Elevated glycolysis is a common metabolic trait of cancer, but what drives such metabolic reprogramming remains incompletely clear. Liu et al. found that ZBTB7A, a member of the POK transcription repressor family, directly binds to the promoter and represses the transcription of critical glycolytic genes. The ZBTB7A locus is frequently deleted in many human tumors. While ZBTB7A-deficient tumors progressed exceedingly fast, they exhibited an unusually heightened sensitivity to glycolysis inhibition.
Keywords: glycolysis, tumor suppressor, ZBTB7A, GLUT3, PFKP, PKM
Abstract
Elevated glycolysis is a common metabolic trait of cancer, but what drives such metabolic reprogramming remains incompletely clear. We report here a novel transcriptional repressor-mediated negative regulation of glycolysis. ZBTB7A, a member of the POK (POZ/BTB and Krüppel) transcription repressor family, directly binds to the promoter and represses the transcription of critical glycolytic genes, including GLUT3, PFKP, and PKM. Analysis of The Cancer Genome Atlas (TCGA) data sets reveals that the ZBTB7A locus is frequently deleted in many human tumors. Significantly, reduced ZBTB7A expression correlates with up-regulation of the glycolytic genes and poor survival in colon cancer patients. Remarkably, while ZBTB7A-deficient tumors progress exceedingly fast, they exhibit an unusually heightened sensitivity to glycolysis inhibition. Our study uncovers a novel tumor suppressor role of ZBTB7A in directly suppressing glycolysis.
Metabolic reprograming is a hallmark of human cancer and accelerated aerobic glycolysis (or the Warburg effect) and is frequently observed in multiple types of cancer (Hanahan and Weinberg 2011). Tumor cells metabolize most glucose into lactate and thus generate abundant glycolytic intermediates as precursors for macromolecular biosynthesis, which enables tumor cells to meet their increased anabolic and energetic demands due to rapid tumor growth (Vander Heiden et al. 2009). A thorough understanding of the molecular mechanisms underlying the Warburg effect may shed new light on tumorigenesis and provide novel strategies for cancer treatment.
Oncogenic pathways have been shown to contribute to the metabolic adaptation of neoplastic cells (Hanahan and Weinberg 2011). Well-known examples include that oncogenic Akt stimulates aerobic glycolysis (Elstrom et al. 2004). While responsible for increased glutaminolysis, Myc also promotes glycolysis through regulating directly or indirectly the expression of glycolytic genes (Dang 2012). The mammalian target of rapamycin complex 1 (mTORC1) enhances glycolysis by stimulating the expression of hypoxia-inducible factor 1a (HIF1a) (Laughner et al. 2001; Düvel et al. 2010). Tumor suppressor p53 favors oxidative phosphorylation by inducing the expression of synthesis of cytochrome c oxidase (SCO2) and the TP53-induced glycolysis and apoptosis regulator (TIGAR) that inhibits glycolysis (Berkers et al. 2013).
Human cancer genomic analysis indicates that glycolytic genes are among the most commonly up-regulated genes in cancer (Altenberg and Greulich 2004; Majumder et al. 2004), implicating a mechanism of transcriptional regulation in the metabolic reprogramming of cancer. MYC and HIF-1 are currently known transcription factors that stimulate the expression of glycolysis genes (Gordan et al. 2007). In addition, nuclear receptors, including estrogen-related receptors, represent another class of transcription factors important in the regulation of glycolysis (Deblois and Giguère 2013). However, it is unclear whether there is any transcription repressor that counterbalances the effect of these oncogenic transcription factors on cellular metabolism.
ZBTB7A—also known as POKEMON (Maeda et al. 2005), FBI (Pessler et al. 1997), LRF (Liu et al. 2004), and OCZF (Kukita et al. 1999)—is a member of the POZ/BTB and Krüppel (POK) family of transcription factors. POK family transcription factors can bind DNA through a Krüppel-like DNA-binding domain and repress transcription by recruiting corepressor complexes through the POZ (poxvirus and zinc finger) domain (Costoya 2007). ZBTB7A is originally known as a proto-oncoprotein due to its ability to suppress the transcription of tumor suppressor gene ARF (Maeda et al. 2005). Overexpression of Zbtb7a in immature mouse T and B lymphoid lineage leads to aggressive lymphomas, consistent with a proto-oncogenic role of ZBTB7A in lymphoma (Maeda et al. 2005). However, the frequent loss of the ZBTB7A gene locus (19p13.3) in many types of human carcinoma (Beroukhim et al. 2010; Zack et al. 2013) argues against it as an oncogene, at least in solid tumors, implicating ZBTB7A as a context-dependent cancer gene. Interestingly, a tumor suppressor role of Zbtb7a was implicated in a recent study that loss of Zbtb7a augmented tumorigenesis of mouse prostate cancer in a Pten-deficient background (Wang et al. 2013), although the mechanism underlying this reported tumor-suppressive activity of ZBTB7A remained incompletely clear.
We report here the discovery of an unexpected function of ZBTB7A in transcriptional suppression of glycolysis. By directly binding to the promoter and repressing the transcription of critical glycolytic genes GLUT3, PFKP, and PKM, ZBTB7A negatively regulates glycolysis. Significantly, ZBTB7A is found frequently down-regulated in many human solid tumors, and, moreover, ZBTB7A-deficient tumors are hypersensitive to the inhibition of glycolysis.
Results
ZBTB7A suppresses glycolysis metabolism
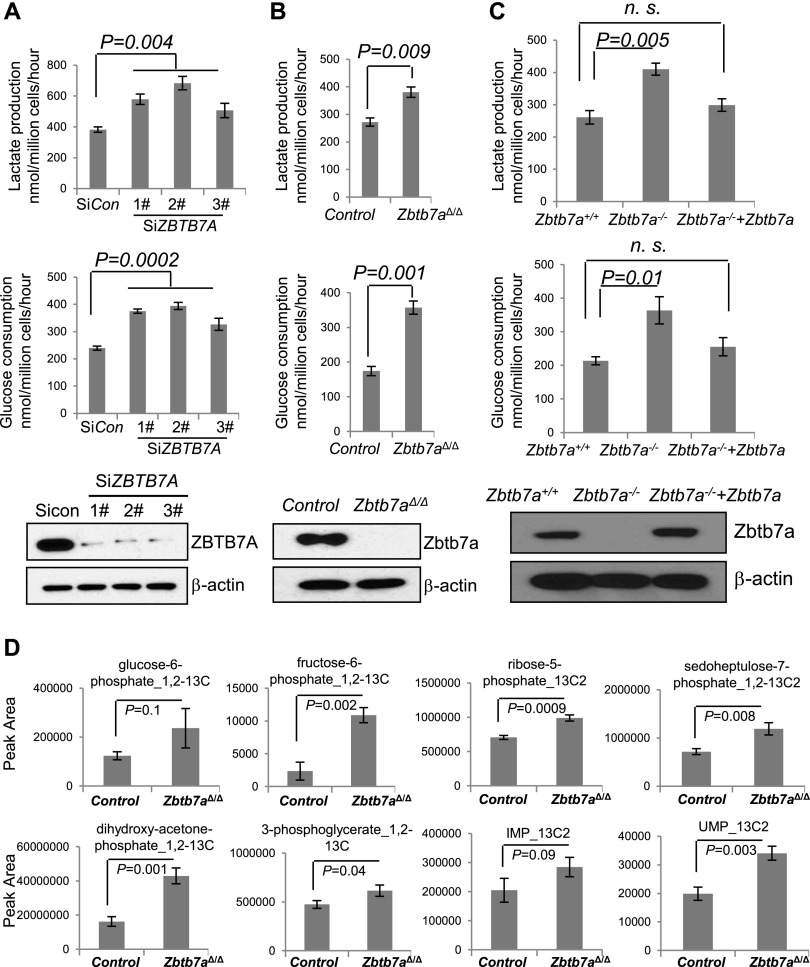
