Abstract
Many human Natural Killer (NK) cells are prevented from killing autologous cells by virtue of inhibitory Killer cell Immunoglobulin-like Receptors (KIR) binding `self' HLA class I molecules. Individual NK cells stably express a selected set of KIR, but it is currently disputed whether the fraction of NK cells expressing a particular inhibitory KIR is influenced by the presence of the corresponding HLA ligand. This issue has been particularly hard to tackle in a statistically meaningful way due to the extreme polymorphism of the KIR and HLA loci, with widely varying affinities for individual KIR and HLA allele combinations. Here, we use a transgenic mouse model to investigate the effect of HLA on KIR repertoire and function. In this model system, a functional interaction between HLA-Cw3 and KIR2DL2 reduced both the surface expression of KIR2DL2 as well as the frequency of KIR2DL2+ cells.
Keywords: Natural Killer cells, transplantation, human, rodent, transgenic mice
Introduction
Under steady state conditions, NK cells are prevented from killing autologous cells by inhibitory NK cell receptors (NKR) binding `self' MHC class I molecules. In humans, this role is fulfilled by multiple inhibitory KIR, binding classical HLA class I molecules (HLA-A, B, −C), and CD94/NKG2A, binding the non-classical HLA-E molecule. While the CD94/NKG2A - HLA-E system is highly conserved, the KIR locus on chromosome 19 and the classical HLA class I locus on chromosome 6 display extensive polymorphism, with only particular KIR and HLA allele products binding each other. KIR3DL1 for example binds HLA-A and −B molecules expressing the Bw4 motif, but the affinity and functionality of this interaction depends on the specific KIR3DL1 and HLA-A/-B alleles involved. Furthermore, individual NK cell receptors are expressed only on a fraction of NK cells, ranging from 0 % to 100 % of NK cells depending on the receptor and the receptor allele. As a result, the NK cell repertoire consists of many NK cell subsets expressing distinct combinations of receptors, and these repertoires differ greatly between individuals.
The mechanisms underlying NK cell tolerance in man have been the subject of intense investigation and are informed by experiments in inbred mouse strains (1). Two non-mutually exclusive mechanisms have been proposed: (a) HLA dictates NKR expression patterns, or (b) HLA modulates NK cell responsiveness depending on the combination of NKR expressed. Initially, based on NK cells randomly cloned from two individuals, it was postulated that every NK cell clone expresses at least one inhibitory NK cell receptor specific for self HLA class I (2). This would ensure NK cell self tolerance and implied a major influence of HLA allotype on KIR repertoire, making responsiveness modulation redundant. Yet, subsequent analyses of the NK cell repertoires of a larger panel of HLA- and/or KIR-identical siblings showed that HLA had only a limited impact on KIR repertoire (3,4). Consistent with his finding, potentially autoreactive NK cells, lacking inhibitory NKR binding autologous HLA class I, were found to constitute a significant fraction (> 10 %) of the mature NK cell repertoire (5,6). These cells were hyporesponsive to stimulation (5), which may explain why they do not appear to kill autologous cells.
Recent developments in multiparameter flowcytometry provided more refined analyses, allowing the simultaneous detection of up to 5 NKR on NK cells at the single-cell level (6–9). The results of these experiments are conflicting. One study in Japanese individuals (n=132) showed an HLA ligand-induced increase in the frequency of NK cells expressing cognate KIR, but this effect was detectable only in the case of high affinity KIR-HLA combinations and neutralized by the presence of additional KIR-HLA interactions (7). This finding was corroborated in Germans (n=150) (6). In contrast, such an effect was undetectable in Swedes (n=44) (9). Rather, the latter study suggested a model in which KIR expression frequencies are genetically hardwired and repertoires low in KIR are buffered by CD94/NKG2A. The overall conclusion from these studies is that much larger studies would be necessary to examine the effect of cognate HLA on the inhibitory KIR repertoire (9). Furthermore, such studies should be based on allele-level KIR genotyping since KIR polymorphism affects the level and frequency of KIR expression (7,8).
To circumvent these issues, we used a KIR and HLA-Cw3 transgenic mouse model on a H-2Kb and H-2Db deficient C57BL/6 background. This mouse is transgenic for an almost intact and fully sequenced KIR B-haplotype (10,11). Similar to humans, KIR genes are expressed stochastically, so that individual KIR genes are expressed only on a subset of NK cells. The absence of H-2Kb and H-2Db eliminated any impact of the endogenous inhibitory interactions between Ly49 and MHC class I. We show that in these mice, a functional interaction between HLA-Cw3 and KIR2DL2 reduces the expression intensity and frequency of KIR2DL2.
