TCC TTC TC-OOO-JTJ TTJ JT-Lys-OO-Lys(TMR)-O |
| bis7 | TMR-OO-Lys-Lys-TCC TTC T-OOO-TJT TJJ T-Lys-Lys |
| S-1TMR | TMR-OO-Lys-Lys-TTT CTC TT-OOO-TTJ TJT TT-Lys-Lys |
| S-2TMR | TMR-OO-Lys-Lys-TTT CTC TT-OOO-TTJ TJT TT-Lys-OO-Lys(TMR)-O |
| S-1Alx | Alexa-OO-Lys-Lys-TTT CTC TT-OOO-TTJ TJT TT-Lys-Lys |
| S-2Alx | Alexa-OO-Lys-Lys-TTT CTC TT-OOO-TTJ TJT TT-Lys-Lys-OO-Lys(Alexa)-O |
The tags include tetramethylrhodamine (TMR) or Alexa 546 fluorophores, which are conjugated to amino-terminal or lysine side-chain amino groups. Fluorophores, Watson-Crick (T+C) strand, and Hoogsteen (T+J) strand are connected with -O- linkers (8-amino-3,6-dioxa-octanoic acid), which form flexible hydrophilic tethers. (T) Thymine; (C) cytosine; (J) pseudoisocytosine. The replacement of C with J in the Hoogsteen strand promotes hybridization of this bisPNA at pH > 6.5 (Kuhn et al. 1999). The lysines introduce positive charges to improve hybridization efficiency (Demidov and Frank-Kamenetskii 2001). The sequences are presented from amino terminus (left) to carboxyl terminus (right). Amino- and carboxy-termini of Watson-Crick strand of PNA (bold) hybridize to 3′- and 5′-termini of DNA target, respectively, whereas amino- and carboxy-termini of Hoogsteen strand of PNA hybridize to 5- and 3′-termini of DNA target, respectively (Nielsen and Egholm 1999).
Figure 3 shows the step-by-step development of the bisPNA H map of λ DNA. This approach uses the central position of one of the targets and differs from the one used in the next section. It was applied to validate DLA and obtain a conversion factor to convert measured lengths into kilobases in this study. Individual DNA molecules, hybridized with tags and backbone stained, were interrogated. Only λ DNA molecules stretched to between 16.25 and 17.25 μm lengths were selected for analysis (within the red square, Fig. 2D). TMR fluorophores of bisPNA tags were excited by green (532 nm) light and detected as photon bursts or spikes (green trace in Fig. 3B). As with the DNA backbone, tags were identified as photon bursts that exceeded a threshold count set by the average and the standard deviation of the background noise (green squares in Fig. 3B). To calculate the location of tags on the DNA backbone, the raw time domain data were converted into distances using the calculated velocity for each DNA event. The molecule in Figure 3B contained two tags bound at the matching sites. The TMR signals at 0.5 and 0.8 msec corresponded to the target locations at 24.4 and 41.6 kb, respectively (Fig. 3, cf. A and B).
To obtain a robust map, tens to thousands of molecular traces were analyzed together. Figure 3C shows a histogram of detected tag locations with respect to the CM of stretched intact DNA molecules. Some DNA molecules traveled headfirst, and others in the opposite orientation. Therefore, three peaks were expected, symmetrical about zero, with the side peaks at one-half of the height of the central peak, because both orientations contribute to the center peak (target position 24.4 kb), whereas the side peaks (target position 41.6 kb) are orientation specific. Each peak was fitted with a Gaussian curve (red lines in Fig. 3C) that allowed peak location to a better accuracy than the measurement resolution (for example, see Lacoste et al. 2000). To eliminate the ambiguity of molecule orientation, the fitted curve for the left peak was mirrored about the position of the central peak. The resulting histogram was then positioned to align the center peak with the CM of the DNA. This fine-tuning step eliminated small day-to-day offsets between the red and green lasers, which was about ±0.4 μm in these measurements. The uncertainty in red/green laser alignment affects the absolute position of the map on the DNA, but it does not influence the relative measurements of the distances between motifs.
We calibrate our measurements comparing measured and genetic distances between the targets. Average geometrical distance between the two bisPNA H peaks was of 5.78 ± 0.27 μm determined using data from five independent experiments (Fig. 3D). This corresponds to the 17.2-kb genetic distance between the target sites, producing a conversion factor of 0.336 ± 0.016 μm/kb = 3.36 ± 0.16 Å/bp. This coefficient was used to convert lengths into kilobases throughout the remainder of this study (see Figs. 4 and 5), as well as to calibrate the x-axis in Figure 3D. This conversion factor is comparable, within experimental uncertainty, to the axial rise of 3.4 Å/bp determined by X-ray diffraction for B-form DNA (Langridge et al. 1960; Sinden 1994). The intertag resolution was 1.5 μm or 5 kb, on the basis of the average full width at half maximum of the fitted Gaussian peaks in Figure 3D.