Transcriptional networks play a crucial role in the regulation of cellular metabolism. Well-known examples are the transcription factors HIF-1 and MYC; when overexpressed, they induce increased expression of glycolytic genes, contributing to elevated aerobic glycolysis (or the Warburg effect) (Dang 1999; Denko 2008; Yeung et al. 2008). It is less clear, however, whether an opposite mechanism or transcription repression may exist to keep glycolytic genes in check. We explored this question by testing ZBTB7A because (1) it is a member of the POK family of transcription repressors (Maeda et al. 2005; Kelly and Daniel 2006; Costoya 2007); (2) recent studies of somatic copy number alterations in human cancers revealed that chromosome 19p13.3, which contains the ZBTB7A gene, is frequently lost in many types of human carcinoma (Beroukhim et al. 2010; Zack et al. 2013); and (3) a marked acidification of the culture medium of ZBTB7A-deficient cells was observed, indicated by a rapid change to yellowish (data not shown). To determine whether this transcription repressor might play any role in the regulation of metabolism, we measured metabolites in the medium of ZBTB7A-deficient and ZBTB7A-proficient cells. Indeed, ZBTB7A-depleted U2OS cells, generated with three independent RNAi sequences (Fig. 1A, bottom), secreted significantly more lactate than control RNAi cells (Fig. 1A, top). Since lactate is produced as the final product of glycolysis, we measured glucose levels and found a marked increase in glucose consumption in ZBTB7A-depleted cells relative to control cells (Fig. 1A, middle). Likewise, Zbtb7a-deleted mouse embryonic fibroblasts (MEFs) exhibited significant increases in glucose consumption and lactate production relative to wild-type controls (Fig. 1B). The ZBTB7A-mediated regulation of glycolysis was further confirmed by restoration of Zbtb7a expression in Zbtb7a-null MEFs, which brought glucose consumption and lactate production back to wild-type levels (Fig. 1C). We substantiated this finding in three additional human cancer cell lines—PC3, HCT116, and M14—where depletion of ZBTB7A with multiple RNAi sequences resulted in induction of glucose consumption and lactate production (Supplemental Fig. S1A–C). The results together suggest a role for ZBTB7A in the attenuation of cellular glycolysis.
Figure 1.
ZBTB7A suppresses glycolytic metabolism in mammalian cells. (A) U2OS cells were transfected with either siControl (Sicon) or three independent ZBTB7A targeting siRNA sequences (SiZBTB7A). (Bottom) The knockdown efficiency was determined by Western analysis. Culture media from an equal number of cells were collected 48 h after transfection for lactate measurement (top) and glucose consumption determination (middle). The numbers are mean ± SD from three independent experiments. (B) Zbtb7a-deleted MEFs (Zbtb7aΔ/Δ MEF) and control MEFs were similarly analyzed for lactate production (top) and glucose consumption (middle). The numbers represent mean ± SD from three independent experiments. (Bottom) Western blot analysis was performed to determine the level of ZBTB7A expression. (C, bottom) The expression of ZBTB7A was restored in Zbtb7a−/− MEFs. Zbtb7a+/+, Zbtb7a−/−, and Zbtb7a−/−+Zbtb7a MEFs were subjected to analysis of lactate production (top) and glucose consumption (middle). Data from three experiments are presented as mean values ± SD. (D) Zbtb7a-deleted and wild-type control MEFs were incubated with [1,2-13C]-glucose for 15 min prior to metabolite extraction and targeted liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. The ratio of 13C-labeled to unlabeled (12C) metabolites was measured by LC-MS/MS and is presented as mean ± SD over four independent samples. Metabolites with P-values for pairwise comparisons are shown.
To further characterize the metabolic changes associated with Zbtb7a deficiency, we used stable isotope labeling with [1,2-13C]-glucose to measure relative flux through glycolysis in Zbtb7a-deleted MEFs (Supplemental Fig. S1D). A 15-min pulse label revealed increased flux through multiple glycolytic intermediates (glucose-6-phosphate, fructose-6-phosphate, dihydroxy-acetone-phosphate, and 3-phosphoglycerate) in Zbtb7a-deleted cells relative to controls (Fig. 1D). Interestingly, flux into intermediates of the pentose phosphate pathway (PPP; ribose-5-phosphate and sedoheptulose-7-phosphate) and the downstream products of purine and pyrimidine metabolism (IMP and UMP) were also increased (Fig. 1D). These data demonstrate that Zbtb7a loss results in the induction of glucose flux into glycolysis and the PPP, consistent with a Zbtb7a-mediated suppression of these metabolic processes.
ZBTB7A suppresses the expression of multiple glycolysis genes
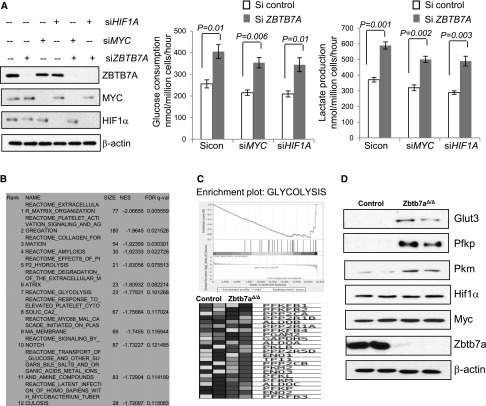
Given that HIF-1α and MYC are key inducers of the glycolytic phenotype in cancer cells (Dang 1999; Denko 2008), we asked whether ZBTB7A-mediated regulation of glycolysis depended on these transcription factors. Comparison of ZBTB7A-deficient and ZBTB7A-proficient cells revealed little difference in the HIF-1α protein abundance (Fig. 2A), and knockdown of HIF-1α had no detectable effect on the induction of glucose consumption and lactate production upon ZBTB7A depletion (Fig. 2A), demonstrating that ZBTB7A regulates glycolysis independent of HIF-1α. Similarly, MYC was also found to be dispensable for ZBTB7A-mediated regulation of glycolysis, as knockdown of MYC had little impact on the increased glycolysis in ZBTB7A-deficient cells (Fig. 2A).
Figure 2.
ZBTB7A suppresses glycolytic metabolism via a MYC- or HIF-1-independent mechanism. (A) U2OS cells were transfected with siRNA targeting HIF-1, MYC, or ZBTB7A. The knockdown efficiency was determined by Western analysis. Forty-eight hours after transfection, the cells were subjected to analysis of lactate production and glucose consumption. The numbers represent mean ± SD from three independent experiments. (B) GSEA of microarray expression data of control and Zbtb7aΔ/Δ MEFs. (C) Genes that were significantly enriched when the levels of expression were compared between control and Zbtb7aΔ/Δ MEFs are shown. (D) Lysates from Zbtb7aΔ/Δ and control MEF were analyzed by Western blot with the indicated antibodies.
