Nonsense-mediated mRNA decay (NMD) controls the quality of eukaryotic gene expression and also degrades physiologic mRNAs. Here, Kurosaki et al. mapped phosphorylated UPF1-binding sites and found them to be enriched on NMD target 3′ UTRs along with SMG5 and SMG7. ATPase/helicase-deficient UPF1 manifests high levels of RNA binding and disregulated hyperphosphorylation. 3′ UTR-associated UPF1 undergoes regulated phosphorylation, providing a binding platform for mRNA-degradative activities.
Keywords: UPF1, ATP-dependent RNA helicase, nonsense-mediated mRNA decay, phosphorylation, RIP-seq of transcriptome-binding sites for phospho-UPF1, mRNP composition
Abstract
Nonsense-mediated mRNA decay (NMD) controls the quality of eukaryotic gene expression and also degrades physiologic mRNAs. How NMD targets are identified is incompletely understood. A central NMD factor is the ATP-dependent RNA helicase upframeshift 1 (UPF1). Neither the distance in space between the termination codon and the poly(A) tail nor the binding of steady-state, largely hypophosphorylated UPF1 is a discriminating marker of cellular NMD targets, unlike for premature termination codon (PTC)-containing reporter mRNAs when compared with their PTC-free counterparts. Here, we map phosphorylated UPF1 (p-UPF1)-binding sites using transcriptome-wide footprinting or DNA oligonucleotide-directed mRNA cleavage to report that p-UPF1 provides the first reliable cellular NMD target marker. p-UPF1 is enriched on NMD target 3′ untranslated regions (UTRs) along with suppressor with morphogenic effect on genitalia 5 (SMG5) and SMG7 but not SMG1 or SMG6. Immunoprecipitations of UPF1 variants deficient in various aspects of the NMD process in parallel with Förster resonance energy transfer (FRET) experiments reveal that ATPase/helicase-deficient UPF1 manifests high levels of RNA binding and disregulated hyperphosphorylation, whereas wild-type UPF1 releases from nonspecific RNA interactions in an ATP hydrolysis-dependent mechanism until an NMD target is identified. 3′ UTR-associated UPF1 undergoes regulated phosphorylation on NMD targets, providing a binding platform for mRNA degradative activities. p-UPF1 binding to NMD target 3′ UTRs is stabilized by SMG5 and SMG7. Our results help to explain why steady-state UPF1 binding is not a marker for cellular NMD substrates and how this binding is transformed to induce mRNA decay.
Nonsense-mediated mRNA decay (NMD) in human cells degrades newly synthesized mRNAs that are aberrant because they contain a premature termination codon (PTC) and thereby have the potential to encode a toxic truncated protein (Karam et al. 2013; Popp and Maquat 2013, 2014; Yamashita 2013). NMD also degrades ∼5%–10% of naturally occurring mRNAs, often as a means of maintaining cellular homeostasis (Huang and Wilkinson 2012; Schweingruber et al. 2013) or regulating developmental processes that include axon guidance, synaptic strength, and neuronal expression (Giorgi et al. 2007; Colak et al. 2013). These functions explain at least in part the intellectual disabilities that typify NMD factor deficiencies (Nguyen et al. 2012, 2013).
In mammals, NMD largely targets newly synthesized mRNAs during a pioneer round of translation (Maquat et al. 2010; Trcek et al. 2013). Depending on the rates at which newly synthesized transcripts are translated and maintain features that are recognized by the NMD machinery, NMD can degrade mRNAs bound at their caps by the cap-binding protein (CBP) heterodimer CBP80 and CBP20 or eukaryotic translation initiation factor 4E (eIF4E) (Ishigaki et al. 2001; Hosoda et al. 2005; Matsuda et al. 2007; Woeller et al. 2008; Sato and Maquat 2009; Durand and Lykke-Andersen 2013; Rufener and Mühlemann 2013). CBP80 promotes NMD (Hosoda et al. 2005; Hwang et al. 2010), as do exon junction complexes (EJCs), which reside ∼20–24 nucleotides (nt) upstream of ∼80% of exon–exon junctions (Le Hir et al. 2000; Saulière et al. 2012; Singh et al. 2012). EJCs often consist of the NMD factors upframeshift 2 (UPF2) and UPF3 or UPF3X (also called UPF3a or UPF3b, respectively) as well as additional proteins (Popp and Maquat 2013, 2014; Yamashita 2013). The essential splicing factor complexed with Cef1 protein 22 (CWC22) interacts with the core EJC constituent eIF4A3 and activates NMD by coupling pre-mRNA splicing to EJC deposition (Alexandrov et al. 2012; Barbosa et al. 2012; Steckelberg et al. 2012).
According to a current model, UPF1 and its phosphatidylinositol 3-kinase-related kinase, suppressor with morphogenic effect on genitalia 1 (SMG1), load onto the termination codon of an NMD target together with the translation termination factors eukaryotic release factor 1 (eRF1) and eRF3, forming the SURF complex (Kashima et al. 2006; Hwang et al. 2010). When an EJC is associated with an exon–exon junction that resides more than ∼50–55 nt downstream from the termination codon so the associated EJC is not removed by the terminating ribosome, UPF1 together with SMG1 bind the EJC (Nagy and Maquat 1998; Le Hir et al. 2000; Lejeune et al. 2003; Yamashita et al. 2009; Hwang et al. 2010). EJC binding occurs via UPF2 and/or one of the UPF3 paralogs and triggers SMG1-mediated UPF1 phosphorylation (Yamashita et al. 2001, 2009; Kashima et al. 2006). Phosphorylated UPF1 (p-UPF1) represses further translation initiation on the NMD target (Isken et al. 2008). p-UPF1 also recruits SMG5, SMG6, and SMG7, which activate multiple and redundant pathways by which NMD targets are degraded and also mediate UPF1 dephosphorylation by protein phosphatase 2A (PP2A) (Anders et al. 2003; Chiu et al. 2003; Ohnishi et al. 2003; Fukuhara et al. 2005; Okada-Katsuhata et al. 2012; Jonas et al. 2013; Loh et al. 2013).
Steady-state UPF1 is largely hypophosphorylated (Chiu et al. 2003; Ohnishi et al. 2003; Isken et al. 2008; Okada-Katsuhata et al. 2012) and found associated with many mRNA 3′ untranslated regions (3′ UTRs), including those that do not belong to NMD targets, and also with long noncoding RNAs (Franks et al. 2010; Hogg and Goff 2010; Hurt et al. 2013; Kurosaki and Maquat 2013; Zünd et al. 2013; Gregersen et al. 2014). Data indicate that translationally active ribosomes remove bound UPF1 from 5′ UTRs, coding regions, and the first ∼35 nt downstream from a termination codon (Hurt et al. 2013; Kurosaki and Maquat 2013; Zünd et al. 2013; Gregersen et al. 2014). Only when a PTC-containing mRNA is compared with its PTC-free counterpart is an NMD target certain to manifest a higher number of and/or affinity for UPF1 molecules bound to its 3′ UTR (Kurosaki and Maquat 2013). Binding is augmented by translation, in particular, interactions between UPF1 and eRF3 during translation termination, and, subsequently, with a 3′ UTR EJC (Kurosaki and Maquat 2013). It remains unknown whether 3′ UTR binding involves hypophosphorylated UPF1 or p-UPF1, the interconversion of which is required for NMD (Isken et al. 2008; Franks et al. 2010; Hogg and Goff 2010; Hurt et al. 2013; Kurosaki and Maquat 2013; Zünd et al. 2013; Gregersen et al. 2014). Furthermore, while data indicate that UPF1 helicase activity remodels NMD targets during the decay process (Franks et al. 2010), assays of steady-state UPF1 binding to cellular RNA cannot differentiate NMD targets from mRNAs that are not NMD targets (Zünd et al. 2013; Gregersen et al. 2014).
To address these issues, we demonstrate using Förster resonance energy transfer (FRET) experiments and also immunoprecipitations of UPF1 from cellular lysates that UPF1 uses ATP hydrolysis as a means to dissociate from cellular RNAs. UPF1 variants (C126S or R843C) lacking RNA-binding ability are hypophosphorylated. However, a UPF1 variant (G495R/G497E) that lacks ATPase and helicase activities but retains the ability to bind RNA accumulates on RNA and becomes hyperphosphorylated, presumably as a consequence of its failure to dissociate. We conclude that ATP binding and hydrolysis generally modulate UPF1–RNA associations, which in turn regulate cellular UPF1 phosphorylation and NMD.
In addition to its quality control function, NMD also regulates many cellular processes, presumably because it controls the levels of many nonmutated cellular transcripts (Mendell et al. 2004; Wittmann et al. 2006; Viegas et al. 2007; Yepiskoposyan et al. 2011; Tani et al. 2012). Genetic depletion studies by design identify both direct and indirect NMD targets, complicating our understanding of how NMD controls these processes. Alternative large-scale efforts to map steady-state UPF1-binding sites to the transcriptome have failed to identify endogenous NMD substrates. Here we map physiologic p-UPF1-binding sites on the cellular transcriptome using deep sequencing (RNA immunoprecipitation [RIP] plus sequencing [RIP-seq]) and show that, unlike UPF1, p-UPF1 binding is a reliable marker of NMD targets. We demonstrate that p-UPF1 binding is accompanied by SMG5 and SMG7 binding, which stabilize p-UPF1 on the 3′ UTRs of NMD targets. While PP2A phosphatase indeed reduces the degree of UPF1 phosphorylation, we unexpectedly found that PP2A is not enriched on NMD targets and does not detectably influence the efficiency of NMD. While other interpretations are possible, it may be that p-UPF1 is dephosphorylated after mRNA decay is initiated. Taken together, our data indicate that p-UPF1 provides the first discriminating mark of NMD targets whose generation is tightly controlled by an ordered series of events that constitute the NMD pathway.
Results
p-UPF1, unlike UPF1, marks cellular NMD targets
Steady-state—i.e., largely hypophosphorylated—UPF1 binding to cellular transcripts cannot be used as an identifier of NMD targets (Zünd et al. 2013; Gregersen et al. 2014; see below). Reasons for this are likely to be multifaceted. Steady-state UPF1 may manifest a sufficient degree of nonspecific binding to RNA to partially mask regulated UPF1 binding to NMD targets (Hurt et al. 2013; Zünd et al. 2013; Gregersen et al. 2014; see below). Additionally, the ability of ribosomes to remove UPF1 from mRNA 5′ UTRs and coding regions leaves 3′ UTRs as the primary sites of UPF1 binding (Hurt et al. 2013; Zünd et al. 2013; Gregersen et al. 2014); however, 3′ UTR length cannot be reliably used to identify NMD targets (Tani et al. 2012; Hurt et al. 2013) because other hallmarks, such as the presence of a 3′ UTR EJC (Hurt et al. 2013; Kurosaki and Maquat 2013), and/or variables, such as how a 3′ UTR folds (Eberle et al. 2008), must be considered.