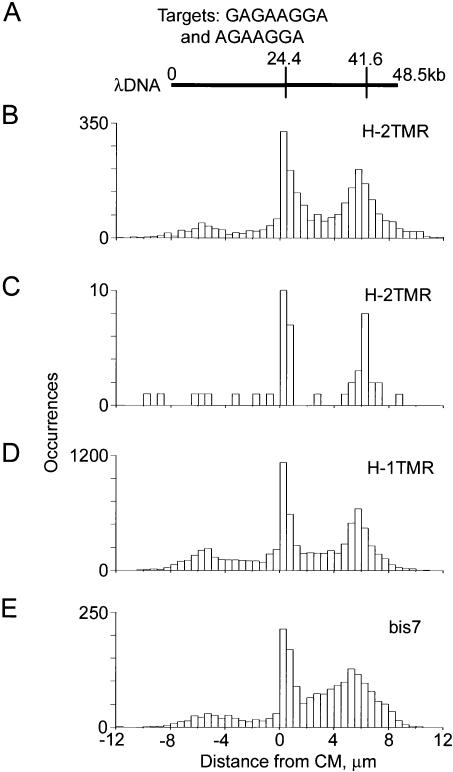
Figure 4.
Theoretical and measured maps of λ phage DNA obtained with H-2TMR, H-1TMR, and bis7 bisPNA tags (Table 1). Motif H has sequence (amino terminus)TCCTTCTC(carboxyl terminus) and motif bis7 has sequence (N)TCCTTCT(C); their corresponding targets are (5′)GAGAAGGA(3′) and (5′)AGAAGGA(3′). (A) Theoretical motif map obtained using either H or bis7. (B–E) Experimental maps obtained with H-2TMR, H-1TMR, and bis7 tags. The data sets used to obtain the maps B, D, and E included 2562, 7794, and 1909 total individual molecular traces, respectively. Map C was obtained using a subset of only 57 molecular traces from the same data set used on map B. These 57 molecules belonged to a very narrow range of burst and linear sizes, representing a small spot within the red box on the graph Figure 2D. All experimental maps are aligned using their centers of molecules (CM, at 0 microns). The center of the theoretical map is aligned with the experimental CM. Theoretical map was scaled using 0.336 μm/kb ratio (see text for details).
Figure 5.
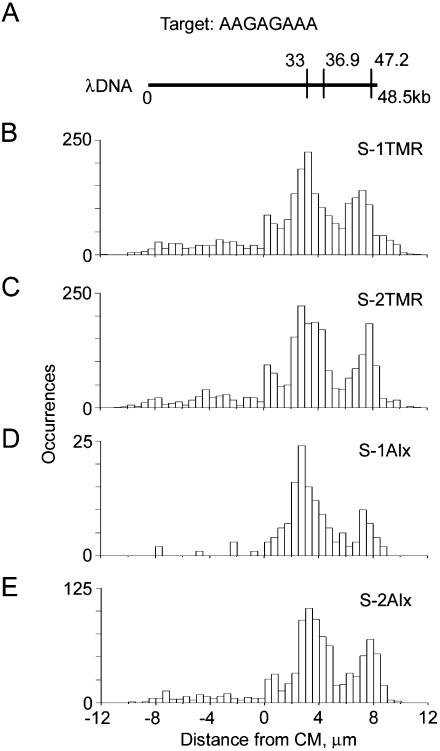
Theoretical and measured maps of λ phage DNA obtained with S-1TMR, S-2TMR, S-1Alx, and S-2Alx bisPNA tags (Table 1). These tags all have the same sequence motif (amino terminus)TTTCTCTT(carboxyl terminus), which recognizes the target (5′)AAGAGAAA(3′). (A) Theoretical map obtained for this motif. (B–E) Experimental maps obtained with S-1TMR, S-2TMR, S-1Alx, and S-2Alx bisPNA tags, respectively. The data sets used to obtain the maps B, C, D, and E included 2279, 2275, 145, and 943 total individual molecular traces, respectively. See Figure 4 caption for details.