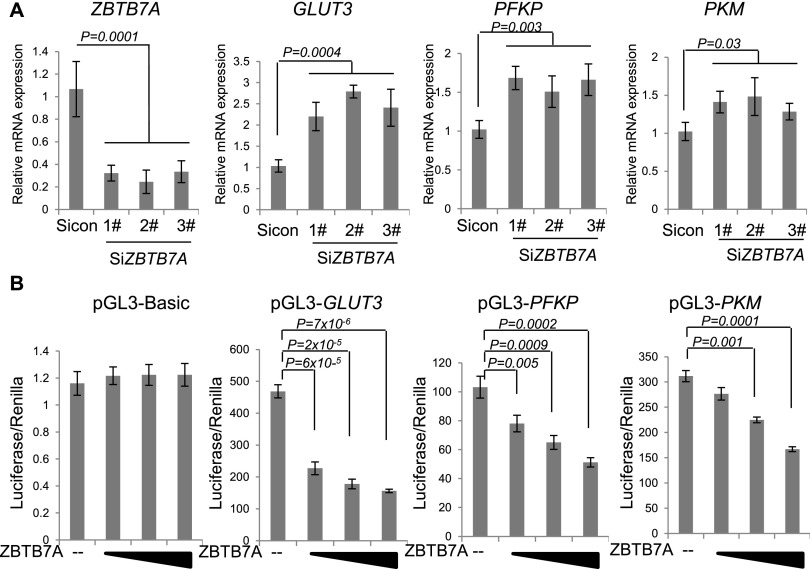
To reveal potential mechanisms of ZBTB7A-mediated control of metabolism, we performed microarray analysis of differentially expressed genes between Zbtb7a-proficient and Zbtb7a-deficient MEFs. Gene set enrichment analysis (GSEA) revealed that metabolic genes were within the top gene clusters enriched in differentially expressed genes (Fig. 2B,C). Among glycolytic genes, the transcript levels of Glut3, Pfkp, and Pkm were significantly increased in Zbtb7a-deficient cells, which was verified via quantitative RT–PCR (qRT–PCR) analysis (Supplemental Fig. S2). Immunoblots revealed a marked increase of the three metabolic proteins in Zbtb7a-deficient cells relative to the control cells (Fig. 2D). As found with ZBTB7A knockdown in U2OS cells, HIF-1α and MYC levels were indistinguishable in Zbtb7a-null MEFs, consistent with a lack of their involvement in ZBTB7A-mediated regulation of the glycolytic genes (Fig. 2D). siRNA-mediated knockdown of ZBTB7A expression with multiple sequences resulted in transcriptional up-regulation of the three glycolytic genes (Fig. 3A). In addition, we created human GLUT3, PFKP, or PKM promoter-driven luciferase reporters to assess the inhibitory effect of ZBTB7A. Indeed, expression of ZBTB7A resulted in a dose-dependent inhibition of the promoter activity of GLUT3, PFKP, and PKM (Fig. 3B). The specific effect of ZBTB7A on these targets was further corroborated by the observation that there was little change in expression from other glycolytic gene promoters, including a number of known HIF-1α target genes (Supplemental Fig. S3). The data together suggest that ZBTB7A suppresses glycolytic metabolism by down-regulation of the expression of key glycolytic genes.
Figure 3.
ZBTB7A suppresses the expression of GLUT3, PFKP, and PKM. (A) U2OS cells expressing Sicon or three independent SiZBTB7A were harvested 48 h after transfection, and mRNA was isolated for qRT–PCR analysis for the expression of ZBTB7A, GLUT3, PFKP, or PKM. Data from four experiments are presented as mean values ± SD with associated P-values. (B) Luciferase reporter assays were performed by cotransfection of pGL3-Basic, GLUT3, PFKP, or PKM with increasing doses of ZBTB7A-expressing plasmid. Luciferase activity was measured 36 h post-transfection. Data from three experiments are presented as mean values ± SD with associated P-values.
ZBTB7A is a bona fide transcriptional repressor of GLUT3, PFKP, and PKM
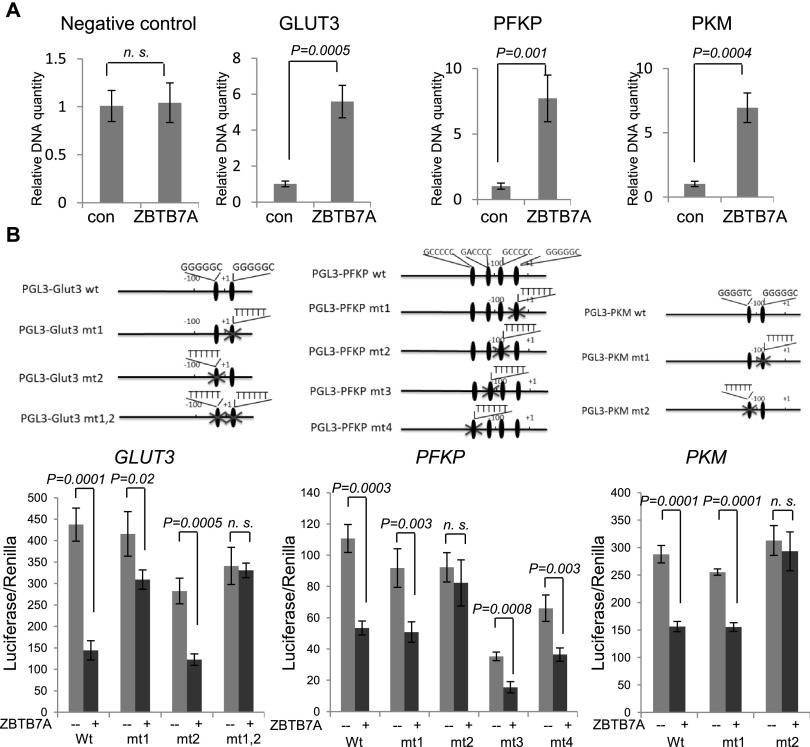
We next investigated the mechanism by which ZBTB7A represses the transcription of the glycolytic genes. It has been reported that POK family proteins can bind to DNA through its zinc finger domain and repress the transcription of target genes by recruiting a corepressor complex to the promoter (Melnick et al. 2002). Therefore, we used chromatin immunoprecipitation (ChIP) to test whether ZBTB7A might repress the expression of these three glycolytic genes via binding to their promoter. The results indicated that ZBTB7A antibody but not control IgG specifically precipitated the promoter sequence of GLUT3, PFKP, or PKM but not control genomic sequences in HeLa cells (Fig. 4A), supporting a direct binding of ZBTB7A to the promoter of the glycolytic genes. To further define ZBTB7A-mediated regulation of glycolytic genes, we analyzed the promoter sequences to identify specific binding sites. ZBTB7A binds to GC-rich DNA sequences (Maeda et al. 2005). Interestingly, the promoters of the three glycolytic genes contain multiple putative ZBTB7A-binding sites; these binding sites were named sites 1–4 based on their distance from the transcription start sites (Supplemental Fig. S4). To identify the binding site, we mutated the putative binding sites individually or in combination within the luciferase reporter constructs (Fig. 4B). As shown in Figure 4B, specific mutation of both predicted ZBTB7A-binding sites in GLUT3, site 2 in PFKP, or site 2 in PKM promoter reporter, respectively, attenuated the ability of ZBTB7A to repress expression from these promoters, whereas mutation of other putative ZBTB7A-binding sites had little effect. The results together establish ZBTB7A as a bona fide transcriptional repressor of GLUT3, PFKP, and PKM.
Figure 4.
ZBTB7A binds to human GLUT3, PFKP, and PKM promoters and directly represses their transcription. (A) ChIP assays were performed in HeLa cells with anti-ZBTB7A antibody or control IgG. The abundance of DNA within the GLUT3, PFKP, or PKM promoter region and minimum protein-binding intragenic DNA were assessed by quantitative real-time PCR with each specific primer. Data from three independent experiments are presented as mean values ± SD. (B, top) The putative ZBTB7A-binding sites within human GLUT3, PFKP, or PKM promoter are shown. Mutation was introduced individually or in combination into each promoter sequence. (Bottom) The luciferase vector pGL3 driven by either the wild-type or mutant promoter was transfected, and luciferase activity was measured. Data from three experiments are presented as mean values ± SD.