Since degradation of NMD targets involves UPF1 phosphorylation (Kashima et al. 2006; Isken et al. 2008), we asked whether assays of p-UPF1 could identify cellular NMD targets by mapping transcriptome-wide p-UPF1-binding sites in vivo. We used an antigen-purified anti-p-UPF1(S1116) antibody to isolate transcripts associated with p-UPF1 from okadaic acid-treated human embryonic kidney 293T (HEK293T) cells. Seven lines of evidence justify using p-UPF1(S1116), which to our knowledge is the only available antibody capable of efficiently immunoprecipitating p-UPF1 (see the Supplemental Material for details), as a highly specific assay for p-UPF1 function during NMD (Supplemental Fig. S1A–G). Under the conditions used, okadaic acid generated an approximately fivefold accumulation of cellular p-UPF1 (Supplemental Fig. S1F).
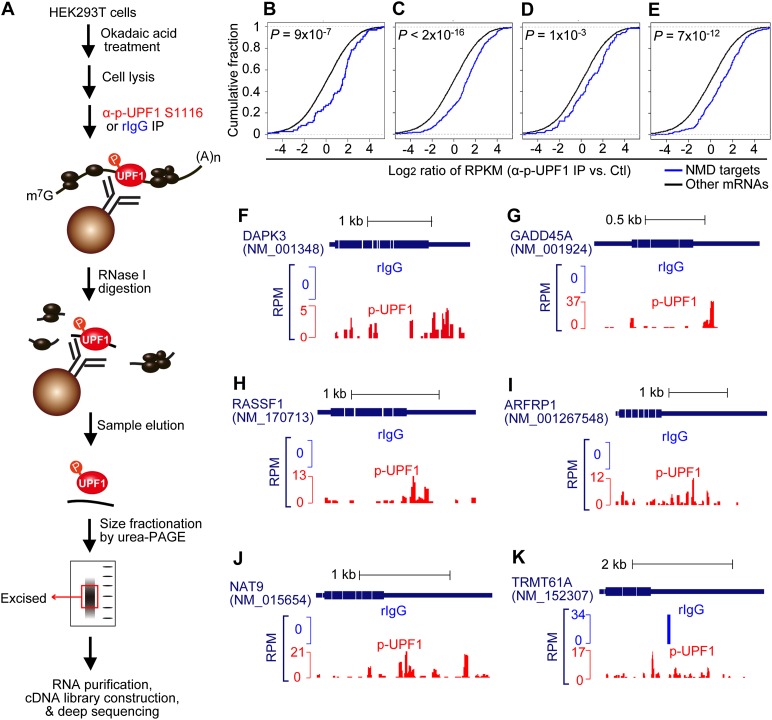
We immunoprecipitated cell lysates using anti-p-UPF1(S1116) or, as a control, rabbit IgG (rIgG) (Fig. 1A) and subsequently generated RNA fragments that were primarily <100 nt using limited RNase I digestion. As a second control (Ctl), cell lysates were generated without immunoprecipitation or RNase I treatment. Immunoprecipitated complexes were eluted from antibody-bound beads using denaturing buffer. Eluted RNA fragments were subjected to denaturing polyacrylamide gel electrophoresis, and those of ∼25–40 nt were purified for cDNA library construction (Fig. 1).
Figure 1.
p-UPF1 provides a useful marker of cellular NMD targets. (A) Scheme used to define HEK293T cell transcriptome-wide p-UPF1-binding sites using RIP-seq (details in the Materials and Methods). Three types of cDNA libraries were made; namely, Ctl, rIgG immunoprecipitation, and rabbit anti-phosphorylated human UPF1(S1116) immunoprecipitation (α-p-UPF1 S1116) (B–E) Cumulative distribution functions (CDFs) of log2 (ratio) of RNA fragment abundance (α-p-UPF1 vs. Ctl). RNA fragment abundance was calculated using RPKM based on exonic regions. Genes detected in both α-p-UPF1 and Ctl samples with ≥20 total reads were used for plotting. P-values were calculated from two biological replicates and were based on the Kolmogorov-Smirnov (K-S) test. (B–E) Target sets are, from left to right, bona fide NMD targets listed in Supplemental Table S1 or putative NMD targets defined by Tani et al. (2012) (group B), Mendell et al. (2004), or Yepiskoposyan et al. (2011). “Other mRNAs” are RefSeq transcripts whose expression was detected in Ctl samples but are not bona fide or putative NMD targets. (F–K) Examples of p-UPF1 association with individual bona fide NMD targets. Amounts of sequencing reads are indicated by numbers of reads per million mapped reads (RPM). As a control, rIgG samples (blue) provide a background with which reads in p-UPF1 samples (red) should be compared.
We obtained 1.6 million to 9.7 million single-end reads from each of the six cDNA libraries; i.e., biological replicates of three libraries. Of these, ∼20%–25% of the reads in Ctl and p-UPF1 samples could be mapped to a unique genomic sequence, in contrast to only ∼2% of the reads in rIgG samples, indicating low nonspecific binding of RNAs to rIgG. We plotted the ratio of RNA fragment abundance (reads per kilobase per million mapped reads [RPKM]) in p-UPF1 samples to those in Ctl samples using what we call bona fide NMD targets. These bona fide NMD targets were defined based on a minimum of two criteria: up-regulation upon UPF1 down-regulation plus a longer half-life upon UPF1 down-regulation (Supplemental Table S1). Results demonstrated that these NMD targets had significantly more p-UPF1 binding compared with other transcripts (P = 9 × 10−7, Kolmogorov–Smirnov test) (Fig. 1B). In contrast, the level of steady-state UPF1 binding to the same bona fide NMD targets using data provided in Gregersen et al. (2014) or Zünd et al. (2013) failed to show significantly more UPF1 binding compared with other transcripts (Supplemental Fig. S1H,I). An enrichment of p-UPF1 binding similar to what we observed for these bona fide NMD targets was obtained using putative NMD targets defined by Tani et al. (2012) (group B), which were defined based on an elongated half-life upon UPF1 down-regulation (Fig. 1C); Mendell et al. (2004) or Yepiskoposyan et al. (2011), which were defined based on an increase in abundance upon UPF1 down-regulation (Fig. 1D,E); or Yepiskoposyan et al. (2011), which were defined based an increase in abundance upon SMG6 or SMG7 down-regulation (Supplemental Fig. S1J,K). However, for unclear reasons, putative NMD targets defined by Viegas et al. (2007) or Wittmann et al. (2006) based on, respectively, UPF1 or UPF2 down-regulation were not enriched in p-UPF1 binding (Supplemental Fig. S1L,M).
Following our global analysis of p-UPF1 binding to the HEK293T cell transcriptome using RIP-seq, we examined the distribution of p-UPF1 on selected bona fide NMD targets. After subtracting background p-UPF1 binding, data revealed that p-UPF1 is enriched on those NMD target 3′ UTRs examined (Fig. 1F–K; data not shown for other NMD targets). This pattern of p-UPF1 binding requires mRNA translation, without which only insignificant levels of p-UPF1 binding are observed (see below).
Cell cycle-regulated histone mRNAs and Staufen-mediated mRNA decay (SMD) targets might also be expected to be enriched in the p-UPF1 RIP-seq (Maquat and Schoenberg and Maquat 2012). While histone cluster 1 H1e (HIST1H1E) and the JUN proto-oncogene SMD target are enriched, HIST1H1C, HIST2H2BF, and HIST1H1D mRNAs as well as SMD targets ADP ribosylation factor 1 and ring finger protein168 mRNAs are only slightly enriched, and other cell cycle-regulated histone mRNAs and SMD targets are not enriched. Possibly, detectable enrichment of histone mRNAs requires cell synchronization and accumulation at the end of S phase, when histone mRNAs are degraded, and detectable enrichment of SMD targets requires a cell type that supports SMD more efficiently than do HEK293T cells (Ricci et al. 2014; our unpublished data).
NMD targets are enriched in p-UPF1, SMG5, and SMG7 but not UPF2, UPF3X, SMG1, SMG6, eRF1, or eRF3
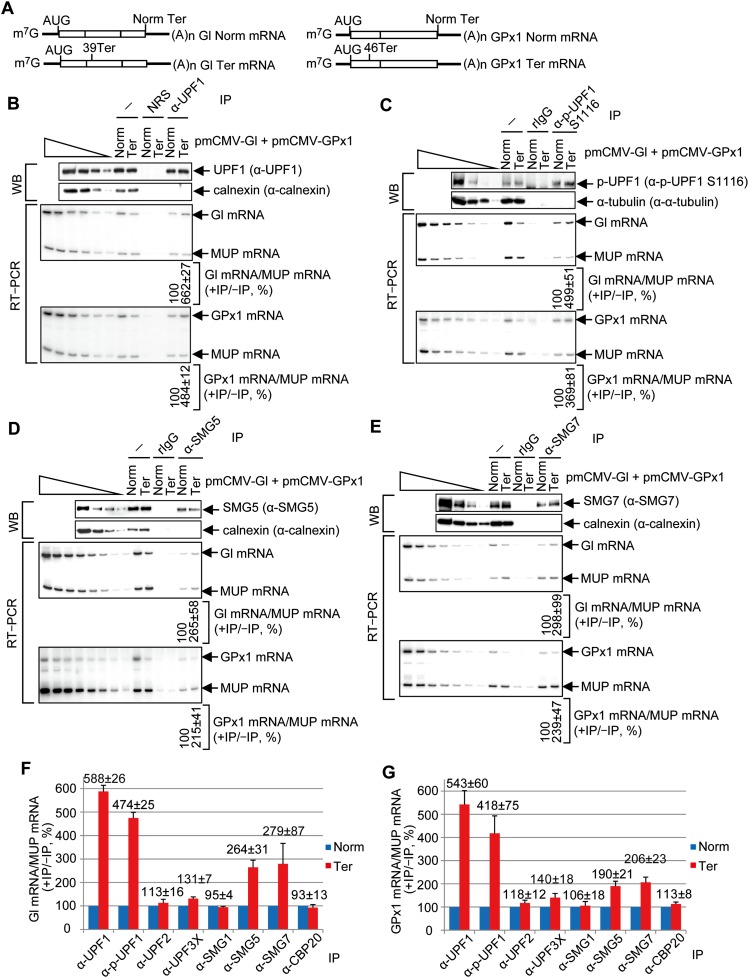
Our finding that p-UPF1 is enriched on NMD targets led us to ask whether other mRNP proteins are likewise enriched. Before assaying other proteins, we initially tested the prediction, deriving from our RIP-seq data (Fig. 1), that p-UPF1 would be enriched on the 3′ UTRs of β-globin (Gl) Ter (i.e., PTC-containing) mRNA and glutathione peroxidase 1 (GPx1) Ter mRNA compared with their corresponding PTC-free counterparts. We prepared lysates of HEK293T cells transiently expressing either Gl Norm mRNA + GPx1 Ter mRNA or Gl Ter mRNA + GPx1 Norm mRNA (Fig. 2A), each together with mRNA that encodes the major urinary protein (MUP) and controls for variations in cell transfection efficiencies and RNA recovery (Kurosaki and Maquat 2013). A fraction of each cell lysate was immunoprecipitated using anti-UPF1, which reacts with all of cellular UPF1 regardless of its phosphorylation status (Supplemental Fig. S1F,G), or anti-p-UPF1(S1116).