Materials and Methods
Mice
Mice transgenic for a KIR B-haplotype and on a mixed (C57BL/6 and CBA) genetic background (11) were back-crossed 8 times onto C57BL/6 (Jackson) mice (12). This KIR B-haplotype has been sequenced in full ((10)) and contains the following intact genes: KIR3DL3*003, KIR2DS2*001, KIR2DL2*003, KIR2DL4*005, KIR3DS1*013, KIR2DL5A*001, KIR2DS5*002, KIR2DS1*002. The presence and integrity of the KIR locus was checked after every backcross by KIR genotyping. C57BL/6 (B6) mice transgenic for genomic HLA-Cw*0304 construct (13) were a kind gift of Eric Vivier (CIML, Marseille, France). To obtain KIR or HLA transgenic mice on a H-2Kb−/− and H-2Db−/− background, KIR+/+, HLA-Cw3+/+ or control B6 mice were crossed with H-2Kb−/−H-2Db−/−β2m−/− (14) mice, also on a B6 background. Mice with the desired phenotype (Kb−Db−β2m+KIR+,Kb−Db−β2m+HLA-Cw3+ or Kb−Db−β2m+) were selected from the F2 and used for further breeding to obtain Kb−/−Db−/−KIR+/+, Kb−/−Db−/−HLA-Cw3+/+ and Kb−/−Db−/− mice. To avoid integration artefacts, mice heterozygous for the KIR and HLA transgenes were used for experiments: KIR+/−, HLACw3+/− and KIR+/−HLA-Cw3+/− mice, all on a Kb−/−Db−/−background. Experiments were approved by the LUMC animal experimental committee, and performed according to local guidelines.
NK cell expression of KIR and Ly49
Mononuclear cells were isolated from spleen using a Ficoll-Hypaque gradient and incubated with fluorescently labelled antibodies. Samples were acquired on a LSRII (BD Biosciences) and analyzed using the FACSDiva software (BD). NK cells were identified as CD3−NK1.1+ cells using a combination of CD3-Pacific Blue (clone 500A2, BD) and NK1.1-PE-Cy7 (clone PK136, BD). For staining of mouse NK receptors, the following FITC-conjugated antibodies were purchased from BD: Ly49A (clone A1), Ly49C/I (clone 5E6), Ly49D (clone 4E5), Ly49G2 (clone 4D11) and NKG2A/C/E (clone 20d5). As B6 NK cells do not express appreciable levels of NKG2C and NKG2E (15), the specificity of the latter antibody was designated as NKG2A. The NKG2A-specific 16a11 antibody (16) conjugated in-house to AlexaFluor647 was also used. KIR2DL4 was also detected using an in-house AlexaFluor647 conjugated antibody (R&D systems). A PE-conjugated antibody to KIR2DL2/KIR2DL3/KIR2DS2 (clone GL183, Coulter Immunotech) was used to detect KIR2DL2/KIR2DS2, since the mouse does not carry the KIR2DL3 gene. HLACw3/GAVDPLLAL tetramers (NIH tetramer facility, etc) were used to selectively stain KIR2DL2, and antibody 1F12 (17) to selectively stain KIR2DS2.
NK responsiveness to crosslinking of Nkrp1c (NK1.1)
Splenic NK cells were stimulated with plate-bound PK136 antibody (specific for NK1.1/Nkrp1c/Klrb1c) for 5 h, with addition of brefeldin A after 1 h, and analyzed for intracellular accumulation of IFNγ (using antibody clone XMG1.2, BD) as described (18,19).
In vivo rejection of target cells
In vivo rejection of CFSE-labelled spleen cells was performed as described (20). Briefly, two different populations of spleen cells, one internal syngeneic control expressing HLA-Cw3 and one lacking HLA-Cw3, were labeled with 0,5 μM and 5 μM CFSE (Invitrogen, Leiden, The Netherlands), respectively, and mixed in a 1:1 ratio. On day 1, 3 and 6 (or 2, 4, 6) after intravenous injection (day 0) of this mixture (cell number), peripheral blood of recipient mice was collected and analyzed by FACS. The relative rejection of HLA-Cw3−/− target cells was calculated as follows: (1-(acquired number of CFSEhigh cells in sample/acquired number of CFSElow cells in sample)/(acquired number of CFSEhigh cells in injection mix/acquired number of CFSElow cells in injection mix)) × 100 %. The HLA-C specific antibody WK4C11 (21) was used to verify HLA-Cw3 expression in donor and recipient mice. In some experiments, NK cells or NKG2A+ cells were depleted by intraperitoneal injection of 200 μg protein A-purified antibody PK136 or 16a11, respectively, in 200 μl PBS on day −4 and day −1.
Results
HLA-Cw3 tetramers stain KIR2DL2
To assess the influence of HLA on KIR expression in a system with minimal genetic variation and minimal influence of endogenous mouse NK cell receptor systems, we compared KIR expression between MHC class I deficient (H-2Kb−/− H-2Db−/−) mice expressing a human KIR locus in the presence or absence of HLA-Cw3. To detect the corresponding inhibitory receptor KIR2DL2, we used HLA-Cw3 tetramers. These should bind KIR2DL2 but not the very similar activating receptor KIR2DS2, since only KIR2DL2 shows measurable binding to HLA-Cw3 (22).
