Hill et al. performed complementary systematic screens in search of new BRCA1 protein-interacting partners. From these results, a new BRCA1 function in the response to transcription-associated DNA damage was detected, specifically in the restart of transcription after UV damage and in preventing or repairing damage caused by stabilized R loops. This new function may be important in BRCA1 tumor suppression, since the expression of several interactors is repeatedly aberrant in both breast and ovarian cancers.
Keywords: BRCA1 interaction screening, DNA damage, transcription
Abstract
BRCA1 is a breast and ovarian tumor suppressor. Given its numerous incompletely understood functions and the possibility that more exist, we performed complementary systematic screens in search of new BRCA1 protein-interacting partners. New BRCA1 functions and/or a better understanding of existing ones were sought. Among the new interacting proteins identified, genetic interactions were detected between BRCA1 and four of the interactors: TONSL, SETX, TCEANC, and TCEA2. Genetic interactions were also detected between BRCA1 and certain interactors of TONSL, including both members of the FACT complex. From these results, a new BRCA1 function in the response to transcription-associated DNA damage was detected. Specifically, new roles for BRCA1 in the restart of transcription after UV damage and in preventing or repairing damage caused by stabilized R loops were identified. These roles are likely carried out together with some of the newly identified interactors. This new function may be important in BRCA1 tumor suppression, since the expression of several interactors, including some of the above-noted transcription proteins, is repeatedly aberrant in both breast and ovarian cancers.
BRCA1 is a breast and ovarian tumor suppressor gene (Miki et al. 1994), and its full-length protein product, p220, is a genome integrity maintenance protein. Its validated functions include, but are not limited to, its role in the repair of double-strand DNA breaks (DSBs) by homologous recombination (HR). However, how and with what protein-binding partners BRCA1 executes its molecular and tumor suppression functions are incompletely understood. Gaining a better understanding of these processes will be beneficial in conceiving new therapies for BRCA1 and BRCA1-like cancers.
Success in gaining a better understanding of the functions of certain proteins has been achieved through systematic mapping of their protein interaction networks (Rual et al. 2005; Stelzl et al. 2005), like that performed for a group of tumor viral oncoproteins (Rozenblatt-Rosen et al. 2012). Focused analysis has identified such BRCA1 and BRCA2 interactors as BARD1 and PALB2 (Wu et al. 1996; Xia et al. 2006), while network analysis (Pujana et al. 2007), an immunoprecipitation-based study (Wang et al. 2000), and a yeast two-hybrid (Y2H) and mass spectrometry (MS)-based analysis of a BRCA1 functional motif have identified yet other BRCA1-interacting proteins (Woods et al. 2012). However, gaps in the BRCA1 network likely remain, given the limited understanding of BRCA1 function.
Therefore, we carried out protein–protein interaction screens using two complementary methodologies to search for new BRCA1-interacting partners (Fig. 1A). A primary goal was to gain evidence of new functions for BRCA1 based on known activities of newly detected interacting proteins (also known as interactors). Another was to identify new cancer genes or therapeutically targetable signal transduction pathways served by newly identified interacting proteins.
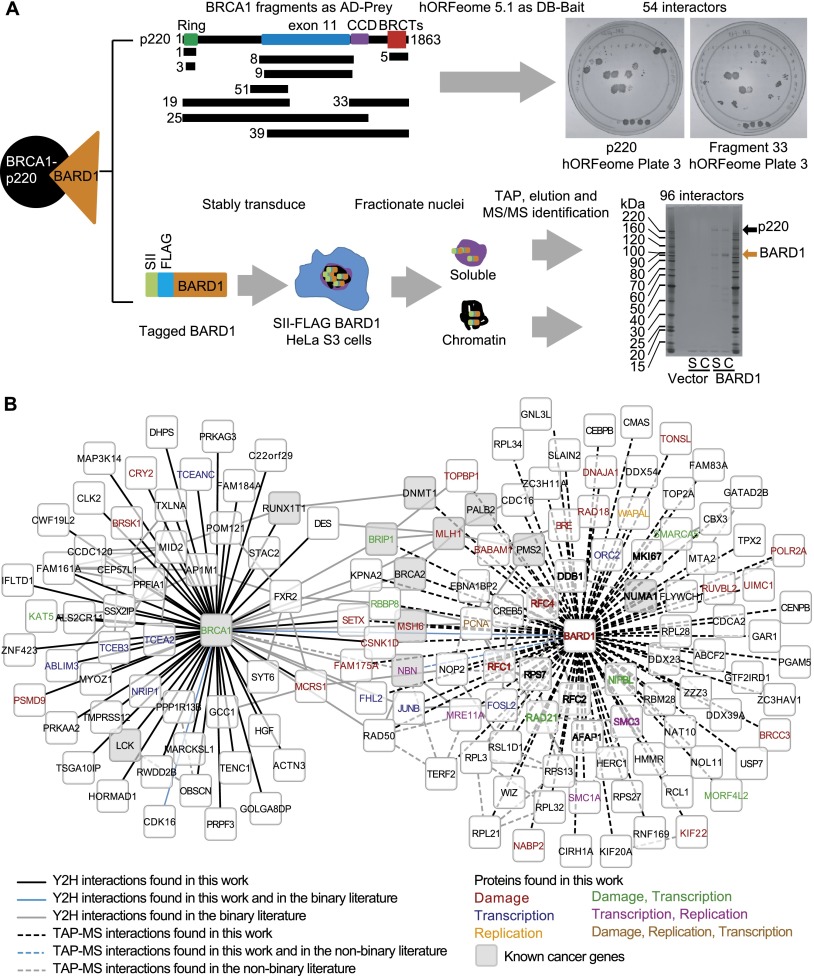
Figure 1.
Bipartite screening effort to identify new protein-interacting partners for BRCA1 (see also Supplemental Figs. S1, S2A–H; Supplemental Tables S1–S5). (A) The Y2H and TAP-MS screening methods used are outlined here. In the Y2H screen, depicted at the top, full-length BRCA1 (p220) and fragments strategically designed to contain specific BRCA1 functional domains (shown in the map) were screened against the human ORFeome version 5.1. The two example plates shown represent full-length BRCA1 and fragment 33 screened against the same 94 members of the ORFeome. The TAP-MS operation sequence is depicted at the bottom. Nuclei were harvested from HeLa S3 cells stably expressing Flag-StrepTactin (SII)-tagged BARD1 or empty vector as a control. The nuclei were fractionated into soluble nuclear and chromatin fractions from which the complexes were tandemly immunoprecipitated. A silver-stained gel depicting a small fraction of one of the three purifications is shown at the bottom of the panel. (S) Soluble fraction; (C) chromatin fraction. (B) This network represents BARD1 and BRCA1 as the central nodes for TAP-MS associations and direct biophysical Y2H interactions, respectively, with all interactors from the screen emanating from them. The code for edge color and style as well as node color is defined in the key below the network.
Results
Bipartite screening effort identifies new BRCA1-interacting partners
The bipartite screening effort involved two complementary approaches. The first consisted of a systematic binary screen using both full-length BRCA1 protein (also known as p220) and strategically designed fragments thereof (Supplemental Table S1) tested against the gene products present in the human ORFeome version 5.1 collection in a high-throughput Y2H format (Dreze et al. 2010). We screened BRCA1 only as prey, since the BRCT domain of this protein acted as an autoactivator. The complementary screen was a tandem affinity purification (TAP) followed by MS analysis (TAP-MS) of BARD1–BRCA1 complexes isolated from the nuclei of HeLa S3 cells (Fig. 1A). BARD1 is the heterodimeric binding partner of BRCA1 and was used as bait because expression of BRCA1 at levels high enough for TAP-MS is toxic to cells.
In the TAP-MS screen, we identified 96 unique interactors (Supplemental Table S2). From the Y2H screen, we identified 54 unique interactors (Supplemental Table S3). Since some of the Y2H interactors bound to multiple overlapping BRCA1 fragments, we were able to map these interactions to specific domains of BRCA1 (Supplemental Fig. S1A; Supplemental Table S3). For example, based on the BRCA1 fragments with which it interacted, we were able to predict that a newly detected BRCA1 interactor, SETX, binds to the C terminus of BRCA1 (Fig. 1A; Supplemental Fig. S1A; Supplemental Table S3). We also predict that the N terminus of SETX is important for it to interact with BRCA1, since only full-length SETX and not a shorter C-terminal fragment interacted with BRCA1 (Supplemental Fig. S1A). Knowing the BRCA1 domain with which proteins interact could be important in predicting the subtype of disease or treatment strategies that can be deployed in association with a BRCA1 mutation that alters a highly specific BRCA1–protein X interaction.
The overall quality of the binary Y2H BRCA1 protein interaction network was evaluated using an empirical framework approach (Venkatesan et al. 2009) in which full-length BRCA1 and all of its Y2H-interacting partners were tested in the orthogonal split Gaussia princeps luciferase complementation assay (GPCA) (Cassonnet et al. 2011). Under conditions where none of our negative control pairs were detected, ∼35% of verified BRCA1 Y2H pairs tested positive in this assay, which is within the expected range of binary assay sensitivity (Braun et al. 2009; Venkatesan et al. 2009). This demonstrates the high quality of the observed Y2H interactions. Coimmunoprecipitation was also performed on a select subset of exogenously overexpressed interactors from both modalities, and we observed frequent coprecipitation with either exogenously overexpressed or endogenous BRCA1 (Supplemental Figs. S2A–H).
In total, 147 interactors were identified in the combined screening effort (Supplemental Table S4). The overlap between the methodologies was small but significant (three common network edges: BARD1, CSNK1D, and SETX; P = 0.002) and within the range expected based on the results of previous dual screens (Rozenblatt-Rosen et al. 2012). The 147 interactors were grouped into a network in which the central nodes are BRCA1 and BARD1 (Fig. 1B). Twenty-five of these interactions had been previously detected as physical interactors in other screening efforts (Wang et al. 2000; Pujana et al. 2007; Woods et al. 2012). The other 122 were novel. In addition, 47 of the hits were identified as potential cancer genes in systematic cancer gene screening efforts (Beroukhim et al. 2010; Rozenblatt-Rosen et al. 2012; T Rolland, M Taşan, B Charloteaux, SJ Pevzner, N Sahni, Q Zhong, S Yi, I Lemmens, C Fontanillo, R Mosca, et al., in prep.), and 12 are present in two large cancer gene lists (the overlap between our 147 hits and these two cancer gene lists was significant; P < 0.001) (Fig. 1B; Supplemental Fig. S1B; Supplemental Table S5; Futreal et al. 2004; Vogelstein et al. 2013).
We queried the gene ontology (GO) term (Ashburner et al. 2000) association of these interactors and found an enrichment (47 of the 147 hits) for proteins involved in DNA damage repair, replication, and transcription among other functions (all highlighted in the network, with the gene symbol being selectively colored to reflect various GO term associations as indicated in the key in Fig. 1B; Supplemental Fig. S1B; Supplemental Table S5). BRCA1 is already known to participate in the first two functions, although the mechanisms by which it operates in these settings are not completely understood. BRCA1 has been indirectly linked to transcription and/or mRNA processing through its interactions with BARD1 and RNA polymerase II (Pol II) (Scully et al. 1997). The functional relevance of the interaction with RNA Pol II is still unclear, but BARD1 has been linked to mRNA processing and transcription-associated damage, thereby suggesting a role for BRCA1 in these processes as well (Kleiman and Manley 1999, 2001; Kleiman et al. 2005).
BARD1 interacts with the polyadenylation factor CstF50 to form a complex that inhibits polyadenylation of mRNA transcripts (Kleiman and Manley 1999). UV- and HU-induced DNA damage leads to an increase in the inhibition of 3′ mRNA processing by this complex (Kleiman and Manley 2001). Although the protein levels do not change after UV exposure, the amount of BRCA1, BARD1, and CstF50 that interacts increases after UV treatment. Moreover, at least the BARD1–CstF50 complex is necessary for the inhibition of 3′ mRNA processing observed after UV damage (Kleiman and Manley 2001). This inhibition of processing likely results from the ubiquitination of active RNA Pol II, as triggered directly or indirectly, by the BRCA1–BARD1 complex and its subsequent degradation (Kleiman et al. 2005). The inhibition of mRNA processing in each of these studies was BARD1-dependent. Nevertheless, the close association between BRCA1 and BARD1 suggests that there may be a role for BRCA1 in this process as well, even if it is simply to enhance the E3 ligase activity of BARD1. In addition, BRCA1 has been speculated to play some unidentified role in transcription and transcription-coupled repair (Scully et al. 1997; Le Page et al. 2000). However, the relevance of the RNA Pol II interaction and the link between BRCA1 and transcription is mechanistically undefined. Therefore, insights into such a BRCA1 function would be valuable.