Figure 2.
p-UPF1, SMG5, and SMG7 preferentially bind to PTC-containing mRNAs compared with their PTC-free counterparts. (A) Diagrams of spliced Gl and GPx1 PTC-free (Norm) and Gl and GPx1 PTC-containing (39 Ter and 46 Ter, respectively) mRNAs. Boxes represent coding regions, vertical lines within boxes show spliced junctions, and horizontal lines denote UTRs. (B) HEK293T cells (8 × 107 per 150-mm dish) were transiently transfected with 2 μg of phCMV-MUP and either 4 μg of pmCMV-Gl Norm + 4 μg of pmCMV-GPx1 Ter or 4 μg of pmCMV-Gl Ter + 4 μg of pmCMV-GPx1 Norm. Immunoprecipitations of lysates were performed using anti-UPF1 (α-UPF1). (Top) Western blotting (WB) before (−) or after immunoprecipitation (IP) using anti-UPF1 or, as a control for nonspecific immunoprecipitation, normal rabbit serum (NRS), where lanes below the wedge analyze serial threefold dilutions of lysate. (Bottom) RT–PCR, where the level of Gl mRNA or GPx1 mRNA before and after immunoprecipitation was normalized to the level of MUP mRNA, the normalized level after immunoprecipitation was calculated as a ratio of the normalized level before immunoprecipitation, and the ratio for Gl Norm mRNA or GPx1 Norm mRNA is defined as 100%. Lanes below the wedge analyze serial twofold dilutions of lysate RNA. (C) As in B, only immunoprecipitation was performed using anti-p-UPF1(S1116) or, as a control, rIgG. (D) As in B, only immunoprecipitation was performed using anti-SMG5. (E) As in B, except immunoprecipitation was performed using anti-SMG7. (F) RT-qPCR of Gl mRNA from samples analyzed in B–E and Supplemental Figure S2. (G) As in F for GPx1 mRNA. All quantitations derive from three to four independently performed experiments and represent the mean plus standard deviations.
Western blotting revealed that for each sample, anti-UPF1 immunoprecipitated comparable amounts of UPF1 (Fig. 2B), and anti-p-UPF1(S1116) immunoprecipitated comparable amounts of p-UPF1(S1116) (Fig. 2C). RT–PCR demonstrated that the levels of Gl and GPx1 Ter mRNAs were, respectively, ∼40% and ∼30% of the corresponding PTC-free mRNAs prior to immunoprecipitation (Fig. 2B,C). As shown previously (Hwang et al. 2010; Kurosaki and Maquat 2013), Gl and GPx1 Ter mRNAs coimmunoprecipitated with UPF1 approximately fivefold to sevenfold more efficiently than did their PTC-free counterparts (Fig. 2B,C; for RT-qPCR data, see Fig. 2F,G). An approximately fivefold to sixfold increase in UPF1 binding to PTC-containing compared with PTC-free mRNA was also found after anti-HA immunoprecipitation of lysates of cells coexpressing HA-tagged MS2 coat protein and either Gl Norm mRNA or Gl Ter mRNA, each harboring six copies of the MS2 coat protein-binding site in its 3′ UTR (Supplemental Fig. S2A,B). Note that here we are comparing UPF1 binding to a PTC-containing transcript with UPF1 binding to its PTC-free counterpart, and the first transcript differs from the latter by having a longer 3′ UTR and a 3′ UTR EJC that cumulatively enhance UPF1 binding (Kurosaki and Maquat 2013). This comparison is distinct from the comparison of cellular mRNAs that are and are not NMD targets—a comparison that failed to show total cell UPF1 enrichment on NMD targets (Zünd et al. 2013; Gregersen et al. 2014) partly because the NMD-inducing features of the bulk of cellular NMD targets are often diverse and undefined.
Anti-p-UPF1(S1116) immunoprecipitations demonstrated that Gl and GPx1 Ter mRNAs coimmunoprecipitated approximately fourfold to fivefold more efficiently with p-UPF1 than did their PTC-free counterparts. Based on the finding that Gl and GPx1 Ter mRNAs coimmunoprecipitated with SMG5 and SMG7 approximately twofold to threefold more efficiently than did their PTC-free counterparts (Fig. 2D-G), at least some of this p-UPF1 appears to be bound by SMG5 and SMG7, each of which preferentially associates with cellular p-UPF1 (Ohnishi et al. 2003; Okada-Katsuhata et al. 2012; Jonas et al. 2013; Loh et al. 2013). We conclude that, under the conditions analyzed, steady-state UPF1 and p-UPF1 preferentially bind PTC-containing NMD targets relative to their PTC-free counterparts to the same order of magnitude.
Like SMG5 and SMG7, UPF2, UPF3X, SMG1, eRF1, and eRF3 form complexes with UPF1 at various stages of the NMD pathway (Serin et al. 2001; Kadlec et al. 2004, 2006; Clerici et al. 2009; Kashima et al. 2006). However, unlike SMG5 and SMG7, each protein was not enriched on PTC-containing relative to PTC-free mRNAs (the slight 1.3–1.4-fold increase observed with UPF3X was not pursued) (Fig. 2F,G; Supplemental Fig. S2C-H). In control experiments, CBP20 was also not enriched on PTC-containing mRNAs (Fig. 2F,G; Supplemental Fig. S2F), as expected. As additional controls, p-UPF1, SMG5, and SMG7, unlike UPF2, UPF3X, SMG1, eRF1, PP2Ac, or CBP20, were enriched on endogenous mRNA for growth arrest and DNA damage-inducible 45A (GADD45A), which is an NMD target (Mendell et al. 2004; Wittmann et al. 2006; Viegas et al. 2007; Tani et al. 2012), compared with endogenous β-actin mRNA, which is not an NMD target (Supplemental Fig. S2I). Under the conditions used, neither PTC-containing nor PTC-free mRNAs detectably coimmunoprecipitated with SMG6 (see the Discussion; data not shown), which also forms a complex with UPF1 (Chiu et al. 2003; Eberle et al. 2009; Franks et al. 2010).
SMG1 is not required for preferential UPF1 binding to PTC-containing vs. PTC-free mRNAs
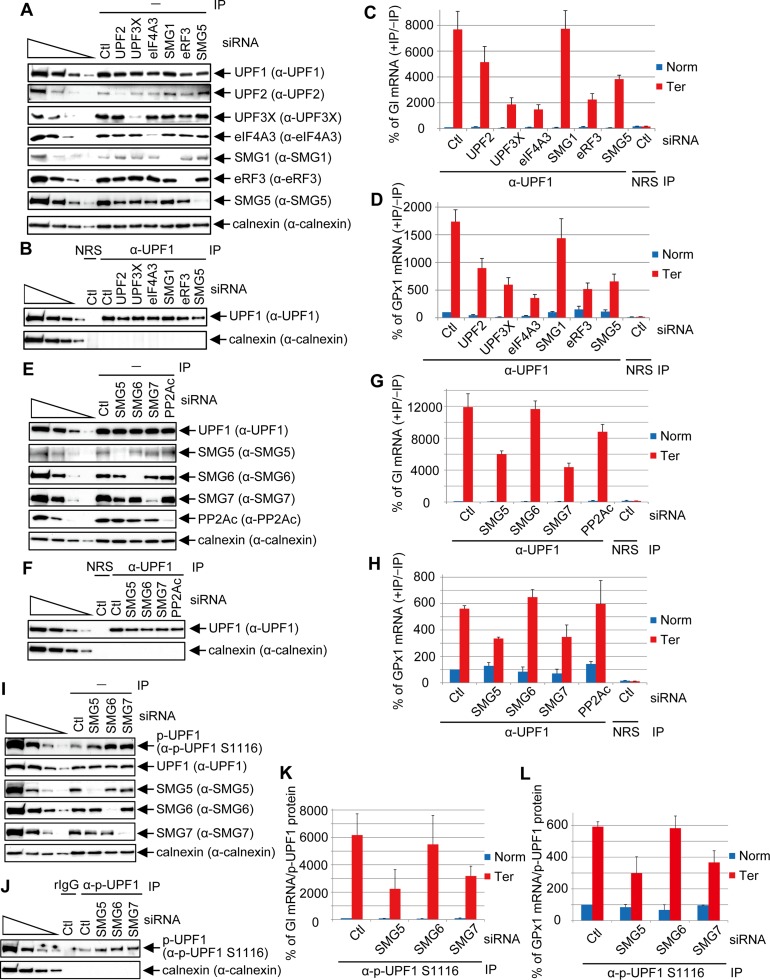
Since anti-UPF1 measures both hypophosphorylated UPF1 and p-UPF1, our finding that UPF1 binding and p-UPF1 binding are enhanced to the same extent on PTC-containing mRNAs does not tell us whether enhanced UPF1 binding occurs before or after SMG1-mediated UPF1 phosphorylation. Furthermore, the degrees to which other mRNP constituents influence UPF1 or p-UPF1 binding have never been characterized. We therefore down-regulated to <15% of their normal levels individual mRNA-associated proteins that are known to form a complex with UPF1 either before or after UPF1 phosphorylation (Fig. 3A,B). We then used RT-qPCR to quantitate the levels of Gl and GPx1 Norm or Ter mRNAs before and after immunoprecipitation using anti-UPF1 or, as a control for nonspecific immunoprecipitation, normal rabbit serum (NRS) (Fig. 3C,D, where NMD efficiencies were much higher than in Fig. 2, making the level of UPF1-binding to PTC-containing vs. PTC-free mRNA higher than in Fig. 2).
Figure 3.