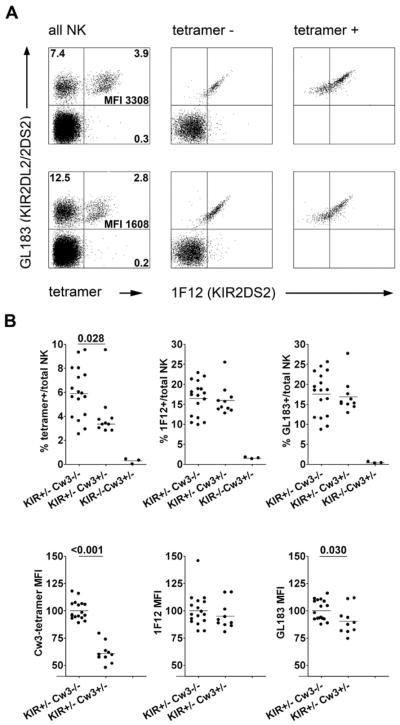
To confirm the specific detection of KIR2DL2 by these tetramers, spleen NK cells were co-stained (Fig. 1A) with HLA-Cw3 tetramer, a KIR2DL2/KIR2DS2-specific antibody (GL183), and a KIR2DS2-specific antibody (1F12). These staining reagents did not interfere with each other's binding (data not shown). The tetramer stained GL183+ but not GL183− NK cells, demonstrating that it did not bind an endogenous mouse receptor or another KIR encoded by the transgene (Fig. 1A). Furthermore, the tetramer bound a subset of GL183+ cells, consistent with the notion that GL183+ cells include both KIR2DL2+ (tetramer+) and KIR2DL2− (tetramer−) cells. The latter population (GL183+tetramer−) cells stained homogeneously with KIR2DS2-antibody 1F12, confirming that the tetramer did not stain KIR2DS2. Finally, tetramer+1F12+ NK cells expressing both KIR2DL2 and KIR2DS2 showed higher staining with GL183 than NK cells expressing KIR2DL2 only (tetramer+1F12−). In conclusion, the HLA-Cw3 tetramer provided a sensitive and specific tool to detect KIR2DL2 expression in the KIR transgenic mice.
Figure 1. The presence of HLA-Cw3 reduces KIR2DL2 expression frequency amd intensity on NK cells from Kb−/−Db−/−KIR+/− mice.
Spleen cells from Kb−/−Db−/− mice transgenic for a KIR B-haplotype and/or HLA-Cw3 were first stained with HLA-Cw3/GAVDPLLAL tetramers and then with antibodies to CD3, NK1.1, KIR2DS2 (1F12) and KIR2DL2/KIR2DS2 (GL183).
(A) Representative dot plots for Kb−/−Db−/−KIR+/− (top) and Kb−/−Db−/−KIR+/−HLA-Cw3+/− (bottom) splenocytes, gated – from left to right - on all NK cells, HLA-Cw3-tetramer-negative or HLA-Cw3-tetramer-positive NK cells. The numbers represent the percentages of cells within each quadrant, as well as the MFI of the tetramer staining (tetramer+ cells only). (B) Summary of four experiments comparing the proportions of NK cells staining with HLA-Cw3-tetramer, KIR2DS2 antibody 1F12 and KIR2DL2/KIR2DS2-antibody GL183 between Kb−/−Db−/−KIR+/−HLA-Cw3−/− (n=17), Kb−/−Db−/−KIR+/−HLA-Cw3+/− (n=10) and Kb−/−Db−/−KIR−/−HLA-Cw3+/− (n=3) mice (top). In each experiment, the mean fluorescence intensity (MFI) values for NK cells staining with these reagents were normalized for the MFI values obtained using Kb−/−Db−/−KIR+/−HLA-Cw3−/− mice before the experiments were pooled (bottom). P values below 0.05 are shown.
HLA-Cw3 reduces the frequency and intensity of NK cell KIR2DL2 expression
We next compared KIR2DL2 expression in the presence or absence of its ligand HLA-Cw3. Consistent with reports in humans, the presence of the HLA-Cw3 transgene reduced the KIR2DL2 mean fluorescence intensity nearly twofold (Fig. 1B), while leaving KIR2DS2 staining unaffected. On average, the frequency of KIR2DL2+ cells within NK cells was also approximately twofold reduced (Fig. 1B), and again no such effect was seen for KIR2DS2. As previously reported for these same KIR transgenic mice on a wild-type C57BL/6 background (12), GL183 (KIR2DL2/KIR2DS2) staining was not significantly different between HLA-Cw3− and HLA-Cw3+ mice. This was due to the fact that the majority of GL183+ NK cells expressed KIR2DS2, while only a minority expressed KIR2DL2 (Fig. 1B). Thus, HLA-Cw3 reduced the frequency as well as the intensity of NK cell KIR2DL2 expression.