Due to imperfect tag chemistry and variability in detection efficiency, both unoccupied targets (false negatives) and spurious signals (false positives) were observed on many molecules. To correct for them, we utilized multiple molecule traces for statistical signal analysis. In general, more bisPNA H false negatives were recorded by DLA than expected from gel band-shift assays; only two-thirds of the detected DNA molecules exhibited one or more tags at any position, in contrast to 89% target-site occupancy from band-shift analysis (see Supplemental material). Most likely, not all bound tags were detected, especially in the periphery of the excitation spots where low excitation energy was insufficient for detecting single-tag fluorophores, but was still enough to detect more fluorescent DNA molecules. Therefore, we summed several hundred molecules to produce histograms of detected tag locations (Figs. 3C, 4, and 5). This approach also diluted the contribution of the false positives that can arise from residual nonreacted tags or from unspecific binding.
Procedure for Obtaining DNA Maps
To obtain maps, we must distinguish between head-first and tail-first orientations of the detected DNA molecules. In this study, we used a procedure, based on the asymmetry of the binding motifs we studied, to obtain the maps presented in Figures 4 and 5. The algorithm oriented each molecular trace before calculation of the sum, so that the half of DNA molecule that emitted more photons in the green channel (data channel 1, which detected emission of tags), was oriented toward the positive axis direction. However, not all molecular traces were oriented properly because of shot noise. Therefore, the maps in Figure 4 have a small ghost peak at –5.6 μm symmetric to the peak at +5.6 μm (41.6 kb). This ghost peak was much smaller than the correct peak, so it did not produce any problem with map interpretation.
Mapping of Different Motifs With Different Tags
To demonstrate that bisPNA tags can be used for DLA mapping and to evaluate the influence of various structural elements on mapping accuracy, we tested a variety of the tags having different sequences, lengths, numbers and types of fluorophores, and numbers of positive charges (Table 1, see Supplemental material for rationale). Three tags were designed in a way that produced maps of similar complexity. However, they differed dramatically in the numbers of SEMM sites within λ DNA, in order to challenge our ability to produce an accurate map. Tag H was the simplest case, as it had only two SEMM sites within λ DNA. In contrast, tag S with its 14 SEMM sites represented a more typical case for an 8-bp sequence motif, and tag bis7 had 37 SEMM sites (one per 1.3 kb on average). At a labeling specificity of 10, which represents the ratio of the proportion of tagged target sites to the proportion of tagged SEMM sites, any given DNA molecule labeled with this tag would have more tagged SEMM sites (two to three) than targets (two or less).
Regardless of the number of SEMM sites and design features, correct maps were obtained with every tag. Measured maps for bisPNAs H and bis7 are shown in Figure 4 and for bisPNA S in Figure 5, with theoretical maps displayed above (Figs. 4A and 5A) for comparison. Each of the observed maps exhibited major peaks easily distinguishable by intensity from other features. These peaks are centered at the expected target positions. For every expected target position, a corresponding peak was found and no other major peaks were observed. The measured maps corresponded directly to target sequences and did not depend on other features of the bisPNA tag designs. These results demonstrate successful mapping of 50 kb-long DNA, using a variety of different bisPNA tags, including those with a different number of charges, as well as a different number and types of fluorophores.
At current labeling specificity, DNA tagged with bis7 bisPNA had more than half of the tags bound to SEMM sites, and these sites were concentrated in the termini-adjacent regions. False positives produced by these SEMM sites complicated the work of the orienting algorithm and resulted in a widening of the measured peak corresponding to the target at 41.6 kb (Fig. 4 E). Regardless of this problem, both major target peaks are clearly visible on the map and centered at the exact positions.
Application of the algorithm, orienting molecular traces using their asymmetry, also makes the central peak of the maps in Figure 4 asymmetric and sharper than the terminal peak. It happens because the tag at 24.4 kb is closer to the CM than current resolution (±2.5 kb), and therefore can be detected both to the left and to the right from CM. However, in the DNA molecules with only this tag detected, the central signal is always oriented by the algorithm to the right. From a different viewpoint, the fact that the central band remains so sharp after summing so many molecular traces proves that our approach of determining the CM position using the backbone intensity profile works very well.
Selecting smaller subsets of molecules belonging to a very narrow range of burst and linear sizes, we estimated that a discernable map can be obtained with fewer than a hundred molecular traces total. One such map obtained with just 57 molecular traces is presented in Figure 4C. This subset was obtained from the same data set as shown in Figure 4B, in which the selected area was only a tiny spot as shown by the red dot within the red box of Figure 2D that includes molecules with lengths of 16.9 ± 0.1 μm and intensities (80 ± 5)*104 photons/sec.