Defective RNA processing or halting of transcription may arise from loss of function of relevant RNA-binding proteins, physiological transcription-modulating sites (e.g., pause sites), or exogenous lesions in the DNA. All can lead to DNA damage. This can occur through multiple mechanisms. One is through the stabilization of R loops, which are normal physiologic structures that consist of the three-stranded nucleic acid structure formed by the nontranscribed DNA strand when a nascent RNA transcript anneals to the transcribed DNA strand, leaving the nontranscribed single strand to bubble out and become susceptible to damage if this structure is stabilized for prolonged periods (Li and Manley 2006; Mischo et al. 2011; Skourti-Stathaki et al. 2011; Aguilera and Garcia-Muse 2012). BRCA1 and BRCA2 have recently been implicated in suppressing the development of R loops, and BRCA2 appears to be important in suppressing the DNA damage associated with them (Bhatia et al. 2014). Transcription-associated damage can also occur through the damaging of DNA opened up for transcription by drugs or other exogenous stressors, the collision of active transcription complexes and replication forks, and/or the collapse of stalled transcription complexes (Paulsen et al. 2009; Aguilera and Garcia-Muse 2012; Helmrich et al. 2013).
BRCA1 has already been linked to transcription through its interaction with RNA Pol II (Scully et al. 1997) and to mRNA processing in its binding to BARD1 (Kleiman and Manley 1999, 2001; Kleiman et al. 2005). Therefore, because of the enrichment of transcription and transcription DNA damage proteins in our network and because it is primarily a DNA damage response protein, we hypothesized that BRCA1 plays a significant role in the prevention or repair of DNA damage associated with transcription arrest and/or RNA processing along with some of the proteins in our BRCA1 interactor data set.
BRCA1 depletion leads to increased sensitivity of cells to transcription arrest
To test the validity of this hypothesis, we asked whether BRCA1 depletion leads to increased cell sensitivity to the development of DNA damage that arises in the setting of transcription arrest induced by certain compounds. We found that depletion of BRCA1 led to increased sensitivity to two compounds, each known to halt transcription and induce DNA damage: 5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside (DRB) (Fig. 2A,B; Supplemental Figs. S2I, S3A; Chodosh et al. 1989) and α-amanitin (Fig. 2C,D; Supplemental Figs. S2I, S3B; Lindell et al. 1970). We verified that treatment of cells with these compounds does cause DNA damage by demonstrating that a 24-h exposure to them leads to an increased number of cells with DNA damage marked by γH2AX foci (Fig. 2B,D). We also demonstrated that the DNA damage caused by α-amanitin is exacerbated in the setting of BRCA1 depletion (Fig. 2E; Supplemental Fig. S2I).
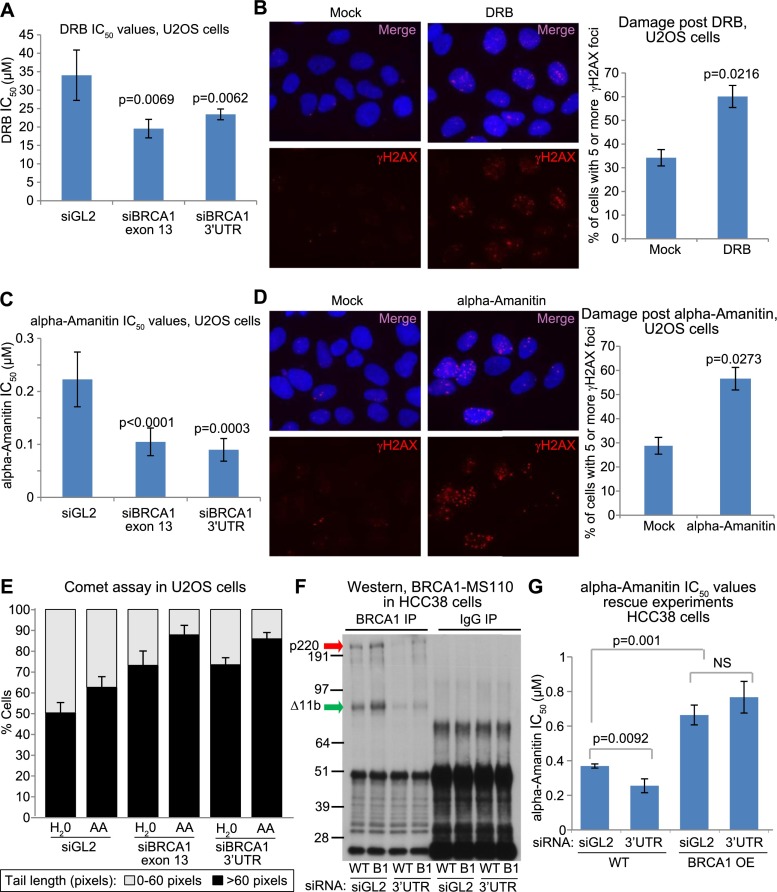
Figure 2.
BRCA1 is required to prevent or repair DNA damage associated with transcription arrest (see also Supplemental Figs. S2I [for siRNA validation], S3A–C). (A) IC50s for DRB sensitivity in U2OS cells depleted of BRCA1 or controls. U2OS cells were transfected with either siGL2 as a control, siBRCA1 exon 13, or siBRCA1 3′ untranslated region (UTR) and tested for sensitivity to varying doses of DRB by colony formation assay. The experiment was repeated a minimum of three times for each siRNA. IC50s were estimated by fitting a nonlinear regression curve to the data from each individual experiment for each siRNA, and the bar graph shows the average of the IC50 values calculated from the replicates for each siRNA. The error bars represent the standard deviation between the IC50s from the multiple experiments. The P-values above the bars (calculated from a two-tailed t-test) indicate the significance of the difference in IC50 values between the siGL2 control and each individual BRCA1 siRNA. (B) In the left panel, representative photos are shown of U2OS cells incubated for 24 h in medium containing either 100 μM DRB or an equivalent volume of ethanol (Mock) and then stained for the classic DNA damage marker γH2AX. This experiment was repeated four times, and the percentage of cells containing five or more γH2AX foci under each condition was counted for each repetition. The bar graph in the right panel represents the average of these percentages for each treatment, and the error bars represent the standard deviation between replicates. The P-value indicates the significance of the difference between the vehicle and DRB treatment as demonstrated by a two-tailed t-test. (C) IC50s for α-amanitin sensitivity in U2OS cells depleted of BRCA1 or controls. U2OS cells were transfected with either siGL2, siBRCA1 exon 13, or siBRCA1 3′ UTR and tested for sensitivity to varying doses of α-amanitin by colony formation assay. The experiment was repeated a minimum of three times for each siRNA. IC50s were estimated by fitting a nonlinear regression curve to the data from each individual experiment for each siRNA, and the bar graph shows the average of the IC50 values calculated from the replicates for each siRNA. The error bars represent the standard deviation between the IC50s from the multiple experiments. The P-values (calculated by a two-tailed t-test) demonstrate the significance of the difference between the IC50s for each BRCA1 siRNA compared with the siGL2 control. (D) In the left panel, representative photos are shown of U2OS cells incubated for 24 h in medium containing either 10 μM α-amanitin or an equivalent volume of ddH2O and then stained for the classic DNA damage marker γH2AX. This experiment was repeated three times, and the percentage of cells containing five or more γH2AX foci under each condition was counted for each repetition. The bar graph in the right panel represents the average of these percentages for each treatment, and the error bars represent the standard deviation between replicates. The P-value (calculated by a two-tailed t-test) indicates the significance of the difference between the vehicle and α-amanitin treatment. (E) Bar graph representing the percentage of cells with two different tail length ranges (0–60 pixels represented as the gray part of the bar, and >60 pixels represented as the black part of the bar) from alkaline comet assays performed on U2OS cells transfected independently with either siGL2, siBRCA1 exon 13, or siBRCA1 3′ UTR and then cultured in medium containing either 0.35 uM α-amanitin (AA) or the equivalent amount of water (H2O) for 24 h. The bars represent the average of the percentages from a minimum of three separate experiments for each siRNA, and the error bars represent the standard deviation between these experiments. P-values that compare the differences in percentages of cells with tail lengths >60 were calculated using a two-tailed t-test. The significant P-values for different comparisons are as follows: siGL2-H2O versus siGL2-AA >60 pixels, P = 0.0106; siBRCA1 exon 13-H2O versus siBRCA1 exon 13-AA >60 pixels, P = 0.0165; siBRCA1 3′ UTR-H2O versus siBRCA1 3′ UTR-AA >60 pixels, P = 0.0096; siGL2-AA versus siBRCA1 exon 13-AA >60 pixels, P = 0.0005; and siGL2-AA versus siBRCA1 3′ UTR-AA >60 pixels, P = 0.0001. (F) Immunoprecipitation-Western blot of wild-type HCC38 (WT) and HCC38+BRCA1 (B1) cDNA lines transfected with siRNAs used in G (siGL2 and siBRCA1 3′ UTR [3′ UTR]). Arrows indicate various BRCA1 isoforms (p220 is full-length BRCA1 and is what we refer to as BRCA1 throughout the text; Δ11b is a shorter isoform lacking exon 11). (G) Wild-type HCC38 cells (WT) or HCC38 cells stably expressing HA-tagged BRCA1 (BRCA1 OE: overexpressed) transfected with either siGL2 or a BRCA1 siRNA targeting its 3′ UTR (3′ UTR) and tested for sensitivity to varying doses of α-amanitin by colony formation assay. The experiment was repeated three times. IC50s were estimated by fitting a nonlinear regression curve to the data from each individual experiment for each siRNA, and the bar graph shows the average of the IC50 values calculated from the replicates for each siRNA. The error bars represent the standard deviation between the IC50s from the three experiments. P-values were calculated using a two-tailed t-test for the differences observed in IC50s and are shown on the bar graph. (The brightness for every photo in B and D was increased by 40% and the contrast was increased by 20% using PowerPoint to overcome the difficulty of converting to PDF. Please note that these images are best viewed in the electronic version of the figure and not on a printed page.)
To ascertain whether this was a BRCA1 depletion-specific effect, we attempted to rescue the α-amanitin sensitivity of cells depleted of BRCA1 by stably expressing an siRNA-resistant BRCA1 cDNA in these cells (Fig. 2F,G; Supplemental Fig. S3C). Rescue was achieved in repeated assays, which suggests that after BRCA1 depletion, the increased α-amanitin sensitivity is in part due to the loss of BRCA1 function. However, other proteins involved in repairing DNA damage may also be associated with transcription arrest.
Taken together, these data support the hypothesis that BRCA1 plays a role in the response to transcription arrest-associated DNA damage. Moreover, BRCA1 may well play a major role in this process, since when the BRCA1 concentration was increased over the baseline level, α-amanitin sensitivity decreased significantly (Fig. 2G, cf. lanes 1 and 3).