SMG5 and SMG7 augment p-UPF1 binding to NMD targets. (A) Western blotting prior to (−) immunoprecipitation (IP) of lysates of HEK293T cells (5 × 106 per well of six-well plate) transiently transfected with 60 pmol of the specified or Ctl siRNA per well and, 1 d later, 0.4 μg of pmCMV Gl Norm or Ter, 0.4 μg of pmCMV-GPx1 Norm or Ter, and 0.2 μg of phCMV-MUP. (B) As in A only after immunoprecipitation using anti-UPF1 (α-UPF1) or, as an immunoprecipitation control, NRS. (C) RT-qPCR of Gl mRNA as in Figure 2F. The ratio for Gl Norm mRNA in Ctl siRNA down-regulation is defined as 100%. (D) RT-qPCR of GPx1 mRNA as in C. (E) As in A using the specified siRNAs. (F) As in B using the specified siRNAs. (G) As in C. (H) As in D. (I) As in A using the specified siRNAs. (J) As in B using the specified immunoprecipitations and siRNAs. (K) Gl mRNA was quantitated as in C but normalized to the level of immunoprecipitated p-UPF1 as exemplified in J. (L) As in K but for GPx1 mRNA. Quantitations derive from three independently performed experiments and represent the mean plus standard deviations.
Analyses of samples after immunoprecipitation revealed that the coimmunoprecipitation (co-IP) of Gl and GPx1 Ter mRNAs with UPF1 was reduced approximately twofold by UPF2 siRNA, approximately threefold to fourfold by UPF3X siRNA, approximately fivefold by eIF4A3 siRNA, or approximately threefold by eRF3 siRNA when compared with their co-IP in the presence of Ctl siRNA (Fig. 3C,D). These findings are consistent with our previous results demonstrating that UPF1 binding to an NMD target 3′ UTR is augmented by the interaction between UPF1 and eRF3 at a PTC and between UPF1 and a 3′ UTR EJC (Kurosaki and Maquat 2013). Our finding that SMG1 siRNA failed to influence UPF1 binding to either NMD target (Fig. 3C,D) may indicate that cellular UPF1 phosphorylation is not required for enhanced UPF1 binding to PTC-containing versus PTC-free mRNAs (see below) and suggests that hypophosphorylated UPF1 initially binds to NMD target 3′ UTRs. In contrast, the co-IP of Gl and GPx1 Ter mRNAs with UPF1 was reduced approximately twofold to threefold by SMG5 siRNA. Considering that SMG5 is enriched on NMD targets (Fig. 2D,F,G), SMG5 may physically stabilize UPF1 binding to NMD targets—in particular the p-UPF1 fraction of cellular UPF1 (see below). In support of these data and the conclusions that we draw from them, similar but less dramatic results were obtained when the co-IP of GADD45A mRNA with UPF1 was quantitated (relative to co-IP of β-actin mRNA with UPF1) in the presence of siRNA to each of the six mRNA-associated proteins (Supplemental Fig. S3A).
SMG5 and SMG7 augment UPF1 and p-UPF1 binding to NMD targets
To further investigate the finding that SMG5 siRNA reduces UPF1 binding to Gl and GPx1 Ter mRNAs, we undertook similar assays of UPF1 binding in the presence of SMG6 siRNA, SMG7 siRNA, or PP2Ac siRNA, the latter of which down-regulates the catalytic subunit of PP2A (Fig. 3E,F). Results revealed that SMG7 but neither SMG6 nor PP2Ac significantly contributes to the preferential binding of UPF1 to PTC-containing mRNAs (Fig. 3G,H) or the GADD45A cellular NMD target (Supplemental Fig. S3B). Since SMG5 and SMG7 form a stable heterodimer that is critical for NMD (Ohnishi et al. 2003; Okada-Katsuhata et al. 2012; Jonas et al. 2013; Loh et al. 2013) and disrupting the SMG5–SMG7 heterodimer greatly reduces SMG5 binding to UPF1 (Loh et al. 2013), our finding that either SMG5 siRNA or SMG7 siRNA reduces UPF1 binding to an NMD target suggests that it is the heterodimer that associates with and enhances the binding of UPF1 to PTC-containing mRNAs.
Given that SMG5 and SMG7 interact with p-UPF1, we next asked whether SMG5 and/or SMG7 enhance(s) p-UPF1 binding to NMD targets. Using lysates of cells in which SMG5, SMG6, or SMG7 was down-regulated to <10% of normal (Fig. 3I), after anti-p-UPF1(S1116) immunoprecipitation (Fig. 3J), SMG5 and SMG7, unlike SMG6, were found to contribute to p-UPF1 binding to Gl and GPx1 Ter mRNAs (Fig. 3K,L). We propose from these results and similar results obtained in assays of cellular GADD45A compared with β-actin mRNA (Supplemental Fig. S3C) that the SMG5–SMG7 heterodimer stabilizes p-UPF1 binding to an NMD target.
DNA oligo-directed RNase H cleavage demonstrates that p-UPF1 binds NMD target 3′ UTRs
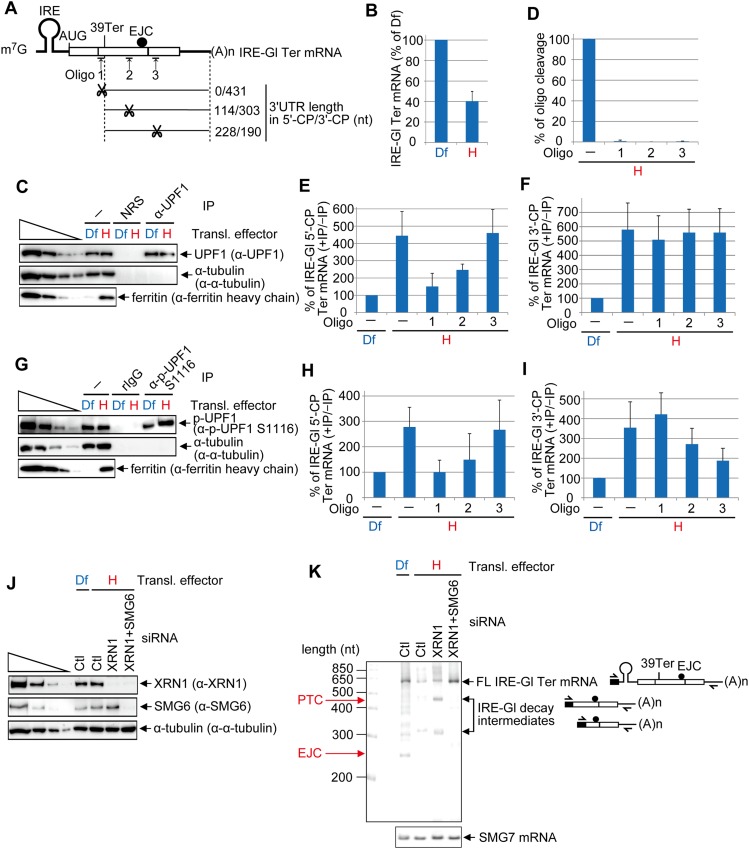
UPF1 binds along the length of the 3′ UTRs of translationally active NMD targets (Hurt et al. 2013; Kurosaki and Maquat 2013; Zünd et al. 2013; Gregersen et al. 2014). Thus, it seemed reasonable that the p-UPF1 that we observed bound to Gl and GPx1 Ter mRNAs (Fig. 2C,F,G) would likewise reflect a translation-dependent 3′ UTR association. To test this hypothesis and examine p-UPF1 distribution along the 3′ UTR, we used HeLa cells that stably express IRE-Gl Ter mRNA (Fig. 4A), which harbors in its 5′ UTR the iron-responsive element (IRE) from ferritin heavy chain mRNA (Thermann et al. 1998; Sato and Maquat 2009; Kurosaki and Maquat 2013). Cells were cultured for 1 d in medium containing the iron chelator deferoxamine mesylate (Df), which represses IRE-Gl mRNA translation, and were subsequently washed and cultured for 8 h in Df or hemin, the latter of which promotes IRE-Gl mRNA translation (Kurosaki and Maquat 2013). Lysates of cells cultured in the presence of hemin were subsequently immunoprecipitated using anti-UPF1 (or, as a control, NRS) or anti-p-UPF1(S1116) (or, as a control, rIgG). After extensive washing, UPF1-bound RNAs were incubated with RNase H in the absence or presence of one of three antisense DNA oligonucleotides (Fig. 4A) while associated with antibody–protein A-agarose beads. By so doing, cleavage products (CPs) that do not associate with UPF1 can be distinguished from those that do. However, this experimental approach does not quantify the amount of bound UPF1. IRE-Gl Ter mRNA 5′-CPs consist of 185, 320, or 434 nt and contain, respectively, 0, 114, or 228 nt of 3′ UTR (where 3′ UTR nucleotide 1 is defined as the nucleotide immediately downstream from Ter) (Fig. 4A). The corresponding 3′-CPs consist of 439, 303, or 190 nt, all of which derive from the 3′ UTR (except for the Oligo 1 3′-CP, which contains 5 nt of coding region plus Ter) (Fig. 4A).
Figure 4.
DNA oligo-directed RNase H cleavage maps p-UPF1 binding to NMD target 3′ UTRs. (A) Diagrams of spliced IRE-Gl PTC-containing (39 Ter) mRNA. Upward-facing arrows indicate a site of DNA oligo-directed RNase H cleavage, and the ratio to the right of scissors specifies the number of 3′ UTR nucleotides in the 5′-CP/number of 3′ UTR nucleotides in the 3′-CP. (B) RT-qPCR of RNA from HeLa cells stably expressing IRE-Gl Ter mRNA (1 × 107 per 150-mm dish) that were cultured in the presence of 100 μM Df or hemin (H) for 8 h. The level of IRE-Gl Ter mRNA was normalized to the level of cellular SMG7 mRNA, and the normalized level in the presence of Df is defined as 100%. (C) Western blotting of lysates of HeLa cells stably expressing IRE-Gl Ter mRNA that were cultured in the presence of Df or hemin for 8 h and subsequently analyzed before (−) or after immunoprecipitation (IP) using the specified antibody (α). (D) RT-qPCR of IRE-Gl Ter mRNA to quantitate the level of cleavage, where the level in the absence (−) of DNA oligo is defined as 100%. (E) RT-qPCR of the 5′-CP of IRE-Gl mRNA essentially as in B except that the RT primer and PCR primer pair were specific for the 5′-CP. (F) As in E, but the RT primer and PCR primer pair were specific for the 3′-CP. (G) As in C, but anti-p-UPF1(S1116) and, as a control, rIgG were used in the immunoprecipitations. (H) As in E. (I) As in F. Quantitations derive from four independently performed experiments and represent the mean plus standard deviations. (J) Western blotting of lysates of HeLa cells stably expressing IRE-Gl Ter mRNA (1 × 107 per 150-mm dish) that were transiently transfected with 450 pmol of Ctl siRNA or XRN1 siRNA or 225 pmol of Ctl siRNA + 225 pmol of XRN1 siRNA and, 2 d later, cultured in the presence of Df or hemin for 8 h prior to lysis. (K) 5′-RACE of full-length (FL) and decay intermediates of IRE-Gl mRNA and RT–PCR of SMG7 mRNA in lysates from J, including diagrams of where each 5′-RACE primer anneals. Diagrams are as in A, with the black box specifying oligo dC generated using terminal deoxnucleotidyl transferase.
