Abstract
The future systematic mapping of variants that confer susceptibility to common diseases requires the construction of a fully informative polymorphism map. Ideally, every base pair of the genome would be sequenced in many individuals. Here, we report 4.75 Mb of contiguous sequence for each of two common haplotypes of the major histocompatibility complex (MHC), to which susceptibility to >100 diseases has been mapped. The autoimmune disease-associated-haplotypes HLA-A3-B7-Cw7-DR15 and HLA-A1-B8-Cw7-DR3 were sequenced in their entirety through a bacterial artificial chromosome (BAC) cloning strategy using the consanguineous cell lines PGF and COX, respectively. The two sequences were annotated to encompass all described splice variants of expressed genes. We defined the complete variation content of the two haplotypes, revealing >18,000 variations between them. Average SNP densities ranged from less than one SNP per kilobase to >60. Acquisition of complete and accurate sequence data over polymorphic regions such as the MHC from large-insert cloned DNA provides a definitive resource for the construction of informative genetic maps, and avoids the limitation of chromosome regions that are refractory to PCR amplification.
The major histocompatibility complex (MHC) is a gene-dense region of the human genome on Chromosome 6p21.31. The complex spans ∼4 Mb and covers >120 expressed genes (The MHC Sequencing Consortium 1999). Forty percent of the expressed loci encode proteins with functions related to immune defense. These include the highly polymorphic class I and class II human leukocyte antigen (HLA) membrane glycoproteins that present peptides for recognition by T lymphocytes.
More than 20,000 papers have been published over the last 30 years describing associations of the human MHC with most autoimmune and some infectious diseases (Warrens and Lechler 1999). However, for most diseases a coherent explanation for their genetic component has not yet emerged. A major limiting factor has been incomplete knowledge of the allelic variation of genes and regions flanking the nine classical HLA loci. More generally, the genetic component of common diseases, such as autoimmune conditions, probably consists of a series of rare and common variants at multiple loci (Dahlman et al. 2002). The search for disease-associated variants of MHC genes, whether non-HLA or even HLA loci themselves, has not been that successful. The mapping of common disease genes with small effects requires a comprehensive knowledge of variation within a genomic region. Then, to distinguish candidates for the causal variant from other polymorphisms associated with the variant via linkage disequilibrium, a detailed association analysis of each allele in a large sample of subjects is required. Finally, an associated variant needs a supporting biological function consistent with the disease phenotype (Ueda et al. 2003). So far, in the vast majority of common disease genetic studies, none of these requirements has been fully met. Even in the intensely studied HLA complex, a contiguous map of allelic variation is not available, partly owing to the inability to PCR-amplify certain DNA segments and the extreme polymorphism of certain genomic regions. The MHC Haplotype Project was designed to overcome this limitation by the cloning and sequencing of BAC clones derived from cell lines homozygous for certain HLA haplotypes (Allcock et al. 2002). Here, we report and compare the first two contiguous haplotype sequences, each spanning 4.75 Mb of the human MHC and chosen because they are common and strongly associated with several autoimmune diseases. The results indicate advantages in using clone-based approaches to obtain complete polymorphism maps.
RESULTS
Two consanguineous, HLA-homozygous cell lines carrying the HLA-A3, B7, Cw7, DR15(DR2) (PGF) and HLA-A1, B8, Cw7, DR3 (COX) haplotypes were selected for study from the 10th International Histocompatibility Workshop panel (Dupont and Ceppellini 1989). The A1, B8, Cw7, DR3 haplotype was chosen because it affects susceptibility to a wide range of diseases and has a northern European frequency in the order of 10%. Relative risks (RR) associated with the haplotype fall across a range from two to four (Price et al. 1999). The B7, DR2 haplotype, again at about a 10% frequency in European populations, provides very significant protection against type 1 diabetes (RR = 0.05) and predisposes to other common diseases such as multiple sclerosis (RR = 2–4; Hall and Bowness 1996; Warrens and Lechler 1999). Contiguous sequences of 4.75 Mb spanning the class I, class II, and class III regions along with much of the extended MHC (from RFP to KIFC1) were obtained from large-insert BAC libraries by shotgun sequencing.
Annotation and Gene Content
From RFP (telomeric boundary) to KIFC1 (centromeric boundary), 162 coding and another 20 transcribed loci were identified as common to both the PGF and COX haplotypes (for details, see Supplemental Table 1). In addition, COX and PGF differed in respect of their HLA-DRB genes (HLA-DRB3 and HLA-DRB5, respectively), and the PGF haplotype had an additional C4A gene (see below). Also, 74 common pseudogenes were observed in both haplotypes.