BRCA1 interacts genetically with transcription-associated proteins
These results prompted a search for links between BRCA1 and transcription-related proteins identified in our screens. To assess the physiological relevance between BRCA1 and previously unknown interactors from our screens, we searched for genetic interactions in cell lines bearing heterozygous mutations (generated by TALENs) (Joung and Sander 2013) in BRCA1 or a given interactor. Verification of mutations and, where relevant, the decreased expression of a protein of interest is shown in Supplemental Figure S4, A–G. We confirmed that all cells that were studied proliferated and cycled at the same rate as wild-type (WT) controls so that no abnormal phenotypes could be attributed to a cell cycle proliferation defect (Supplemental Fig. S4H). We also attempted to determine whether the BRCA1+/− cells manifested a BRCA1 functional defect so that any phenotypes observed in this cell line and not in the WT/WT control cells could be attributed to decreased BRCA1 protein levels. The WT/WT parent line and BRCA1+/− lines both contain a single copy of a well-known HR reporter stably integrated into their genomes (Weinstock et al. 2006). This reporter consists of two fragments of a GFP sequence, one of which contains an I-SceI recognition site. When the cells are transfected with I-SceI, the single site on the reporter is cleaved, generating a DSB. If the cell is competent to perform HR, the break will be repaired in such a way that a full-length functional GFP cDNA is generated, and GFP is produced. This allows the number of GFP-positive cells to be quantified and used as a reporter of the capacity of the cell line to perform HR. We infected the WT/WT and BRCA1+/− cell lines with a control and multiple BRCA1 shRNAs and performed this HR assay. As would be expected, both cell lines had a reduced HR capacity upon BRCA1 depletion (Supplemental Fig. S4D). The BRCA1+/− cell line yielded a significantly lower percentage of GFP-positive cells than the WT/WT line even when treated with the control shRNA, however, indicating that the BRCA1+/− line was defective in HR, likely due to its markedly lower BRCA1 protein level compared with the WT/WT line. The presence of a BRCA1-associated functional defect in the BRCA1+/− line indicates that other phenotypes, such as genetic interactions, observed in this line are likely related to its decreased level of BRCA1 protein compared with the control.
Searching for genetic interactions in such heterozygous mutant lines as a means of validating novel interactions is a reasonable approach given that a recent study in cells heterozygous for mutations in the BRCA1-binding partner PALB2 demonstrated that PALB2 heterozygosity can be associated with phenotypes that have mechanistic and biological implications (Nikkila et al. 2013).
Thus, we posed the question of whether a BRCA1 heterozygous cell line reveals synthetic lethality upon depletion of a BRCA1-interacting protein of interest. Positive results would reflect a breakdown in the physiological interaction between these proteins.
Genetic interaction between BRCA1 and an interactor from the screen was demonstrated by colony formation assays performed on WT/WT and BRCA1+/− cell lines transfected with a control siRNA (siGL2) or with two siRNAs targeting the interactor of interest. If gene-specific depletion led to fewer colonies in the BRCA1+/− line than the WT/WT control, that would reflect a genetic interaction between BRCA1 and the interactor gene of interest.
By colony formation assay, we detected a genetic interaction between BRCA1 and SETX, an RNA helicase that prevents R-loop-associated damage (Fig. 3A; Supplemental Fig. S2M; Mischo et al. 2011; Skourti-Stathaki et al. 2011). BRCA1 and SETX co-occupy sites on selected meiotic chromosomes (XY body), where the possibility of R-loop involvement for both proteins in this setting exists (Becherel et al. 2013). We also detected genetic interactions between BRCA1 and each of two transcription elongation factors: TCEANC (Fig. 3B; Supplemental Fig. S2N) and TCEA2 (Fig. 3C; Supplemental Fig. S2O). TCEA2 is a member of a family of proteins that assists RNA Pol II in traversing pause sites and certain transcription-arresting sites (Wind and Reines 2000). It also engages in responses to DNA damage/errors that arise during transcription (Wind and Reines 2000). In contrast, the BRCA1-interacting and transcription-elongating protein TCEB3 failed this test, indicating that not all core transcription factors that directly interact with BRCA1 do so in a genetically definable manner.
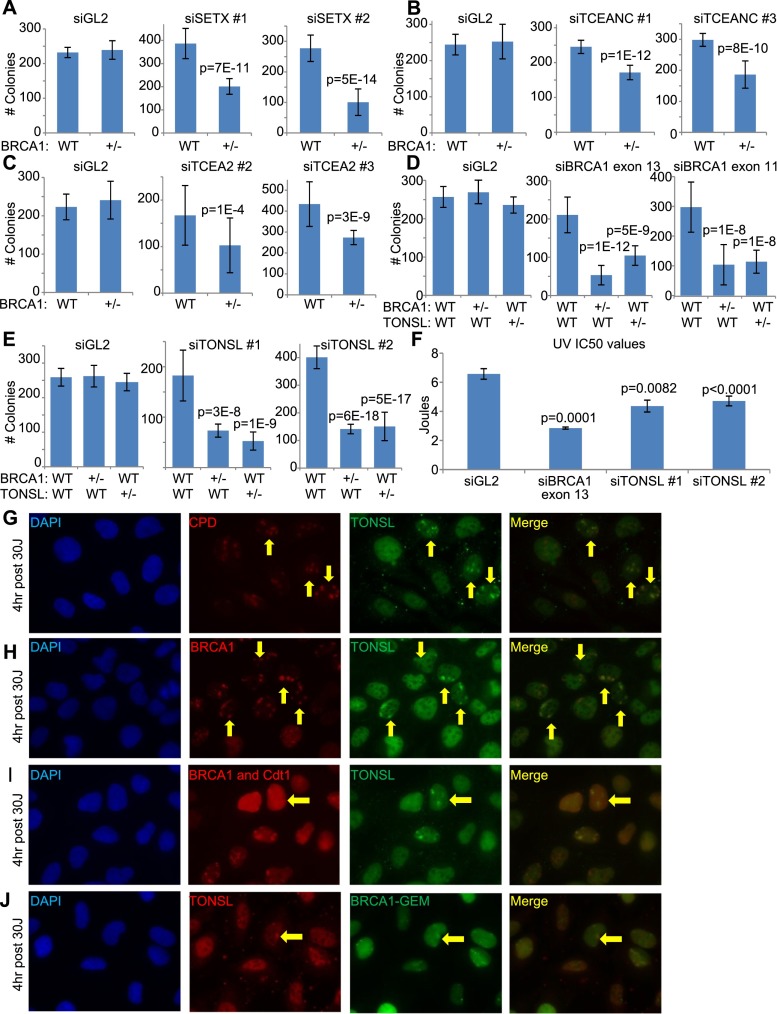
Figure 3.
BRCA1 interacts genetically with various transcription-associated proteins, one of which has not been previously linked with transcription or transcription-associated DNA damage (see also Supplemental Figs. S2I,J,M,N,O [for siRNA validation], S3D, S4, S5). (A–C) WT/WT (WT) and BRCA1+/− (+/−) U2OS cell lines were transfected with a control (siGL2) and multiple gene-specific siRNAs to search for synthetic lethality upon codepletion of specific gene products together with BRCA1. The average number of colonies from at least three separate experiments is shown for the WT/WT and BRCA1+/− cell lines transfected with either siGL2 or two different siRNAs targeting either SETX (A), TCEANC (B), or TCEA2 (C). The error bars represent the standard deviation between the replicates of the experiment, and the P-values (calculated by a two-tailed t-test on a union of all replicates of the repetitions of the experiments) indicate the significance of the difference in colony number between heterozygous mutants and the WT/WT line for the gene-specific siRNA transfections. Due to differences in colony-forming efficiency with each siRNA, the siGL2 results can only be compared with siGL2 and so forth. (D) The average number of colonies from three separate experiments is shown for WT/WT, BRCA1+/−, and TONLS+/− U2OS cell lines transfected with either siGL2, siBRCA1 exon 13, or siBRCA1 exon 11. The plots are designed as described above. (E) The average number of colonies from three separate experiments is shown for WT/WT, BRCA1+/−, and TONLS+/− U2OS cell lines transfected with siGL2, siTONSL #1, or siTONSL #2, plotted as described above. (F) UV IC50s for U2OS cells depleted of TONSL or BRCA1 compared with controls are shown. U2OS cells were transfected with siRNAs, plated at a suitable density for colony formation, treated with varying doses of UV, and later stained and counted. The experiment was repeated four times. The IC50 was estimated by fitting a nonlinear regression curve to the data from each individual experiment for each siRNA, and the bar graph shows the average value calculated from the four replicates for each siRNA. The error bars represent the standard deviation between the four experiments. The P-values (calculated by a two-tailed t-test) indicate the significance of the difference between the siGL2 values and the three individual gene-specific siRNAs. (G,H) Representative immunofluorescence photos of U2OS cells treated with 30 J UV-C through micropores and stained for TONSL and cyclobutane pyrimidine dimers (CPDs are a marker of UV damage) (G) or TONSL and BRCA1 (H) 4 h after treatment. Yellow arrows indicate examples of cells in which there is either CPD–TONSL or BRCA1–TONSL costaining. (I,J) U2OS cells stably expressing either G1-specific Cdt1-RFP (I, red) or S-phase-specific Geminin-GFP (J, green) degrons were irradiated with 30 J through micropores and, 4 h later, stained for BRCA1 and TONSL. Normally, Geminin and Cdt1 are stably expressed in S phase and G1, respectively, and are rapidly degraded upon exit from these phases (Sakaue-Sawano et al. 2008). Tagging them or their isolated degrons with fluorescent markers has been a useful way to mark cells being studied by immunofluorescence in specific phases of the cell cycle. In this method, the fluorescent tag is only expressed in the phase of the cell cycle in which the degron is inactive (Sakaue-Sawano et al. 2008). Yellow arrows indicate phase-specific cells in which TONSL localized in UV micropores. (Please note that these images are best viewed in the electronic version of the figure and not on a printed page.)
These results reinforce the view that BRCA1 responds to transcription-based DNA damage, since it influences or is influenced by these transcription-engaging proteins. Since some of the BRCA1 interactors with which genetic interactions were detected perform different biochemical activities related to this function, one can speculate that BRCA1 performs its roles in this process through a number of biochemical routes, depending on the specific functionality of its interacting partner proteins.
Genetic interaction between TONSL and BRCA1
We also tested another interactor and potential transcription protein, TONSL, which was identified as an interactor in the BARD1 TAP-MS analysis. We focused on TONSL for multiple reasons. First, it was identified in Sleeping Beauty transposon tumorigenesis screens in mice (Supplemental Fig. S1B; Supplemental Table S5), signifying it as a candidate cancer gene (Starr et al. 2009). In these screens, the Sleeping Beauty transposon was used to insert loss-of-function and gain-of-function mutations into mouse genomes in such a way that a mutated gene was easily identified, and the mice were followed for cancer development, thereby allowing for the rapid identification of new cancer genes in the context of an animal model (Collier and Largaespada 2007). In addition, complexes of overexpressed, tagged TONSL and endogenous BRCA1 concentrated in chromatin, consistent with them exerting genome-associated functions (Supplemental Fig. S2B). In this regard, TONSL is already known to participate in the repair of collapsed or stalled replication forks (Duro et al. 2010; O’Connell et al. 2010; O’Donnell et al. 2010; Piwko et al. 2010), which is also a known BRCA1 function (Pathania et al. 2011; Schlacher et al. 2012). However, as shown below, we found that TONSL may also participate in the response to transcription-associated damage.
Genetic interaction between BRCA1 and TONSL was demonstrated by colony formation assays performed on WT/WT, BRCA1+/−, and TONSL+/− cell lines (Supplemental Fig. S4A–G) transfected with a control siRNA (siGL2), two siRNAs targeting BRCA1 (Fig. 3D; Supplemental Fig. S2I), or two siRNAs targeting TONSL (Fig. 3E; Supplemental Fig. S2J). Compared with WT/WT controls, the BRCA1+/− and TONSL+/− lines were more sensitive to depletion of their respective wild-type residual proteins. This shows that the decreased baseline levels of each of these proteins in the heterozygotes led to a gene-specific effect on colony formation, thereby validating the specificity of genetically observed phenotypes in these lines. This evidence is supported by the demonstration of a BRCA1 HR defect in the BRCA1+/− cell line (Supplemental Fig. S4D).
TONSL and BRCA1 localize to UV-induced DNA damage sites
Since TONSL and BRCA1 both participate in the repair of stalled replication forks (Duro et al. 2010; O’Connell et al. 2010; O’Donnell et al. 2010; Piwko et al. 2010; Pathania et al. 2011; Schlacher et al. 2012), we asked whether a BRCA1–TONSL complex is engaged in this repair. To test this possibility, we searched for TONSL localization in UV irradiation-associated nuclear territories (micropores) (Polo et al. 2006) where BRCA1 localizes and where its only known function thus far is in the repair of stalled replication forks (Pathania et al. 2011). Thus, cells were exposed to UV micropore irradiation and then immunostained for TONSL and BRCA1 up to 24 h thereafter. Like BRCA1, TONSL localized in micropores (Fig. 3G,H; Supplemental Fig. S5). However, TONSL first appeared there 4–8 h after irradiation, which is long after BRCA1 had concentrated at these sites (Fig. 3G,H; Supplemental Fig. S5; Pathania et al. 2011).