Mapping the sequence motif AAGAGAAA includes extra challenges such as multiple SEMM sites, two close targets at 33.0 and 36.9 kb, and the third target site, at 47.2 kb, which is so close to the end that it could be accurately mapped only with completely stretched DNA molecules (Fig. 5). The termini-adjacent regions were much more prone to fluctuations related to incompletely stretched DNA because of the stem-and-flower conformation. We measured maps using different designs of tag S with different numbers of positive charges, different types, and numbers of fluorophores. Correct maps were obtained with every tag tested (Fig. 5). A small peak was visible on some maps in the middle of the DNA at ∼0.5 μm. This originated from a cluster of four close SEMM sites between 23 and 26 kb of λ DNA. Regardless of finite tagging specificity, which was between 7 and 12 (see Supplemental material), this central peak was still lower than the peaks at the target positions. With current resolution of 5 kb, we were not able to resolve the central doublet (33 and 36.9 kb).
DISCUSSION
The study is, in essence, a proof of principle of Direct Linear Analysis as a new technology for DNA analysis. The two key components of DLA are (1) a microfluidic system for flowing and stretching single DNA molecules so that they can be read rapidly in a linear fashion and (2) fluorescence-based detection that allows the reading of fluorescent tags on single molecules of long DNA. Its combination of features, namely single-molecule sensitivity, analysis of very long DNA molecules, and high throughput makes DLA an attractive approach for many genomic applications. Single-molecule sensitivity eliminates the need for DNA amplification, and could be particularly valuable for discerning haplotypes (Ding and Cantor 2003; Mitra et al. 2003). The ability to analyze very long DNA molecules preserves the higher-order information in the genome, such as the sequence of linkage disequilibrium blocks. For this, DNA fragments should be sufficiently long to include two or more LD blocks, which are tens to hundreds of kilobases long (Patil et al. 2001; Gabriel et al. 2002; Weiss and Clark 2002). Furthermore, because we analyze large DNA fragments at a rate of 30–60 million bp/min, the throughput of DLA has the potential to address microbial and human genomics.
The post and funnel microfluidic device used in this study allowed us to efficiently stretch λ DNA so that >50% of the DNA molecules were routinely stretched to 15 μm or longer (i.e., ≥90% of the contour length). Our data analysis allowed rapid identification and selection of a population of stretched molecules that proved suitable for mapping. Due to stem-and-flower considerations, in cases in which a target site was located within 1 μm of the termini (e.g., at 47.2 kb, Fig. 5A), only molecules >16 μm long were selected for analysis, which represented 10%–20% of all λ DNA molecules. Although the fluidic system required high flow rates (>10μm/msec) to achieve stretching, a noteworthy feature of the system was the ability to simultaneously localize single fluorophore tags along the backbone.
bisPNAs were used to tag sequence-specific sites within the λ genome. Although DLA analyzes individual DNA molecules, traces of 50–5000 molecules were summed to obtain more reliable maps. This served to reduce false-negative and false-positive signals resulting from imperfect labeling and detection of individual molecules. Typically, about 1000 stretched molecules were sufficient for a good quality map. Thus, even in cases in which 5000 total DNA molecules are observed to obtain traces of 1000 stretched molecules, it took only 1.4 min to obtain a reliable map of the λ genome at a typical velocity of 10 μm/msec. This time also accounts for dilution needed to avoid overlapping, because of which, DNA molecules were detected only 10% of the time.
By evaluating the bisPNA tagging of known sites within λ DNA, we were able to assess the resolution and accuracy of DLA for mapping. The minimal distances resolved in the presented experiments were 10.3 and 8.5 kb, which were between 36.9 and 47.2 kb targets and between the middle of SEMM cluster at 24.5 kb and the target at 33.0 kb, respectively (Fig. 5). The resolution of the system cannot be better than a peak width (Hecht and Zajac 1979), which is about 5 kb for a single bisPNA site (e.g., Fig. 3D). The peak width is determined principally by molecule velocity and detection frequency. At a typical velocity of 10 μm/msec and detection frequency of 10 kHz, one data bin encompasses 1 μm or 3 kb of stretched DNA. Improvements in resolution could be attained by reducing molecule velocity, which would reduce throughput, or by increasing sampling frequency, which would require brighter fluorescent tags. Nonetheless, resolution is fundamentally limited by the diffraction limit of optical detection, which is 0.23 μm or 0.68 kb according to 0.61λ/NA for wavelength λ = 532 nm and a numeric aperture NA = 1.4 (Hecht and Zajac 1979). Of note, two tags labeled with different colors can be both detected, regardless of the separating distance. In this simultaneous analysis, which is compatible with our current system, the resolution is not limited by the peak widths because the tags are detected simultaneously.