In a comparison of the official reference MHC gene sequence (The MHC Sequencing Consortium 1999; Mungall et al. 2003), which is a composite of many haplotypes, to the PGF and COX haplotype sequences, one major difference was observed. Previously, the intronless pseudogene PPP1R2P1 was observed to have a frameshift mutation. However, in both the PGF and COX haplotypes, a full-length open reading frame was found. Expressed transcripts of this locus have been reported (Wu and Moses 2001). Screening of 18 random genomic samples revealed that nine were homozygous for the continuous ORF, seven were heterozygous, and two were homozygous for the frameshift, consistent with Hardy-Weinberg equilibrium and a frameshift gene frequency of ∼0.31, suggesting this may be a functional gene in many individuals.
The gene numbers in this study differ substantially from the publication of the first composite MHC sequence (The MHC Sequencing Consortium 1999) for two reasons. Firstly, our criteria for gene annotation (see Methods) filtered several pseudogenes from the annotation. For example, many HCG pseudogenes within the MHC class I region were not annotated because no protein homology to the translated genomic sequence was identified. In addition, many of the previously identified P5-1 pseudogenes are highly homologous to HERV repeat elements (Kulski and Dawkins 1999) and were, therefore, annotated accordingly by RepeatMasker. Secondly, evidence for certain MHC genes has only recently been acquired, for example, the C6orf15 (STG), PSORS1C1 (SEEK1), and PSORS1C2 (SPR1) genes identified in a psoriasis susceptibility gene search (Oka et al. 1999).
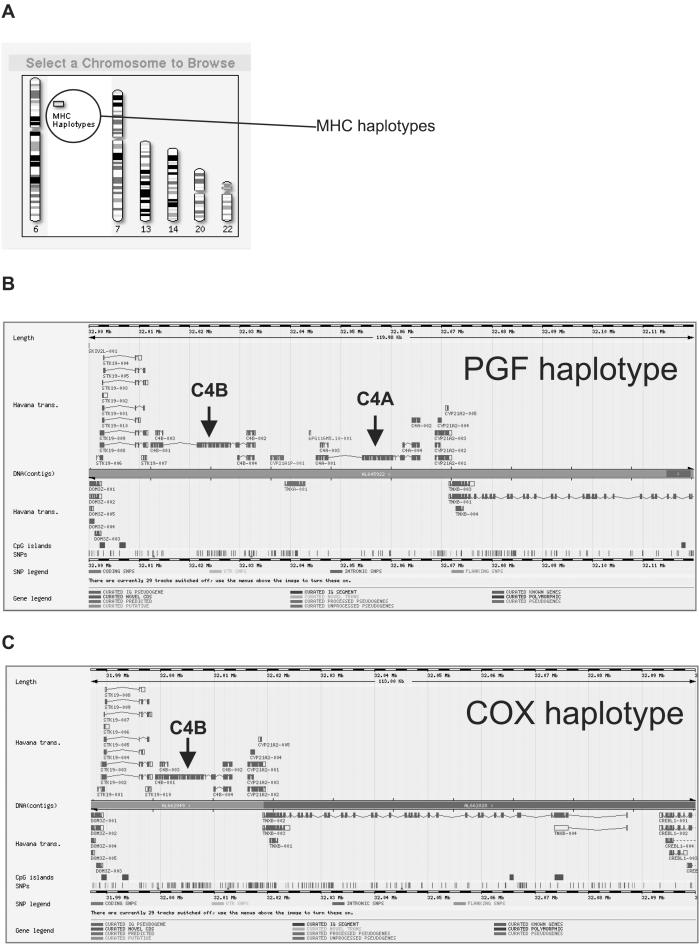
The annotation of the PGF and COX haplotypes is available as a general resource through the VEGA database (http://vega.sanger.ac.uk/; Vertebrate Genome Annotation database). All annotation is curated manually, thereby providing accuracy far greater than is currently possible through in silico methods, and is updated upon the release of new evidence (Ashurst and Collins 2003). Structural information for genes and SNP data are all accessible through the database. An example showing differences between PGF and COX around the C4 loci (see below) is shown in Figure 1.
Figure 1.
Representation of the PGF and COX haplotypes in the VEGA genome annotation browser. (A) Title screen showing the availability of annotated MHC haplotypes. (B,C) Manually curated gene structures in the RCCX region (see Fig. 3 and accompanying text). The PGF haplotype has two copies of the gene for complement component C4 (C4A and C4B), whereas COX has only one (C4B).
