K-Ras activation with a CC10(Scgb1a1)-CreER driver leads to lung adenocarcinoma in a subset of alveolar type II cells and hyperplasia in the bronchioalveolar duct region. Xu et al. find that Notch inhibition strongly inhibits adenocarcinoma formation but promotes squamous hyperplasia in the alveoli. In contrast, activation of Notch leads to widespread Sox2+, Sox9+, and CC10+ papillary adenocarcinomas throughout the bronchioles. Sox2 binds to NOTCH1 and NOTCH2 regulatory regions and reduces Notch1 and Notch2 transcripts. This study shows that the cell of origin of K-Ras-induced tumors depends on levels of Sox2 expression affecting Notch signaling.
Keywords: Notch, Sox2, K-Ras, lung cancer
Abstract
Cell type-specific conditional activation of oncogenic K-Ras is a powerful tool for investigating the cell of origin of adenocarcinomas in the mouse lung. Our previous studies showed that K-Ras activation with a CC10(Scgb1a1)-CreER driver leads to adenocarcinoma in a subset of alveolar type II cells and hyperplasia in the bronchioalveolar duct region. However, no tumors develop in the bronchioles, although recombination occurs throughout this region. To explore underlying mechanisms, we simultaneously modulated either Notch signaling or Sox2 levels in the CC10+ cells along with activation of K-Ras. Inhibition of Notch strongly inhibits adenocarcinoma formation but promotes squamous hyperplasia in the alveoli. In contrast, activation of Notch leads to widespread Sox2+, Sox9+, and CC10+ papillary adenocarcinomas throughout the bronchioles. Chromatin immunoprecipitation demonstrates Sox2 binding to NOTCH1 and NOTCH2 regulatory regions. In transgenic mouse models, overexpression of Sox2 leads to a significant reduction of Notch1 and Notch2 transcripts, while a 50% reduction in Sox2 leads to widespread papillary adenocarcinoma in the bronchioles. Taken together, our data demonstrate that the cell of origin of K-Ras-induced tumors in the lung depends on levels of Sox2 expression affecting Notch signaling. In addition, the subtype of tumors arising from type II cells is determined in part by Notch activation or suppression.
Lung adenocarcinoma is a heterogeneous disease, both histologically and genetically. While much progress has been made in therapeutically targeting specific driver mutations (EGFR, ALK, and ROS) (Lynch et al. 2004; Takeuchi et al. 2012), K-RAS mutant tumors remain difficult to treat effectively. Farnesyl transferase inhibitors have failed (Johnson and Heymach 2004), and MEK inhibitors have proven ineffective (Adjei et al. 2008), partly because they block only one pathway downstream from the oncogenic kinase. Synthetic-lethal approaches identify genes that, when inhibited, are lethal to K-RAS-activated but not wild-type cells (Luo et al. 2009; Scholl et al. 2009). However, these have not resulted in effective therapies (Luo et al. 2009). One reason for these failures may be the inadequate consideration of the cellular context of K-RAS activation. We previously demonstrated that in the mouse lung, K-RasG12D induces adenocarcinoma formation preferentially in a subset of type II cells in the alveoli, even when the oncogenic mutation is induced throughout the bronchiolar airways (Xu et al. 2012). Others have subsequently corroborated this cell of origin specificity but have not identified underlying mechanisms (Mainardi et al. 2014; Sutherland et al. 2014). Here, we address this question by identifying molecular differences between bronchiolar cells that are not transformed and alveolar cells that are tumorigenic when K-RasG12D is induced. Specifically, we focus on the role of Notch signaling in controlling this differential response.
The Notch signaling pathway plays an important role in lung development and cell fate decisions (Tsao et al. 2009; Rock et al. 2011; Guha et al. 2012; Morimoto et al. 2012). Mounting data implicate alteration of the Notch signaling pathway in lung cancer development in both humans and mice (Westhoff et al. 2009; Eliasz et al. 2010; Allen et al. 2011; Osanyingbemi-Obidi et al. 2011; Wang et al. 2011; Yang et al. 2011; Maraver et al. 2012; Licciulli et al. 2013). Notch receptors transduce signals in response to ligands on neighboring cells, regulating lineage selection and developmental patterning. To activate Notch transcriptional targets, ligands (Jagged1, Jagged2, and Delta1–3) bind to receptors (Notch1–4), leading to two proteolytic cleavages at the membrane. The second of these, mediated by γ-secretase, frees the Notch intracellular domain (ICD), which translocates to the nucleus and forms a DNA-binding complex with coactivators, including Mastermind-like (MAML). This transcriptional complex activates downstream effectors, including Hes1, Hes5, Hey1, Hey2, and Hey5. Recent studies (Wang et al. 2011; Cancer Genome Atlas Research Network 2012) have demonstrated that inhibitory alterations of Notch signaling are frequent in human squamous non-small cell lung cancer (NSCLC).
In this study, we show that inhibition of Notch signaling strongly abrogates alveolar K-Ras-induced adenocarcinoma formation, while activation of Notch in bronchiolar cells allows K-Ras-induced tumorigenesis. Further investigation demonstrates that NOTCH is a direct target of the SOX2 transcription factor in human cells and that Sox2 overexpression leads to transcriptional repression of Notch1 and Notch2 in mice. Genetically reducing the level of Sox2 by 50% in CC10-CreER; Lox-Stop-Lox (LSL)-K-RasG12D mice allows bronchiolar tumor formation. Together, these data identify Notch signaling as a key determinant of both tumor initiation and tumor progression in K-Ras-activated respiratory epithelial cells.