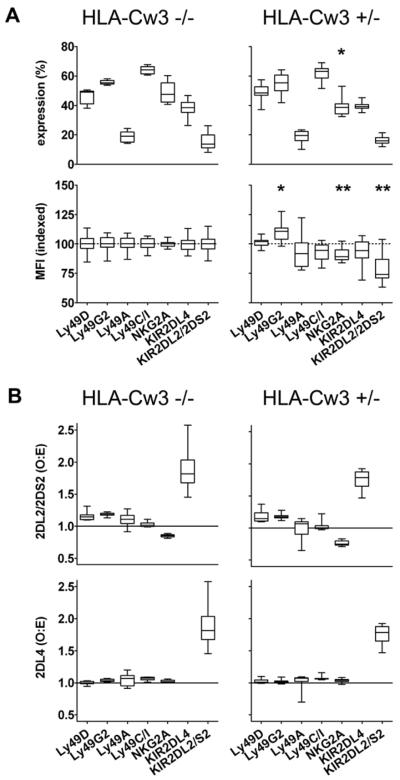
The introduction of HLA-Cw3 did not alter the expression frequencies of mouse Ly49 receptors (Fig. 2A), but was associated with a slight, but significant, reduction in the expression frequency and intensity of NKG2A (Fig. 2A), an inhibitory mouse receptor for the MHC class Ib molecule Qa-1. Using another NKG2A-specific antibody (16a11, ref. 16) in an additional set of 17 Kb−/−Db−/−KIR+/−HLA-Cw3−/− and 10 Kb−/−Db−/−KIR+/−HLA-Cw3+/− mice, this HLA-Cw3 effect on NKG2A expression frequency (p=0.0005) and intensity (p=0.0024) was confirmed (data not shown). The expression of KIR2DL4, a (putative) HLA-G or bacterial CpG receptor also encoded by the KIR transgene, was unaffected by HLA-Cw3. In general, individual KIR and Ly49 genes are expressed largely independently of other KIR and Ly49 genes. Yet, in our mice KIR2DL4 was preferentially co-expressed on the same cell with KIR2DL2/KIR2DS2, and the opposite was true for NKG2A and KIR2DL2/KIR2DS2 (Fig. 2B). These co-expression biases were unaffected by the presence of HLA-Cw3. In summary, HLA-Cw3 did not influence expression of mouse Ly49 receptors or another non-HLA-Cw3-specific KIR encoded by the transgene, but did significantly reduce NKG2A expression frequency and intensity.
Figure 2. The presence of HLA-Cw3 reduces mouse NKG2A expression frequency and intensity on NK cells from Kb−/−Db−/−KIR+/− mice.
Spleen cells from Kb−/−Db−/−KIR+/− mice transgenic (HLA-Cw3+/−) or not (HLA-Cw3−/−) for HLA-Cw3 were stained with antibodies to CD3, NK1.1, KIR2DL2/2DS2 and KIR2DL4 in combination with antibodies directed against mouse Ly49 receptors or NKG2A.
(A) Frequency of surface expression of individual mouse (Ly49s, NKG2A) or human (KIR2DL4, KIR2DL2/KIR2DS2) NK receptors by spleen NK (CD3−NK1.1+) cells from Kb−/−Db−/−KIR+/−HLA-Cw3−/− and Kb−/−Db−/−KIR+/−HLA-Cw3+/− mice. The mean fluorescence intensity (MFI) values for NK cells positive for individual receptors were normalized for the MFI values obtained using Kb−/−Db−/−KIR+/−HLA-Cw3−/− mice as in Fig. 1B.
(B) Co-expression of transgenic KIR2DL2/KIR2DS2 (top) or KIR2DL4 (bottom) with endogenous mouse NK receptors was quantified in terms of deviation from the `product rule' (2). O represents the observed frequency of cells co-expressing KIR2DL2/KIR2DS2 or KIR2DL4 and a particular mouse NK receptor among NK cells, and E represents the product of the individual expression frequencies of these human and mouse receptors on NK cells, i.e. the expected frequency of cells expressing these receptors. Co-expression values for KIR2DL2/KIR2DS2 and KIR2DL4 were also calculated.
Data are from 8 Kb−/−Db−/−KIR+/−HLA-Cw3−/− and 8 Kb−/−Db−/−KIR+/−HLA-Cw3+/− mice. Horizontal bars represent median values, boxes extend from the 25th to the 75th percentile and whiskers represent the total range of the measurements. For each receptor, the results from HLA-Cw3+/− compared to HLA-Cw3−/− mice were compared using a 2-sided Student's t-test, without correcting for multiple comparisons: * indicates a p value <0.05, ** a p value <0.005.
HLA-Cw3 increases responsiveness of KIR2DL2/KIR2DS2+ NK cells
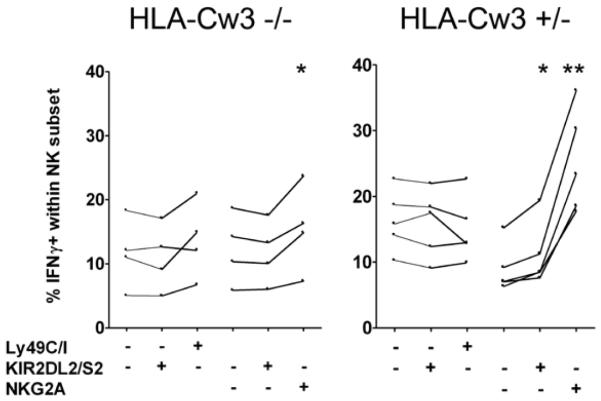
NK cells expressing inhibitory receptors that bind endogenous MHC class I are more responsive to activating stimuli than NK cells that do not express such `useful' inhibitory receptors (5,18,19). Hence, the presence of HLA-Cw3 might also influence the potency of NK cells expressing a HLA-Cw3-specific inhibitory receptor. In mice lacking both mouse and human MHC class Ia molecules (Kb, Db and HLA-Cw3), NK cells were poorly responsive to NK1.1-crosslinking irrespective of the receptors they carried (Fig. 3). In the presence of HLA-Cw3, KIR2DL2/KIR2DS2+ NK cells produced slightly more IFNγ than KIR2DL2/KIR2DS2− NK cells. In contrast, the responsiveness of NKG2A+ NK cells was greatly increased in the presence of HLA-Cw3, suggesting that NKG2A+ NK cells are also educated by HLA-Cw3 in this model system.