Therefore, one manifestation of the BRCA1–TONSL genetic interaction may well operate at sites of UV-induced damage, where both proteins are normally engaged in complex DNA damage responses (Duro et al. 2010; O’Connell et al. 2010; O’Donnell et al. 2010; Piwko et al. 2010; Pathania et al. 2011). Indeed, as one might expect for a protein involved in a UV damage response, cells depleted of TONSL became more sensitive to UV irradiation than controls but not as sensitive as when BRCA1 was depleted (Fig. 3F; Supplemental Figs. S2I,J, S3D). Since TONSL did not localize at micropores in every cell in which BRCA1 was so localized, it is possible that colocalization occurs only during a limited cell cycle interval, which may offer a clue as to the function of the BRCA1–TONSL complex.
To test this possibility, detection of TONSL and BRCA1 in micropores was assessed in cells expressing cell cycle-dependent reporters that are degraded along with their fluorescent tags as soon as cells exit the phase for which the reporters are specific (Mechali and Lutzmann 2008; Sakaue-Sawano et al. 2008). The results showed that TONSL concentrates in these damage sites during both S and G1 (Fig. 3I,J). The S-phase localization supports the suggested role for TONSL in repair of stalled and collapsed replication forks, but the G1-phase localization does not. However, a putative role in the response to transcription-associated DNA damage is possible.
Indeed, the list of known TONSL interactors includes proteins that function in transcription and likely operate in both S and G1—in particular, both members of the FACT complex: SSRP1 and SUPT16H (Duro et al. 2010; O’Connell et al. 2010; O’Donnell et al. 2010; Piwko et al. 2010). The FACT complex supports transcription elongation through regions of chromatin by facilitating alteration and accurate replacement of nucleosome structure (Reinberg and Sims 2006). This allows transcriptional progression through chromatin regions without permanently disrupting epigenetic markers (Reinberg and Sims 2006). In addition, the FACT complex has recently been implicated in multiple aspects of transcription-associated DNA damage control and the resolution of collisions between replication and transcription complexes (Dinant et al. 2013; Herrera-Moyano et al. 2014). Such functions could occur in either G1 or S.
TONSL and BRCA1 function at sites of transcription-associated damage together with the FACT complex
Thus, we searched for evidence linking FACT, TONSL, and BRCA1 at UV damage sites. We first asked whether TONSL localization at micropores is dependent on BRCA1 or FACT. We performed UV micropore assays with and without BRCA1, SSRP1, SUPT16H, or TONSL depletion (Figs. 4A; Supplemental Figs. S2I–L, S6A,B). While SSRP1, SUPT16H, and TONSL depletion had no effect on BRCA1 micropore localization (Supplemental Fig. S6A), BRCA1, SSRP1, or SUPT16H depletion led to fewer cells exhibiting TONSL in micropores (Fig. 4A). Under the conditions used, none of these depletions caused significant cell cycle arrest (Supplemental Fig. S6B). Thus, BRCA1, SSRP1, and SUPT16H expression may be necessary for TONSL localization at these damage sites in cells in multiple phases of the cell cycle.
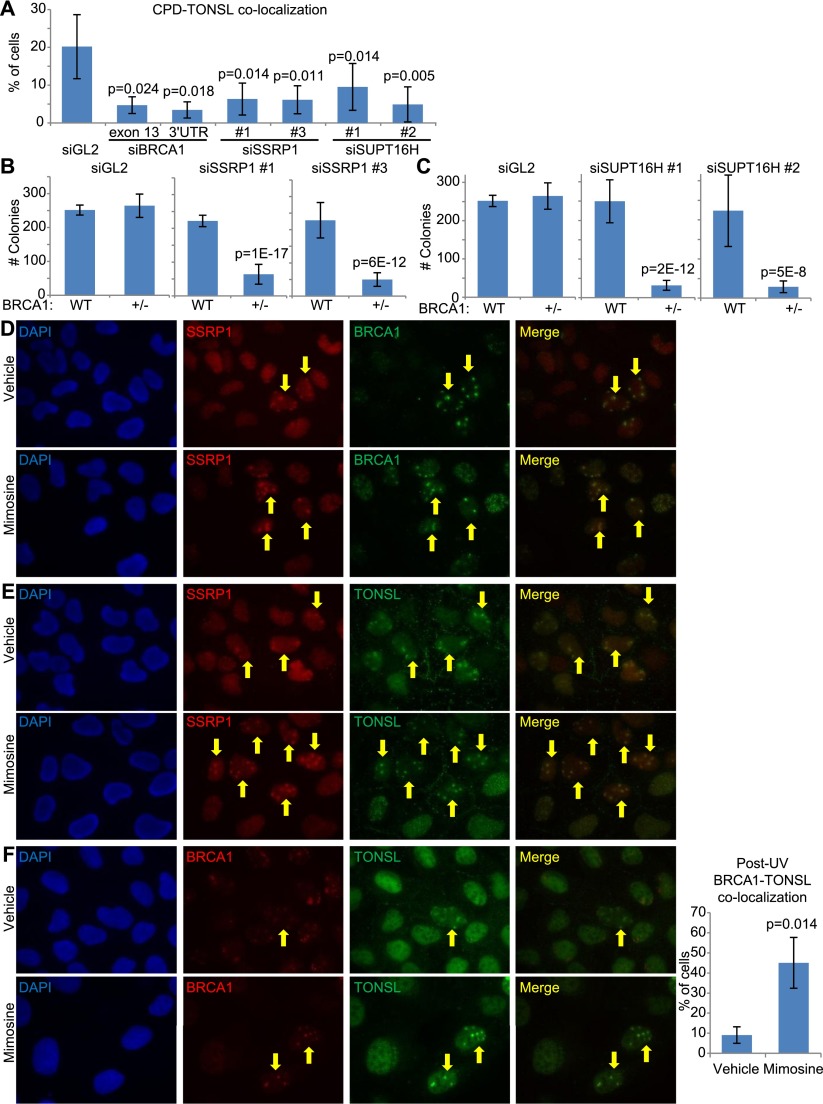
Figure 4.
BRCA1, TONSL, and FACT interact functionally at sites of UV damage (see also Supplemental Figs. S2I,J,K,L, S4, S6). (A) The bar graph represents the average number of U2OS cells containing TONSL in CPD-positive micropores from four separate experiments in which cells were transfected with a control siRNA (siGL2) or one of two different BRCA1-, SSRP1-, or SUPT16H-specific siRNAs. After transfection, the cells were irradiated with 30 J UV and stained 4 h later for BRCA1, CPDs, and TONSL. The error bars represent the standard deviation between the counts from the four experiments. The P-values (calculated by a two-tailed t-test) demonstrate the significance of the difference between the siGL2 values and the various gene-specific siRNA values. (B,C) The average number of colonies from three separate experiments is shown for the WT/WT (WT) and BRCA1+/− (+/−) U2OS cell lines transfected with either siGL2 or two different siRNAs targeting either SSRP1 (B) or, independently, SUPT16H (C). The plots are designed as described in Figure 3. (D–F) U2OS cells were incubated in medium containing 0.5 mM mimosine or an equivalent volume of vehicle for 24 h and then irradiated with 30 J through micropores, allowed to recover for 4 h, and costained for SSRP1 and BRCA1 (D), SSRP1 and TONSL (E), and TONSL and BRCA1 (F). Arrows mark some of the cells where colocalization is observed. The BRCA1–TONSL costaining in F was repeated four times, and the number of cells in which TONSL and BRCA1 colocalized in micropores in either vehicle- or Mimosine-containing medium was counted each time. The bar graph in the right panel of F shows the average percentage of cells containing colocalizing BRCA1 and TONSL under both conditions. The bars are the averages from four experiments, and the error bars represent the standard deviation between experiments. The P-value (calculated using a two-tailed t-test) indicates the significance of the difference between vehicle- and Mimosine-treated cells. (Please note that these images are best viewed in the electronic version of the figure and not on a printed page.)
Given the influence of FACT on TONSL and the link between TONSL and BRCA1, we wondered whether there was also a link between FACT and BRCA1. Thus, we tested for a genetic interaction between BRCA1 and either SSRP1 (Fig. 4B; Supplemental Fig. S2K) or SUPT16H (Fig. 4C; Supplemental Fig. S2L), again using the colony formation assay. We found that the BRCA1+/− line produced significantly fewer colonies than the WT/WT line when transfected with siRNAs specific for either SSRP1 or SUPT16H. Therefore, we conclude that BRCA1 interacts genetically with TONSL and the TONSL partners SSRP1 and SUPT16H.
Given this association, the question arose of whether either member of the FACT complex also localizes to UV-induced damage sites with BRCA1 and, if so, in which phase of the cell cycle this occurs. We searched for SSRP1 and SUPT16H in micropores with either TONSL or BRCA1 in both asynchronous cells and cells arrested in G1 by mimosine, a plant amino acid that arrests cells in G1 (Fig. 4D–F; Supplemental Fig. S6C). We failed to detect SUPT16H in micropores, but SSRP1 colocalized with both BRCA1 (Fig. 4D) and TONSL (Fig. 4E) in these UV-irradiated territories in both asynchronous and G1-arrested cells. We also observed that TONSL colocalizes with BRCA1 in both asynchronous and G1-arrested cells and that the percentage of cells containing BRCA1 and TONSL colocalizing micropores increases significantly in G1-arrested cells, indicating that an important function of the BRCA1–TONSL–FACT complex is likely manifest in G1, which is consistent with the prospect of transcription-associated damage being dealt with by such a complex in G1 (Fig. 4F).
Given the influence of FACT and BRCA1 on TONSL localization, the genetic interactions between FACT and BRCA1 and between TONSL and BRCA1 and the colocalization of FACT, BRCA1, and TONSL at UV damage sites in G1-arrested cells, we speculated that, in addition to aiding in the repair of stalled replication forks, TONSL and BRCA1 cofunction with FACT at sites of UV damage. There, one might speculate that they (1) prevent transcription through such sites, (2) contribute to its restart or repair, and/or (3) resolve R-loop-associated damage that has occurred due to the stabilization of these structures at arrested transcription complexes. These hypotheses are supported by existing evidence. First, after UV damage, the FACT complex interacts with RNA Pol II pS5 and certain transcription-coupled nucleotide excision repair (NER) proteins and participates in transcription restart (Dinant et al. 2013). Second, the FACT complex is important in preventing DNA damage associated with the stabilization of R loops, some of which is caused by collisions between transcription and replication complexes (Herrera-Moyano et al. 2014).
BRCA1 localizes at sites of post-UV transcription-associated DNA damage
To determine whether the UV sites at which BRCA1, TONSL, and FACT colocalize contain transcription-associated DNA damage, we asked whether actively transcribing RNA Pol II (RNA Pol II pS5) colocalizes with the DNA damage marker γH2AX and BRCA1 after UV treatment. We were not able to perform micropore assays because RNA Pol II pS5 does not concentrate in these large territories at levels greater than those detected in an unirradiated nucleus. Thus, we performed whole-cell UV irradiation, since RNA Pol II pS5 forms distinct foci in this setting when cells are appropriately fixed (Espinosa et al. 2003). In preparation for these assays, we validated the specificity of our BRCA1 and RNA Pol II pS5 antibodies by various means, since RNA Pol II detection requires harsher fixation conditions than micropore immunofluorescence staining (Supplemental Fig. S7A–E).