Results
Notch signaling is required for K-Ras-induced lung adenocarcinoma
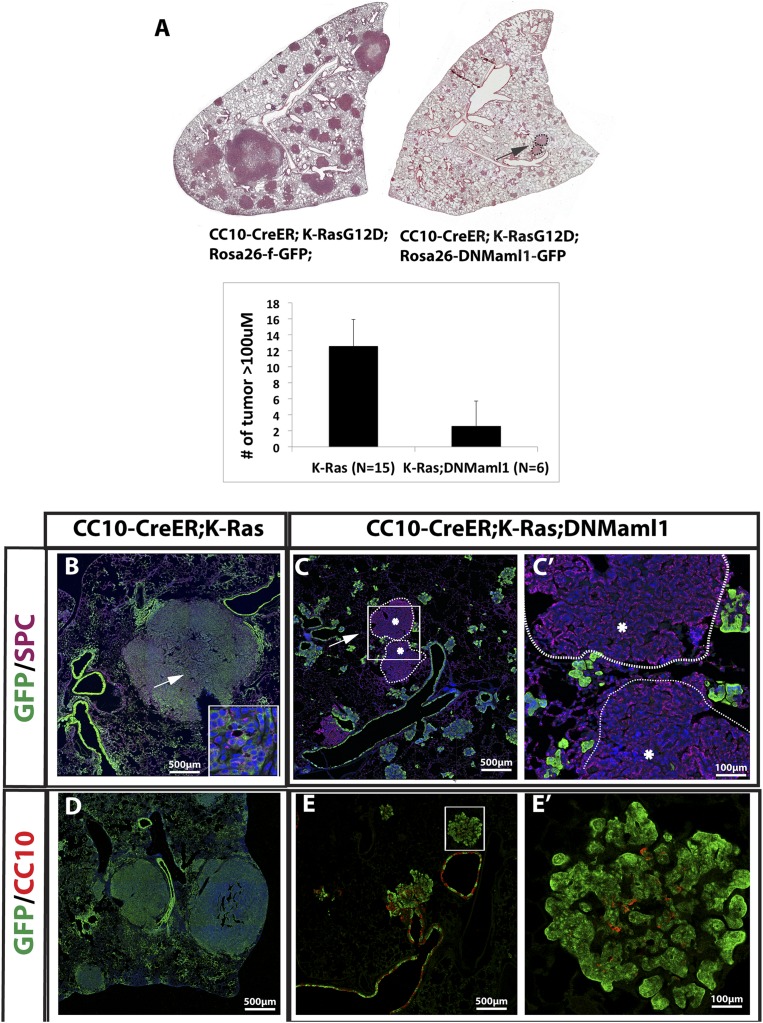
Involvement of the Notch pathway in K-Ras-induced tumors has been controversial. Using mouse models of K-RasG12D activated by inhaled adenoviral Cre recombinase, groups have published conflicting data regarding its necessity for in vivo lung tumorigenesis (Osanyingbemi-Obidi et al. 2011; Maraver et al. 2012). In order to determine whether Notch activation is necessary for K-Ras-induced tumors in the subset of alveolar type II cells that expresses CC10+, we exploited a conditional Rosa26-DNMaml1-GFP allele encoding a dominant-negative Mastermind-like protein (DNMaml1) tagged with GFP (Tu et al. 2005). Recombination of this allele results in the expression of a competitive inhibitor of the Rpbj coactivator Maml that is required for transcriptional activation of Notch targets (Nam et al. 2006). We generated CC10-CreER; LSL-K-RasG12D; Rosa26-DNMaml1-GFP (hereafter, CC10-CreER; K-Ras; DNMaml1) mice and used CC10-CreER; LSL-K-RasG12D; Rosa26-f-GFP (CC10-CreER; K-Ras) mice as controls. At 15 wk after four doses of tamoxifen, the lungs of control CC10-CreER; K-Ras mice contain widespread adenocarcinomas and adenomas (Fig. 1A, left panel). In contrast, CC10-CreER; K-Ras; DNMaml1 lungs (Fig. 1A, right panel) contain mostly small clusters of hyperplastic GFP+ columnar cells in the alveoli and bronchioalveolar duct junction (BADJ), very few adenomas >100 μm in diameter, and no adenocarcinomas (Fig. 1A). Quantification reveals 12.6 ± 2.6 lesions >100 μm in diameter per left lobe in CC10-CreER; K-Ras mice and only 2.8 ± 0.8 lesions of this size in the CC10-CreER; K-Ras; DNMaml1 mice (P < 0.05). The adenomas and adenocarcinomas in CC10-CreER; K-Ras mice are GFP+ Sftpc+ (Fig. 1B). In contrast, the cells in alveolar hyperplasia/small adenomas in CC10-CreER; K-Ras; DNMaml1 mice are GFP+ Sftpc− (Fig. 1C). Significantly, the very rare larger adenomas (>100 μm) that do develop in experimental lungs are GFP-negative (Fig. 1C,C′), demonstrating activation of the K-Ras allele only and escape from recombination of the DNMaml1 allele. Immunohistochemistry for CC10 shows that the tumorigenic cells in CC10-CreER; K-Ras mice lose CC10 expression when K-Ras is activated (Fig. 1D). This is consistent with our previous report (Xu et al. 2012). However, although the majority of the GFP+ cells in CC10-CreER; K-Ras; DNMaml1 mice are CC10-negative (Fig. 1E,E′), some cells in the hyperplasia/small adenomas continue to express CC10. Thus, in our model, active Notch signaling is required for K-RasG12D-induced adenocarcinoma formation in the subset of alveolar type II cells that express CC10.
Figure 1.
Genetic inhibition of Notch signaling significantly reduces K-Ras-induced lung adenocarcinoma formation. (A) Sections of left lobes of CC10-CreER; LSL-K-RasG12D; Rosa26-f-GFP (CC10-CreER; K-Ras) (left) and CC10-CreER; LSL-K-RasG12D; Rosa26-LSL-DNMaml1-GFP (CC10-CreER; K-Ras; DNMaml1) (right) mice 15 wk after tamoxifen injection, stained with H&E. Note that the lung expressing both activated K-Ras and DNMaml1 has significantly fewer large tumors (>100 μm), which is quantified in the bottom panel. (B) Section of CC10-CreER; K-Ras mouse lung stained with DAPI, GFP (green), and Sftpc (purple). The inset shows higher magnification of tumor cells (arrow). Note that tumors are GFP+Sftpc+. (C) Section of CC10-CreER; K-Ras; DNMaml1 mouse lung stained with DAPI, GFP (green), and Sftpc (purple). Note that widespread GFP+ hyperplastic clusters are Sftpc−. Rare larger solid adenomas (arrow) are GFP− Sftpc+ (white dashed circles with asterisk), indicating that these rare large adenomas escape DNMaml1 expression. (C′) Higher magnification of the boxed area in C. (D) Section of CC10-CreER; K-Ras mouse lung stained with DAPI, GFP (green), and CC10 (red) showing that tumors are CC10−. (E) Section of CC10-CreER; K-Ras; DNMaml1 mouse lung stained with GFP and CC10. (E′) Higher magnification of the white boxed area in E demonstrating that hyperplastic clusters/small adenomas contain a majority of cells that are GFP+ alone.