Figure 3. The presence of HLA-Cw3 increases the responsiveness of KIR2DL2/2DS2+ as well as NKG2A+ NK cells.
Spleen cells from Kb−/−Db−/−KIR+/−HLA-Cw3−/− (HLA-Cw3−/−) and Kb−/−Db−/−KIR+/−HLA-Cw3+/− (HLA-Cw3+/−) mice were stimulated with plate-bound anti-NK1.1 for 5 hours, and accumulation of intracellular IFNγ in NK cells (CD3−DX5+) was subsequently analyzed by flowcytometry. NK cells were subdivided according to their expression of KIR2DL2/KIR2DS2, of which the inhibitory KIR2DL2 binds HLA-Cw3, and their expression of inhibitory mouse receptors binding mouse MHC class I. Ly49C and Ly49I both bind H-2Kb (absent from the mice) and NKG2A binds Qa-1. Data are from 2 experiments using a total of 4 Kb−/−Db−/−KIR+/−HLA-Cw3−/− and 5 Kb−/−Db−/−KIR+/−HLA-Cw3+/− mice. Lines connect data from individual mice. For each receptor combination, the results were compared between cells expressing a particular receptor (e.g. KIR2DL2/S2+NKG2A−) and those lacking that receptor (e.g. KIR2DL2/S2−NKG2A−), using a paired 2-sided Student's t-test, without correcting for multiple comparisons: * indicates a p value <0.05, ** a p value <0.005.
KIR-dependent rejection of `missing self' HLA-Cw3 in vivo
To test whether KIR and HLA mediated missing self recognition in these mice, we analyzed the rejection of Kb−/−Db−/− spleen cells by KIR and HLA transgenic Kb−/−Db−/− mice (Fig. 4). Missing self rejection was tested using an in vivo assay based on differential labeling of donor cells using the CFSE dye (20). Mixed CFSEhigh Kb−/−Db−/− and control CFSElo Kb−/−Db−/−HLA-Cw3+/− spleen cells were injected intravenously into Kb−/−Db−/−KIR+/−HLA-Cw3+/− or control Kb−/−Db−/−KIR−/−HLA-Cw3+/− mice. Both types of recipient mice swiftly rejected about 80 % of HLA-Cw3-negative Kb−/−Db−/− target cells (Fig 4A). The presence of the KIR transgene only marginally improved rejection, showing that KIR were not necessary for rejection.
Figure 4. In KIR and HLA-Cw3 transgenic Kb−/−Db−/− mice, KIR and NKG2A contribute to the rejection of Kb−/−Db−/− grafts.
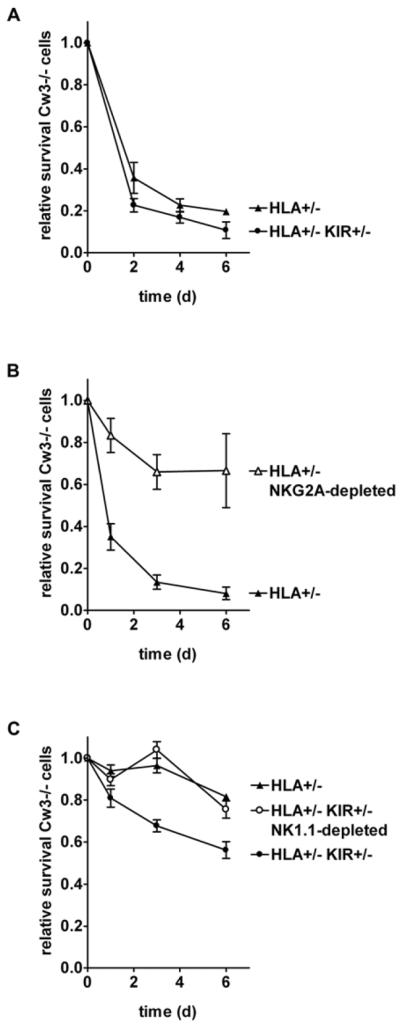
Kb−/−Db−/−KIR+/−HLA-Cw3+/− (HLA+/− KIR+/−) or control Kb−/−Db−/−KIR−/−HLA-Cw3+/− (HLA+/−) mice were injected intravenously with mixed CFSE-labeled Kb−/−Db−/− (CFSEhi) and control Kb−/−Db−/−HLA-Cw3+/− (CFSElo) spleen cells. The relative survival of CFSEhi (Cw3−/−) cells in peripheral blood, normalized for the CFSEhi/CFSElo ratio in the injected cells, was tracked in (A) non-depleted Kb−/−Db−/−KIR+/−HLA-Cw3+/− (n=4) or control Kb−/−Db−/−KIR−/−HLA-Cw3+/− (n=3) mice, in (B) non-depleted (n=4) versus 16a11(NKG2A)-depleted (n=3) Kb−/−Db−/−KIR−/−HLA-Cw3+/− mice and in (C) 16a11-depleted Kb−/−Db−/−KIR+/−HLA-Cw3+/− (n=5), 16a11-depleted Kb−/−Db−/−KIR−/−HLA-Cw3+/− (n=5), and in 16a11- and PK136(NK1.1)-depleted Kb−/−Db−/−KIR+/−HLA-Cw3+/− (n=5) mice.