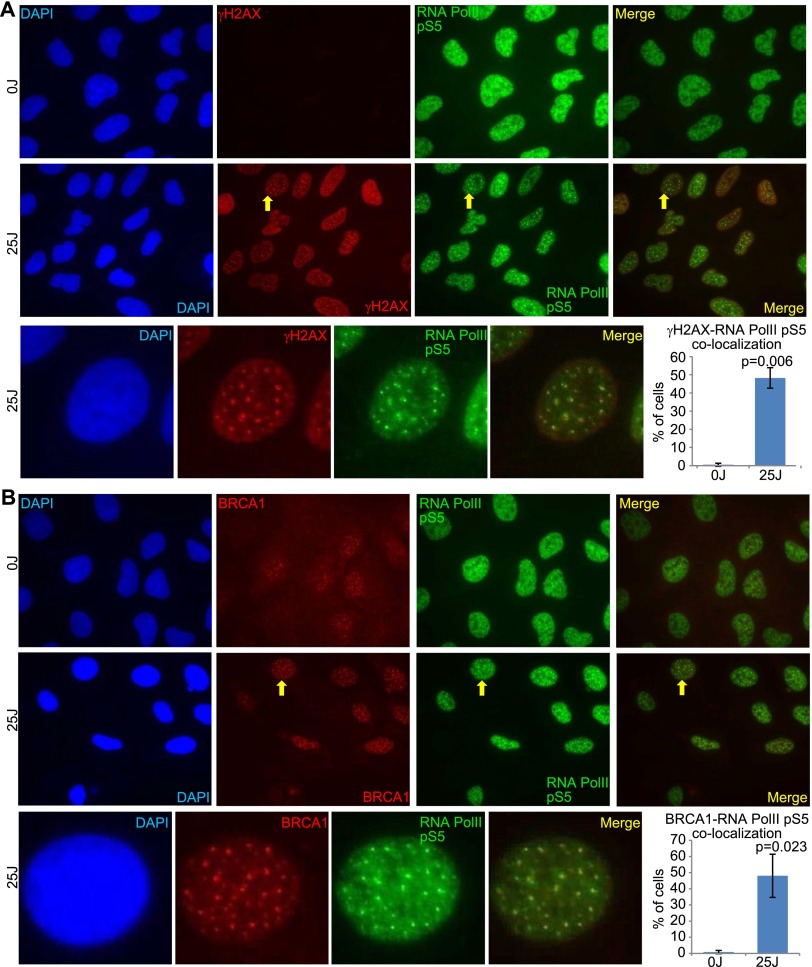
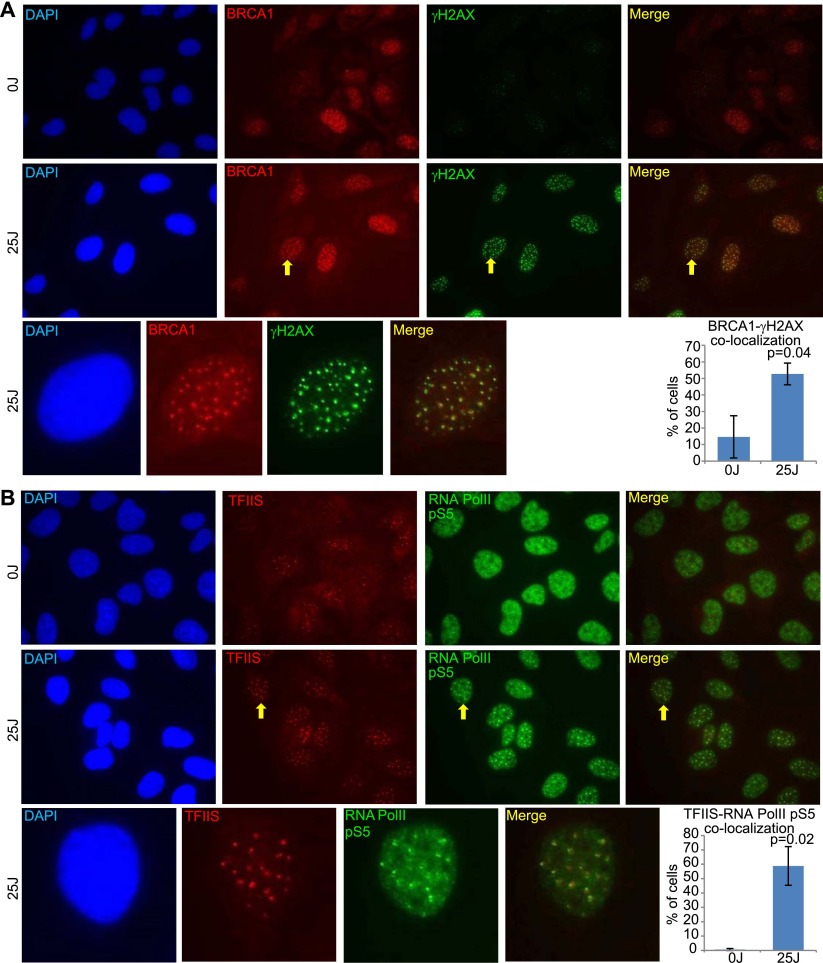
The results showed that active RNA Pol II forms an increased number of distinct foci after UV treatment (Figs. 5, 6B). These foci costained with the DNA damage marker γH2AX as well as with BRCA1 after UV treatment, and the percentages of cells containing foci in which there was colocalization between active RNA Pol II and these two proteins were greatly increased after UV treatment (Fig. 5A,B). BRCA1 also colocalized with γH2AX foci in a greater percentage of cells after UV treatment (Fig. 6A). Given the large increase in cells containing foci in which there is colocalization between RNA Pol II pS5 and γH2AX, between BRCA1 and γH2AX, and between RNA Pol II pS5 and BRCA1, it is likely that RNA Pol II pS5, BRCA1, and γH2AX are colocalizing in a significant fraction of the same cells after UV exposure.
Figure 5.
Active RNA polymerase and BRCA1 colocalize in distinct foci after UV damage (see also Supplemental Figs. S7, S8). (A,B) U2OS cells were exposed to 0 J or 25 J (whole-cell irradiation, without micropore filters), fixed with methanol–acetic acid 4 h later, and costained for either active RNA polymerase (RNA Pol II pS5) and γH2AX (A) or RNA Pol II pS5 and BRCA1 (B). One representative photo of a field of cells is shown for each staining after 0 J and 25 J of UV, and one representative cell from the 25-J photo in which there is significant colocalization between the costained proteins has been cut out and magnified using PowerPoint at the bottom of the panel. Yellow arrows point to the magnified cell in the 25-J field of cells photos. In addition, at the right of the magnified photos is a bar graph showing the average percentage of cells containing three or more colocalizing foci for the stained protein pair in that panel. A minimum of 200 cells were counted for each treatment in each experiment. The bars in each bar graph represent the average percentages from three separate experiments for each staining pair, and the error bars represent the standard deviation between experiments. The P-values (calculated by a two-tailed t-test) represent the significance of the difference between the 0-J and 25-J counts. In the bar graph in A, the 0-J value is extremely low compared with the 25-J value and is difficult to see in the bar graph. The average percent for 0 J in A was 0.49, with a standard deviation of 0.84. The 0-J value in the bar graph in B is also very low when compared with the 25-J value. The average percent for 0 J in B was 0.82, with a standard deviation of 1.03. (The brightness for every photo in the figure has been increased by 40% using PowerPoint to overcome the difficulty of converting to PDF. Please note that these images are best viewed in the electronic version of the figure and not on a printed page.)
Figure 6.
BRCA1 and TFIIS localize to sites of UV-induced transcription-associated DNA damage (see also Supplemental Figs. S7, S8). (A,B) U2OS cells were treated with 0 J or 25 J (whole-cell irradiation, without micropore filters), fixed with methanol-acetic acid 4 h later, and costained for either γH2AX and BRCA1 (A) or RNA Pol II pS5 and TFIIS (B). One representative photo of a field of cells is shown for each staining after 0-J and 25-J irradiation, and one representative cell from the 25-J photo in which there is significant colocalization between the costained proteins has been cut out and magnified using PowerPoint below the two fields. Yellow arrows point to the magnified cell in the 25-J photos. In addition, at the right of the magnified photos is a bar graph showing the average percentage of cells containing three or more colocalizing foci for the staining pair in that panel. A minimum of 200 cells was counted for each treatment in each experiment. The bars in each bar graph represent the average percentages from three separate experiments for each staining pair, and the error bars represent the standard deviation between experiments. The P-values (calculated by a t-test) represent the significance of the difference between the 0-J and 25-J counts. The 0-J value in the bar graph in B is very low and difficult to see compared with the 25-J value. The average percent for 0 J in B was 0.64, with a standard deviation of 0.73. (The brightness for each photo in the figure has been increased by 40% using PowerPoint to overcome the difficulty of converting to PDF. Please note that these images are best viewed in the electronic version of the figure and not on a printed page.)
The colocalization of BRCA1 and RNA Pol II at sites of UV DNA damage fits with previous results indicating that the BARD1–BRCA1 complex contributes to the degradation of RNA Pol II after UV damage (Kleiman et al. 2005). However, the persistence of these structures 4 h post-irradiation suggests that causing degradation of RNA Pol II at sites of UV damage is not the only role for BRCA1 at such sites. It suggests that, in the setting of irreparable damage, BRCA1–BARD1 promotes the degradation of RNA Pol II to allow for repair at that site, while in the setting of repairable damage, BRCA1 may aid in overcoming the damage and restarting transcription, both proposals to be studied further. Furthermore, both directly and indirectly, UV irradiation contributes to the development of multiple forms of DNA damage as well as transcription arrest. Conceivably, BRCA1 plays specific roles in the relevant responses to these perturbations.
To test whether there are ssDNA stretches and thus R loops, collapsed transcription complexes, or perhaps collapsed replication forks that had collided with a transcription fork at these damage sites, we stained for colocalization of BRCA1 and phosphorylated RPA, which is known to coat ssDNA at such sites (Supplemental Fig. S8A). We found that the number of cells containing BRCA1 and pRPA colocalized at foci greatly increased after UV treatment (Supplemental Fig. S8A). This implies that stabilized R loops, fork collapse, and/or collisions are forms of transcription-associated DNA damage that are likely confronted by BRCA1 at these sites.
One possibility is that, upon recognition of a stalled transcription complex, BRCA1 recruits other proteins to assist its restart. One of the interactors detected in our screen, TCEA2, is a member of the TFIIS family of transcription factors that is responsible for assisting RNA polymerase in overcoming various obstacles encountered during transcription (Wind and Reines 2000). Although no available TCEA2 antibodies worked with the relevant fixation protocol, an antibody directed at a closely related isoform, TCEA1 (commonly referred to as TFIIS and referred to as TFIIS in the figures and other parts of this study), did operate in this staining protocol (Supplemental Fig. S7F–H). Moreover, after UV irradiation, there was a significant increase in the percentage of cells containing foci in which RNA Pol II pS5 and TFIIS foci colocalized (Fig. 6B), and the same was true for TFIIS and γH2AX (Supplemental Fig. S8B). Thus, like BRCA1, TFIIS also localized at post-UV transcription-associated damage foci. Given the increased numbers of cells after UV treatment that contain foci in which TFIIS and BRCA1 colocalize with either γH2AX or active RNA Pol II, it is likely that BRCA1 and TFIIS also colocalize in at least a fraction of these transcription-associated damage foci.
We were unable to test TCEANC, SETX, TONSL, SSRP1, or SUPT16H for RNA Pol II pS5 colocalization, since we lacked antibodies that function in this fixation protocol. Nevertheless, the above-noted evidence further supports the hypothesis that BRCA1 functions at sites of transcription-associated damage and the notion that a BRCA1–TONSL–FACT complex functions at these sites. Given the colocalization of BRCA1 with active RNA Pol II at post-UV damage structures, it is likely that a BRCA1–TONSL–FACT complex participates in some part of the recognition or repair of these structures. This is because these proteins all localize to sites of UV damage in G1-arrested cells where transcription and not DNA replication is in progress and because FACT has already been implicated in such a function (Dinant et al. 2013).
Furthermore, this evidence, along with the above-noted TFIIS staining, implies that BRCA1 can participate in the response to various forms of transcription-associated DNA damage or stalling and that it does so in different ways by interacting with different protein-binding partners, such as TFIIS or TONSL–FACT.
BRCA1 participates in various processes at sites of transcription-associated DNA damage
Given the localization of BRCA1 at sites of transcription-associated damage and the multiple functions of its new interactors that could be occurring at these sites, we set out to determine what functional roles BRCA1 exhibits in this setting.