Inhibition of Notch in K-RasG12D; CC10-expressing alveolar cells leads to a Sox2+Krt5+Trp63+ squamous phenotype
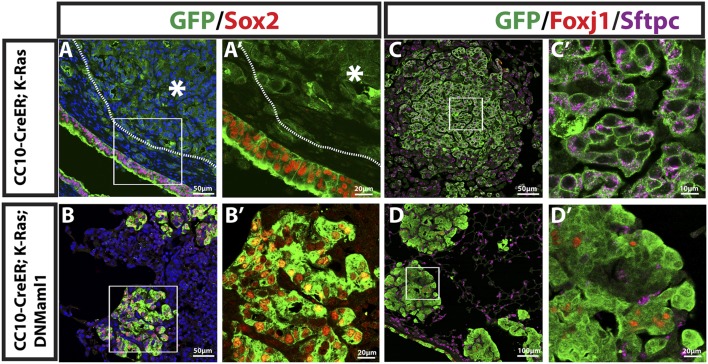
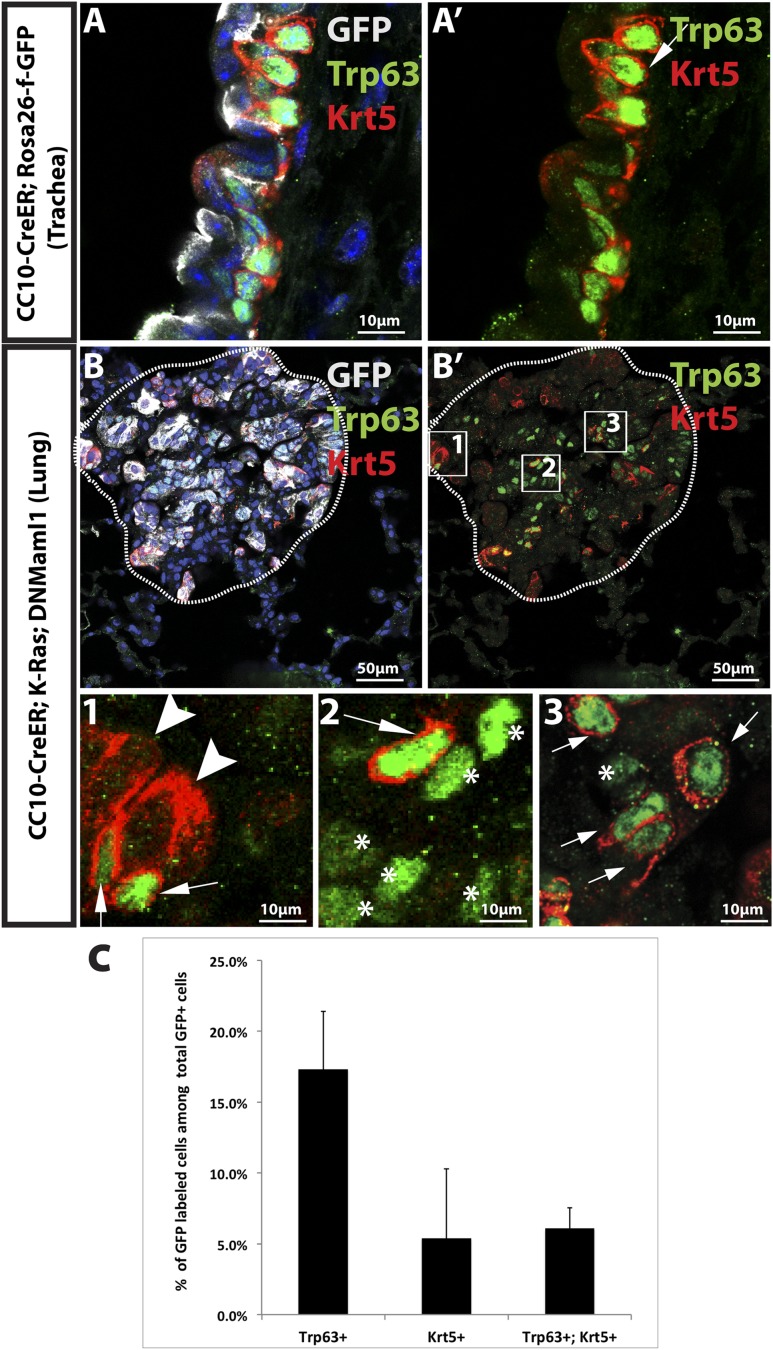
Recently, it was demonstrated that γ-secretase inhibition abrogates K-Ras-induced tumor formation in the mouse lung (Maraver et al. 2012). However, while Notch ablation caused inhibition of large tumors in our CC10-CreER; K-Ras; DNMaml1 mice, clusters of hyperplastic cells and small adenomas persisted. To further characterize these lesions, we performed immunostaining for lung epithelial markers. As shown in Figure 2, A and A′, consistent with our previous report (Xu et al. 2012), the majority of lung adenomas and lung adenocarcinomas in the alveolar region of CC10-CreER; K-Ras mice do not express the transcription factor Sox2. However, expression of DNMaml1 leads to clear expression of Sox2 in all GFP-positive cells (Fig. 2B,B′). Moreover, Foxj1, a marker for ciliated airway epithelial cells, is not seen in lung adenomas and adenocarcinomas in CC10-CreER; K-Ras mice (Fig. 2C,C′) but is expressed in some Sox2+ cells in the hyperplastic lesions in CC10-CreER; K-Ras; DNMaml1 mice (Fig. 2D,D′). To further test whether hyperplastic lesions express other markers of cell types normally present in the Sox2+ proximal airways of the mouse lung, we stained for Trp63 and Krt5. These proteins are normally expressed only in basal cells of the pseudostratified epithelium of the mouse trachea (Fig. 3A,A′) and proximal, extralobar bronchi. They are also markers for squamous carcinoma in the human lung. As shown in Figure 3, B and B′, some cells in the hyperplastic lesions in the alveoli of CC10-CreER; K-Ras; DNMaml1 mice express Trp63 alone (17.3%), Krt5 alone (5.4%), or both Trp63 and Krt5 (6.1%) (Fig. 3C). These expression patterns are never seen in the alveoli of control lungs (data not shown). These results indicate that Notch inhibition with resultant Sox2 up-regulation drives a squamous phenotype in cells that were originally K-RasG12D-expressing type II cells. These Sox2-high/Trp63+/Krt5+ cells do not progress to larger tumors at the times of sacrifice in this study.
Figure 2.
Inhibition of Notch signaling leads to high Sox2 and Foxj1 expression in CC10-expressing alveolar cells. (A,B), Mouse lung sections stained with DAPI, GFP (green), and Sox2 (red). (A) Sections of CC10-CreER; K-Ras mouse lung. Sox2 expression is seen in airway epithelial cells but not tumor (white dashed line and asterisk). (A′) Higher magnification of boxed region in A. (B) Sections of CC10-CreER; K-Ras; DNMaml1 mouse lung. Hyperplasia/small adenoma with high Sox2 expression in all GFP+ cells. (B′) Higher magnification of the boxed region in B. (C,D) GFP (green), Foxj1(red), and Sftpc (purple) staining for the indicated mouse lung sections 15 wk after tamoxifen treatment. (C) Tumor cells in CC10-CreER; K-Ras mouse lung are GFP+Sftpc+Foxj1−. (C′) Higher magnification of the boxed region in C. (D) GFP+Foxj1+ cells in some alveolar hyperplastic lesions in CC10-CreER; K-Ras; DNMaml1 mouse lung. Note that all GFP+ cells are Sftpc−. (D′) Higher magnification of the boxed region in D.
Figure 3.
Inhibition of Notch in K-RasG12D; CC10-expressing cells leads to hyperplasia with squamous features. (A,B) GFP (gray),Trp63 (green), and Krt5 (red) staining in mouse lung sections of the indicated genotypes 15 wk after tamoxifen treatment. (A) The section including CC10-CreER; Rosa26-f-GFP mouse trachea shows normal Trp63 and Krt5 staining in basal cells. (A′) Trp63 and Krt5 channels only. (B) Representative image of GFP-labeled hyperplastic lesion in CC10-CreER; K-Ras; DNMaml1 mouse lung expressing Trp63 and Krt5. (B′) Trp63 and Krt5 channels only. (1,2,3) Higher magnification of boxed regions in B′ labeled with each number. Arrows show Trp63+Krt5+ cells. Asterisks show Trp63+Krt5− cells. The arrowhead shows Trp63−Krt5+ cells. (C) Bar graph showing the percentage of cells in each lesion. Fifteen lesions from each mouse and a total of two mice was analyzed (n = 2).