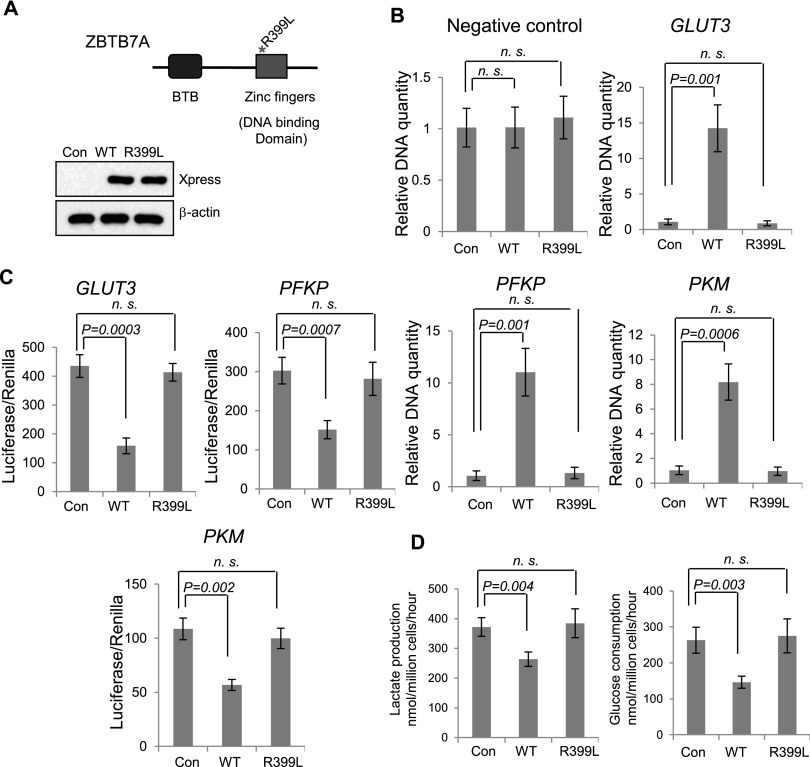
We also generated a zinc finger (R399L) mutant of ZBTB7A to additionally test the requirement of promoter binding in ZBTB7A-mediated transcriptional repression of glycolytic genes (Fig. 5A, top). When comparable protein levels were expressed (Fig. 5A, bottom), in contrast to wild-type ZBTB7A, the R399L mutant was defective in binding to the promoter of GLUT3, PFKP, or PKM, as indicated by ChIP assay (Fig. 5B). The promoter-driven luciferase reporter assays revealed that, unlike the wild-type counterpart, ZBTB7A (R399L) failed to suppress the promoter activity (Fig. 5C). Consistently, this zinc finger mutant was no longer capable of suppressing glycolysis, as indicated by its inability to inhibit lactate production and glucose consumption (Fig. 5D). The data together indicate that ZBTB7A suppresses glycolysis via directly binding to the promoter and repressing the expression of these glycolytic genes.
Figure 5.
ZBTB7A zinc finger mutant R399L is defective in binding to the glycolysis gene promoters and unable to repress glycolysis. (A, top) Domain architecture of ZBTB7A protein and the design of ZBTB7A zinc finger mutant R399L. Western blot analysis was performed to determine the expression of the R399L mutant and wild-type ZBTB7A. Both the R399L mutant and wild-type ZBTB7A were tagged with Xpress. (B) Xpress-tagged wild-type ZBTB7A (WT), R399L mutated ZBTB7A (R399L), and control Xpress vector were transfected in U2OS cells, and ChIP was performed with Xpress Ab. Primers specific to GLUT3, PFKP, or PKM promoter as in Figure 4A were used to determine DNA abundance with quantitative real-time PCR. Data shown are average values of three experiments, with error bars indicating mean ± SD. (C) Luciferase assays as described in Figure 4B were carried out to determine the ability of ZBTB7A R399L, with its wild-type counterpart as a control, to suppress the activity of GLUT3, PFKP, and PKM promoters, respectively. The data shown are average values of three experiments, with error bars indicating mean ± SD. (D) Plasmids encoding wild-type or R399L ZBTB7A were expressed in U2OS cells. Glucose consumption and lactate production were measured. Lactate produced and glucose consumed per 106 cells in a 1-h period of three experiments are shown. Error bars represent mean ± SD.
Genetic loss of ZBTB7A in multiple types of human cancer
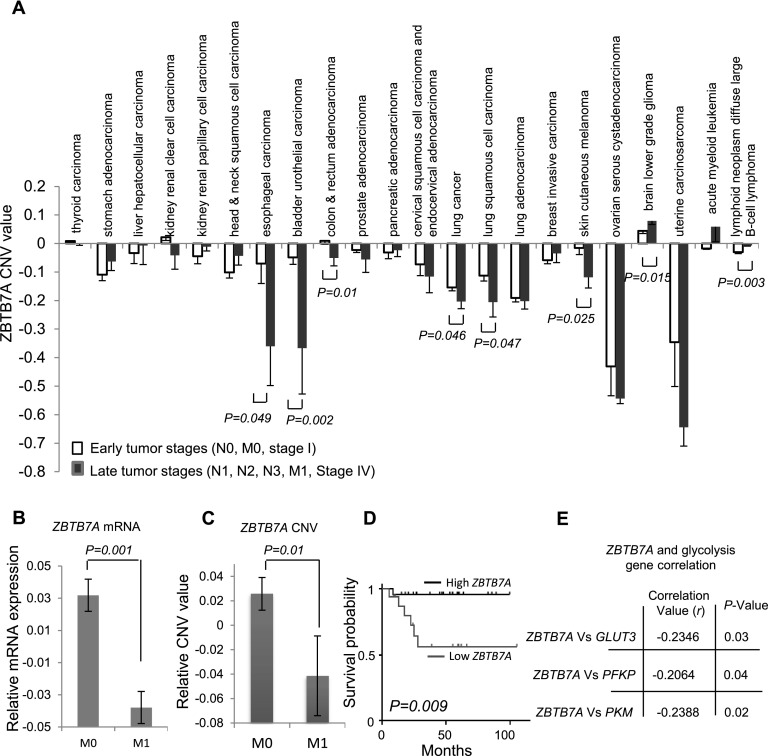
Given its role in controlling aerobic glycolysis, we sought to determine whether ZBTB7A plays a role in cancer by mining The Cancer Genome Atlas (TCGA) databases. In agreement with one of the genes localized within chromosome 19p13.3, which is frequently lost in human cancers (Beroukhim et al. 2010; Zack et al. 2013), a significant decrease of ZBTB7A copy number variation (CNV) was seen in many types of human carcinoma, especially at the late stage of tumors (Fig. 6A). Using colon carcinoma as a representative, we conducted further analysis and discovered a close association of reduced mRNA level and CNV of ZBTB7A (Supplemental Fig. S5A), implicating chromosome loss as an important mechanism for ZBTB7A down-regulation. Interestingly, both ZBTB7A transcripts (Fig. 6B) and CNV (Fig. 6C) were significantly lower in the late stage relative to the early stage of tumors. There was also a significant correlation of ZBTB7A expression with patient outcomes, as lower levels of ZBTB7A transcripts (Fig. 6D) and CNV (Supplemental Fig. S5B) were associated with poorer patient survival. Moreover, in line with our findings of ZBTB7A-mediated suppression of specific glycolytic genes, the expression of GLUT3, PFKP, and PKM showed an inverse correlation with the expression of ZBTB7A in human colon cancer specimens (Fig. 6E; Supplemental Fig. S5C). Together, the results from human cancer genome data analysis implicate ZBTB7A as a tumor suppressor and its loss as a novel genetic event contributing to the elevated expression of glycolytic genes and altered metabolic program, at least in colon cancer cells.
Figure 6.