Variation Between the PGF and COX Haplotype Sequences
Of several alignment procedures tested, cross_match gave the most accurate detection of PGF-COX variations (see Methods) and was, therefore, used for this analysis. Table 1 summarizes all variations between the PGF and COX haplotypes. Across the 4.75-Mb regions, a total of 18,414 variations were observed. Of these, 16,013 were SNPs. The mean density of variations within intergenic, non-repeat DNA was 4.48 variations per kilobase. Pseudogene sequences and interspersed repeats displayed similar variation densities, whereas lower variation densities were observed in regions of exonic UTR (2.59/kb), intronic DNA (2.87/kb), and coding regions (1.41/kb). Subdivision of the coding SNPs into the types of codon changes they introduce (Table 2) revealed that the nine classical MHC genes had 127 nonsynonymous codon changes compared with 48 synonymous codon changes. Of the nonsynonymous changes, 59 were nonconservative, consistent with balancing selection acting on these genes (Hughes and Nei 1988, 1989). In contrast, the pooled set of “other genes” had the expected ratio of synonymous to nonsynonymous substitutions at 78:67, of which 36 were conservative.
Table 1.
Variations Between the PGF and COX MHC Haplotypes
| Context | SNPs | Indels | Total | bp | kb-1 |
|---|---|---|---|---|---|
| Codinga,b | 341 | 8 | 349 | 247,604 | 1.41 |
| Exonic UTR | 308 | 26 | 334 | 128,976 | 2.59 |
| Intronic | 2552 | 472 | 3024 | 1,052,077 | 2.87 |
| Pseudogenic | 460 | 33 | 493 | 107,079 | 4.60 |
| Microsatellitec | 262 | 230 | 492 | 28,788 | 17.09 |
| Interspersed repeatsd | 3616 | 719 | 4335 | 1,092,788 | 3.97 |
| Other intergenic | 8474 | 913 | 9387 | 2,097,517 | 4.48 |
| Total | 16,013 | 2401 | 18,414 | 4,754,829 | 3.87 |
| kb-1 | 3.37 | 0.51 | |||
| Transversions | 5189 | ||||
| Transitions | 10,824 |
Variations are classified according to type and position.
Coding variations were in 320 codons as follows: 297 with one SNP; 21 with two SNPs; two with three SNPs. Codon changes due to these variations are shown in Table 2.
Indels within coding regions did not disrupt the reading frame at the end of the exon containing them: either by being multiples of 3 nt, or by pairing with another indel that had an opposite effect on the reading frame.
Microsatellites were annotated by the Tandem Repeats Finder (Benson 1999) as ≥7 copies of a ≥2-mer.
Interspersed repeats were annotated by RepeatMasker as LINEs or SINEs.
Table 2.
Codon Changes Due to Coding SNPs Between PGF and COX
| Classical MHC genes | Other genes | Total | |
|---|---|---|---|
| Synonymous | 48 | 78 | 126 |
| Nonsynonymous | |||
| Conservative | 68 | 36 | 104 |
| Nonconservativea | 59 | 31 | 90 |
| Total | 175 | 145 | 320 |
Distribution of the 320 codon changes.
Nonconservative variations were defined as amino acid changes that gave a negative score in a BLOSUM 62 matrix. Classical MHC genes include HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DRA1, HLA-DQB1, HLA-DQA1, HLA-DPB1, HLA-DPA1; other genes include all other coding genes.
The standard measure of heterozygosity (π), the per nucleotide difference between any two haplotypes throughout the genome, has been estimated to lie between 4 × 10–4 and 9 × 10–4 (Li and Sadler 1991; Wang et al. 1998; Cargill et al. 1999; Halushka et al. 1999; Altshuler et al. 2000; Sachidanandam et al. 2001; Venter et al. 2001). The level of SNP variation between COX and PGF, 3.4 × 10–3, is, therefore, fourfold to ninefold higher than estimates for genome-wide heterozygosity. In comparing our statistic for PGF/COX variations to π, there is a slight bias caused by our selection of two HLA-disparate haplotypes. However, the high population frequencies of each of these HLA haplotypes suggest that these levels of heterozygosity are common for the MHC.