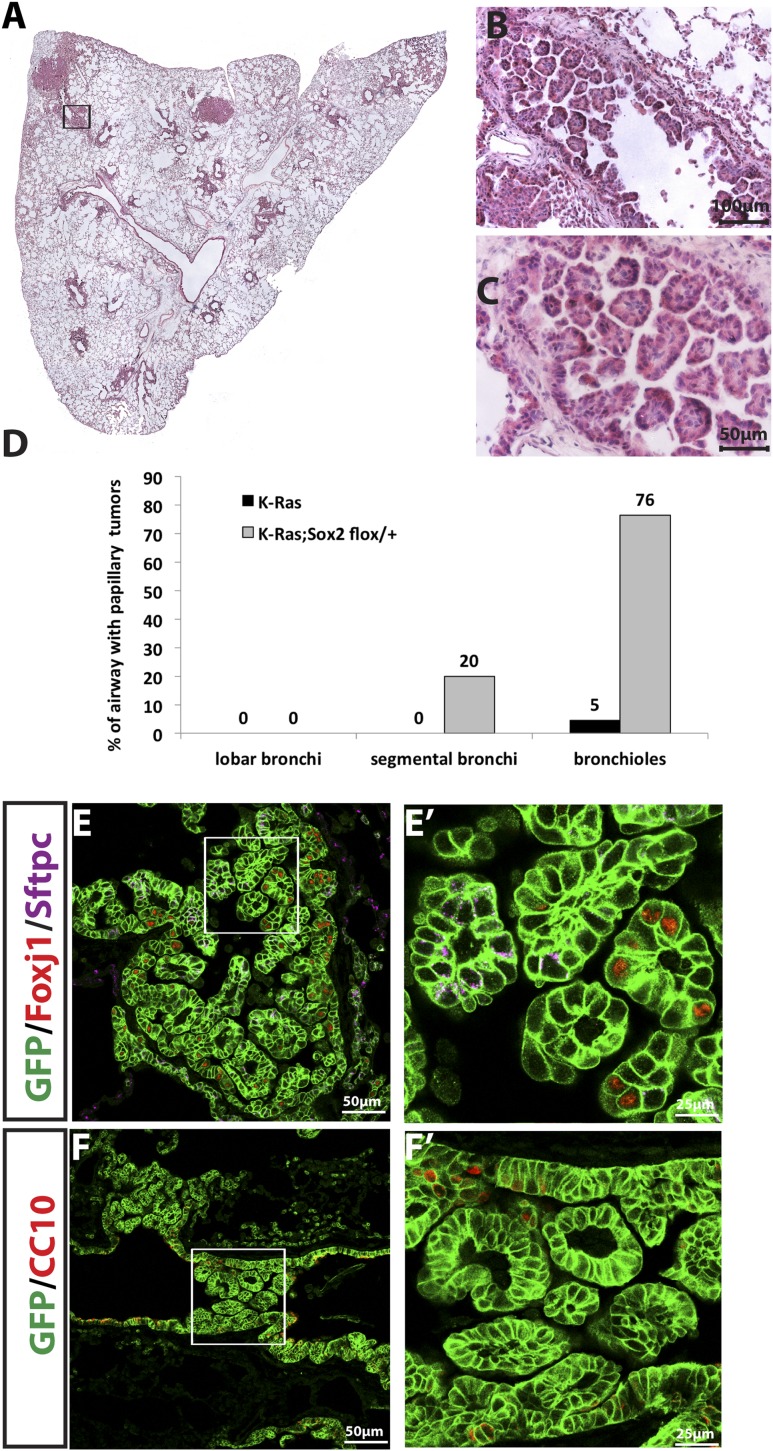
Overexpression of Notch1 ICD promotes papillary adenocarcinoma tumor formation in CC10+ airway cells that express K-RasG12D
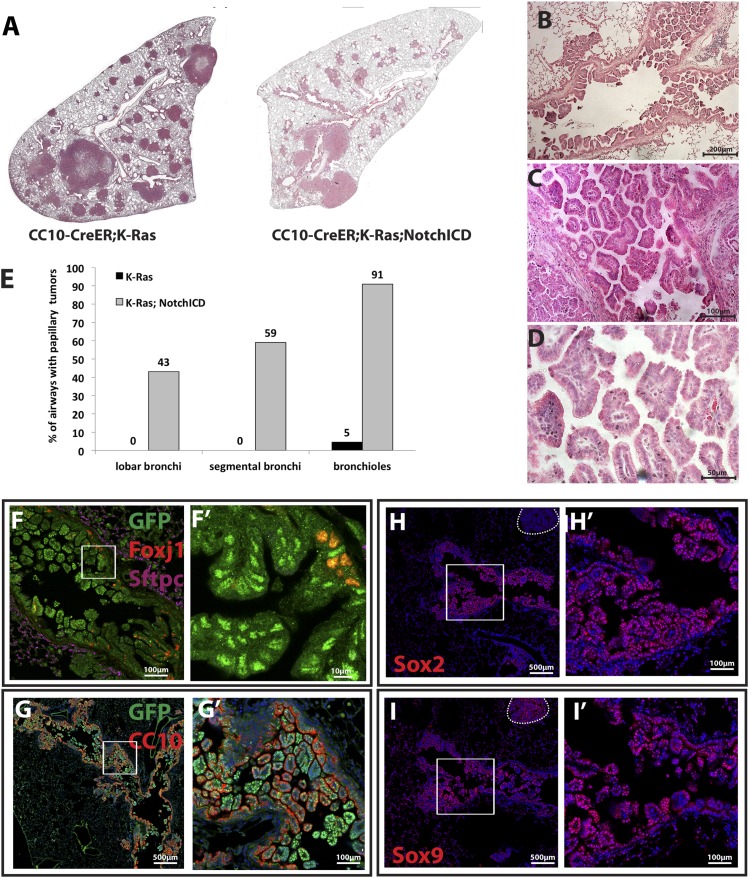
Because Notch inhibition suppresses K-Ras-induced tumors in the alveoli, we reasoned that Notch activation might allow or promote K-Ras-induced tumor formation in CC10+ airway cells. In order to determine whether bronchiolar cells form tumors when Notch is activated in conjunction with K-Ras, we made use of the Rosa26-NotchICD-GFP allele (Murtaugh et al. 2003). Following Cre-mediated recombination, this allele results in the constitutive expression of the ICD of Notch1 that is sufficient for ligand-independent, cell-autonomous Notch activation as well as nuclear GFP. We treated adult CC10-CreER; LSL-K-RasG12D; Rosa26-NotchICD-GFP (CC10-CreER; K-Ras; NotchICD) mice with tamoxifen. CC10-CreER; LSL-KrasG12D; Rosa26-f-GFP (CC10-CreER; K-Ras) mice of the same age were controls. After 15 wk, we assessed the lung tumor burden of both groups. Strikingly, we observed the widespread appearance of papillary lung adenocarcinomas in airways of CC10-CreER; K-Ras; NotchICD mice (Fig. 4A, right panel). In contrast, such tumors are very rare in CC10-CreER; K-Ras control mice (Fig. 4A, left panel). We quantified tumors per airway and found that CC10-CreER; K-Ras; NotchICD mice had significantly higher numbers of airway adenocarcinomas than CC10-CreER; K-Ras mice (Fig. 4E; Supplemental Table S1). Thus, taken together, our loss- and gain-of-function Notch allele studies demonstrate that Notch signaling activity determines tumor location/cell of origin when K-RasG12D is induced.
Figure 4.
Overexpression of Notch transforms tumor-resistant K-RasG12D+ airway CC10+ cells to form papillary adenocarcinomas. (A) Section of CC10-CreER; K-Ras (left) and CC10-CreER; K-Ras; NotchICD (right) mouse left lung lobes 15 wk after tamoxifen injection stained with H&E. (B–D) Higher magnification shows all airways affected with tumors (B) with papillary morphology (C) and mitoic figures (D). (E) Bar graph shows the percentage of the indicated airways with tumors. (F–I) Immunostaining of mouse lung sections of CC10-CreER; K-Ras; NotchICD mice. (F) GFP (green), Foxj1 (red), and Sftpc (purple) staining. Note that GFP+ papillary tumors are Sftpc−. (F′) Higher magnification of the area in F marked with a box. In the airway, GFP+Foxj1+ cells are significantly reduced, and almost all papillary tumor cells are GFP+Foxj1−. (G) GFP (green) and CC10 (red) staining demonstrates widespread CC10 expression in almost all airway epithelial cells and papillary tumor cells. (G′) Higher magnification of the boxed area in G. (H,I). Sox2 (H) and Sox9 (I) staining from adjacent consecutive sections of CC10-CreER; K-Ras; NotchICD mouse lung sections. (H) Sox2 is expressed in all airway papillary tumor cells but not in alveolar tumors (dashed line in H). (H′) Higher magnification of the boxed area in H. (I) All airway papillary tumor cells express Sox9. Sox9 is also expressed in one alveolar tumor (dashed line in I). Note that same tumor is negative for Sox2 (dashed line in H). (I′) Higher magnification of the boxed area in I.