Genetic loss of ZBTB7A in multiple types of human cancer. (A) CNV values of ZBTB7A in early stages (N0, M0, and stage I) and late stages (N1, N2, N3, M1, and stage IV) of multiple types of human cancer were downloaded from the TCGA SNP array data set. Average CNV values and associated SEM and P-values are shown. Expression levels of ZBTB7A mRNA (B) and relative copy number variation (CNV) values (C) in human colorectal cancer at M0 (without distant metastasis) or M1 (with distant metastasis) stage are shown. Error bars represent mean ± SEM. (D) Kaplan-Meier overall survival curves of colorectal cancer patients with high or low ZBTB7A mRNA expression. (E) The correlation of mRNA levels of ZBTB7A with GLUT3, PFKP, or PKM in human colorectal cancer.
ZBTB7A-deficient cancer cells display increased dependency on glycolysis
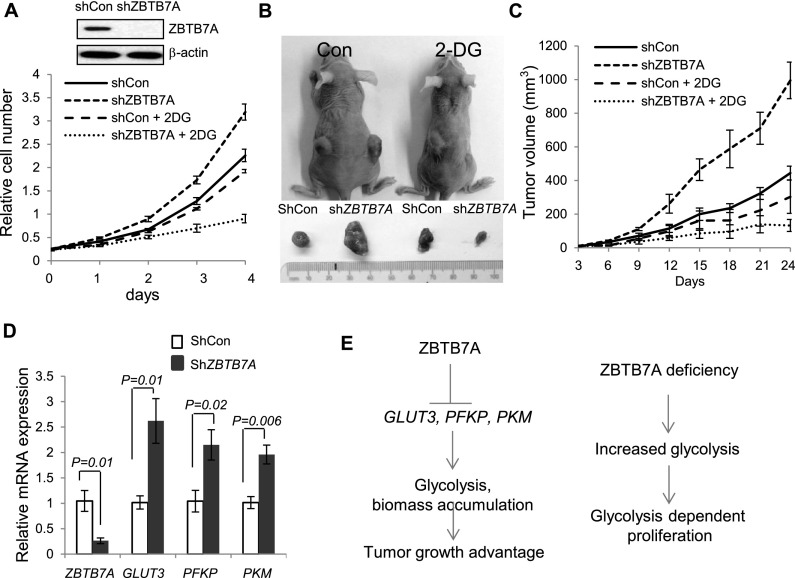
Considering the importance of glycolytic metabolism in supporting cancer cell proliferation (Vander Heiden et al. 2009; Cairns et al. 2011; Lunt and Vander Heiden 2011), we examined whether the enhanced glycolysis in ZBTB7A-deficient cells might affect the rate of cell proliferation. Indeed, shRNA-mediated knockdown of ZBTB7A in colon carcinoma cells HCT116 resulted in a considerable increase in cell proliferation relative to cells expressing control shRNAs (Fig. 7A). Importantly, ZBTB7A-deficient cells exhibited an increased dependence on glycolytic metabolism. Treatment with the glycolytic inhibitor 2-deoxy-D-glucose (2-DG) slowed markedly the proliferation of cells lacking ZBTB7A while having a relatively milder impact on the control cells (Fig. 7A). To complement the result obtained from shRNA experiments, we expressed ZBTB7A in SW48 cells, a human colon carcinoma cell line expressing a relatively low endogenous level of ZBTB7A. Significantly, the introduction of ZBTB7A expression in SW48 colon cancer cells was associated with a marked inhibition of proliferation (Supplemental Fig. S6A) and of the expression of glycolytic genes (Supplemental Fig. S6B), supporting a tumor suppressor role for ZBTB7A.
Figure 7.
ZBTB7A suppresses human colon carcinoma cell growth by inhibiting glycolytic metabolism. (A) Cell growth curve of HCT116 cells stably expressing shCon or shZBTB7A construct in the absence or presence of 2 mM 2-DG. Data from three independent experiments are presented as mean values ± SD. The efficiency of knockdown by shZBTB7A was determined by Western blot. (B) Representative images of tumor-bearing mice implanted with HCT116 cells expressing shZBTB7A or shCon that were treated with or without 2-DG. (C) Tumor volume was measured in mice implanted with shCon or shZBTB7A-expressing HCT116 cells. Data shown are mean ± SD of five mice. (D) Tumor tissues from the indicated mice were dissected, and mRNA was isolated. The mRNA levels of ZBTB7A, GLUT3, PFKP, or PKM were determined by qRT–PCR. Data from three experiments are presented as mean ± SD. (E) Working model for the role of ZBTB7A in regulating glycolysis and cell proliferation.
To determine the relevance of ZBTB7A-mediated regulation of cellular metabolism for tumorigenesis, we generated xenograft tumors with these HCT116 cell lines in mice. HCT116 cells proficient and deficient in ZBTB7A were subcutaneously implanted pairwise into opposite flanks, and tumor development was monitored. In agreement with the in vitro data, ZBTB7A loss greatly enhanced tumor growth, resulting in tumor volume at day 24 that was approximately twice that derived from control cells expressing ZBTB7A (Fig. 7B,C). Upon resection, the tumor tissues were analyzed to ascertain the difference in ZBTB7A and glycolytic gene expression. The reduced levels of ZBTB7A expression in ZBTB7A shRNA-expressing tumors was associated with significantly increased expression of GLUT3, PFKP, and PKM compared with controls (Fig. 7D), consistent with ZBTB7A-mediated suppression of the glycolytic genes. To determine the dependency of ZBTB7A-deficient tumor growth on glycolytic metabolism, mice were treated with vehicle or 2-DG. Remarkably, treatment of the mice with 2-DG was associated with strong inhibition of tumor growth of shZBTB7A-expressing cells, while control cells were considerably less affected by 2-DG treatment and grew into readily detectable tumors (Fig. 7B,C). These results indicate an enhanced dependency of ZBTB7A-deficient tumors on glycolysis, rendering them exceedingly vulnerable to this inhibitor of glycolysis (Fig. 7E).
Discussion
Elevated aerobic glycolysis metabolism (or the Warburg effect) is commonly observed in cancers (Hanahan and Weinberg 2011). In a sharp contrast to oncogenic transcription factors HIF-1α and MYC that induce the expression of many genes key to glycolysis (Dang 1999; Denko 2008; Yeung et al. 2008), we demonstrate a ZBTB7A-mediated transcriptional repression of glycolytic genes, uncovering a novel transcriptional repression mechanism in negative regulation of cellular glycolysis in mammalian cells. The frequent loss of ZBTB7A in many human tumors may represent a previously unrecognized genetic basis of the unique metabolic phenotype of cancer cells.
Cancer cell proliferation depends on enhanced glycolysis (Vander Heiden et al. 2009). The suppression of increased glycolytic metabolism may represent a novel mechanism of tumor suppression. We demonstrate that ZBTB7A negatively regulates glycolysis. Reduced expression of ZBTB7A caused a marked increase in glycolysis, as evidenced by the observation that knocking down the expression of ZBTB7A in multiple carcinoma cell lines resulted in a robust induction of glycolysis. This ZBTB7A-dependent suppression of glycolysis was further validated with Zbtb7a-null MEFs that displayed highly elevated glycolysis, and, importantly, restoration of Zbtb7a expression in Zbtb7a-null MEFs was sufficient to bring the increased rate of glycolysis back to the wild-type level. In the context of tumor cells, introduction of ZBTB7A in SW48 colon carcinoma cells was also associated with suppression of glycolysis and proliferation, implicating a tumor suppressor function of ZBTB7A. Of interest was the striking glycolytic phenotype of ZBTB7A-deficient cells in the absence of any apparent involvement of HIF-1α and MYC, suggesting a novel mechanism of glycolysis regulation. In line with this notion was our identification of GLUT3, PFKP, and PKM as novel target genes of ZBTB7A. Multiple lines of evidence were presented to establish ZBTB7A as a bona fide transcription repressor of these three glycolytic genes.