For mapping purposes, once tags are resolved, that is, located >5 kb apart, the distance between them can be measured with considerably greater accuracy using Gaussian interpolations. This approach has been used to determine positions of fluorophores with accuracy down to 10 nm (Lacoste et al. 2000). In our case, the accuracy is determined not only by the number of detected fluorophores, but also by reproducibility of stretching and was ±0.27 μm or ±0.8 kb (Fig. 3D). To determine positions of the tagged sites in the genome, the pattern of the detected tags should be aligned on the DNA backbone profile. The accuracy of this alignment was ±0.4 μm or ±1.2 kb, and resulted from an offset between the red and green lasers. Thus, distances between distant motif tags could be reproducibly measured at ±0.8 kb, and these tags could be localized on the DNA backbone with an accuracy of ±2 kb.
Two major strategies can be envisioned for mapping genomes using DLA, labeling a repeating motif and using the pattern of the motif distribution along the DNA or labeling unique site(s) within a genome. Herein, we demonstrated the applicability of short bisPNAs for tagging sequence motifs as short as 8, or even 7 bp. By determining the number and location of such repeating motifs within a target DNA molecule, a highly informative and generally unique map can be obtained. Alternatively, labeling of unique sites within a genome or DNA fragment can be achieved, for example, with fluorescent conjugates of long runs of single-strand DNA oligonucleotides (20 bp or longer) or DNA mimics like PNAs (Nielsen and Egholm 1999) or LNAs (Braasch and Corey 2001), which form more stable complexes, so the target sequences can be shorter.
Traditional gel-based restriction mapping provides high resolution with high-density map information (Brown 1999). However, unlike DLA, this technique does not directly provide an ordered map of the restriction sites. Furthermore, because a restriction map must be derived from independent measurements of many fragments, the length measurement error accumulates during reconstruction and therefore decreases the accuracy of determining the positions of restriction sites. Moreover, in extreme cases, insufficient accuracy of fragment lengths can result in incorrect reconstruction of maps (Thayer et al. 1999). Mapping of genomes includes measurements of long DNA fragments, which are analyzed with pulsed-field gel electrophoresis (Ho and Monaco 1995). Its accuracy depends upon the fragment size and is about ±1 kb for 50–100-kb fragments (Birren and Lai 1993). The minimal amount of dsDNA that can be detected in a gel with fluorescent staining is about 25 pg (Haugland 2002) or 5*105 copies of 50-kb DNA, generally obligating one to an amplification step. Finally, restriction mapping to identify genomic variations is inherently limited by the repertoire of available enzymes, and hence, the sites they target. Thus, traditional restriction mapping is cumbersome, relatively time and cost expensive, requires DNA amplification, and fails to provide haplotype information from diploid sources of DNA. Therefore, alternative approaches would be valuable.
One such technique, optical mapping, can provide ordered restriction maps from a small population of long individual DNA molecules (Aston et al. 1999a,b). In this approach, long DNA is stretched, usually by molecular combing, and bound to a treated glass surface. DNA is then cleaved with a restriction endonuclease, stained, and individual molecules are imaged using a high-power fluorescence microscope. The restriction sites in the extended molecules appear as visible gaps. The most striking use of this optical mapping technique was the genome mapping of the pathogenic Escherichia coli strain O157:H7 (Lim et al. 2001), in which DNA was randomly broken into fragments between 150 and 2900 kb. To construct a map, data from 840 fragments, which were digested with XhoI and NheI endonucleases, were obtained. Fragment sizes were determined to an accuracy of about 2.1 kb. The process from image acquisition to image analysis and map making was automated.
As for every technique, optical mapping has its intrinsic difficulties. For successful stretching and immobilization of DNA, the glass surface must be modified to an optimal charge density, depending on the size of DNA molecules to be stretched (Aston et al. 1999a; Taylor et al. 2000). However, as this procedure is highly variable, surfaces must be assessed after every preparation (Aston et al. 1999a). The derivatized glass surfaces are labile and can be stored in treatment solution for only a few weeks (Aston et al. 1999a). Moreover, DNA samples can undergo nonspecific photolysis catalyzed by DNA-bound intercalators (Akerman and Tuite 1996). Finally, restriction endonucleases are less efficient on immobilized DNA than on DNA in solution and may not cleave all sites (Aston et al. 1999b).
DLA technology exhibits notable advantages over optical mapping. First, in place of a derivatized surface, DNA is stretched in a microfluidic chip, which can be manufactured in bulk and deliver reproducible results. Because fluidic stretching of DNA occurs on the fly and does not involve manual manipulation, it has very high throughput. Photodamage of DNA is not relevant in our case, because DNA relaxation time (Ladoux and Doyle 2000) is longer than the measurement time. Therefore, the trace of a DNA backbone is uninterrupted regardless of photo-induced nicks. Furthermore, we perform the tagging reaction in solution rather than on immobilized DNA; hence, all targets are equally accessible. In conclusion, Direct Linear Analysis of DNA represents a novel sequence-specific structural mapping technology with broad applications in genomics, genetics, and other DNA sequence- and structure-based research.