Since the presence of HLA-Cw3 affected NK cell NKG2A expression levels as well as the frequency and functionality of NKG2A+ NK cells, we next tested whether these cells contributed to rejection (Fig. 4B). In Kb−/−Db−/−HLA-Cw3+/− mice, depletion of NKG2A+ cells before and during the experiment indeed greatly reduced the rejection of Kb−/−Db−/− cells. To isolate the effect of KIR on rejection, the fate of injected Kb−/−Db−/− cells was compared between NKG2A-depleted Kb−/−Db−/−HLA-Cw3+/− mice having or lacking the KIR transgene (Fig. 4C). Rejection was significantly greater in the mice carrying the KIR transgene (Fig. 4C), but only approximately half that of non-depleted mice (Fig. 4B). Additional depletion of NK cells in these KIR transgenic mice reduced rejection to the level of control KIR-less mice, supporting the idea that all KIR-dependent rejection in NKG2A-depleted mice was mediated by NK cells. In conclusion, the mouse CD94/NKG2A receptor dominated the `missing HLA' response in KIR and HLA transgenic mice, and only upon depletion of NKG2A+ NK cells did KIR-mediated rejection become apparent.
Discussion
We used a humanized mouse model to investigate the effect of HLA on KIR repertoire and function. In this MHC class I-deficient (Kb−/−Db−/−) model system, the presence of HLA-Cw3 reduced both the surface expression of KIR2DL2 as well as the proportion of KIR2DL2+ cells. In addition, HLA-Cw3 influenced the expression frequency and intensity of NKG2A. In line with these observations, both KIR and NKG2A contributed to the rejection of `missing self' target cells lacking HLA-Cw3.
Studies on human NK cell repertoires in most cases showed no HLA effect on KIR expression frequencies (3,4,7,9), except in very specific circumstances. For example, in individuals homozygous for specific inhibitory KIR binding their ligand with high affinity (KIR2DL1 or KIR3DL1*001/KIR3DL1*015/KIR3DL1*020) the presence of ligand was associated with increased frequencies of NK cells expressing these receptors, but only in the absence of too many additional inhibitory KIR-ligand interactions (6,7). A similar, albeit less pronounced, effect was observed for KIR2DL3 and C1 (6). These effects were detected in individuals homozygous for KIR A-haplotypes, characterized by the absence of KIR2DL2, KIR2DL5 and most activating receptors (KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5, KIR3DS1). In Caucasoids, such A-homozygous individuals make up less than half of the population.
KIR repertoires in individuals carrying KIR B-haplotypes have been more difficult to study, mainly due to the fact that antibodies specific for inhibitory KIR crossreact with activating KIR present in B- but not A-haplotypes. Group 1 HLA-C effects on KIR2DL2 expression frequencies have been particularly difficult to detect, not only because the available antibodies crossreact with the activating KIR2DS2, nearly always present on the same haplotype, but also because KIR2DL2 also binds group 2 HLA-C alleles (23). These problems were circumvented in our humanized mice, since we were able to use HLA-C tetramers to detect specifically KIR2DL2, and since we compared mice lacking or having an HLA-C group 1 allele. In these mice, the presence of HLA-Cw3 decreased the frequency of KIR2DL2+ NK cells. Thus, in this model system with a genetically homogeneous background, the presence of an HLA ligand clearly did influence the expression frequency of the corresponding inhibitory KIR
In the Kb−/−Db−/− mice transgenic for KIR and HLA-Cw3, NKG2A+ cells contributed to the rejection of `missing HLA-Cw3'. In line with this finding, the introduction of HLA-Cw3 in KIR transgenic Kb−/−Db−/− mice affected the expression of NKG2A, as well as the responsiveness of NKG2A+ NK cells. Importantly, Kb−/−Db−/− mice transgenic for HLA-Cw3 only already rejected Kb−/−Db−/− target cells, and this rejection was inhibited by antibody-mediated depletion of NKG2A+ cells. NKG2A is an inhibitory receptor binding Qa-1, whose surface expression in C57BL/6 mice largely depends on its loading with the H-2Db leader peptide AMAPRTLLL. The very similar HLA-Cw3 leader peptide VMAPRTLIL also binds Qa-1b and thereby induces a functional ligand for CD94/NKG2A in Kb−/−Db−/− mice (24,25). Thus, the recognition by CD94/NKG2A of HLA-Cw3 leader peptides bound to Qa-1b likely contributes to the education, selection and function of the NK cell repertoire in HLA-Cw3 transgenic Kb−/−Db−/− mice, irrespective of the presence of KIR. Therefore, NKG2A may also have contributed to the rejection of `missing HLA-Cw3' targets in the KIR2DL3 and HLA-Cw3 transgenic Kb−/−Db−/− mice described by Sola et al. (26).