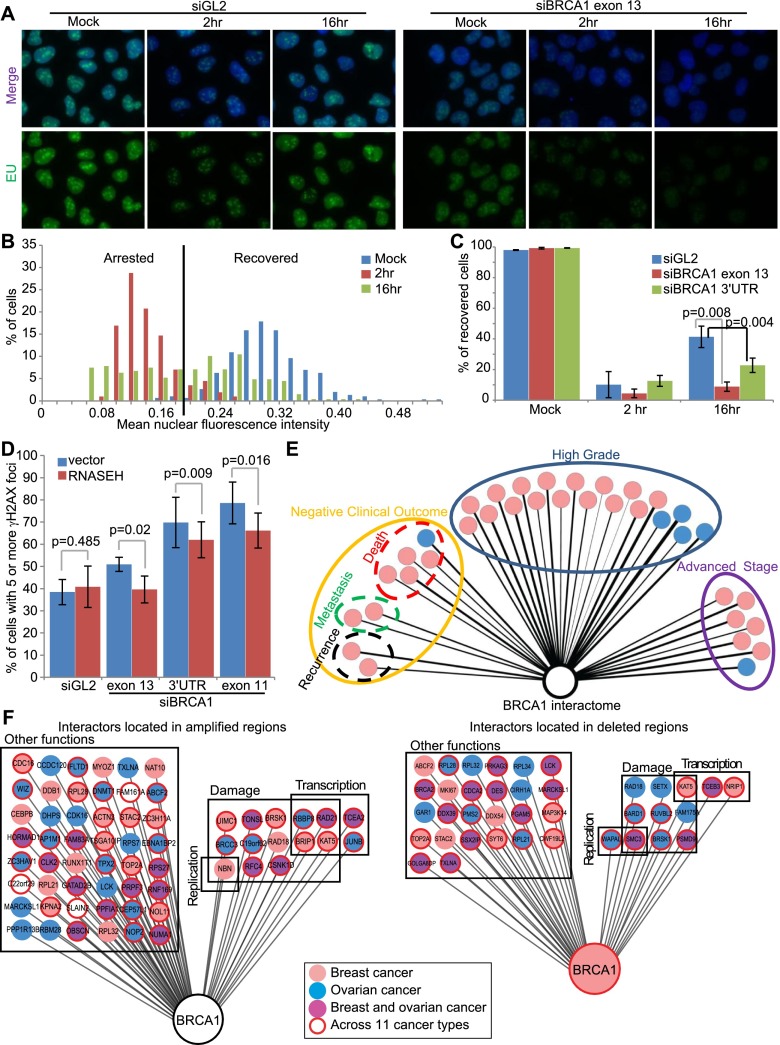
One group has already shown that at least one member of the FACT complex is important in supporting the restart of transcription after UV damage (Dinant et al. 2013). Thus, we asked whether BRCA1 is also involved in this function and found that depletion of BRCA1 leads to a slower recovery from UV-associated transcription arrest than is observed in control cells (Fig. 7A–C). This fits with data indicating that, after UV damage, a BARD1–CstF50 complex prevents aberrant 3′ mRNA processing, likely through signaling for the degradation of RNA Pol II by the BRCA1–BARD1 complex (Kleiman and Manley 2001; Kleiman et al. 2005). Perhaps the BRCA1–BARD1 complex triggers RNA Pol II degradation or transcriptional restart, depending on the severity of the damage. It is likely that a BRCA1–FACT complex and either a BRCA1–TONSL–FACT complex or, given the known function of TFIIS and its localization to UV-induced transcriptional damage, a BRCA1–TFIIS complex are recruited to sites of arrested transcription to aid in damage repair and/or transcription restart at these sites.
Figure 7.
BRCA1 functions in multiple ways at sites of transcription arrest, and its transcription-associated damage repair function is relevant in breast and ovarian cancer development and progression (see also Supplemental Figs. S2I [for siRNA validation], S7I; Supplemental Table S6). (A–C) The role of BRCA1 in transcription restart following UV damage was measured and compared in control siRNA and BRCA1 siRNA (either siBRCA1 exon 13 or siBRCA1 3′ UTR) transfected U2OS cells by measuring RNA synthesis using 5-ethinyl uridine (EU) uptake over a 2-h labeling period (Dinant et al. 2013). siRNA transfected cells were exposed to an equivalent air exposure (mock) or 8 J, labeled with EU for 2 h either immediately (air sample; referred to as “mock” in the figures) or 2 or 16 h after UV treatment, fixed, and stained. Photographs were taken of the stained cells for each treatment at each time point, and at least 300 nuclei were analyzed for their mean fluorescent intensity using CellProfiler for each treatment at each time point. Representative photos of the siGL2 cells and one of the siBRCA1-treated sets of cells are shown in A at each time point. The merged Hoechst-EU staining is shown at the top, and the EU staining alone is shown at the bottom. A histogram depicting the distribution of mean nuclear EU intensities from one representative experiment for siGL2 is shown in B. The arrested cells, appearing at the left, revealed a lower EU-staining intensity than the recovered cells at the right. The black line denotes two standard deviations below the mean of the mock distribution. Irradiated cells at the right of this line are considered to have recovered to normal transcriptional levels, whereas those at the left are considered to be transcriptionally arrested (cf. siGL2-2 h and siGL2-16 h with siGL2-Mock, and siBRCA1-2 h and siBRCA1-16 h with siBRCA1-Mock). The bar graph in C shows the percentage of cells for each siRNA that has recovered based on the above calculations at a given time point. Results are the average of three experiments, and the error bars represent the standard deviation between experiments. P-values were calculated using a two-tailed t-test. (D) To test the possibility that BRCA1 is involved in preventing or repairing R-loop-associated DNA damage, U2OS cells were transfected with either siGL2 or one of three different BRCA1-specific siRNAs (targeting exon 13, exon 11, or the 3′ UTR) on days 1 and 2, transfected with either empty vector or RNASEH1 on day 3, and then fixed 24 h later. The cells were costained for γH2AX and RNASEH1. For vector+siRNA transfected cells, the number of cells containing five or more γH2AX foci was counted from the whole population, and for the RNASEH1+siRNA transfected cells, the number of RNASEH1-expressing cells containing five or more γH2AX foci was counted. A minimum of 150 cells was counted for each treatment in each experiment. This experiment was repeated five times for each siRNA, and the average percentage of cells containing five or more γH2AX foci for each siRNA with either vector or RNASEH1 expression was calculated from these five experiments and is represented by the bars in the bar graph shown here comparing the vector and RNASEH1 values for each siRNA. The error bars represent the standard deviation between the five replicates, and the P-values (calculated by a two-tailed t-test) above the paired counts represent the statistical significance of the difference between the vector and RNASEH counts for each individual siRNA. (E) A network representing overlap between genes in the BRCA1 interactome and previously reported gene expression profiling results obtained from breast and ovarian tumors. The expression of genes detected as BRCA1-interacting protein partners (BRCA1 Interactome) was queried in various gene expression profiling data sets in Oncomine Concepts Map for both breast and ovarian cancers. Each group of nodes encircled by different colors represents a molecular concept with which the BRCA1 interactome is associated, and the individual nodes represent significantly associated data sets within that particular concept with a Q-value ≤0.01 (the majority are overexpression data sets). The width of each edge reflects the number of overlapping genes between the BRCA1 interactome and the linked data set—the wider the edge, the greater the overlapping gene number. Node color represents the tumor type analyzed in the linked data set. (Pink) Breast; (blue) ovarian. (F) Networks representing the genes from the BRCA1 interactome that were found to be in either amplified (left panel) or deleted (right panel) regions in a statistically significant way (P < 0.05 and Q < 0.25) in a TCGA (The Cancer Genome Atlas) data set for breast cancers or ovarian cancers or across 11 different cancers. The nodes are grouped according to association with select GO terms, and the key to node color is noted below the networks.
UV arrest of transcription could lead to damage by multiple mechanisms, one of which is stabilization of R loops and subsequent evolution of single-strand breaks at these sites into DSBs. This mechanism of damage is particularly interesting because several of the new BRCA1 interactors identified in this screen are involved in preventing this kind of damage, including SETX and at least one member of the FACT complex (Mischo et al. 2011; Skourti-Stathaki et al. 2011; Herrera-Moyano et al. 2014). In addition, BRCA1 and BRCA2 depletion have recently been linked to an increase in the number of DNA:RNA hybrids (Bhatia et al. 2014).
Thus, to address whether this is a type of DNA damage that BRCA1 contributes to repairing or overcoming at sites of transcription arrest, we tested whether overexpressing RNASEH1, an enzyme that specifically digests DNA:RNA hybrids such as those underpinning R loops, decreases the amount of DNA damage marked by γH2AX foci that is observed after BRCA1 depletion. We found that RNASEH1 overexpression caused a small decrease in the number of cells with γH2AX foci for multiple different BRCA1 siRNAs compared with an empty vector (Fig. 7D; Supplemental Fig. S7I). This fits with the fact that BRCA1 depletion leads to an increase in the abundance of DNA:RNA hybrids (Bhatia et al. 2014) and suggests that BRCA1 contributes to either preventing or repairing DNA damage associated with the persistence of R loops.
Importantly, the decreases observed in γH2AX foci after RNASEH1 expression, while not large, were statistically significant and reproducible. This is not surprising, since BRCA1 likely deals with a variety of forms of DNA damage that lead to γH2AX foci, only some of which are related to transcription and R loops. Therefore, only a small percentage of the damage that BRCA1 deals with is likely to be caused by R-loop persistence/abnormal stabilization. The increase in DNA:RNA hybrids observed upon BRCA1 depletion was also observed to be small (Bhatia et al. 2014). This supports the idea that suppressing or repairing DNA damage associated with these structures is only one of the molecular functions of BRCA1.
Novel BRCA1 interactors identified in our screen exhibit cancer links with therapeutic implications
The question of whether any of our interactors or the new roles for BRCA1 in the response to transcription-associated DNA damage are clinically relevant was also addressed. Oncomine Concepts Map (Rhodes et al. 2007) was used to search for associations between our results and those of prior gene expression profiling studies performed on breast and ovarian cancers. BRCA1 tumor suppression largely extends to these two organs.
The results show that our 147 screening hits are enriched for proteins that are significantly overexpressed or underexpressed in tumors associated with negative clinical outcomes, high-grade pathology, and advanced clinical stage (Fig. 7E). This suggests that a number of interactors exhibit altered expression in clinically aggressive tumors and may therefore contribute to disease progression and lack of therapeutic benefit.
In keeping with these observations, we also asked whether any of our interactors are encoded by genes that map to amplified or deleted regions in breast tumors or ovarian tumors or across a set of 11 cancer types that have been systematically analyzed (Zack et al. 2013). Indeed, 90 out of 147 of our hits were located within these genomic regions (Fig. 7F; Supplemental Table S6). Sixty-three hits were localized to amplified regions; 15 of these 63 proteins are involved in transcription, replication, and/or DNA damage. Thirty-nine additional hits mapped to deleted regions, and 12 of these hits are known to engage in one or more of these same functions. Moreover, some of the genes that we demonstrated as being genetically associated with BRCA1 are located in these regions, including TONSL, TCEA2, and SETX. These data are particularly interesting with respect to TONSL, since it was found in a Sleeping Beauty screen, thereby implying that it affects tumorigenesis, at least in animal models (Supplemental Fig. S1B; Supplemental Table S5).
One caveat associated with these data is that the regions identified here are large and contain multiple genes. Hence, there is no guarantee that a BRCA1 interactor gene of interest operates as a tumor suppressor or an oncogene in these regions. However, this concern is lessened by the fact that 12 of our hits are already known to be cancer-associated (Fig. 1B; Supplemental Fig. S1B; Supplemental Table S5; Futreal et al. 2004; Vogelstein et al. 2013). Therefore, these data suggest that a number of the hits from our BRCA1 interactome screens and the new functional role proposed for BRCA1 participate in human tumorigenesis.
Discussion
The complementary protein interaction screening approaches used here have led to the generation of a more complete BRCA1 protein interaction network. Knowledge of the anatomy of this network has made possible genetic tests of the physiological relevance of certain interactions. From the results of these analyses, the relevance of the function of certain BRCA1-interacting proteins to BRCA1 function was demonstrated, and a combined role for BRCA1 and these interacting proteins in the response to transcription-associated DNA damage was detected.