METHODS
Preparation and Characterization of Tagged DNA Samples
λ Phage DNA (accession #NC_001416) was purchased from Promega Corp. To evaluate admixtures of degraded DNA, the samples were analyzed by pulsed-field gel electrophoresis (CHEF Mapper, Bio-Rad). A ChemiGenius bioimaging system with GeneSnap and GeneTools software (Syngene) was used for photometric analysis of the gels.
The procedure for λ DNA tagging and for control of specificity and efficiency of the hybridization with electrophoretic band-shift assay is described in detail in the Supplemental material.
For DLA, DNA samples were stained with TOTO-3 (Molecular Probes) at the measurement concentration of 120 pg/μL. Solutions of DNA were incubated at room temperature for 2–4 h at base pair to dye molar ratios between 10:1 and 5:1. On the one hand, we wanted to have our DNA stained as close to saturation as possible to have the dye evenly spaced along DNA backbone (Larsson et al. 1994; Carlsson et al. 1995); the saturating ratio for homologous YOYO-1 dye was estimated to be 4:1 bp per dye molecules (Larsson et al. 1994). The even distribution was needed for accurate determination of CM of DNA molecules. On the other hand, we avoided staining of DNA at higher proportion of the intercalator, because this decreased the number of detected DNA molecules probably facilitating their adsorption onto the internal chip surfaces and promoting DNA aggregation (see also Akerman and Tuite 1996). Cyanine dyes are easily adsorbed out of aqueous solutions onto surfaces (Haugland 2002); therefore, the dissolved TOTO-3 moiety could be considerably depleted because of the adsorption onto the reaction vessel walls, and the degree of DNA staining was even lower than calculated. Lower than saturating amount of the dye also explains why the measured lengths of completely stretched DNA were similar to the contour length of unstained molecules (16.5 μm for unstained λ DNA). The contour length of λ DNA saturated with intercalating dye can be considerably longer than that of dye-free DNA. For example, Perkins at al. (1995) observed up to 22 μm for λ DNA saturated with TOTO-1, a homologous cyanine dye. Similar extension was expected for λ DNA saturated with TOTO-3 in our experiments. However, only a minor proportion (<2%) of λ DNA molecules longer than 18 μm was detected in our experiments (Fig. 2A). Another effect that can potentially impact in the observed DNA length is incomplete stretching of the terminal regions in the stem-and-flower conformation. However, this effect did not contribute >0.9 μm, because the S tag at 47.2 kb (97.3% of total DNA length) was found at the correct position.
Experimental Setup
The experimental setup was built around an Eclipse E800 fluorescence microscope with a PlanApo 100× objective lens (Nikon Corp.). One diode-pumped solid-state laser (532 nm wavelength, mod. #100GLS, QED) and two He-Ne ion lasers (633 nm, model #31-2140-000, Coherent) were used. Each beam provided up to 20 mW at the excitation point. Optical filters and dichroic mirrors were obtained from Chroma Technology Corp. The microscope was equipped with two V-IBS beamsplitter modules for multicolor detection, one with a dichroic mirror to separate TMR and TOTO-3 fluorescence, and another with a beamsplitter to separate signals from the excitation spots ExII and ExIII (Fig. 1B). The spectral band of detection in data channel 1 was centered at 590 nm and had a width of 50 nm. The centers and widths of spectral bands were 665 nm and 50 nm, respectively, in data channels 2 and 3. Fluorescence was delivered by optical-fiber to photon-counting modules with avalanche photodiodes (SPCM-AQR-12-FC, PerkinElmer Optoelectronics). Signals from all three data channels were directed to a computer with a PCI-6602 interface board (National Instruments). Custom software was used for collecting and processing of the data. Data were collected at 10 kHz sampling frequency.
The microfluidic chip structures were etched in fused silica by electron beam lithography and reactive ion etching (DuPont Photomasks), diced and drilled by Ceramic Grinders, and sealed by glass cover (0.15-mm thick) with KASIL-2130 (PQ Corp.). The depth of the chip cavity was 1 μm. The distances between the posts' axes were 2.5 μm, and the sides of the square posts were 1 μm.