Papillary adenocarcinomas in CC10+; K-RasG12D airway cells overexpressing Notch express CC10 and both Sox2 and Sox9
To further characterize the airways of CC10-CreER; K-Ras mice with and without NotchICD, we immunostained lung sections with antibodies to Sftpc, Foxj1, CC10, Krt5, and Trp63. The columnar epithelial cells lining the airways in control CC10-CreER; K-Ras mice have an increased number of GFP+Foxj1+ cells 15 wk after tamoxifen injection compared with CC10-CreER; Rosa26-f-GFP mice (Supplemental Fig. S1). The GFP+Foxj1+ cells in CC10-CreER; K-Ras mice express acetylated tubulin and do not incorporate BrdU (data not shown). This phenotype mimics the effect of inhibiting Notch during lung development, which leads to terminal differentiation of CC10+ cells into Foxj1+ ciliated cells when a proliferative signal is encountered (Rock et al. 2011; Guha et al. 2012). In contrast, consistent with high Notch promoting secretory airway phenotypes, most of the GFP+ tumor cells in the papillary adenocarcinomas of the CC10-CreER; K-Ras; NotchICD mice are CC10+ and Foxj1−. They are also negative for Sftpc (Fig. 4F,F′,G,G′), Krt5, and Trp63 (data not shown).
To further analyze the phenotype of the papillary lung adenocarcinomas in CC10-CreER; K-Ras; NotchICD mice, we stained sections for Sox2 and Sox9. Both transcription factors are expressed in the embryonic lung, but Sox9 is normally down-regulated at birth (Chang et al. 2013). In contrast to the alveolar adenocarcinomas in CC10-CreER; K-Ras mice, which are Sox2− and Sox9+ (data not shown), the papillary adenocarcinomas in CC10-CreER; K-Ras; NotchICD mice express both Sox2 and Sox9, as judged by staining of adjacent sections (Fig. 4H,H′,I,I′). These data demonstrate that Notch activation leads to expression of Sox9, a distal embryonic lung marker that is often expressed in human lung tumors (Jiang et al. 2010; Zhou et al. 2012).
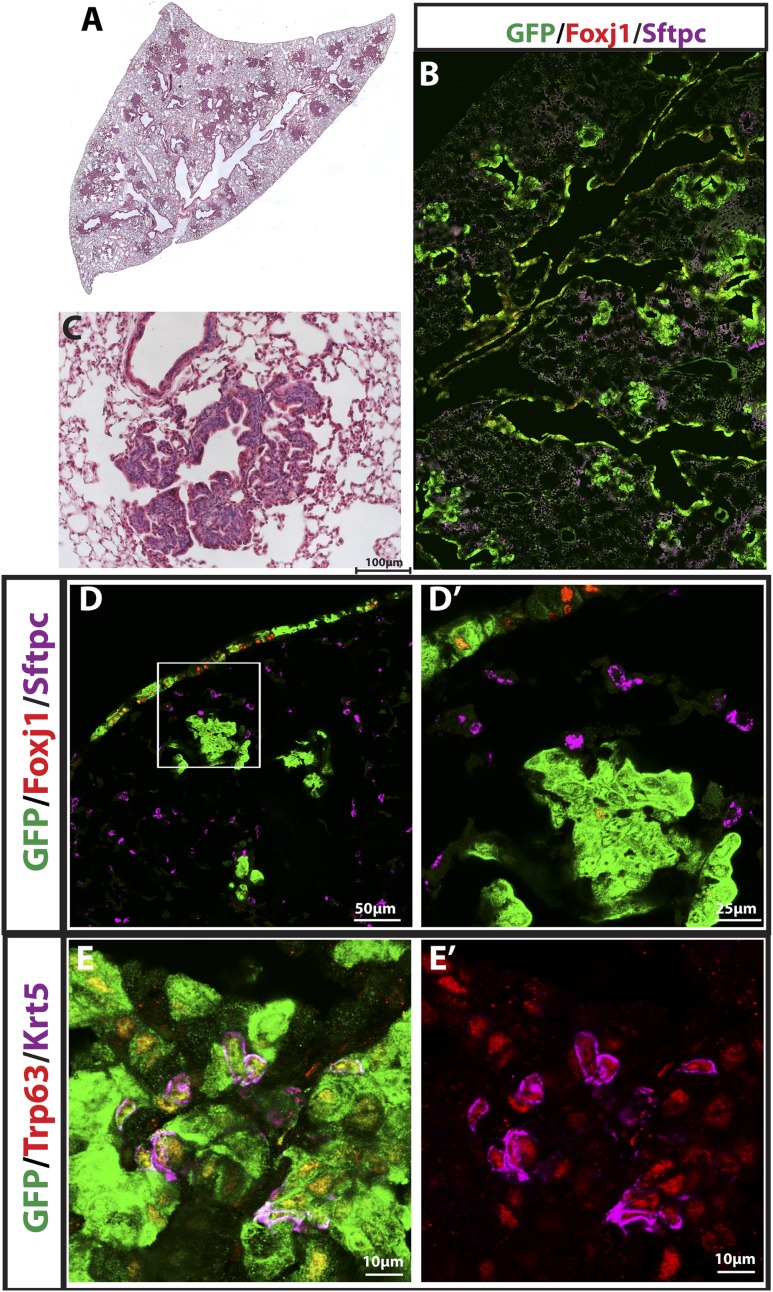
Reduction of Sox2 expression in CC10+; K-RasG12D airway cells leads to CC10+ papillary adenocarcinoma
Notch is activated only in tumor-forming type II cells when K-Ras is induced (Maraver et al. 2012). We next asked what regulates the Notch pathway in these cells. Sox2, a high-mobility box transcription factor, is normally expressed in epithelial cells of the proximal trachea, bronchi, and bronchioles but not in the distal alveolar region of the lung (Tompkins et al. 2009). Because of this expression pattern and because it binds a Notch1 intergenic region in the retina (Taranova et al. 2006), we reasoned that Sox2 limits Notch transcription in airway cells. We previously generated a Rosa26-LSL-CAG-Sox2-IRES-GFP (LSL-Sox2) mouse line (Lu et al. 2010). When CC10-CreER; LSL-Sox2 mice are administered tamoxifen, adenocarcinoma with squamous features develops in the alveoli in about half of the mice. In order to identify potential mechanistic effectors of Sox2, we isolated RNA from GFP+ cells present in CC10-CreER; LSL-Sox2 (n = 7) and CC10-CreER; Rosa26-f-GFP (n = 4) lungs 16 wk after tamoxifen treatment. Microarray analysis was performed using Affymetrix 430.2 chips. We found that Hes1, Notch1, and Notch2 are significantly down-regulated when Sox2 is overexpressed (14.4-fold, 18.9-fold, and 15.3-fold, respectively) (Supplemental Table S2; Supplemental Fig. S2A). In addition, we performed chromatin immunoprecipitation (ChIP) using a SOX2 antibody in SOX2-overexpressing H520 human lung cancer cells. The resulting DNA was sequenced, and significant peaks were noted at the promoter region of HES1 and NOTCH1 and in an intergenic region of NOTCH2 (Supplemental Fig. S2B; Supplemental Table S2). Taken together, these analyses suggest that Sox2 directly binds regulatory sequences of Hes1, Notch1, and Notch2, leading to suppression of transcription.