In control experiments performed before immunoprecipitation, Df (but not hemin) increased the level of IRE-Gl Ter mRNA prior to RNase H-mediated cleavage (i.e., inhibited NMD) (Fig. 4B) and also blocked the production of ferritin (Fig. 4C,G). Cleavage was complete as indicated by the loss of full-length IRE-Gl Ter mRNA (Fig. 4D). Relative to Df, hemin enhanced by approximately fourfold to sixfold or approximately threefold to fourfold the amount of uncleaved IRE-Gl Ter mRNA that coimmunoprecipitated with UPF1 or p-UPF1(S1116), respectively (Fig. 4E,F,H,I), consistent with enhanced binding of p-UPF1 to an NMD target relative to its PTC-free counterpart requiring a translation termination event that triggers NMD. Since the same conclusion can be drawn for UPF1 (Kurosaki and Maquat 2013), there is likely to be a precursor–product relationship between NMD target-bound UPF1 and p-UPF1.
In the presence of hemin, 5′-CPs of IRE-Gl Ter mRNA containing 0, 114, or 228 nt of 3′ UTR coimmunoprecipitated with, respectively, ∼1.5-fold, ∼2.5-fold, or ∼4.5-fold more UPF1 than did uncleaved mRNA in the presence of Df (Fig. 4E), whereas each of the corresponding 3′-CPs containing 431, 303, or 190 nt of 3′ UTR coimmunoprecipitated with fivefold more UPF1 than did uncleaved mRNA in the presence of Df (Fig. 4F). As concluded previously (Kurosaki and Maquat 2013), these results are consistent with UPF1 removal from the 5′ UTR, coding region, and first ∼35 nt of the 3′ UTR by translating ribosomes (see also below). Furthermore, there is at least one UPF1 molecule bound to each 3′-CP.
p-UPF1 binding also increased with increased 5′-CP length to levels that were, respectively, approximately onefold, ∼1.5-fold, or approximately threefold the level of binding to uncleaved mRNA exposed to Df (Fig. 4H), while p-UPF1 binding decreased with decreasing 3′-CP length to levels that were, respectively, approximately fourfold, approximately threefold, or approximately twofold (Fig. 4I). Therefore, the probability of p-UPF1 binding along the Gl Ter mRNA 3′ UTR decreases as the distance from the termination codon increases. A diminishing distribution of p-UPF1 relative to distance downstream from the termination codon appears to typify the 3′ UTRs of some (Fig. 1F,H,K) but not all (Fig. 1G,I,J) cellular NMD targets.
It has been previously shown that SMG6 cleaves NMD targets in the vicinity of their PTCs (Gatfield and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009). Given its preferential binding to p-UPF1, SMG6 endonuclease presumably cleaves NMD targets after it is recruited by p-UPF1 (Chiu et al. 2003; Okada-Katsuhata et al. 2012; see below). Thus, we next asked whether p-UPF1-binding sites coincide with sites of endonucleolytic cleavage. To define these sites, we used 5′-RACE to map the 5′ ends of IRE-Gl Ter mRNA 3′-CPs in lysates of cells exposed to one or more siRNAs and, subsequently, Df or hemin. The 5′-to-3′ exonuclease XRN1 was down-regulated (Fig. 4J) so as to allow SMG6-generated 3′-CPs to accumulate (Eberle et al. 2009; Franks et al. 2010). As controls, (1) both XRN1 and SMG6 were down-regulated (Fig. 4J) to inhibit the formation of SMG6-generated 3′-CPs, and (2) control siRNA (Fig. 4J) was used to allow SMG6-generated 3′-CPs to be degraded by XRN1 and thus fail to accumulate (Eberle et al. 2009; Franks et al. 2010). As expected, full-length IRE-Gl Ter mRNA was readily detected when cells were exposed to Df alone, which inhibits IRE-Gl Ter mRNA translation and thus NMD, or hemin after XRN1 + SMG6 siRNAs, which allows for IRE-Gl Ter mRNA translation but inhibits the decay step (Fig. 4K). In contrast, the level of IRE-Gl Ter mRNA was reduced when cells were exposed to hemin after either control siRNA or XRN1 siRNA (Fig. 4K). Consistent with previous data that mapped SMG6 cleavage sites, the majority of decay intermediates that were unique to cells treated with hemin after XRN1 siRNA exposure mapped to near the PTC and ∼35-nt upstream of the EJC (Fig. 4K); i.e., they were roughly coincident with where p-UPF1 binds (Fig. 4I). This suggests that p-UPF1-binding sites overlap with SMG6 cleavage sites.
UPF1 recognition of NMD targets requires its ATPase and helicase activities
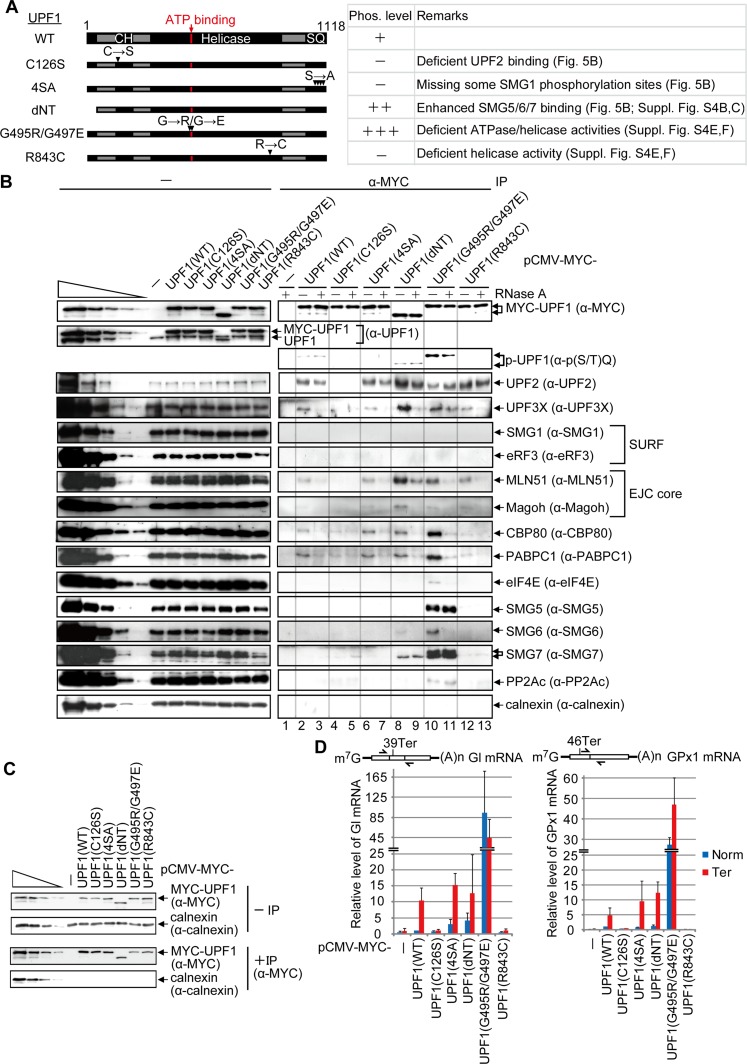
Neither SMG1 siRNA nor PP2A siRNA detectably affect the differential binding of UPF1 to NMD targets compared with their PTC-free counterparts (Fig. 3; Supplemental Fig. S3), consistent with UPF1 phosphorylation occurring after UPF1 binding to an NMD target. To further examine how and when UPF1 phosphorylation occurs during NMD, we constructed a series of UPF1 variants and analyzed their (1) phosphorylation status using anti-p(S/T)Q, (2) relative binding to PTC-containing versus PTC-free mRNAs, and (3) effects on mRNP composition. Lysates were generated from HEK293T cells transiently expressing MYC alone; MYC-UPF1(WT), which is primarily hypophosphorylated (Chiu et al. 2003; Ohnishi et al. 2003; Isken et al. 2008; Okada-Katsuhata et al. 2012); or one of five MYC-UPF1 variants (Fig. 5A) that have been shown to inhibit NMD (Sun et al. 1998; Kashima et al. 2006; Isken et al. 2008). Their relative phosphorylation status was found to be MYC-UPF1(G495R/G497E) >> MYC-UPF1(dNT) > MYC-UPF1(WT) > MYC-UPF1(C126S) ≈ MYC-UPF1(4SA) ≈ MYC-UPF1(R843C) (Fig. 5A,B, lanes 2–13; Supplemental Fig. S4A).
Figure 5.
UPF1 recognition of NMD targets requires ATPase and helicase activities. (A) Diagrams of UPF1 variants denoting the CH region and the region containing the four serine–glutamine (SQ) SMG1 phosphorylation sites. Horizontal arrows denote amino acid changes at positions specified by downward-facing arrowheads. (B) HEK293T cells (8 × 107 per 150-mm dish) were transiently transfected with 0.6 μg of pmCMV-MYC-UPF1(WT), 2 μg of MYC-UPF1(C126S), 1 μg of MYC-UPF1(4SA), 0.5 μg of MYC-UPF1(dNT), 4 μg of MYC-UPF1(G495R/G497E), 0.8 μg of MYC-UPF1(R843C), or, as a negative control, 0.5 μg of pmCMV-MYC. Cell lysates were treated with (+) or without (−) RNase A and subsequently immunoprecipitated using anti-MYC (α-MYC). Western blotting before (−) or after (α-MYC) immunoprecipitation. For samples after immunoprecipitation, fivefold less immunoprecipitate was loaded in the analysis of MYC-UPF1 compared with the analysis of other proteins. (C) HEK293T cells were transiently transfected as described in B except that transfections included 0.5 μg of phCMV-MUP and 1 μg each of pmCMV-Gl and pmCMV-GPx1, either Norm or Ter. Cell lysates were immunoprecipitated using anti-MYC and analyzed by Western blotting. (D) RT-qPCR of full-length Gl or GPx1 mRNA from samples in C. The level of Gl or GPx1 mRNA before immunoprecipitation was normalized to the level of MUP mRNA, and, given the comparable immunoprecipitation efficiencies, the normalized level of Gl Norm or GPx1 Norm mRNA after immunoprecipitation that coimmunoprecipitated with MYC-UPF1(WT) is defined as 1. Diagrams are of Gl mRNA or GPx1 mRNA as in Figure 2A, and the arrows specify PCR primer positions. Quantitations derive from two to three independently performed experiments and represent the mean plus standard deviations.