Given the fact that cancer cells depend on aerobic glycolysis to support proliferation (Vander Heiden et al. 2009), ZBTB7A-mediated suppression of glycolysis would need to be lost for tumor growth or progression. Indeed, analysis of a human cancer genome database uncovered that the ZBTB7A gene locus is frequently deleted in many types of human carcinoma. Consistent with a ZBTB7A-dependent repression of GLUT3, PFKP, and PKM, mining TCGA data of human colon carcinoma revealed an inverse correlation between the expressions of ZBTB7A and its three target glycolytic genes. Moreover, lower levels of ZBTB7A expression correlated with a later stage of cancer and poor patient survival, in line with a tumor suppressor role for ZBTB7A. In agreement with the importance of glycolytic metabolism in tumorigenesis, ZBTB7A deficiency-induced glycolysis was associated with a remarkable increase in cancer cell proliferation in vitro and tumor growth in mice. Of therapeutic potential is the observation that ZBTB7A-deficient cancer cells exhibited a significantly increased dependency on glycolytic metabolism, which resulted in a heightened sensitivity to the inhibition of glycolysis. 2-DG is already in clinical trials for the treatment of certain solid tumors either alone or in combination with other therapies (Aghaee et al. 2012; Raez et al. 2013). Our data suggest that ZBTB7A can be used as an important therapeutic marker for the use of inhibitors of glycolysis for cancer treatment.
In summary, we uncovered ZBTB7A as a novel transcriptional repressor directly suppressing key glycolytic genes and influencing metabolic flux through glycolysis and the PPP pathway. The significance of ZBTB7A-mediated inhibition of glycolysis is highlighted by the finding that the gene locus of ZBTB7A is frequently lost in human cancers, implicating that ZBTB7A is a tumor suppressor that, upon down-regulation, provides a novel genetic mechanism driving the Warburg effect in cancer cells to support tumor growth. The unusual susceptibility of ZBTB7A-deficient tumors to glycolysis inhibitors promises a very attractive therapeutic opportunity to effectively target those tumors with inhibitors of glycolysis.
Materials and methods
Cell culture
MEF, human fibroblast, 293T, M14, SW48, U2OS, and MCF7 cells were cultured in DMEM (Corning, Cellgro) plus 10% fetal bovine serum (FBS) (Gibco), 100 U/mL penicillin G, and 100 μg/mL streptomycin (Corning, Cellgro). A549 and H1299 were cultured in RPMI (Corning, Cellgro) plus 10% FBS, 100 U/mL penicillin G, and 100 μg/mL streptomycin. HCT116 was cultured in McCoy’s 5A (Corning, Cellgro) plus 10% FBS (Gibco), 100 U/mL penicillin G, and 100 μg/mL streptomycin. All cells were cultured in a 37°C, 5% CO2 incubator.
Antibodies and reagents
Anti-ZBTB7A antibody (Santa Cruz Biotechnology, 13E9), anti-GLUT3 antibody (Abcam, ab41525), anti-PFKP antibody (LifeSpan BioSciences, 1D6), anti-PKM antibody (Cell Signaling, C103A3), anti-MYC antibody (Cell Signaling, D84C12), anti-HIF1α antibody (BD, 610958), and anti-β-actin antibody (Sigma, AC-15) were used for Western blot; anti-ZBTB7A antibody (Bethyl Laboratories, A300-549A) and anti-Xpress antibody (Invitrogen) were used for ChIP.
Glucose uptake and lactate production measurement
Cells (3 × 104) were seeded into 24-well plates. The next day, cells were washed twice with culture medium, and fresh culture medium was added. Five hours later, cell culture medium was collected and frozen in −80°C, and the cell numbers in each well were counted using a hemocytometer. Glucose and lactate concentration in the collected cell culture medium was measured using Colorimetric kit (Biovision) according to the attached protocol.
Metabolomics analysis
The metabolomics flux studies were performed as described previously (Düvel et al. 2010; Yuan et al. 2012). Briefly, cells in 10-cm plates (∼70% confluency) were washed once with glucose-free DMEM and then incubated in DMEM containing a 10 mM 1:1 mixture of D-[1,2-13C]-glucose (Sigma) and unlabeled D-glucose (Sigma) for 15 min. Metabolites were extracted on dry ice with 5 mL of 80% methanol. The extract was dried down under nitrogen and resuspended in 80 μL of water just prior to the liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis in the Beth Israel Deaconess Medical Center Mass Spectrometry facility.
Microarray analysis
Arf−/− Zbtb7aFlox/Flox MEFs were transduced with MSCV-PIG-Cre or empty control vector retroviruses for 2 d at passage 2. After selection with puromycin for 2 d, total RNAs were purified using the RNAeasy minikit (Qiagen) and treated with RNase-free DNase set (Qiagen). RNAs from two independent experiments were labeled and hybridized using Affymetrix GeneChip HT Mouse Genome 430 arrays by the microarray core facility of Dana-Farber Cancer Institute. GSEA was conducted using GSEA software (Subramanian et al. 2007). Reactome pathways were used as the gene set database. Normalized enrichment score (NES) and false discovery rate (FDR) Q-values were calculated using the GSEA protocol.
Plasmid mutation
ZBTB7A-binding sites in human GLUT3, PFKP, and PKM promoter regions and R399L in Xpress-tagged human ZBTB7A were mutated with the Q5 site-directed mutagenesis kit (New England Biolabs). Mutation primers were designed using NEBaseChanger software provided by the Q5 mutagenesis kit. Mutation PCR, kinase–ligase–DpnI (KLD) enzyme treatment, and transformation were performed according to the manufacturer’s instructions. All of the mutations were confirmed by plasmid sequencing.
Luciferase reporter assay
293 cells were cultured in 48-well plates, and transfection was started when cells reached 50% confluence. Twenty nanograms of pGL3-GLUT3, PFKP, or PKM vector; 10 ng of pRL-CMV Renillar luciferase reporter; and varying doses of pcDNA3.1 control or pcDNA3.1-ZBTB7A expression plasmid were cotransfected into 293 cells. After 36 h, the firefly (pGL3) and Renilla luciferase activity was measured with the dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions. The ratio of firefly luciferase/Renilla luciferase represents the relative activity of different promoters.
Lentiviral shRNA-mediated knockdown
293T cells were seeded in 10-cm tissue culture plates, and plasmid transfection was performed when the cells reached 70% confluence. Prior to transfection, 293T cell culture medium was changed to antibiotic-free DMEM with 10% FBS. Ten micrograms of shZBTB7A plasmid (Sigma, TRCN0000137332), 7.5 μg of psPAX2 packaging plasmid, and 2.5 μg of pMD2.G envelope plasmid were cotransfected using 20 μL of Lipofectamine 2000 transfection reagent (Invitrogen). The next day after transfection, the transfection medium was changed to fresh complete culture medium. The virus supernatant was collected every 12 h, filtered through a 0.45-μm filter, and frozen at −80°C until further usage. The virus titer was determined by serial dilution assay using 3T3 cells. For lentiviral transfection, 5 × 105 cells were seeded in 10-cm dishes and incubated with virus at a multiplicity of infection (MOI) of ∼1 for 12 h in the presence of 8 μg/mL polybrene (Sigma). Seventy-two hours later, the cells were selected with 1 μg/mL puromycin (Sigma) for 3 d. The ployclonal cells after puromycin selection were used in downstream experiments. The sequence of shRNA targeting ZBTB7A was CCGGGCTGGACCTTGTAGATCAAATCTCGAGATTTGATCTACAAGGTCCAGCTTTTTTG.