Based on these findings, we reasoned that reducing the level of Sox2 in bronchial/bronchiolar epithelial cells would release the inhibition of Notch signaling when K-RasG12D is activated. Because Sox2 is required for CC10 cell survival (Tompkins et al. 2009), and CC10-CreER; LSL-K-RasG12D; Sox2flox/flox mice die early due to abnormal airways (Supplemental Fig. S3), we generated CC10-CreER; LSL-K-RasG12D; Sox2flox/+ (CC10-CreER; K-Ras; Sox2flox/+) mice, which have 50% of the wild-type Sox2 level (Supplemental Fig. S4). At 12 wk after tamoxifen, CC10-CreER; K-Ras; Sox2flox/+ mice already developed airway papillary adenocarcinomas (Fig. 5A–D). To characterize these lesions, we immunostained sections for Foxj1, CC10, and Sox2. As described earlier, control CC10-CreER; K-Ras mice have a single layer of GFP+ epithelial cells lining the airways 15 wk after tamoxifen injection. In contrast, airways in CC10-CreER; K-Ras; Sox2flox/+ mice contain papillary adenocarcinoma cells. Most tumor cells that extend into the airway lumen in these mice are Foxj1−, and a few GFP+Foxj1+ cells remain in the normal airway epithelial layer (Fig. 5E,E′,F,F′). This result suggests that a reduction in Sox2 levels can allow tumor-resistant CC10+ airway cells to initiate tumors when K-Ras is induced.
Figure 5.
Reduction of Sox2 in CC10+ airway cells leads to papillary adenocarcinoma formation. (A, right) Sections of CC10-CreER; LSL-K-RasG12D; Sox2 flox/+ mouse lungs 12 wk after tamoxifen injection stained with H&E. (B,C) Higher magnification of papillary airway tumors. (D) Bar graph demonstrating percentage of airways with tumors in CC10-CreER; LSL-K-RasG12D mice and CC10-CreER; LSL-K-RasG12D; Sox2 flox/+ lungs. (E,E′,F,F′, right) Sections of CC10-CreER; LSL-K-RasG12D; Sox2 flox/+ mouse lungs stained with the indicated markers.
To ask whether Sox2 overexpression suppresses tumor formation in K-RasG12D-expressing type II cells, we created a CC10-CreER; LSL-K-RasG12D; LSL-Sox2 (CC10-CreER; K-Ras; LSL-Sox2) mouse line and administered tamoxifen at 6 wk after birth. As Figure 6 reveals, these mice have markedly smaller alveolar tumors than CC10-CreER; K-Ras mice at similar ages. Immunohistochemical analysis of these alveolar hyperplastic regions demonstrates positive staining for the squamous markers Krt5 and Trp63. All hyperplastic cells were noted to be strongly GFP+ (Sox2 label). These data demonstrate that overexpression of Sox2 is tumor-suppressive for large adenocarcinomas in this model.
Figure 6.
Overexpression of Sox2 inhibits K-Ras-induced lung adenocarcinoma formation and leads to hyperplasia/adenoma with squamous features. (A) Sections of CC10-CreER; K-Ras; LSL-Sox2 mouse lungs 20 wk after tamoxifen injection stained with H&E. (B) Higher magnification of hyperplasia in A. (C) GFP (green), Foxj1 (red), and Sftpc (purple) staining of mouse lung sections. Note the GFP+ hyperplasia but absence of GFP+ adenoma or adenocarcinoma. (D) GFP (green), Foxj1 (red), and Sftpc (purple) staining of hyperplasia. Alveolar hyperplasia is GFP+ Sftpc−. (D′) Higher magnification of the white box in D. (E) GFP (green), Trp63 (red), and Krt5 (purple) staining. Note the GFP+ Trp63+ Krt5+ cells. (E′)Trp63 and Krt5 channels only.
Discussion
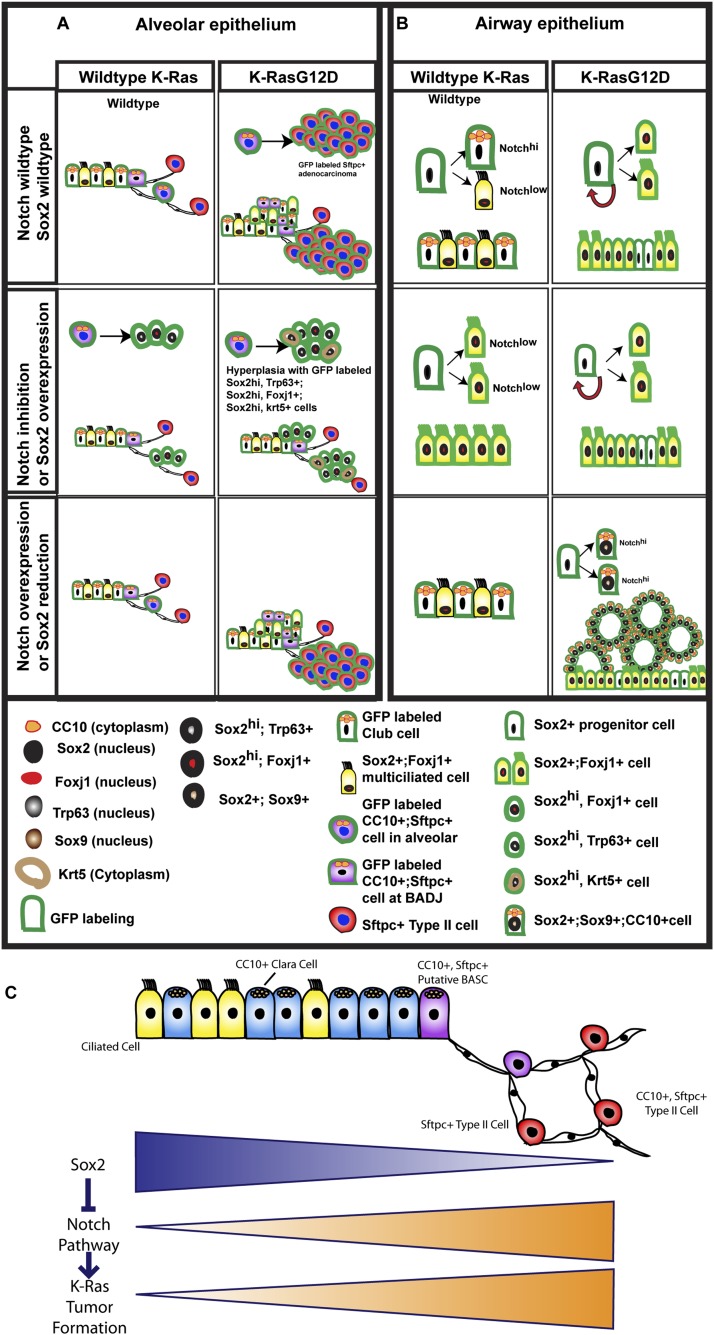
Collectively, our data provide further insights into the pivotal roles of Sox2 and Notch signaling in K-Ras-driven lung tumors. As the model in Figure 7A summarizes, in the alveoli, CC10+Sftpc+ cells in which K-Ras has been activated progress to lung adenocarcinoma, and this progression requires the ability to activate Notch signaling. When we block Notch signaling, adenocarcinoma formation is inhibited, and only hyperplastic clusters/small adenomas are found. These cells express proximal lung epithelial markers and do not progress to adenocarcinomas over the course of this study. In the airways (Fig. 7B), K-Ras-activated CC10+ cells differentiate into Foxj1+ ciliated cells and are resistant to tumor formation. However, when Notch is activated or Sox2 is down-regulated, the K-Ras-activated CC10+ cells can form CC10+Foxj1− papillary adenocarcinomas.
Figure 7.
Schematic summary of genetic interaction between Notch, Sox2, and K-Ras in lung tumor initiation and phenotype in the alveoli (A) and airway (B). (C) Model for some of the genetic interactions that determine tumor initiation in the epithelium of the mouse lung. (Top line) Known proximal–distal gradient of Sox2 expression. (Bottom lines) Proposed distal–proximal gradients of potential for Notch pathway activation and K-Ras tumor initiation.