Upon the deletion of NKG2A+ cells, KIR-dependent rejection of `missing HLA-Cw3' by KIR and HLA-Cw3 transgenic Kb−/−Db−/− mice was considerably greater than we observed previously in KIR and HLA-Cw3 transgenic mice on a wild-type C57BL/6 background (12). This is reminiscent of the experiments of Johansson et al., who found that the education of NK cells by weak Ly49 ligands was attenuated by the presence of strong Ly49 ligands (27). Hence, in the transgenic mice on a wild-type C57BL/6 background, the presence of the strong interaction between Ly49C and H-Kb may have obscured the detection of `missing HLA-Cw3'. In agreement with the differences in KIR-mediated rejection, the presence of KIR2DL2/KIR2DS2 on NK cells did not detectably increase their responsiveness in mice on a C57BL/6 background (12), but on a Kb−/−Db−/− background a small but significant increase in responsiveness was detected. Since only a third of KIR2DL2/KIR2DS2+ NK cells expressed KIR2DL2, this weak response may have been due to dilution rather than poor responsiveness of KIR2DL2+ NK cells. As in our hands HLA-Cw3 tetramer stains did not work in combination with intracellular IFNγ staining, we were unable to discriminate between these possibilities.
In heavily T-cell depleted haploidentical stem cell transplantations of AML patients, the absence of donor KIR ligands from the patient was associated with graft-versus-host NK cell alloreactivity, with improved engraftment and with reduced relapse rates (28,29). In analogy with this finding, the engraftment of sublethally irradiated mice transplanted with fully MHC-mismatched bone marrow was greatly improved by the infusion of alloreactive donor NK cells (29), suggesting that the beneficial effects in human transplantation were mediated by donor NK cells. We performed a series of experiments to test whether we could reproduce these results in our humanized mouse model. In our hands, the infusion of 1–2 million IL-15 activated and NKG2A-depleted donor NK cells did not improve engraftment of a suboptimal dose of KIR and HLA-Cw3 transgenic Kb−/−Db−/− bone marrow in sublethally irradiated Kb−/−Db−/− mice, compared to control NKG2A-depleted NK cells from KIR but not HLA-Cw3 transgenic Kb−/−Db−/− mice (data not shown). This may be due to the low frequency of NK cells able to detect missing HLA-Cw3 (< 5 % of total NK are KIR2DL2+) as well as weaker alloreactivity mediated by KIR2DL2 compared to Ly49 receptors. Therefore, to obtain beneficial effects of alloreactive NK cells in the clinic, the infusion of large numbers of NK cells recognizing a strong KIR-ligand mismatch may be required.
Our data show that in a setting with minimal variation in genetic background, a functional interaction between the products of a single inhibitory KIR allele and a single HLA allele reduces both the surface expression levels of that KIR as well as the fraction of NK cells expressing it. This finding supports the idea that HLA influences the human NK cell receptor repertoire.
Acknowledgements
The authors are grateful to the NIH tetramer facility for providing HLA-Cw3 tetramers and to Michel Mulders, Suzanne van Duikeren and Arie Boon for assistance with animal experiments.
Abbreviations used in this paper
- KIR
Killer Immunoglobulin Receptor(s)
- B6
C57BL/6
Footnotes
This work was supported by Landsteiner grant 0515 (to J.v.B.)
References
- 1.Hoglund P, Brodin P. Current perspectives of natural killer cell education by MHC class I molecules. Nat. Rev. Immunol. 2010;10:724–734. doi: 10.1038/nri2835. [DOI] [PubMed] [Google Scholar]
- 2.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D'Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 3.Gumperz JE, Valiante NM, Parham P, Lanier LL, Tyan D. Heterogeneous phenotypes of expression of the NKB1 natural killer cell class I receptor among individuals of different human histocompatibility leukocyte antigens types appear genetically regulated, but not linked to major histocompatibililty complex haplotype. J. Exp. Med. 1996;183:1817–1827. doi: 10.1084/jem.183.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shilling HG, Young N, Guethlein LA, Cheng NW, Gardiner CM, Tyan D, Parham P. Genetic control of human NK cell repertoire. J. Immunol. 2002;169:239–247. doi: 10.4049/jimmunol.169.1.239. [DOI] [PubMed] [Google Scholar]
- 5.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagne F, Ugolini S, Vivier E. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Schonberg K, Sribar M, Enczmann J, Fischer JC, Uhrberg M. Analyses of HLA-C-specific KIR repertoires in donors with group A and B haplotypes suggest a ligand-instructed model of NK cell receptor acquisition. Blood. 2011;117:98–107. doi: 10.1182/blood-2010-03-273656. [DOI] [PubMed] [Google Scholar]
- 7.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson S, Fauriat C, Malmberg JA, Ljunggren HG, Malmberg KJ. KIR acquisition probabilities are independent of self-HLA class I ligands and increase with cellular KIR expression. Blood. 2009;114:95–104. doi: 10.1182/blood-2008-10-184549. [DOI] [PubMed] [Google Scholar]