While the overlap in hits that we detected in our two BRCA1-focused screens was small (three out of 147 interactors), it is not surprising, since the two methods ask different questions, and each has different limitations. The Y2H screen can detect only direct interactions that do not require post-translational modifications of either interactor and is limited to the library with which one is screening (in our case, a library of ∼15,000 human ORFs). BRCA1 itself undergoes many post-translational modifications, including but not limited to multiple phosphorylations, which are important in mediating protein–protein interactions (Manke et al. 2003; Yu et al. 2003; Greenberg 2008; Huen et al. 2010). In contrast to the Y2H method, the TAP-MS method detects interactions that may be indirect among members of large protein complexes and may rely on either post-translational modifications or perhaps linking structures like mammalian chromatin to facilitate their interactions. The Y2H could therefore miss certain interactions detected in a TAP-MS screen because those interactions may require post-translational modifications, be indirect, involve a protein or isoform of a protein not represented in the ORFeome library that we screened, or be dependent on the complex containing both BRCA1 and BARD1. On the other hand, detection of an interactor in the TAP-MS method is dependent on the stability of the interaction and the abundance of the interacting proteins and therefore could miss interactions that would be detectable by Y2H. These limitations contribute to the lack of overlap between the screens but also point out the potential complementarity of performing a dual screen.
Demonstrating the physiological relevance of interactions identified in a screen can be challenging. Here, we used a genetic assay to test whether the candidate interactors showed synthetic lethality with BRCA1 depletion. Because complete depletion of BRCA1 can lead to cell cycle arrest (Xu et al. 2001), it is especially difficult to perform mechanistic studies on several of its functions. However, the BRCA1+/− lines used in our experiments cycled normally and thus made it easier to study certain functions through genetic manipulations, allowing physiologically relevant insights to be made. Similar methods could be extended to other systems in the future.
The results of our screens and of the validation of multiple candidate interactors have uncovered a new role for BRCA1 in the prevention and/or repair of DNA damage associated with transcription arrest. Imputed functions of TONSL and known functions of the TONSL-binding FACT complex and of SETX, TCEANC, and TCEA2—all proteins engaged in transcription and/or RNA processing and all proteins that genetically interact with BRCA1—represent new processes through which BRCA1 participates in the prevention and/or repair of DNA damage. That BRCA1 is engaged in transcription-associated damage control is supported as a concept by these results and by BRCA1- and BARD1-linked phenomena observed by others (Scully et al. 1997; Kleiman and Manley 1999, 2001; Le Page et al. 2000; Kleiman et al. 2005; Becherel et al. 2013; Yuce and West 2013; Bhatia et al. 2014). Moreover, data reported here now strongly suggest that BRCA1 responds to DNA damage at sites of stalled or defective transcription to either aid in transcription restart and/or resolve certain damaged structures at these sites. These findings are also consistent with the hypothesis that BRCA1 can perform different functions at these sites, depending on the type of damage and on the particular interacting proteins (TONSL–FACT, TCEA2, etc.) that are corecruited with it.
For example, both the FACT complex and SETX are involved in preventing R-loop stabilization (Mischo et al. 2011; Skourti-Stathaki et al. 2011; Herrera-Moyano et al. 2014). We showed that BRCA1 is required to prevent or repair DNA damage associated with R loops in an RNASEH1 rescue assay (Fig. 7D). This is supported by data from others showing that BRCA1 depletion leads to accumulation of DNA:RNA hybrids (Bhatia et al. 2014). We showed that BRCA1 is important in not only resolving these structures but also preventing or repairing the DNA damage caused by their persistence. It is conceivable that BRCA1 accompanies RNA Pol II, with which it is known to interact (Scully et al. 1997), to prevent stalling and repair damage at R loops. Alternatively, BRCA1 might be recruited to RNA Pol II that collects at pause sites or sites of DNA damage and recruits SETX or the FACT complex to these sites to prevent R-loop stabilization or repair R-loop-associated damage.
The FACT complex has multiple other functions as well. The best understood function of FACT is in rearranging chromatin to allow transcription elongation to proceed through chromatin without disturbing its epigenetic architecture (Reinberg and Sims 2006), and FACT has now been implicated in transcription restart after and potential repair of UV-induced, transcription-associated damage (Dinant et al. 2013). In addition to detecting a role for BRCA1 (like that of FACT) in preventing R-loop-associated damage, we also found that BRCA1 accelerates the restart of transcription after UV damage (again, like the FACT complex). This fits with the demonstration by others that BARD1 is important in preventing 3′ mRNA processing after UV damage and that BRCA1–BARD1 complexes promote RNA Pol II degradation after UV damage (Kleiman and Manley 1999, 2001; Kleiman et al. 2005). Conceivably, a BRCA1–TONSL–FACT complex is recruited to active RNA Pol II in such a setting. If there is associated damage or an obstacle in the DNA that can be overcome, then BRCA1, possibly together with FACT and TONSL, aids in restarting transcription. If the DNA damage is too severe, then BARD1–BRCA1 may signal for the degradation of RNA Pol II and aid in the repair of the damage. It will be interesting in the future to see whether BRCA1 and its role as a ubiquitin ligase and/or its functional interaction with PARP1 are linked to the regulation of FACT activity at sites of DNA damage, especially in the setting of transcription-associated DNA damage. FACT is known to be regulated by a complex parsylation signaling cascade in the setting of DNA damage (Huang et al. 2006; Heo et al. 2008).
In keeping with this theme, the interactor TCEA2 is a member of the TFIIS family of transcription elongation factors. TCEA1 (also known as TFIIS or S-II) is the most studied member of this family. TCEA1 promotes transcription elongation-associated RNA transcript cleavage as well as the elimination of transcript errors when RNA Pol II encounters various endogenous and exogenously induced DNA obstacles (Wind and Reines 2000). While TCEA2 was originally identified as a testes- and ovary-specific version of TFIIS (Wind and Reines 2000), it is also expressed in certain somatic cells (Supplemental Fig. S2O). TCEA2 and TCEA1 are homologous for a functional domain that is critical for TFIIS transcription elongation function (Wind and Reines 2000), and both stimulate RNA Pol II in the same way (Wind and Reines 2000). Since BRCA1 and TCEA1 (labeled TFIIS in the figures) colocalized at UV-induced sites of transcription-associated damage, it is likely that the closely related and BRCA1-interacting TCEA2 operates similarly with BRCA1. These hypotheses fit with our observation that BRCA1 depletion is associated with increased sensitivity to the halting of transcription.
The possibility that BRCA1 responds to transcription-associated damage by multiple mechanisms may mean that there are multiple steps in these repair/response pathways that could be targets for mutation and possibly even therapy. The Cancer Genome Atlas (TCGA) data cited earlier (Fig. 7F) are consistent with the notion that the function of multiple transcription-associated proteins is altered across different types of cancers. For example, TCEA2 is amplified across 11 different tumor types, including breast and ovarian cancers (Fig. 7F). One wonders whether this is a manifestation of compensation for ongoing broadly based threats to transcription procession, given the functional interaction of TCEA2 with BRCA1. If this were the case, increasing transcription-associated damage or blocking TCEA2 involvement in such tumors might evolve as part of a rational therapeutic strategy for these cancers.
In considering the therapeutic relevance of the BRCA1 interaction network, HCC38, the cells studied in the α-amanitin rescue experiments, represent a sporadic breast cancer cell line in which the BRCA1 promoter is methylated (Xu et al. 2010). This results in these cells expressing less BRCA1 than normal (Xu et al. 2010). Such a deficiency makes it possible that HCC38 harbors defects in BRCA1 function (Xu et al. 2010). Not only was the HCC38 α-amanitin sensitivity that was induced by BRCA1 depletion rescued after overexpressing BRCA1 cDNA (Fig. 2G), there was also decreased sensitivity to α-amanitin in the BRCA1 cDNA-expressing line compared with naïve HCC38 cells (Fig. 2G, cf. lanes 1 and 3). This suggests that promoter methylation-associated BRCA1 depletion, a relatively common event in sporadic BRCA1+/+ breast and ovarian cancers (Press et al. 2008; Dworkin et al. 2009), can give rise to chronically enhanced sensitivity to α-amanitin or other transcription inhibitors. If so, deficient BRCA1-driven, transcription-associated DNA damage control might prove to be a specific molecular vulnerability or even a therapeutic target in this setting.
There may already be an example of clinical success in targeting this BRCA1-driven transcription-associated damage control in BRCA1 mutant and sporadic basal-like breast tumors. The latter are, in part, BRCA1 mutant breast cancer phenocopies. Multiple clinical trials and retrospective studies of BRCA1 mutant (Silver et al. 2010) and sporadic basal-like breast tumors (Sirohi et al. 2008; Chew et al. 2009; Silver et al. 2010; Staudacher et al. 2011; Hurley et al. 2013) reveal signs of efficacy of the cross-linking agent cisplatin. Cisplatin cross-links cause DNA damage by blocking the procession of transcription or DNA replication (Todd and Lippard 2009). BRCA1 is known to participate in the repair of stalled replication forks, and loss of this function could at least in part explain the therapeutic efficacy of cisplatin in these tumors (Pathania et al. 2011; Schlacher et al. 2012). However, data shown here suggest that a block of BRCA1 participation in transcription-associated damage control could also contribute to cisplatin efficacy in such tumors. Indeed, cisplatin is also known to cause DNA damage and cell death through transcription arrest (Todd and Lippard 2009). Thus, it is reasonable to speculate that cisplatin efficacy in these tumors is further enhanced by eliciting a defect in BRCA1-driven, transcription-associated DNA damage control.
In conclusion, gaining a better understanding of how BRCA1 functions at sites of transcription-associated DNA damage with the interactors identified here and likely other proteins yet to be detected may also prove to be of therapeutic interest. It could provide opportunities to generate biomarkers useful in identifying subsets of breast and ovarian tumors that are susceptible to therapy that targets this pathway. In addition, understanding the methods of BRCA1-dependent and BRCA1-independent repair of transcription-associated damage may be important in other diseases as well. Transcription occurs constantly even after terminal differentiation, such as in neuronal cells and cardiac myocytes (Zhang et al. 2008). Thus, one might speculate that transcription-associated DNA damage plays a role in the pathogenesis of certain diseases in these tissues.
Materials and methods
A detailed explanation of all procedures and statistical analysis performed is included in the Supplemental Material.
Dual-protein interaction screen
The Y2H screen was carried out as previously described (Dreze et al. 2010) using full-length BRCA1 and strategically designed BRCA1 fragments as prey. The TAP-MS analysis was performed on BARD1–BRCA1 complexes purified from the chromatin and soluble nuclear fractions of the nuclei of HeLa S3 cells stably expressing Flag-StrepTactin-tagged BARD1 or empty vector. More details are available in the Supplemental Material.
Tissue culture and transfection
HeLa S3 cells and wild-type and TALEN-induced heterozygotes of U2OS cells were cultured in DMEM containing 10% FBS and 1% penicillin/streptomycin (Pen/Strep). HCC38 cells were cultured in American Type Culture Collection RPMI 1640 (catalog no. 30-2001) with 10% FBS and 1% PenStrep. All siRNA transfections were carried out using Lipofectamine RNAiMax (Life Technologies). More details are available in the Supplemental Material.
Immunofluorescence
UV treatment and micropore staining were carried out as previously described (Polo et al. 2006; Pathania et al. 2011). For UV micropore immunofluorescence, cells were fixed in 3% paraformaldehyde and permeabilized with 0.5% Triton X-100. For SSRP1 staining, the cells were permeabilized first. For RNA Pol II staining (Figs. 5, 6), methanol–acetic acid fixation was used, and the staining was carried out as described previously (Espinosa et al. 2003). More details are available in the Supplemental Material.
Transcription restart assay
UV treatment was performed as previously described (Pathania et al. 2011). U2OS cells transfected with various siRNAs were exposed to either 8J or an equivalent time of air exposure and allowed to recover for various amounts of time before being incubated in medium containing 1 mM 5-ethinyl uridine (EU). The cells were fixed 2 h after addition of EU and stained for measurement of post-UV transcriptional restart using a Life Technology RNA synthesis labeling kit (catalog no. C10329) as described (Dinant et al. 2013). Analysis of nuclear mean fluorescent intensity of the cells was performed using CellProfiler (Carpenter et al. 2006; Kamentsky et al. 2011). A minimum of 300 cells was analyzed for each siRNA at each time point in all three replicates of the experiment. More details are available in the Supplemental Material.