EJC-interacting UPF2 coimmunoprecipitated in a largely RNase A-insensitive fashion with MYC-UPF1(WT) and all variants except for MYC-UPF1(C126S) (Fig. 5B, lanes 2–13), which harbors a point mutation in the cysteine–histidine-rich (CH) domain that disrupts UPF2 binding (Kashima et al. 2006). Thus, the interaction of UPF2 with UPF1, which is known to be direct, is independent of UPF1 phosphorylation. Core EJC constituents metastatic lymph node 51 (MLN51) and human homolog of Drosophila melanogaster magonashi (Magoh) as well as the more peripherally associated UPF3X were not detectable in the immunoprecipitation of MYC-UPF1(C126S), indicating dependence on UPF2 for their co-IP with UPF1 (Fig. 5B, lanes 4,5). UPF2, UPF3X, MLN51, Magoh, CBP80, poly(A)-binding protein C1 (PABPC1), SMG6, and SMG7 coimmunoprecipitated more efficiently with MYC-UPF1(dNT) than with MYC-UPF1(WT) (Fig. 5B, lanes 2,3,8,9). Although these SMG6 data contradict a report that localized SMG6 binding to the N terminus of UPF1 (Okada-Katsuhata et al. 2012), we corroborated our results with the demonstration that MYC-UPF1(dNT) coimmunoprecipitates with both HA-SMG5 and HA-SMG6 (Supplemental Fig. S4B,C). That MYC-UPF1(dNT) is phosphorylated at a level that exceeds that of MYC-UPF1(WT) is consistent with its increased co-IP with SMG5, SMG6, and SMG7 (Fig. 5B, lanes 2,3,8,9; Supplemental Fig. S4B,C).
SMG5, SMG6, and SMG7 indeed interact most efficiently with p-UPF1 in a partially (SMG6) or entirely (SMG5 or SMG7) RNase A-insensitive manner, as indicated by assays of hyperphosphorylated MYC-UPF1(G495R/G497E) (Fig. 5B, lanes 10,11). The weak to nonexistent co-IP of eIF4E with MYC-UPF1(WT) and all variants but MYC-UPF1(G495R/G497E), which manifests a high degree of nonspecific binding to RNA (see below), agrees with the bulk of NMD targeting CBP80–CBP20-bound mRNAs (Maquat et al. 2010; Trcek et al. 2013 and references therein).
As expected, MYC-UPF1(WT) exhibited ∼10-fold and approximately fivefold more binding to, respectively, Gl and GPx1 Ter mRNAs compared with their PTC-free counterparts (Fig. 5C,D). The failure of MYC-UPF1(C126S) to detectably coimmunoprecipitate with Gl Ter or GPx1 Ter mRNA above the level of binding to, respectively, Gl Norm or GPx1 Norm mRNA (Fig. 5C,D) is consistent with its failure to coimmunoprecipitate with CBP80 or PABPC1 (Fig. 5B, lanes 4,5) and indicates that UPF2 binding to UPF1 is important for NMD target recognition (Fig. 3C,D). MYC-UPF1(4SA), which includes S1116A, resembled MYC-UPF1(WT) in its interactions with not only mRNP proteins (Fig. 5B, lanes 2,3,6,7) but also PTC-containing and PTC-free mRNAs (Fig. 5C,D), providing further evidence that UPF1 phosphorylation occurs on Ter mRNAs (Fig. 5C,D). MYC-UPF1(dNT) still distinguished PTC-containing and PTC-free mRNAs (Fig. 5C,D); however, its abnormal interactions with mRNP proteins, including NMD factors (Fig. 5B, lanes 8,9), suggests that the UPF1 N terminus contributes to proper mRNP configuration.
Consistent with the enhanced co-IP of CBP80 and PABPC1 with MYC-UPF1(G495R/G497E) (Fig. 5B, lanes 10,11), Gl and GPx1 Norm and Ter mRNAs coimmunoprecipitated more efficiently with MYC-UPF1(G495R/G497E) than with MYC-UPF1(WT) (Fig. 5C,D). Given that MYC-UPF1(G495R/G497E) fails to support NMD (Isken et al. 2008) and its binding to mRNA was not significantly enhanced by the presence of a PTC in either Gl or GPx1 mRNA (Fig. 5C,D), we suggest that binding is largely nonspecific. In support of this idea, the abnormally elevated amounts of SMG5, SMG6, and SMG7 that coimmunoprecipitated with MYC-UPF1(G495R/G497E) were incapable of degrading mRNA. Assays of Escherichia coli-produced human UPF1 (Supplemental Fig. S4D) revealed that the G495R/G497E mutations inhibit UPF1 ATPase and helicase activities (Supplemental Fig. S4E,F).
FRET analyses showed that UPF1 is efficiently released from RNA in the presence of ATP compared with ADP or the nonhydrolyzable ATP analog AMPPNP or ADP-Bex (Supplemental Fig. S4G–I). This was determined using human UPF1(295–914) that was site-specifically modified by Sortase labeling (Popp et al. 2007) so that its C terminus harbors a Cy5 dye (Supplemental Fig. S4G,H) and generates a FRET signal when the Cy5 dye is in close proximity to a donor Cy3-labeled 50-nt RNA (Supplemental Fig. S4I). Hence, ATP hydrolysis, rather than simply ATP binding, leads to dissociation of UPF1 from RNA, in keeping with the hypothesis that UPF1 is a helicase that translocates along RNA in a 5′-to-3′ direction (Bhattacharya et al. 2000; Chamieh et al. 2008; Chakrabarti et al. 2011; Shigeoka et al. 2012; Park et al. 2013). We suggest that the large decrease in FRET observed in the presence of ATP is due to movement of UPF1 away from the donor fluorophore attached to RNA and/or dissociation of UPF1 from the 3′ end of donor-labeled RNA (see Supplemental Fig. S4J for stop–flow kinetic FRET analyses).
As expected from the failure of mRNP constituents to detectably coimmunoprecipitate with hypophosphorylated MYC-UPF1(R843C) (Fig. 5B, lanes 12,13), Gl and GPx1 Norm and Ter mRNAs also failed to detectably coimmunoprecipitate (Fig. 5C,D). Thus, the observed interaction between MYC-UPF1(R843C) and EJC constituents likely occurs while not bound to RNA. The R843C mutation inhibits UPF1 helicase but not ATPase activity (Supplemental Fig. S4E,F). Just as MYC-UPF1(G495R/G497E) is hyperphosphorylated because it fails to dissociate from RNA, MYC-UPF1(R843C) is hypophosphorylated because it fails to detectably bind RNA. Thus, the R843C mutation apparently uncouples ATPase activity from RNA binding.
We conclude that, under the conditions used, UPF1 binds both PTC-containing and PTC-free mRNAs indiscriminately in the absence of ATP and requires the ability to cleave ATP to recognize an NMD target. The enhanced binding of UPF1 to PTC-containing relative to PTC-free mRNAs requires that UPF1 have an intact CH domain, which allows interaction with EJCs, and also possess helicase activity, which requires ATP hydrolysis (Weng et al. 1998; Bhattacharya et al. 2000; Cheng et al. 2007; Chamieh et al. 2008; Chakrabarti et al. 2011; Shigeoka et al. 2012; Park et al. 2013).
PP2Ac depletion up-regulates the level of p-UPF1 without inhibiting NMD
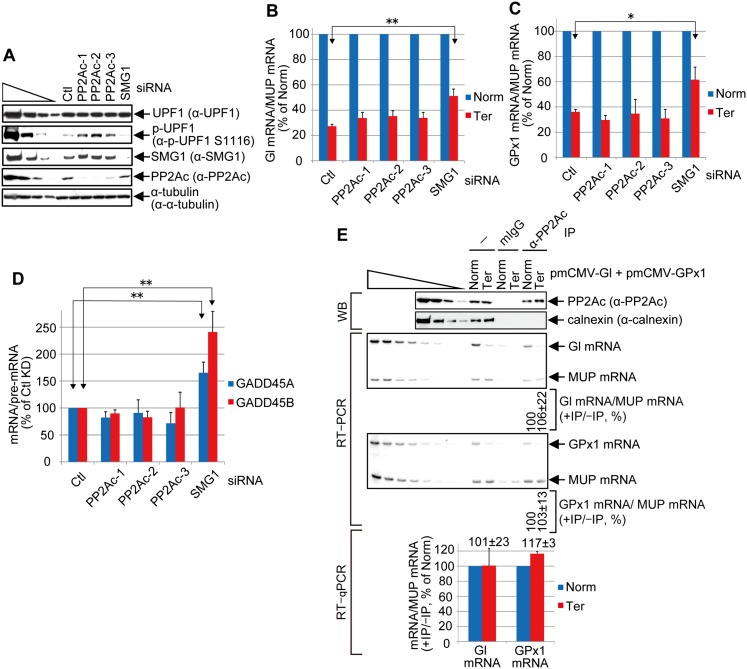
Current models indicate that PP2A is responsible for dephosphorylating p-UPF1 during NMD despite the absence of data showing that PP2A directly contributes to mRNA-degradative activities and/or restores UPF1 to its largely hypophosphorylated steady state (Anders et al. 2003; Chiu et al. 2003; Ohnishi et al. 2003; Kashima et al. 2006). To assay for PP2A function in NMD, three individual siRNAs targeting PP2Ac were separately used to down-regulate the level of PP2Ac to <10%–25% of normal without affecting the level of cellular UPF1 but increasing the level of cellular p-UPF1 approximately twofold to fourfold (Fig. 6A). Remarkably, no PP2Ac siRNA inhibited the NMD of Gl or GPx1 Ter mRNA (Fig. 6B,C) or endogenous NMD targets GADD45A or GADD45B mRNA (Fig. 6D). In striking contrast, down-regulating SMG1 to ∼10% of normal, which decreased the level of cellular p-UPF1 ∼2.5-fold, efficiently inhibited the NMD of the two reporter and the two endogenous NMD targets (Fig. 6B–D). Furthermore, immunoprecipitations using anti-PP2Ac revealed that PP2Ac is not enriched on Gl or GPx1 Ter mRNA (Fig. 6E) or GADD45A mRNA (Supplemental Fig. S2I). While it is possible that, despite siRNA-mediated down-regulation, residual levels of PP2Ac were sufficient to fully support NMD and/or that another phosphatase functionally replaced PP2Ac, PP2Ac may only indirectly function in NMD via its role in p-UPF1 recycling to a hypophosphosphorylated status. At least under the experimental conditions tested, the level of hypophosphorylated UPF1 did not limit UPF1 phosphorylation and thus NMD.
Figure 6.