There are at least two subtypes of type II cells in the alveoli (a majority that are CC10−Sftpc+ and a minority that are CC10+Sftpc+). Our current results from CC10-CreER-driven models indicate that inhibition of Notch signaling or overexpression of Sox2 can inhibit K-Ras-activated lung adenocarcinoma formation originating from CC10+Sftpc+ cells. Preliminarily, Sftpc-CreER; LSL-K-RasG12D; LSL-Sox2 mice also have markedly abrogated tumor formation (data not shown). Further analysis of these mice will be the focus of another study. Our data are consistent with a previous report (Sutherland et al. 2014) that lung adenocarcinoma formation requires reduced Sox2 expression.
A previous study suggested that genetic or γ-secretase inhibition of the Notch pathway blocks tumor growth in codon 12 mutant Kras mice (Maraver et al. 2012). However, in humans, missense and nonsense mutations in Notch ligands and receptors lead to squamous cell tumors in the lung (Wang et al. 2011; Cancer Genome Atlas Research Network 2012). This is consistent with the development of alveolar hyperplasia with squamous markers that we observed with Notch inhibition in the current study. Because of this, we believe that γ-secretase inhibitors should be used with great caution in patients with lung adenocarcinoma. Because γ-secretase inhibition blocks all Notch receptors, a future direction will be to target individual Notch receptors and Notch targets as potential therapeutic strategies.
CC10+ airway papillary adenocarcinomas in CC10-CreER; K-Ras; Sox2 flox/+ and CC10-CreER; K-Ras; NotchICD mice, unlike alveolar tumors, do not express Sftpc, a marker for type II cells that is expressed in the majority of lung adenocarcinomas. In humans, K-RAS mutant tumors are strictly alveolar (Cooper et al. 1997), although our data would predict that K-RAS mutant tumors may arise from bronchiolar cells if NOTCH signaling is also activated (or if SOX2 is inactivated). This will be a focus of further study with human lung adenocarcinoma samples.
Terminal differentiation of CC10+ airway cells into Foxj1+ ciliated cells when K-RasG12D is induced is similar to the effect of inhibition of Notch in lung development. As the airway cells proliferate during airway development, low levels of Notch signaling result in the differentiation of progenitors into Foxj1+ ciliated cells (Guha et al. 2012; Morimoto et al. 2012). This emphasizes the importance of cellular context in determining the phenotype of tumors formed in response to oncogene activation. Future therapeutic opportunities may be identified if the specific factors that drive differentiation in cells proliferating in response to K-Ras induction can be delineated.
K-Ras mutant lung adenocarcinomas in humans may be divided into K-Ras-dependent and K-Ras-independent subsets. K-Ras-dependent tumors are epithelial and may respond better to MEK inhibition, while K-Ras-independent tumors are mesenchymal and may be more resistant to treatment (Singh et al. 2009; Corcoran et al. 2013). These distinctions do not correlate with genetic mutations but are epigenetic. In fact, exogenous administration of TGF-β leads to loss of K-Ras dependence. Interestingly, Sox2 overexpression in the respiratory epithelium leads to induction of TGF-β targets (Tompkins et al. 2011). In addition to lung cancer, Notch signaling is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma (Mazur et al. 2010). It would be interesting to test whether altered Sox2 expression plays a role in K-Ras-driven pancreatic adenocarcinoma development. A future direction will be to determine whether Sox2 is inhibiting K-Ras-induced tumor formation in additional ways that are distinct from Notch inhibition.
In summary, our data support a general model depicted in Figure 7C. According to this model, K-Ras activation of type II cells leads to adenocarcinoma because these cells are able to sustain or maintain Notch signaling when K-Ras is activated. Sox2, which is present in airway cells but only at very low levels in type II cells, suppresses the expression of Notch receptors at the transcriptional level, and this in turn inhibits K-Ras-induced tumor formation.
Materials and methods
Mice
The CC10-CreER; LSL-K-RasG12D; Rosa26R-f-GFP and Rosa26-LSL-CAG-Sox2-IRES-GFP mouse lines have been described previously (Rawlins et al. 2009; Xu et al. 2012). Rosa26-LSL-DNMaml-GFP mice (Tu et al. 2005) were kindly provided by Warren Pear (University of Pennsylvania, Philadelphia, PA). Rosa26-Notch1ICD-GFP mice (Murtaugh et al. 2003) were from The Jackson Laboratories. Sox2 flox/flox mice (Taranova et al. 2006) were generated by Larysa Pevny (University of North Carolina, Chapel Hill, NC) and kindly provided by Barry Stripp (Cedars Sinai, Los Angeles, CA). All animal experiments were approved by the Duke Medicine Institutional Animal Care and Use Committee.
Tamoxifen administration
A 20 mg/mL tamoxifen (Sigma) solution was made by dissolving tamoxifen in Mazola corn oil. Six-week-old to 8-wk-old mice were injected with 0.25 mg per gram of body weight tamoxifen every other day for four doses.
Histology and immunohistochemistry
For immunohistochemistry, lungs were perfused with PBS and then inflated and fixed with 4% PFA overnight. Fixed lungs were washed three times with PBS, gradually dehydrated in ethanol, and embedded in paraffin. Paraffin sections (7 μm) were stained with the following primary antibodies: chicken anti-GFP (1:400; Aves Laboratories, GFP1020), rabbit anti-proSftpc (1:500; Millipore, AB3786), goat-anti CC10 (1:10,000; kindly provided by Barry Stripp), rabbit-anti CC10 (1:10,000; Barry Stripp), mouse anti-Foxj1 (1:200; eBioscience, 14-9965), rabbit anti-Sox2 (1:1000; Seven Hills Bioreagents, WRAB-Sox2), mouse anti-Trp63 (1:200; Santa Cruz Biotechnology), rabbit-anti-Krt5 (1:500; Covance), rabbit anti-Krt8 (1:200; Developmental Studies Hybridoma Bank), and rabbit anti-Sox9 (1:1000; Millipore). Alexa-fluor-coupled secondary antibodies (Invitrogen) were used at 1:400 dilution. All images were captured on a Leica Sp2 laser scanning confocal microscope.
ChIP/sequencing
Approximately 50 million NCI-H520 (American Type Culture Collection, HTB-182) cells were fixed with 1% PFA and then subjected to ChIP with an anti-SOX2 antibody (Cell Signaling, catalog no. 5024) using the Cell Signaling SimpleChIP enzymatic ChIP kit (Cell Signaling, catalog no. 9003) as per the manufacturer’s instructions. Input and SOX2-immunoprecipitated chromatin samples were sequenced at the Duke University Genome Sequencing and Analysis Core. Approximately 31 million input and 26 million SOX2-immunoprecipitated 36-base-pair reads were acquired. Reads were mapped to human genome assembly GRCh37/hg19 using Bowtie via the Galaxy online analysis toolkit (http://galaxyproject.org) (Goecks et al. 2010). SOX2-binding sites were then predicted using Model-Based Analysis of ChIP-Seq (MACS) version 1.4.1 (Zhang et al. 2008).
Microarray analysis
CC10-CreER; LSL-Sox2 mice and CC10-CreER; Rosa26-f-GFP mice were sacrificed 20 wk after tamoxifen injection. Lungs were perfused with 10 mL of PBS and lavaged four times with 1 mL of PBS and 0.2 μM EGTA. One milliliter of 4 U/mL Elastase solution (Worthington Biochemical) was instilled in the airways and incubated for 5 min at 37°C followed by three 0.5-mL instillations for 5 min each. Lung lobes were then minced and incubated with DNase I solution for 10 min at 37°C. Cells were passed through a 100-μm cell strainer, and red blood cells (RBCs) were lysed using RBC lysing buffer (eBioscience, Inc.). Isolated cells were resuspended at 1 × 106 cells in 100 μL of Hanks’ balanced saline solution, 10 mM HEPES, and 2% fetal bovine serum. Propidium iodide (PI) was used for dead cell discrimination. GFP-positive cells were sorted and collected on FACS Vantage SE. RNA was then extracted using Qiagen protocols. The RNA was reverse-transcribed, and the cDNA was hybridized to Affymetrix 420 mouse microarray chips in the Duke Medicine microarray facility. The resulting CEL files were rma-normalized in Bioconductor in the R environment. Differential expression was carried out using the Limma package with multiple comparisons controlled by the method of Benjamini and Hochberg.