The single molecule detection capability was assessed using very diluted (1–100 pM) solutions of TMR (data channel 1) and Cy5 (data channels 2 and 3) fluorophores (Eigen and Rigler 1994). Depending on flow velocity and excitation laser power, between 10 and 150 photons were detected per single fluorophore. The time width of detection of a single fluorophore never exceeded a single bin, even at 20-kHz detection frequency (0.05 msec/bin). Background noise depends on the excitation intensity and was between 6 and 30 counts per bin in our measurements.
Acknowledgments
We thank John L. Harris, Eric A. Nalefski, Jenny E. Rooke, and Michael A. Shia for critically reviewing this manuscript, Kevin M. Phillips for electrophoretic analysis of the DNA sample, and James Ku for the preparation of Figure 1, A and B. We thank Lance Gleich and Parris S. Wellman for construction of the experimental setup and Robert H. Austin, John F. Marko, and Jonas Tegenfeldt for fruitful discussions. This research was partially funded by NASA grant NAS2-00001 and NSF grant DMI-0213876.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1635204.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Akerman, B. and Tuite, E. 1996. Single- and double-strand photocleavage of DNA by YO, YOYO and TOTO. Nucleic Acids Res. 24: 1080–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston, C., Hiort, C., and Schwartz, D.C. 1999a. Optical mapping: An approach for fine mapping. Methods Enzymol. 303: 55–73. [DOI] [PubMed] [Google Scholar]
- Aston, C., Mishra, B., and Schwartz, D.C. 1999b. Optical mapping and its potential for large-scale sequencing projects. Trends Biotech. 17: 297–302. [DOI] [PubMed] [Google Scholar]
- Birren, B. and Lai, E. 1993. Pulsed field gel electrophoresis: A practical guide. Academic Press, Inc., San Diego, CA.
- Braasch, D.A. and Corey, D.R. 2001. Locked nucleic acid (LNA): Fine-tuning the recognition of DNA and RNA. Chem. Biol. 8: 1–7. [DOI] [PubMed] [Google Scholar]
- Brown, T.A. 1999. Genomes. John Wiley & Sons Inc., New York.
- Bustamante, C., Marko, J.F., Siggia, E.D., and Smith, S. 1994. Entropic elasticity of λ-phage DNA. Science 265: 1599–1600. [DOI] [PubMed] [Google Scholar]
- Bustamante, C., Smith, S.B., Liphardt, J., and Smith, D. 2000. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 10: 279–285. [DOI] [PubMed] [Google Scholar]
- Carlsson, C., Jonsson, M., and Akerman, B. 1995. Double bands in DNA gel electrophoresis caused by bis-intercalating dyes. Nucleic Acids Res. 23: 2413–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, H.-P., Spence, C., Scherer, A., and Quake, S.R. 1999. A microfabricated device for sizing and sorting DNA molecules. Proc. Natl. Acad. Sci. 96: 11–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov, V.V. and Frank-Kamenetskii, M.D. 2001. Sequence-specific targeting of duplex DNA by peptide nucleic acids via triplex strand invasion. Methods 23: 108–122. [DOI] [PubMed] [Google Scholar]
- Ding, C. and Cantor, C.R. 2003. Direct molecular haplotyping of long-range genomic DNA with M1-PCR. Proc. Natl. Acad. Sci. 100: 7449–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen, M. and Rigler, R. 1994. Sorting single molecules: Application to diagnostics and evolutionary biotechnology. Proc. Natl. Acad. Sci. 91: 5740–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foquet, M., Korlach, J., Zipfel, W., Webb, W.W., and Craighead, H.G. 2002. DNA fragment sizing by single molecule detection in submicrometer-sized closed fluidic channels. Anal. Chem. 74: 1415–1422. [DOI] [PubMed] [Google Scholar]
- Gabriel, S.B., Schaffner, S.F., Nguyen, H., Moore, J.M., Roy, J., Blumenstiel, B., Higgins, J., DeFelice, M., Lochner, A., Faggart, M., et al. 2002. The structure of haplotype blocks in the human genome. Science 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- Haugland, R.P. 2002. Nucleic acid detection and genomics technology. In Handbook of fluorescent probes and research products (ed. J. Gregory), pp. 265–352. Molecular Probes, Inc., Eugene, OR.
- Hecht, E. and Zajac, A. 1979. Optics. Addison-Wesley Publishing Company, Reading, MA.
- Ho, M.F. and Monaco, A.P. 1995. PFGE in physical mapping. In Pulsed field gel electrophoresis: A practical approach (ed. A.P. Monaco), pp. 21–43. IRL Press, at Oxford University Press, New York.