Evidence that PP2Ac may dephosphorylate p-UPF1 after mRNA decay initiates. (A) Western blotting of lysates of HEK293T cells (5 × 106 per each six-well plate) transiently transfected with 60 pmol of the specified siRNA and, 1 d later, 0.4 μg of pmCMV-Gl Norm or Ter, 0.4 μg of pmCMV-GPx1 Norm or Ter, and 0.2 μg of phCMV-MUP. (B) Using samples from A, RT-qPCR of Gl Norm or Ter mRNA as described in Figure 2F for Gl mRNA before immunoprecipitation. (C) As in B but analyzed as in Figure 2G for GPx1 mRNA before immunoprecipitation. (D) Using samples from A, RT-qPCR of endogenous NMD targets GADD45A and GADD45B mRNAs. mRNA levels were normalized to the level of the corresponding pre-mRNA using PCR primers described in Supplemental Table S2. Quantitations in B–D derive from three independently performed experiments and represent the mean plus standard deviations as statistically analyzed using the two-tailed t-test. (*) P < 0.05; (**) P < 0.01. (E) As in Figure 2, B, F, and G, but using anti-PP2Ac or mouse IgG (mIgG) in the immunoprecipitations. Two to three independently performed experiments represent the mean plus standard deviations.
Discussion
Here, we establish using transcriptome-wide deep sequencing of cellular RNAs that p-UPF1 provides the first reliable marker of cellular NMD targets (Fig. 1; Supplemental Fig. S1). A complete inventory of the cellular mRNAs targeted by NMD has been lacking for a number of reasons: UPF1 binding does not provide a marker of cellular NMD targets (Zünd et al. 2013; Gregersen et al. 2014), direct NMD targets form a collection with myriad incompletely defined cis-acting NMD triggers (uORFs, 3′ UTR EJCs, 3′ UTR length, etc.), and experimentally down-regulating NMD factors by nature identifies both direct and indirect NMD targets. Thus, our finding that p-UPF1 serves as the discriminating mark of direct cellular NMD targets is essential toward elucidating how NMD regulates a multitude of cellular processes. Our results indicate that SMG5 and SMG7 binding should also identify direct NMD targets (Fig. 2; Supplemental Fig. S2I). Our finding a lack of UPF2, UPF3X, SMG1, eRF1, and eRF3 enrichment on NMD targets (Fig. 2; Supplemental Fig. S2) suggests, but certainly does not prove, that (1) SURF forms only transiently on NMD targets, (2) the peripheral EJC constituents UPF2 and UPF3X spend comparable time on NMD targets and their PTC-free counterparts, and (3) SMG1-mediated UPF1 phosphorylation occurs through transient interactions of SMG1 with NMD target-bound UPF1. Notably, either p-UPF1 or UPF1 can be used as an NMD target marker when analyses are confined to 3′ UTRs provided that (1) a PTC-containing mRNA is compared with its PTC-free counterpart, or (2) an mRNA that terminates translation normally and has a long 3′ UTR that triggers NMD is compared with the same mRNA with a short 3′ UTR that fails to trigger NMD (Figs. 2, 4, 5; Hogg and Goff 2010; Hurt et al. 2013; Kurosaki and Maquat 2013; Zünd et al. 2013; Gregersen et al. 2014).
We also examine the role of ATP hydrolysis on UPF1 binding to RNA. While it was known that UPF1 switches between ATP- and ADP-bound states, how the cycle of ATP hydrolysis is involved in NMD remained unresolved. Early biochemical data suggested that ATP binding significantly decreases the affinity of UPF1 to RNA (Weng et al. 1998; Bhattacharya et al. 2000; Cheng et al. 2007; Chamieh et al. 2008; Chakrabarti et al. 2011). Here, we demonstrate using ensemble FRET and immunoprecipitations of UPF1 variants from cellular lysates that UPF1 dissociation from RNA requires the binding and hydrolysis of ATP (Fig. 5; Supplemental Fig. S4G–J). When expressed in HEK293T cells, neither MYC-UPF1(R843C), which manifests ATPase but not helicase activity, nor MYC-UPF1(G495R–G497E), which manifests neither ATPase nor helicase activity, can discriminate between mRNAs that are and are not NMD targets (Fig. 5). Since MYC-UPF1(G495R/G497E) cannot dissociate from nonspecific interactions with RNA, it coimmunoprecipitates with three to four orders of magnitude more RNA than does MYC-UPF1(R843C), which fails to detectably bind RNA (Fig. 5). Furthermore, since MYC-UPF1(G495R/G497E) is hyperphosphorylated, whereas MYC-UPF1(R843C) is hypophosphorylated (Fig. 5), and since UPF1 phosphorylation occurs on RNA (Fig. 5), we conclude that prolonged UPF1 binding to RNA results in UPF1 hyperphosphorylation.
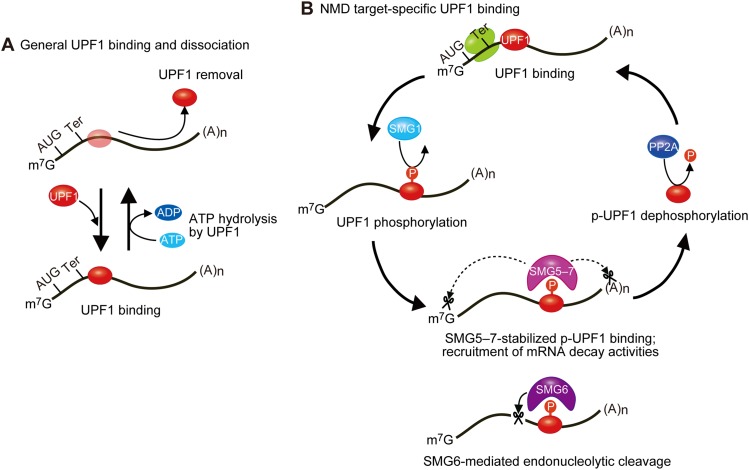
Taken together, our findings unveil the following model in which a number of important new links have been added (Fig. 7A). Steady-state hypophosphorylated UPF1 associates and dissociates from mRNAs that are and are not NMD targets nonspecifically in a mechanism that depends on ATP binding and hydrolysis (Fig. 5; Supplemental Fig. S4G–J). Translating ribosomes also regulate the distribution of UPF1 along mRNAs to remove nonspecifically bound UPF1 from the 5′ UTRs and coding regions and specifically recruit UPF1 to the 3′ UTRs of NMD targets (Figs. 1–4; Hurt et al. 2013; Kurosaki and Maquat 2013; Zünd et al. 2013; Gregersen et al. 2014). Data indicate that UPF1 moves along the mRNA 3′ UTR in a 5′-to-3′ direction (Bhattacharya et al. 2000; Chamieh et al. 2008; Chakrabarti et al. 2011; Shigeoka et al. 2012; Park et al. 2013), facilitating mRNA scanning (Shigeoka et al. 2012).
Figure 7.
Model for the dynamics of UPF1 binding to cellular mRNAs. (A) Steady-state UPF1 binds and hydrolyzes ATP as a means to dissociate from nonproductive mRNA binding; e.g., binding that does not lead to UPF1 phosphorylation and the subsequent p-UPF1-mediated recruitment of degradative activities and mRNA decay. UPF1 binding to translationally active mRNAs is largely restricted to 3′ UTRs at least in part because translating ribosomes remove UPF1 from 5′ UTRs and coding regions. (B) Regulated UPF1 phosphorylation by transiently associating SMG1 requires UPF1 recognition of a termination codon as one that triggers NMD; e.g., as one situated sufficiently upstream of a 3′ UTR EJC (not shown). p-UPF1 recruits SMG6 and/or SMG5−SMG7, which directly (solid arrow) or indirectly (dotted arrows) trigger(s) mRNA decay, respectively. While the binding of SMG6 is sufficiently transient to be undetectable, SMG5–SMG7 stabilize p-UPF1 binding to NMD target 3′ UTRs. It may be that PP2A returns p-UPF1 to a dephosphorylated state after mRNA decay is initiated.
In later steps (Fig. 7B), NMD targets are marked as such by regulated SMG1-mediated UPF1 phosphorylation (Fig. 6A; Supplemental Fig. S1G), which is enhanced by cap-bound CBP80–CBP20 and UPF factors bound to an EJC that is situated downstream from the PTC (Figs. 3, 5; Kashima et al. 2006; Hwang et al. 2010; Franks et al. 2010). Notably, UPF1 phosphorylation occurs while UPF1 is bound to the 3′ UTR via a transient association with SMG1 (Figs. 2, 5, 6; Supplemental Fig. S1G). How many UPF1 molecules load onto a 3′ UTR and which are converted to p-UPF1 molecules remain to be fully understood. p-UPF1 recruits the endonuclease SMG6 and also SMG5–SMG7, the latter of which stabilizes p-UPF1 binding to an NMD target 3′ UTR (Figs. 2, 3, 5) and recruits mRNA degradative activities (Lejeune et al. 2003; Ohnishi et al. 2003; Fukuhara et al. 2005; Eberle et al. 2009; Okada-Katsuhata et al. 2012; Jonas et al. 2013; Loh et al. 2013). Although NMD targets cannot be detected in immunoprecipitations of SMG6 (data not shown), they are cleaved by SMG6 (Fig. 4J,K). Thus, it appears that SMG6 spends relatively little time on an NMD target prior to its decay, although other explanations are possible. Provided that experimental results using PP2Ac siRNAs are not confounded by levels of residual PP2Ac that are sufficient for NMD or the activity of a phosphatase that functionally substitutes of PP2Ac, our data indicate that p-UPF1 dephosphorylation occurs after the initiation of mRNA decay (Fig. 6). The ratio of UPF1 to p-UPF1 that moves along the mRNA 3′ UTR to facilitate mRNP remodeling and decay (Franks et al. 2010) remains to be determined.
Taken together, our data elucidate how UPF1 ATPase and helicase activities contribute to the identification of NMD targets, which are marked by regulated UPF1 phosphorylation. Accordingly, UPF1 phosphorylation is tightly controlled by the remaining NMD factors.