Pathologic analysis
Tumor sections were reviewed by Dr. Thomas Sporn of the Department of Pathology, Duke Medicine.
Tumor analysis
For each mouse, three longitudinal sections of the left lung lobe (600 μm, 800 μm, and 1200 μm from the surface) containing the main axial bronchial pathway were stained with H&E. Tumor size and number were scored using ImageJ software. Airway tumors were assessed by ≥30% of airway space covered by tumor cells, and airways were categorized by lobar bronchi, segmental bronchi, and bronchiole.
Acknowledgments
We thank Jing Zhang for mouse husbandry, Dr. Tim Reddy in the Duke Institute for Genome Science and Policy for advice with bioinformatics, and Henry Yen for technical assistance. This work was supported by Howard Hughes Medical Institute Early Career Grant, Department of Defense Promising Young Investigator Grant, National Lung Cancer Partnership Research Grant, and Thoracic Surgery Foundation for Research and Education Research Grant.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.243717.114.
References
- Adjei AA, Cohen RB, Franklin W, Morris C, Wilson D, Molina JR, Hanson LJ, Gore L, Chow L, Leong S, et al. 2008. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol 26: 2139–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TD, Rodriguez EM, Jones KD, Bishop JM. 2011. Activated Notch1 induces lung adenomas in mice and cooperates with Myc in the generation of lung adenocarcinoma. Cancer Res 71: 6010–6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2012. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489: 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DR, Martinez Alanis D, Miller RK, Ji H, Akiyama H, McCrea PD, Chen J. 2013. Lung epithelial branching program antagonizes alveolar differentiation. Proc Natl Acad Sci 110: 18042–18051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CA, Carby FA, Bubb VJ, Lamb D, Kerr KM, Wyllie AH. 1997. The pattern of K-ras mutation in pulmonary adenocarcinoma defines a new pathway of tumour development in the human lung. J Pathol 181: 401–404 [DOI] [PubMed] [Google Scholar]
- Corcoran RB, Cheng KA, Hata AN, Faber AC, Ebi H, Coffee EM, Greninger P, Brown RD, Godfrey JT, Cohoon TJ, et al. 2013. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell 23: 121–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasz S, Liang S, Chen Y, De Marco MA, Machek O, Skucha S, Miele L, Bocchetta M. 2010. Notch-1 stimulates survival of lung adenocarcinoma cells during hypoxia by activating the IGF-1R pathway. Oncogene 29: 2488–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Galaxy T. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha A, Vasconcelos M, Cai Y, Yoneda M, Hinds A, Qian J, Li G, Dickel L, Johnson JE, Kimura S, et al. 2012. Neuroepithelial body microenvironment is a niche for a distinct subset of Clara-like precursors in the developing airways. Proc Natl Acad Sci 109: 12592–12597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SS, Fang WT, Hou YH, Huang SF, Yen BL, Chang JL, Li SM, Liu HP, Liu YL, Huang CTet al. 2010. Upregulation of SOX9 in lung adenocarcinoma and its involvement in the regulation of cell growth and tumorigenicity. Clin Cancer Res 16: 4363–4373 [DOI] [PubMed] [Google Scholar]
- Johnson BE, Heymach JV. 2004. Farnesyl transferase inhibitors for patients with lung cancer. Clin Cancer Res 10: 4254s–4257s [DOI] [PubMed] [Google Scholar]
- Licciulli S, Avila JL, Hanlon L, Troutman S, Cesaroni M, Kota S, Keith B, Simon MC, Pure E, Radtke F, et al. 2013. Notch1 is required for Kras-induced lung adenocarcinoma and controls tumor cell survival via p53. Cancer Res 73: 5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, Onaitis MW. 2010. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS ONE 5: e11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. 2009. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- Mainardi S, Mijimolle N, Francoz S, Vicente-Duenas C, Sanchez-Garcia I, Barbacid M. 2014. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc Natl Acad Sci 111: 255–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraver A, Fernandez-Marcos PJ, Herranz D, Canamero M, Munoz-Martin M, Gomez-Lopez G, Mulero F, Megias D, Sanchez-Carbayo M, Shen J, et al. 2012. Therapeutic effect of γ-secretase inhibition in KrasG12V-driven non-small cell lung carcinoma by derepression of DUSP1 and inhibition of ERK. Cancer Cell 22: 222–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur PK, Einwachter H, Lee M, Sipos B, Nakhai H, Rad R, Zimber-Strobl U, Strobl LJ, Radtke F, Kloppel G, et al. 2010. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc Natl Acad Sci 107: 13438–13443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M, Nishinakamura R, Saga Y, Kopan R. 2012. Different assemblies of Notch receptors coordinate the distribution of the major bronchial Clara, ciliated and neuroendocrine cells. Development 139: 4365–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. 2003. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci 100: 14920–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. 2006. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124: 973–983 [DOI] [PubMed] [Google Scholar]
- Osanyingbemi-Obidi J, Dobromilskaya I, Illei PB, Hann CL, Rudin CM. 2011. Notch signaling contributes to lung cancer clonogenic capacity in vitro but may be circumvented in tumorigenesis in vivo. Mol Cancer Res 9: 1746–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, Hogan BL. 2009. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4: 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Gao X, Xue Y, Randell SH, Kong YY, Hogan BL. 2011. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 8: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S, et al. 2009. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137: 821–834 [DOI] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. 2009. A gene expression signature associated with ‘K-Ras addiction’ reveals regulators of EMT and tumor cell survival. Cancer Cell 15: 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland KD, Song JY, Kwon MC, Proost N, Zevenhoven J, Berns A. 2014. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci 111: 4952–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, et al. 2012. RET, ROS1 and ALK fusions in lung cancer. Nat Med 18: 378–381 [DOI] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. 2006. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev 20: 1187–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins DH, Besnard V, Lange AW, Wert SE, Keiser AR, Smith AN, Lang R, Whitsett JA. 2009. Sox2 is required for maintenance and differentiation of bronchiolar Clara, ciliated, and goblet cells. PLoS ONE 4: e8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins DH, Besnard V, Lange AW, Keiser AR, Wert SE, Bruno MD, Whitsett JA. 2011. Sox2 activates cell proliferation and differentiation in the respiratory epithelium. Am J Respir Cell Mol Biol 45: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J, Cardoso WV. 2009. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 136: 2297–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L, Fang TC, Artis D, Shestova O, Pross SE, Maillard I, Pear WS. 2005. Notch signaling is an important regulator of type 2 immunity. J Exp Med 202: 1037–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, Leigh IM, Collisson EA, Gordon PB, Jakkula L, et al. 2011. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci 108: 17761–17766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff B, Colaluca IN, D’Ario G, Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G, Viale G, et al. 2009. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci 106: 22293–22298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Rock JR, Lu Y, Futtner C, Schwab B, Guinney J, Hogan BL, Onaitis MW. 2012. Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proc Natl Acad Sci 109: 4910–4915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, Alvarez CA, Moreira DC, Creighton CJ, Gregory PA, et al. 2011. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest 121: 1373–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CH, Ye LP, Ye SX, Li Y, Zhang XY, Xu XY, Gong LY. 2012. Clinical significance of SOX9 in human non-small cell lung cancer progression and overall patient survival. J Exp Clin Cancer Res 31: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]