RNASEH1 DNA damage rescue assay
This assay was performed as described previously (Herrera-Moyano et al. 2014). U2OS cells were seeded on coverslips on day 1, transfected with siRNAs on days 2 and 3, transfected with pcDNA3-empty vector or pcDNA3-RNASEH1 on day 4, and fixed and stained for γH2AX and RNASEH1 staining 24 h after the day 4 transfection. The numbers of cells containing greater than five γH2AX foci were counted from the whole population of vector transfected cells and only from RNASEH1-expressing cells in the RNASEH1 transfected population. More details are available in the Supplemental Material.
Competing interest statement
No authors have declared any conflicts of interest with the work reported here. J.K.J. has no financial conflict with the work reported in this manuscript but discloses the following: J.K.J. has a financial interest in Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. J.K.J. is a consultant for and holds equity in Gengine, a company seeking to commercialize genome-editing technologies.
Acknowledgments
We thank members of the Livingston laboratory and Dana-Farber Cancer Institute Center for Cancer Systems Biology for helpful discussions and technical advice, Myles Brown and Min Ni for helpful discussion regarding Oncomine Concepts Map analysis, Xialu Li for the gift of the pcDNA3-RNASEH1 plasmid, and Shailja Pathania for the gift of the pLX304-HA-Myc-BRCA1 plasmid. S.J.H. was supported initially by Department of Defense Breast Cancer Research Program Fellowship W81XWH-08-1-0748 and subsequently by National Cancer Institute (NCI) Fellowship 1F30CA167895-01. This work was supported by grants from the Breast Cancer Research Foundation, the Susan G. Komen Foundation for the Cure (SAC110022), an NCI SPORE grant (P50CA089393) in breast cancer research to the Dana-Farber/Harvard Cancer Center, and R01 (R01CA136512) and P01 (P01CA080111) grants from the NCI to D.M.L. D.R., S.Q.T., and J.K.J. were supported by a National Institutes of Health (NIH) Director’s Pioneer Award (DP1 GM105378) and the Jim and Ann Orr Massachusetts General Hospital Research Scholar Award. J.A.M. and G.A. were supported by a National Institute of Neurological Disorders and Stroke P01 grant (P01NS047572), the Dana-Farber Cancer Institute Strategic Research Initiative, the Susan Smith Center for Women’s Cancers, and The Honorable Tina Brozman Foundation. This work was also supported by National Human Genome Research Institute grant R01HG001715 awarded to M.V. and D.E.H., and NCI grant U54CA112962 subaward to M.V. S.J.H. and D.M.L. conceived the project. S.J.H., D.M.L., and D.E.H. wrote the paper with contributions to the Materials and Methods from T.R., G.A., X.X., S.G., and M.S.O. S.J.H. performed the experiments, with contributions from G.A., A.D., Y.J., and N.S. Data analysis was performed by S.J.H., with contributions from T.R., G.A., T.I.Z., M.S.O., X.X., S.G., and T.H. D.M.L., D.E.H., M.V., J.A.M., R.B., K.M.M., S.G., and J.K.J. advised on various technologies. D.R., S.Q.T., and J.K.J. designed and generously provided the TALEN plasmids. A.P.C. provided the BRCA1 cDNA-expressing HCC38 cell line.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.241620.114.
References
- Aguilera A, Garcia-Muse T. 2012. R loops: from transcription byproducts to threats to genome stability. Mol Cell 46: 115–124 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherel OJ, Yeo AJ, Stellati A, Heng EY, Luff J, Suraweera AM, Woods R, Fleming J, Carrie D, McKinney K, et al. 2013. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet 9: e1003435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. 2014. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511: 362–365 [DOI] [PubMed] [Google Scholar]
- Braun P, Tasan M, Dreze M, Barrios-Rodiles M, Lemmens I, Yu H, Sahalie JM, Murray RR, Roncari L, de Smet AS, et al. 2009. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods 6: 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassonnet P, Rolloy C, Neveu G, Vidalain PO, Chantier T, Pellet J, Jones L, Muller M, Demeret C, Gaud G, et al. 2011. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat Methods 8: 990–992 [DOI] [PubMed] [Google Scholar]
- Chew HK, Doroshow JH, Frankel P, Margolin KA, Somlo G, Lenz HJ, Gordon M, Zhang W, Yang D, Russell C, et al. 2009. Phase II studies of gemcitabine and cisplatin in heavily and minimally pretreated metastatic breast cancer. J Clin Oncol 27: 2163–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh LA, Fire A, Samuels M, Sharp PA. 1989. 5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J Biol Chem 264: 2250–2257 [PubMed] [Google Scholar]
- Collier LS, Largaespada DA. 2007. Transposons for cancer gene discovery: Sleeping Beauty and beyond. Genome Biol 8: S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, Theil AF, van Cappellen WA, Kimura H, Bartek J, et al. 2013. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell 51: 469–479 [DOI] [PubMed] [Google Scholar]
- Dreze M, Monachello D, Lurin C, Cusick ME, Hill DE, Vidal M, Braun P. 2010. High-quality binary interactome mapping. Methods Enzymol 470: 281–315 [DOI] [PubMed] [Google Scholar]
- Duro E, Lundin C, Ask K, Sanchez-Pulido L, MacArtney TJ, Toth R, Ponting CP, Groth A, Helleday T, Rouse J. 2010. Identification of the MMS22L–TONSL complex that promotes homologous recombination. Mol Cell 40: 632–644 [DOI] [PubMed] [Google Scholar]
- Dworkin AM, Huang TH, Toland AE. 2009. Epigenetic alterations in the breast: implications for breast cancer detection, prognosis and treatment. Semin Cancer Biol 19: 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM. 2003. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell 12: 1015–1027 [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. 2004. A census of human cancer genes. Natl Rev 4: 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg RA 2008. Recognition of DNA double strand breaks by the BRCA1 tumor suppressor network. Chromosoma 117: 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Nudler E, Tora L. 2013. Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol 20: 412–418 [DOI] [PubMed] [Google Scholar]
- Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. 2008. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell 30: 86–97 [DOI] [PubMed] [Google Scholar]
- Herrera-Moyano E, Mergui X, Garcia-Rubio ML, Barroso S, Aguilera A. 2014. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev 28: 735–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Chen WH, Chang YL, Wang HT, Chuang WT, Lee SC. 2006. Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucleic Acids Res 34: 2398–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, Chen J. 2010. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 11: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J, Reis IM, Rodgers SE, Gomez-Fernandez C, Wright J, Leone JP, Larrieu R, Pegram MD. 2013. The use of neoadjuvant platinum-based chemotherapy in locally advanced breast cancer that is triple negative: retrospective analysis of 144 patients. Breast Cancer Res Treat 138: 783–794 [DOI] [PubMed] [Google Scholar]
- Joung JK, Sander JD. 2013. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. 2011. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27: 1179–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL. 1999. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science 285: 1576–1579 [DOI] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL. 2001. The BARD1–CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 104: 743–753 [DOI] [PubMed] [Google Scholar]
- Kleiman FE, Wu-Baer F, Fonseca D, Kaneko S, Baer R, Manley JL. 2005. BRCA1/BARD1 inhibition of mRNA 3′ processing involves targeted degradation of RNA polymerase II. Genes Dev 19: 1227–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Page F, Randrianarison V, Marot D, Cabannes J, Perricaudet M, Feunteun J, Sarasin A. 2000. BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesion in human cells. Cancer Res 60: 5548–5552 [PubMed] [Google Scholar]
- Li X, Manley JL. 2006. Cotranscriptional processes and their influence on genome stability. Genes Dev 20: 1838–1847 [DOI] [PubMed] [Google Scholar]
- Lindell TJ, Weinberg F, Morris PW, Roeder RG, Rutter WJ. 1970. Specific inhibition of nuclear RNA polymerase II by α-amanitin. Science 170: 447–449 [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639 [DOI] [PubMed] [Google Scholar]
- Mechali M, Lutzmann M. 2008. The cell cycle: now live and in color. Cell 132: 341–343 [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66–71 [DOI] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikkila J, Christin Parplys A, Pylkas K, Bose M, Huo Y, Borgmann K, Rapakko K, Nieminen P, Xia B, Pospiech H, et al. 2013. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat Commun 4: 2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell BC, Adamson B, Lydeard JR, Sowa ME, Ciccia A, Bredemeyer AL, Schlabach M, Gygi SP, Elledge SJ, Harper JW. 2010. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L–NFKBIL2 complex required for genomic stability. Mol Cell 40: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell L, Panier S, Wildenhain J, Tkach JM, Al-Hakim A, Landry MC, Escribano-Diaz C, Szilard RK, Young JT, Munro M, et al. 2010. The MMS22L–TONSL complex mediates recovery from replication stress and homologous recombination. Mol Cell 40: 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania S, Nguyen J, Hill SJ, Scully R, Adelmant GO, Marto JA, Feunteun J, Livingston DM. 2011. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol Cell 44: 235–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwko W, Olma MH, Held M, Bianco JN, Pedrioli PG, Hofmann K, Pasero P, Gerlich DW, Peter M. 2010. RNAi-based screening identifies the Mms22L–Nfkbil2 complex as a novel regulator of DNA replication in human cells. EMBO J 29: 4210–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Roche D, Almouzni G. 2006. New histone incorporation marks sites of UV repair in human cells. Cell 127: 481–493 [DOI] [PubMed] [Google Scholar]
- Press JZ, De Luca A, Boyd N, Young S, Troussard A, Ridge Y, Kaurah P, Kalloger SE, Blood KA, Smith M, et al. 2008. Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al. 2007. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet 39: 1338–1349 [DOI] [PubMed] [Google Scholar]
- Reinberg D, Sims RJ 3rd. 2006. De FACTo nucleosome dynamics. J Biol Chem 281: 23297–23301 [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Tomlins SA, Mahavisno V, Kasper N, Varambally R, Barrette TR, Ghosh D, Varambally S, Chinnaiyan AM. 2007. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia 9: 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, et al. 2012. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487: 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. 2005. Towards a proteome-scale map of the human protein–protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132: 487–498 [DOI] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51–BRCA1/2. Cancer Cell 22: 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Anderson SF, Chao DM, Wei W, Ye L, Young RA, Livingston DM, Parvin JD. 1997. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci 94: 5605–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, et al. 2010. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol 28: 1145–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi B, Arnedos M, Popat S, Ashley S, Nerurkar A, Walsh G, Johnston S, Smith IE. 2008. Platinum-based chemotherapy in triple-negative breast cancer. Ann Oncol 19: 1847–1852 [DOI] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42: 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O’Sullivan MG, Matise I, Dupuy AJ, et al. 2009. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 323: 1747–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudacher L, Cottu PH, Dieras V, Vincent-Salomon A, Guilhaume MN, Escalup L, Dorval T, Beuzeboc P, Mignot L, Pierga JY. 2011. Platinum-based chemotherapy in metastatic triple-negative breast cancer: the Institut Curie experience. Ann Oncol 22: 848–856 [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. 2005. A human protein-protein interaction network: a resource for annotating the proteome. Cell 122: 957–968 [DOI] [PubMed] [Google Scholar]
- Todd RC, Lippard SJ. 2009. Inhibition of transcription by platinum antitumor compounds. Metallomics 1: 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, et al. 2009. An empirical framework for binary interactome mapping. Nat Methods 6: 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. 2013. Cancer genome landscapes. Science 339: 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 14: 927–939 [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. 2006. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol 409: 524–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wind M, Reines D. 2000. Transcription elongation factor SII. BioEssays 22: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods NT, Mesquita RD, Sweet M, Carvalho MA, Li X, Liu Y, Nguyen H, Thomas CE, Iversen ES Jr, Marsillac S, et al. 2012. Charting the landscape of tandem BRCT domain-mediated protein interactions. Sci Signal 5: rs6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. 1996. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet 14: 430–440 [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22: 719–729 [DOI] [PubMed] [Google Scholar]
- Xu X, Qiao W, Linke SP, Cao L, Li WM, Furth PA, Harris CC, Deng CX. 2001. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet 28: 266–271 [DOI] [PubMed] [Google Scholar]
- Xu J, Huo D, Chen Y, Nwachukwu C, Collins C, Rowell J, Slamon DJ, Olopade OI. 2010. CpG island methylation affects accessibility of the proximal BRCA1 promoter to transcription factors. Breast Cancer Res Treat 120: 593–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J. 2003. The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
- Yuce O, West SC. 2013. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol Cell Biol 33: 406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. 2013. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]