- Kuhn, H., Demidov, V.V., Nielsen, P.E., and Frank-Kamenetskii, M.D. 1999. An experimental study of mechanism and specificity of peptide nucleic acid (PNA) binding to duplex DNA. J. Mol. Biol. 286: 1337–1345. [DOI] [PubMed] [Google Scholar]
- Lacoste, T.D., Michalet, X., Pinaud, F., Chelma, D.S., Alivisatos, A.P., and Weiss, S. 2000. Ultrahigh-resolution multicolor colocalization of single fluorescent probes. Proc. Natl. Acad. Sci. 97: 9461–9466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoux, B. and Doyle, P.S. 2000. Stretching tethered DNA chains in shear flow. Europhys. Lett. 52: 511–517. [Google Scholar]
- Langridge, R., Marvin, D.A., Seeds, W.E., Wilson, H.R., and Hamilton, L.D. 1960. The molecular configuration of deoxyribonucleic acid. II. Molecular models and their Fourier transforms. J. Mol. Biol. 2: 38–64. [Google Scholar]
- Larsson, A., Carlsson, C., Jonsson, M., and Albinsson, B. 1994. Characterization of the binding of the fluorescent dyes YO and YOYO to DNA by polarized light spectroscopy. J. Amer. Chem. Soc. 116: 8459–8465. [Google Scholar]
- Lim, A., Dimalanta, E.T., Potamousis, K.D., Yen, G., Apodoca, J., Tao, C., Lin, J., Qi, R., Skiadas, J., Ramanathan, A., et al. 2001. Shotgun optical maps of the whole Escherichia coli O157:H7 genome. Genome Res. 11: 1584–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manneville, S., Cluzel, P., Viovy, J.-L., Chatenay, D., and Caron, F. 1996. Evidence for the universal scaling behaviour of a freely relaxing DNA molecule. Europhys. Lett. 36: 413–418. [Google Scholar]
- Marko, J.F. and Siggia, E.D. 1995. Stretching DNA. Macromolecules 28: 8759–8770. [Google Scholar]
- Mitra, R.D., Butty, V.L., Shendure, J., Williams, B.R., Housman, D.E., and Church, G.M. 2003. Digital genotyping and haplotyping with polymerase colonies. Proc. Natl. Acad. Sci. 100: 5926–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, P.E. and Egholm, M. 1999. An introduction to PNA. In Peptide nucleic acids. Protocols and applications. (eds. P.E. Nielsen and M. Egholm), pp. 1–19. Horizon Scientific Press, Norfolk, UK.
- Olive, D.M. and Bean, P. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37: 1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, N., Berno, A.J., Hinds, D.A., Barrett, W.A., Doshi, J.M., Hacker, C.R., Kautzer, C.R., Lee, D.H., Marjoribanks, C., McDonough, D.P., et al. 2001. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294: 1719–1723. [DOI] [PubMed] [Google Scholar]
- Perkins, T.T., Smith, D.E., Larson, R.G., and Chu, S. 1995. Stretching of a single tethered polymer in a uniform flow. Science 268: 83–87. [DOI] [PubMed] [Google Scholar]
- Perkins, T.T., Smith, D.E., and Chu, S. 1997. Single polymer dynamics in an elongational flow. Science 276: 2016–2021. [DOI] [PubMed] [Google Scholar]
- Rigler, R., Mets, U., Widengren, J., and Kask, P. 1993. Fluorescence correlation spectroscopy with high count rate and low background: Analysis of translational diffusion. Eur. Biophys. J. 22: 169–175. [Google Scholar]
- Shi, M.M. 2002. Technologies for individual genotyping: Detection of genetic polymorphisms in drug targets and disease genes. Am. J. Pharma. 2: 197–205. [PubMed] [Google Scholar]
- Sinden, R.R. 1994. DNA structure and function. Academic Press, San Diego, CA.
- Smith, S.B., Cui, Y., and Bustamante, C. 1996. Overstretching B-DNA: The elastic response of individual double-stranded and single stranded DNA molecules. Science 271: 795–799. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, A., and French, L. 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 10: 1772–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl, G. 1997. The physics of polymers. Springer, Berlin, Germany.
- Taylor, J.R., Fang, M.M., and Nie, S. 2000. Probing specific sequences on single DNA molecules with bioconjugated fluorescent nanoparticles. Anal. Chem. 72: 1979–1986. [DOI] [PubMed] [Google Scholar]
- Thayer, E.C., Olson, M.V., and Karp, R.M. 1999. Error checking and graphical representation of multiple-complete-digest (MCD) restriction-fragment maps. Genome Res. 9: 79–90. [PMC free article] [PubMed] [Google Scholar]
- Weiss, K.M. and Clark, A.G. 2002. Linkage disequilibrium and the mapping of complex human traits. Trends Genet. 18: 19–24. [DOI] [PubMed] [Google Scholar]