- 10.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belkin D, Torkar M, Chang C, Barten R, Tolaini M, Haude A, Allen R, Wilson MJ, Kioussis D, Trowsdale J. Killer cell Ig-like receptor and leukocyte Ig-like receptor transgenic mice exhibit tissue- and cell-specific transgene expression. J. Immunol. 2003;171:3056–3063. doi: 10.4049/jimmunol.171.6.3056. [DOI] [PubMed] [Google Scholar]
- 12.Van Bergen J, Thompson A, Retiere C, Trowsdale J, Koning F. Cutting edge: killer Ig-like receptors mediate “missing self” recognition in vivo. J. Immunol. 2009;182:2569–2572. doi: 10.4049/jimmunol.0804042. [DOI] [PubMed] [Google Scholar]
- 13.Dill O, Kievits F, Koch S, Ivanyi P, Hammerling GJ. Immunological function of HLA-C antigens in HLA-Cw3 transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 1988;85:5664–5668. doi: 10.1073/pnas.85.15.5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grigoriadou K, Menard C, Perarnau B, Lemonnier FA. MHC class Ia molecules alone control NK-mediated bone marrow graft rejection. Eur. J. Immunol. 1999;29:3683–3690. doi: 10.1002/(SICI)1521-4141(199911)29:11<3683::AID-IMMU3683>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 15.Vance RE, Jamieson AM, Raulet DH. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J. Exp. Med. 1999;190:1801–1812. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vance RE, Jamieson AM, Cado D, Raulet DH. Implications of CD94 deficiency and monoallelic NKG2A expression for natural killer cell development and repertoire formation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:868–873. doi: 10.1073/pnas.022500599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David G, Morvan M, Gagne K, Kerdudou N, Willem C, Devys A, Bonneville M, Follea G, Bignon JD, Retiere C. Discrimination between the main activating and inhibitory killer cell immunoglobulin-like receptor positive natural killer cell subsets using newly characterized monoclonal antibodies. Immunology. 2009;128:172–184. doi: 10.1111/j.1365-2567.2009.03085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieve self-tolerance without expressing inhibitory receptors specific for self MHC molecules. Blood. 2005;105:4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, French AR, Sunwoo JB, Lemieux S, Hansen TH, Yokoyama WM. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 20.Oberg L, Johansson S, Michaelsson J, Tomasello E, Vivier E, Karre K, Hoglund P. Loss or mismatch of MHC class I is sufficient to trigger NK cell-mediated rejection of resting lymphocytes in vivo - role of KARAP/DAP12-dependent and -independent pathways. Eur. J. Immunol. 2004;34:1646–1653. doi: 10.1002/eji.200424913. [DOI] [PubMed] [Google Scholar]
- 21.Zoet YM, Eijsink C, Bohmova R, Witvliet MD, Kardol MJ, Franke ME, Claas FH, Mulder A, Doxiadis II. Single-antigen-expressing cell lines are excellent tools for detecting human leukocyte antigen-C-reactive antibodies in kidney transplant recipients. Transplantation. 2005;79:1268–1272. doi: 10.1097/01.tp.0000161246.33019.b6. [DOI] [PubMed] [Google Scholar]
- 22.Vales-Gomez M, Reyburn HT, Erskine RA, Strominger J. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J. Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 24.Kurepa Z, Hasemann CA, Forman J. Qa-1b binds conserved class I leader peptides derived from several mammalian species. J. Exp. Med. 1998;188:973–978. doi: 10.1084/jem.188.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sivakumar PV, Gunturi A, Salcedo M, Schatzle JD, Lai WC, Kurepa Z, Pitcher L, Seaman MS, Lemonnier FA, Bennett M, Forman J, Kumar V. Cutting edge: expression of functional CD94/NKG2A inhibitory receptors on fetal NK1.1+Ly-49- cells: a possible mechanism of tolerance during NK cell development. J. Immunol. 1999;162:6976–6980. [PubMed] [Google Scholar]
- 26.Sola C, Andre P, Lemmers C, Fuseri N, Bonnafous C, Blery M, Wagtmann NR, Romagne F, Vivier E, Ugolini S. Genetic and antibody-mediated reprogramming of natural killer cell missing-self recognition in vivo. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12879–12884. doi: 10.1073/pnas.0901653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson S, Johansson M, Rosmaraki E, Vahlne G, Mehr R, Salmon-Divon M, Lemonnier F, Karre K, Hoglund P. Natural killer cell education in mice with single or multiple major histocompatibility complex class I molecules. J. Exp. Med. 2005;201:1145–1155. doi: 10.1084/jem.20050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruggeri L, Mancusi A, Capanni M, Urbani E, Carotti A, Aloisi T, Stern M, Pende D, Perruccio K, Burchielli E, Topini F, Bianchi E, Aversa F, Martelli MF, Velardi A. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood. 2007;110:433–440. doi: 10.1182/blood-2006-07-038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, Posati S, Rogaia D, Frassoni F, Aversa F, Martelli MF, Velardi A. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]