Nude mouse subcutaneous transplantation assay and 2-DG administration
shCon and shZBTB7A HCT116 cells (1 × 106) were subcutaneously injected into the left and right flanks of nude mice (nu/nu, female, 6–8 wk old; Charlies River Laboratories), respectively. One day after HCT116 cell injection, the mice were randomly assigned into two groups: One group of mice received 2g/kg 2-DG (dissolved in PBS), and the other group of mice received PBS by intraperitoneal injection every other day. The size of the tumor was measured with a caliper every 3 d. Tumor volume was calculated by using the formula V = (π /6)(d1 × d2)3/2. At the end point, mice were euthanatized, tumors from each mouse were excised, and total RNA was extracted with TRIzol reagents. All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the Harvard School of Public Health.
Human cancer genomics analysis
ZBTB7A CNV and mRNA values in multiple types of human cancers (which include thyroid carcinoma, stomach adenocarcinoma, liver hepatocellular carcinoma, kidney clear cell carcinoma, head and neck squamous cell carcinoma, esophageal carcinoma, bladder urothelial carcinoma, colon and rectum adenocarcinoma, prostate adenocarcinoma, pancreatic adenocarcinoma, cervical squamous cell carcinoma, endocervical adenocarcinoma, lung cancer, breast invasive carcinoma, skin cutaneous melanoma, ovarian serous cystadenocarcinoma, brain lower-grade glioma, uterine carcinosarcoma, acute myeloid leukemia, and lymphoid neoplasm diffuse large B-cell lymphoma) were downloaded from TCGA data sets. Kaplan-Meier survival curve analysis was performed using GraphPad Prism software. “ZBTB7A low” represents the samples with the lowest 30% of ZBTB7A expression, while “ZBTB7A high” represents the samples with the highest 30% of ZBTB7A expression. The data for ZBTB7A expression and colorectal cancer prognosis come from study GSE29623. Correlation analysis was performed using GraphPad Prism software. Data set GSE29623 was used for correlation analysis between ZBTB7A and glycolysis genes GLUT3, PFKP, and PKM in human colorectal cancer. TCGA data set was used for ZBTB7A mRNA and CNV correlation in human colorectal cancer. Pearson’s R-values were generated to determine the degree of correlation. Linear regression analysis was also performed using GraphPad Prism software, where the P-value designates the degree to which the regression is nonzero.
Immunoblot
Cells were lysed in buffer (50 mM Tris, at pH 8.0, 150 mM NaCl, 0.5% NP-40). Protein concentrations of the lysates were measured by Bradford assay. The lysates were then resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
Cell proliferation assay
Cells were seeded in 12-well plates at a density of 104 per well and then left to grow for 4 d in the presence or absence of 2 mM 2-DG (Sigma). Cells were fixed by paraformaldehyde at each time point and stained with crystal violet. After extensive washing, crystal violet was resolubilized in 10% acetic acid and quantified at 595 nm as a relative measure of cell number as described previously (Carnero et al. 2000).
Real-time PCR to quantify gene expression
Total RNA was extracted with TRIzol reagents (Invitrogen) according to the provided protocol. One microgram of total RNA was reversed transcribed with iScript cDNA synthesis kit (Bio-Rad). Real-time qPCR was performed using diluted cDNA, SYBR Green JumpStart Taq ReadyMix (Sigma), and appropriate primers in StepOnePlus real-time PCR system (Applied Biosystems). 18s was used as an endogenous control for normalization. The primer sequences for the genes were as follows: HuZBTB7A-5, GCTTGGGCCGGTTGAATGTA; HuZBTB7A-3, GGCTGTGAAGTTACCGTCGG; Hu18s-5, CAGCCACCCGAGATTGAGCA; Hu18s-3, TAGTAGCGACGGGCGGTGTG; HuGLUT3rt-5, GCTGGGCATCGTTGTTGGA; HuGLUT3rt-3, GCACTTTGTAGGATAGCAGGAAG; HuPKMrt-5, ATGTCGAAGCCCCATAGTGAA; HuPKMrt-3, TGGGTGGTGAATCAATGTCCA; HuPFKPrt-5, GACCTTCGTTCTGGAGGTGAT; HuPFKPrt-3, CACGGTTCTCCGAGAGTTTG; HuHIF1Art-5, ATCCATGTGACCATGAGGAAATG; HuHIF1Art-3, TCGGCTAGTTAGGGTACACTTC; HuMYCrt-5, GGCTCCTGGCAAAAGGTCA; HuMYCrt-3, CTGCGTAGTTGTGCTGATGT; Mszbtb7a-5, CTTTGCGACGTGGTGATTCTT; Mszbtb7a-3, CGTTCTGCTGGTCCACTACA; Ms18s-5, AGGGGAGAGCGGGTAAGAGA; Ms18s-3, GGACAGGACTAGGCGGAACA; MsGlut3rt-5, ATGGGGACAACGAAGGTGAC; MsGlut3rt-3, GTCTCAGGTGCATTGATGACTC; MsPkmrt-5, GCCGCCTGGACATTGACTC; MsPkmrt-3, CCATGAGAGAAATTCAGCCGAG; Mspfkprt-5, GAAACATGAGGCGTTCTGTGT; Mspfkprt-3, CCCGGCACATTGTTGGAGA; MsHif1art-5, GGGGAGGACGATGAACATCAA; MsHif1art-3, GGGTGGTTTCTTGTACCCACA; MsMycrt-5, CCCTATTTCATCTGCGACGAG; and MsMycrt-3, GAGAAGGACGTAGCGACCG.
ChIP
ChIP was performed as described previously (Nelson et al. 2006). Briefly, protein–DNA complexes were cross-linked for 10 min at room temperature with 1% formaldehyde added directly into the culture medium. The reaction was stopped by the addition of glycine (final concentration 0.125 mol/L) and incubated for 5 min with gentle rocking. The cells were washed with PBS and buffer (10 mM Tris at pH 8.0, 10 mM EDTA, 0.5 mM EGTA, 0.25% Triton-X-100), suspended in 200 μL of lysis buffer (1.1% Triton-X-100, 4 mM EDTA, 40 mM Tris at pH 8.1, 300mM NaCl), and submitted to sonication to produce small DNA fragments (200–1000 base pairs). Chromatin was precleared and immunoprecipitated with the indicated antibody. Precipitated DNA and protein complexes were reverse-cross-linked, proteins were digested with proteinase K (Fermentas), and DNA fragments were purified with a QIAquick PCR purification kit (Qiagen). The purified DNAs were amplified by real-time qPCR using StepOnePlus (Applied Biosystems) and SYBR Green JumpStart Taq ReadyMix (Sigma). Primers to quantify the abundance of human Glut3, PFKP, and PKM promoters were as follows: GLUT3CHIP-5, CCCCTGAAGCAATCTTGTGATC; GLUT3CHIP-3, AAAAACCCAGGGTGGAGAGAG; PFKPCHIP-5, TCATCTCTAGAGCCCCCAAC; PFKPCHIP-3, GTGTGGGCAGGAGCATCTAC; PKMCHIP-5, ATGGGGTGTCTTTTCTTGGC; and PKMCHIP-3, TTTCCGTTCAGCTCAGTCTCC. Negative control primers were NegCHIP-5, ATGGTTGCCACTGGGGATCT; and NegCHIP-3, TGCCAAAGCCTAGGGGAAGA.