Materials and methods
Cell transfections and lysis
HEK293T cells or HeLa cells stably expressing IRE-Gl Ter mRNA (Kurosaki and Maquat 2013) were propagated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum. DMEM for HeLa cell growth additionally contained 100 μM hemin (Sigma) or Df (Sigma) as described before (Kurosaki and Maquat 2013). When specified, cells were treated with 100 nM wortmannin (Caymen Chemicals) or 200 nM okadaic acid (Sigma) or transiently transfected with 20–30 nM control siRNA (Ambion) or an experimental siRNA (Dharmacon) (Supplemental Table S3) using Lipofectamine RNAiMAX (Life Technologies) and/or plasmid DNAs using Lipofectamine 2000 (Life Technologies). Cell lysates were prepared using hypotonic gentle lysis buffer (10 mM Tris at pH 7.4, 10 mM NaCl, 10 mM EDTA, 0.5% [w/w] Triton X-100) (Kurosaki and Maquat 2013) with protease inhibitor cocktail (Roche). Protein was analyzed after the addition of NaCl to 150 mM, and RNA was extracted and purified using TRIzol reagent (Life Technologies).
p-UPF1-bound RIP and fragmentation for cDNA library construction
HEK293T cells (24 × 107 in three 150-mm dishes) were incubated in 200 nM okadaic acid for 3 h. Cellular RNAs bound by p-UPF1 were immunoprecipitated using anti-p-UPF1(S1116) [Millipore, anti-phospho-Upf1(Ser1127)] and Dynabeads protein A magnetic beads (Life Technologies). The RNA in bead-bound RNA–p-UPF1(S1116) complexes was digested to primarily <100 nt by incubation for 30 min at 4°C with 1 U/μL RNase I (Life Technologies); for RNA size estimations after RNase I digestion, ∼200 ng of RNA was radiolabeled using γ32PATP (Perkin Elmer) and T4 polynucleotide kinase (New England Biolabs) and subsequently visualized using a Typhoon 9410 variable mode imager (GE Healthcare) after electrophoresis in 6M urea–15% polyacrylamide. To prepare RNA for cDNA library construction, after extensive washing, bound complexes were eluted using immunoprecipitation elution buffer (Ishigaki et al. 2001), and RNA fragments were separated in 6M urea–15% polyacrylamide in parallel with a DynaMarker prestain marker for small RNA (BioDynamics Laboratory). Small-range RNAs (∼25–40 nt) were excised from 6 M urea–15% polyacrylamide and agitated overnight at 25°C in RNA extraction buffer (20 mM Tris, 300 mM sodium acetate, 2 mM EDTA, 0.2% [v/v] SDS). RNAs were eluted from 6 M urea–15% polyacrylamide gel using a Coaster Spin-X column (Corning) and further purified using TRIzol reagent followed by ethanol precipitation. In parallel, control immunoprecipitations using rIgG were performed. Additional control samples were prepared without immunoprecipitation.
cDNA library construction for RIP-seq
Purified 25- to 40-nt RNA fragments were treated with recombinant shrimp alkaline phosphatase (New England Biolabs) to remove 3′ phosphates and were subsequently phosphorylated at 5′-hydroxyl groups using T4 polynucleotide kinase (New England Biolabs). Phosphorylated RNA fragments were purified with RNeasy mini columns (Qiagen). A 3′ adenylated adapter was ligated to the phosphorylated RNA fragments using truncated T4 RNA ligase (New England Biolabs). An RT primer was annealed to the adapted RNAs to prevent adapter self-ligation, followed by 5′ RNA adapter ligation using T4 RNA ligase (New England Biolabs). After RT of adapter-ligated RNAs, cDNAs were amplified using 15 PCR cycles. Amplified cDNAs were purified in 8% polyacrylamide, and the quality and quantity of cDNAs were assessed using an Agilent Bioanalyzer and qPCR. cDNAs were then sequenced using the Illumina HiSeq 2500 platform.
Computational analysis of RIP-seq data
The 3′ adapter sequence was first removed, and reads with a length of <15 nt were discarded. Reads were mapped to the human genome (hg19) using Bowtie2 (local mode). Reads with a mapping quality score (MAPQ) of ≥10 were selected for further analysis. mRNA abundance was measured using RPKM based on exonic regions of RefSeq sequences. The 3′ ends of genes in HEK293 cells were defined using the 3′READS method (Hoque et al. 2013).
For plasmid constructions, immunoprecipitations, Western blotting, RT–PCR, RT-qPCR, 5′-RACE, mass spectrometry, in vitro UPF1 helicase and ATPase activity assays, protein production, solid-phase peptide synthesis, and GGGC(Cy5) Sortase-compatible probe construction, Sortase labeling of hUPF1(295–914)-Srt, or FRET measurements and data analyses, see the Supplemental Material.
Accession number
The sequencing data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE60045.
Acknowledgments
We thank Galina Pavlencheva for isolating human UPF1(115–915) variants from E. coli; Mohammad Kayedkhordeh for preliminary kinetic data; Elie Farah for preparing fluorescently labeled RNAs; Akio Yamashita and Shigeo Ohno for pSR-HA-UPF1 4SA and anti-p-UPF1 S1078, S1096 (7H1); Jungwook Hwang for pCMV-MYC-UPF1(4SA); Jens Lykke-Andersen for pcDNA3-Gl Norm-MS2bs; Oliver Mühlemann for pcDNA3-HA-SMG6; Elisa Izaurralde for pEYFP-C1-SMG5 and pEYFP-N1-SMG7; David Liu and Brent Dorr for the SrtA 5° construct; Manny Ares for the T7 polymerase expression vector; and Eric Spooner and the Whitehead Institute Mass Spectrometry Facility. This work was supported by National Institutes of Health R01 GM59614 to L.E.M., GM099719 to D.N.E., and GM084089 to B.T. M.W.-L.P. is a Howard Hughes Medical Institute Post-doctoral Fellow of the Damon Runyon Cancer Research Foundation, DRG-2119-12.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.245506.114.
References
- Alexandrov A, Colognori D, Shu M-D, Steitz JA. 2012. Human spliceosomal protein CWC22 plays a role in coupling splicing to exon junction complex deposition and nonsense-mediated decay. Proc Natl Acad Sci 109: 21313–21318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders KR, Grimson A, Anderson P. 2003. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J 22: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa I, Haque N, Fiorini F, Barrandon C, Tomasetto C, Blanchette M, Le Hir H. 2012. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat Struct Mol Biol 19: 983–990 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. 2011. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 41: 693–703 [DOI] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H. 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15: 85–93 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. 2007. Structural and functional insights into the human Upf1 helicase core. EMBO J 26: 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S, Serin G, Ohara O, Maquat LE. 2003. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mourão A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S. 2009. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J 28: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak D, Ji S-J, Porse BT, Jaffrey SR. 2013. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell 153: 1252–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Lykke-Andersen J. 2013. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat Struct Mol Biol 20: 702–709 [DOI] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O. 2008. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 6: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J. 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143: 938–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E. 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578 [DOI] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. 2007. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell 130: 179–191 [DOI] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. 2014. MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell 54: 573–585 [DOI] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. 2013. Analysis of alternative cleavage and polyadenylation by 3 ′ region extraction and deep sequencing. Nat Methods 10: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda N, Kim YK, Lejeune F, Maquat LE. 2005. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol 12: 893–901 [DOI] [PubMed] [Google Scholar]
- Huang L, Wilkinson MF. 2012. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA 3: 807–828 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E. 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB. 2013. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 23: 1636–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE. 2010. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell 39: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617 [DOI] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JWB, Maquat LE. 2008. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Weichenrieder O, Izaurralde E. 2013. An unusual arrangement of two 14-3-3-like domains in the SMG5–SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes Dev 27: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J, Izaurralde E, Cusack S. 2004. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 11: 330–337 [DOI] [PubMed] [Google Scholar]
- Kadlec J, Guilligay D, Ravelli RB, Cusack S. 2006. Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA 12: 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam R, Wengrod J, Gardner LB, Wilkinson MF. 2013. Regulation of nonsense-mediated mRNA decay: implications for physiology and disease. Biochim Biophys Acta 1829: 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. 2006. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. 2013. Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc Natl Acad Sci 110: 3357–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12: 675–687 [DOI] [PubMed] [Google Scholar]
- Loh B, Jonas S, Izaurralde E. 2013. The SMG5–SMG7 heterodimer directly recruits the CCR4–NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev 27: 2125–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Tarn W-Y, Isken O. 2010. The pioneer round of translation: features and functions. Cell 142: 368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Hosoda N, Kim YK, Maquat LE. 2007. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat Struct Mol Biol 14: 974–979 [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. 1998. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 23: 198–199 [DOI] [PubMed] [Google Scholar]
- Nguyen LS, Jolly L, Shoubridge C, Chan WK, Huang L, Laumonnier F, Raynaud M, Hackett A, Field M, Rodriguez J, et al. . 2012. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol Psychiatry 17: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LS, Kim H-G, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y, Layman LC, Shaffer LG, Gécz J. 2013. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet 22: 1816–1825 [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12: 1187–1200 [DOI] [PubMed] [Google Scholar]
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. 2012. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 40: 1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Gleghorn ML, Maquat LE. 2013. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci 110: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW-L, Maquat LE. 2013. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet 47: 139–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Maquat LE. 2014. The dharma of nonsense-mediated mRNA decay in Mammalian cells. Mol Cells 37: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. 2007. Sortagging: a versatile method for protein labeling. Nat Chem Biol 3: 707–708 [DOI] [PubMed] [Google Scholar]
- Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE, Ashar-Patel A, Peng L, Moore MJ. 2014. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol 21: 26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufener SC, Mühlemann O. 2013. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol 20: 710–717 [DOI] [PubMed] [Google Scholar]
- Sato H, Maquat LE. 2009. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin β. Genes Dev 23: 2537–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulière J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonquèze O, Audic Y, Paillard L, Roest Crollius H, Le Hir H. 2012. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol 19: 1124–1131 [DOI] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE. 2012. Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13: 246–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. 2013. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta 1829: 612–623 [DOI] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. 2001. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol 21: 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Kato S, Kawaichi M, Ishida Y. 2012. Evidence that the Upf1-related molecular motor scans the 3′-UTR to ensure mRNA integrity. Nucleic Acids Res 40: 6887–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. 2012. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151: 750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckelberg A-L, Boehm V, Gromadzka AM, Gehring NH. 2012. CWC22 connects pre-mRNA splicing and exon junction complex assembly. Cell Reports 2: 454–461 [DOI] [PubMed] [Google Scholar]
- Sun X, Perlick HA, Dietz HC, Maquat LE. 1998. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci 95: 10009–10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, Akimitsu N. 2012. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 9: 1370–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. 1998. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17: 3484–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Sato H, Singer RH, Maquat LE. 2013. Temporal and spatial characterization of nonsense-mediated mRNA decay. Genes Dev 27: 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. 2007. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the nonsense mediated decay pathway. Nucleic Acids Res 35: 4542–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. 1998. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA 4: 205–214 [PMC free article] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jäck H. 2006. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol 26: 1272–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeller CF, Gaspari M, Isken O, Maquat LE. 2008. NMD resulting from encephalomyocarditis virus IRES-directed translation initiation seems to be restricted to CBP80/20-bound mRNA. EMBO Rep 9: 446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A 2013. Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay. Genes Cells 18: 161–175 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev 15: 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. . 2009. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 23: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Mühlemann O. 2011. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17: 2108–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zünd D, Gruber AR, Zavolan M, Mühlemann O. 2013. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol 20: 936–943 [DOI] [PubMed] [Google Scholar]