Promoter cloning
Primers for human GLUT3, GLUT1, HK1, PFKP, PFKL, PKM, ENO1, HIF1A, GAPDH, and IDHA promoter cloning were as follows: GLUT3pro-3, ATGCCTCGAGGTCTCGCTGGGATCATGACTATTTA; GLUT3pro-5, ATGCGGTACCCAGAATTCAGAATGGGTGATATGGC; GLUT1pro-3, ATGCAAGCTTGAGGGCATTCGCTGTGTGACTCAG; GLUT1pro-5, ATGCGAGCTCCACCAACCAGTAAATGAGAACTTCC; HK1pro-5, ATGCGGTACCGAGAGCCACCGGAGCTGGTGACTC; HK1pro-3, ATGCAAGCTTCCTGGCGAGCCGTGGTCCTCCGG; PFKPpro-3, ATGCAAGCTTCGGGAGTCGTCCGCGTCCATGGC; PFKPpro-5, ATGCGGTACCGATCCACTCTCCGTCCGTCCTTC; PFKLpro-5, ATGCGAGCTCCATGCCCGGCTAATTTTTGTATTTT; PFKLpro-3, ATGCAAGCTTCTTCTCCAGGTCCACCGCGGCCATG; PKMpro-5, ATGCGGTACCTAAAGGGACCAGGAAAGACTTACAG; PKMpro-3, ATGCAAGCTTCCGCGGGCGCAGTCACCTTCAGGA; ENO1pro-3, ATGCAAGCTTCTCCGTCACGTACTCCGAGTCCC; ENO1pro-5, ATGCGGTACCTCTTACAGACCAGCAGTAATGTTGA; HIF1Apro-3, ATGCAAGCTTCAGCGTCAGGGGGCGGGCAAGGG; HIF1Apro-5, ATGCGGTACCAGTCATTTATACCTATTTGCAAATGCT; GAPDHpro-3, ATGCAAGCTTCATTCATTTCCTTCCCGGTTGCAAC; GDPDHpro-5, ATGCGGTACCGGTTTCTATAAATTGAGCCCGCAG; LDHApro-5, ATGCGGTACCGCTGTAATCCCATCTTCTCAGGAG; and LDHApro-3, ATGCAAGCTTGCCAGACAACCGACCGGCAGACTG.
Genomic DNA extracted from normal human fibroblasts was used as the template for PCR. PFKP and PFKL promoters were amplified using PrimeSTAR HS with GC buffer (Takara), and other promoters were amplified with Q5 high-fidelity DNA polymerase (New England Biolabs). GLUT3 PCR fragment was digested with KpnI and XhoI; HK1, HIF1A, GAPDH, ENO1, LDHA, PKM, and PFKP PCR fragments were digested with KpnI and HindIII; and GLUT1 and PFKL PCR fragments were digested with SacI and HindIII and then cloned into pGL3-Basic vector (Promega). Restriction enzymes were purchased from New England Biolabs. The resulting pGL3-GLUT3, PFKP, PKM, GLUT1, HIF1A, HK1, PFKL, ENO1, GAPDH, and IDHA vectors were confirmed by sequencing.
siRNA-based gene knockdown
siRNA target human ZBTB7A was synthesized by Sigma. The sequences of three ZBTB7A targeting siRNAs were as follows: ZBTB7A siRNA-1, CGCUCAUGGACUUCGCCUA; ZBTB7A siRNA-2, CAGACAAGACCUUAAAUGA; ZBTB7A siRNA-3, CGCUCACCGCGCUCAUGGA; MYC siRNA, CGUCCAAGCAGAGGAGCAA; and HIF1A siRNA, GAUUAACUCAGUUUGAACU.
For siRNA transfection, 3 × 105 cells were reverse-transfected with a mixture of siRNA and Lipofectamine RNAiMAX reagent (Invitrogen) in a 35-cm dish according to the manufacturer’s protocol.
Statistical analysis
Log-rank tests were used for Kaplan-Meier survival analysis. Unpaired Student’s t-test was used for the comparisons of the means, and error bars represent the standard deviation (SD) unless otherwise stated. The statistical significances between data sets were expressed as P-values, and P < 0.05 was considered statistically significant.
Acknowledgments
We are grateful to current and former members of the Yuan laboratory for experimental support, advice, and helpful discussions. This work was supported in part by the Morningside Foundation and grants from National Institutes of Health/National Cancer Institute (R01CA085679, RO1CA167814, and RO1CA125144).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.245910.114.
References
- Aghaee F, Pirayesh Islamian J, Baradaran B. 2012. Enhanced radiosensitivity and chemosensitivity of breast cancer cells by 2-deoxy-d-glucose in combination therapy. J Breast Cancer 15: 141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenberg B, Greulich KO. 2004. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 84: 1014–1020 [DOI] [PubMed] [Google Scholar]
- Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. 2013. Metabolic regulation by p53 family members. Cell Metab 18: 617–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. 2011. Regulation of cancer cell metabolism. Nat Rev Cancer 11: 85–95 [DOI] [PubMed] [Google Scholar]
- Carnero A, Hudson JD, Price CM, Beach DH. 2000. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat Cell Biol 2: 148–155 [DOI] [PubMed] [Google Scholar]
- Costoya JA 2007. Functional analysis of the role of POK transcriptional repressors. Brief Funct Genomic Proteomic 6: 8–18 [DOI] [PubMed] [Google Scholar]
- Dang CV 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol 19: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 2012. MYC on the path to cancer. Cell 149: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblois G, Giguère V. 2013. Oestrogen-related receptors in breast cancer: control of cellular metabolism and beyond. Nat Rev Cancer 13: 27–36 [DOI] [PubMed] [Google Scholar]
- Denko NC 2008. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer 8: 705–713 [DOI] [PubMed] [Google Scholar]
- Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. 2010. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39: 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, et al. 2004. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res 64: 3892–3899 [DOI] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. 2007. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell 12: 108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Kelly KF, Daniel JM. 2006. POZ for effect—POZ-ZF transcription factors in cancer and development. Trends Cell Biol 16: 578–587 [DOI] [PubMed] [Google Scholar]
- Kukita A, Kukita T, Ouchida M, Maeda H, Yatsuki H, Kohashi O. 1999. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood 94: 1987–1997 [PubMed] [Google Scholar]
- Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. 2001. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1α (HIF-1α) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol 21: 3995–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CJ, Prazak L, Fajardo M, Yu S, Tyagi N, Di Cesare PE. 2004. Leukemia/lymphoma-related factor, a POZ domain-containing transcriptional repressor, interacts with histone deacetylase-1 and inhibits cartilage oligomeric matrix protein gene expression and chondrogenesis. J Biol Chem 279: 47081–47091 [DOI] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. 2011. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 27: 441–464 [DOI] [PubMed] [Google Scholar]
- Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. 2005. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature 433: 278–285 [DOI] [PubMed] [Google Scholar]
- Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, Manola J, Brugarolas J, McDonnell TJ, Golub TR, et al. 2004. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med 10: 594–601 [DOI] [PubMed] [Google Scholar]
- Melnick A, Carlile G, Ahmad KF, Kiang CL, Corcoran C, Bardwell V, Prive GG, Licht JD. 2002. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol Cell Biol 22: 1804–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 1: 179–185 [DOI] [PubMed] [Google Scholar]
- Pessler F, Pendergrast PS, Hernandez N. 1997. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol Cell Biol 17: 3786–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raez LE, Papadopoulos K, Ricart AD, Chiorean EG, Dipaola RS, Stein MN, Rocha Lima CM, Schlesselman JJ, Tolba K, Langmuir VK, et al. 2013. A phase I dose-escalation trial of 2-deoxy-D-glucose alone or combined with docetaxel in patients with advanced solid tumors. Cancer Chemother Pharmacol 71: 523–530 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. 2007. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics 23: 3251–3253 [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Lunardi A, Zhang J, Chen Z, Ala U, Webster KA, Tay Y, Gonzalez-Billalabeitia E, Egia A, Shaffer DR, et al. 2013. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat Genet 45: 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung SJ, Pan J, Lee MH. 2008. Roles of p53, MYC and HIF-1 in regulating glycolysis—the seventh hallmark of cancer. Cell Mol Life Sci 65: 3981–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Breitkopf SB, Yang X, Asara JM. 2012. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc 7: 872–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. 2013. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]