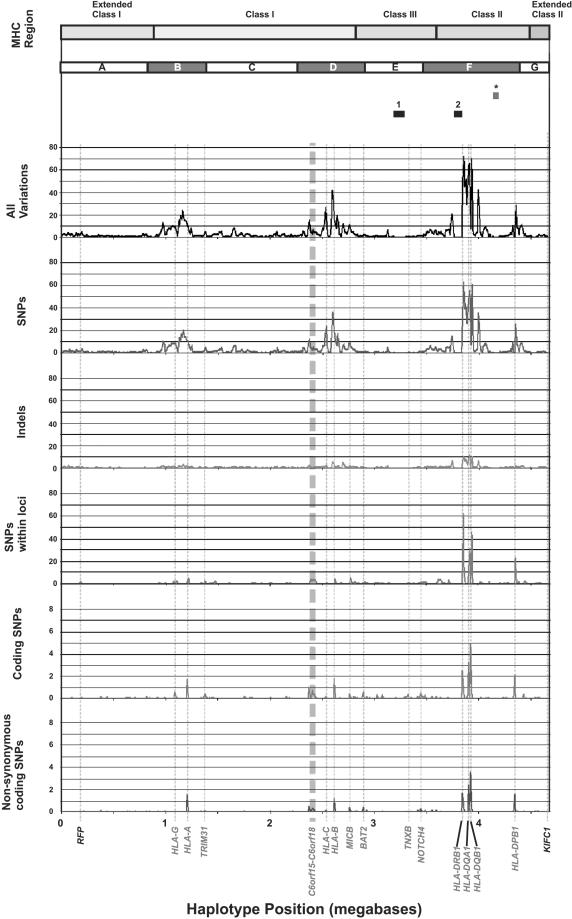
To analyze further the variations between the PGF and COX haplotypes, we plotted variation density against genomic position (Fig. 2). As expected, much of the variation concentrated in peaks overlying the classical class I and class II loci. This was most apparent when only SNPs occurring within defined loci (all genomic sequence from the 5′-start of a gene to the 3′-end) were considered. Some smaller peaks corresponded to genes other than the classical class I and II loci, consistent with independent selection for variation. For example, we observed a peak telomeric of HLA-C from CDSN to POU5F1, the borders of which correspond very well with the region shown to be important in susceptibility to psoriasis (Balendran et al. 1999; Oka et al. 1999; Nair et al. 2000). Balancing or overdominant selection has been demonstrated for the peptide-binding domains of HLA loci (Hughes and Nei 1988, 1989) and most likely explains the concentrated variation we observe around them. Most of the variation accounting for these peaks is noncoding and may be caused by “hitch-hiking” with the selected amino-acid changing variants (Maynard-Smith and Haigh 1974). At high resolution, the peaks of variation were not uniform and were interrupted by regions of similarity. One example is the TAP1 to HLA-DMB region (labeled with * in Fig. 2). This region lies between two recombination hot spots and shows a high level of linkage disequilibrium (Jeffreys et al. 2001), suggesting that genetic exchange by recombination resulted in the PGF and COX haplotypes having highly similar sequences through this region.
Figure 2.
Positional distribution of variations between COX and PGF MHC sequences. MHC sequences were divided into 10-kb bins, and variations were calculated in each bin. Results are expressed as variations per 1 kb. A locus is defined as all genomic DNA between the 5′-start of a gene to the 3′-end. Boundaries of the class I, II, and III regions are shown. The positions of genes RFP and KIFC1 that define the ends of the MHC haplotype sequencing project are indicated in black. Other genes with five or more SNPs between the haplotypes are labeled in blue. HLA-C is also labeled. Regions of interest are labeled above the graph. Regions 1 and 2 are the RCCX module and the HLA-DRB region, respectively. (*) The TAP to HLA-DMB region, which shows little variation between haplotypes. Blue bars B, D, and F label regions in which variation is thought to be classical MHC class I and class II gene-associated.
The distribution of insertions and deletions (indels) throughout the MHC correlates well with the distribution of SNPs, showing increased density around the classical class I and class II loci. This correlation indicates that the same genetic pressures give rise to the patterns of both forms of variation.
To determine whether the high degree of variation observed between PGF and COX was purely due to the regions surrounding classical class I and class II genes, we subdivided the sequence comparison into those regions with classical class I/II associated peaks (Fig. 2, blue bars B, D, and F), and other regions (bars A, C, E, and G). As expected, the SNP variations per nucleotide were highest for the HLA-A-associated region (region B), 5.49 × 10–3; for the HLA-B- and -C-associated region (D), 5.23 × 10–3; and for the HLA-D-associated region (F), 7.79 × 10–3. On the other hand, the SNP variation observed in the non-class I/class II-associated regions was indistinguishable from that observed else-where in the genome. For regions A, C, E, and G, the per nucleotide SNP variation was 9.1 × 10–4, 13.7 × 10–4, 5.6 × 10–4, and 8.9 × 10–4, respectively. These values are only slightly greater than, if not within, the estimated figures for genome-wide heterozygosity, π, between 4 × 10–4 and 9 × 10–4. The region with the least SNP variation between PGF and COX, region E, extends across the MHC class III region from BAT4 to AGPAT1. The class III region has an extremely high gene density (The MHC Sequencing Consortium 1999; Xie et al. 2003) and is bordered by the highly variable class I and class II regions. One explanation for these lower levels of variation is that the selective effects at the class I and class II loci and accompanying hitch-hiking do not extend into such regions of the MHC. Alternatively, the net effect of both hitch-hiking and purifying selection could result in a figure for heterozygosity close to the genome-wide average.
Complex Polymorphic Regions
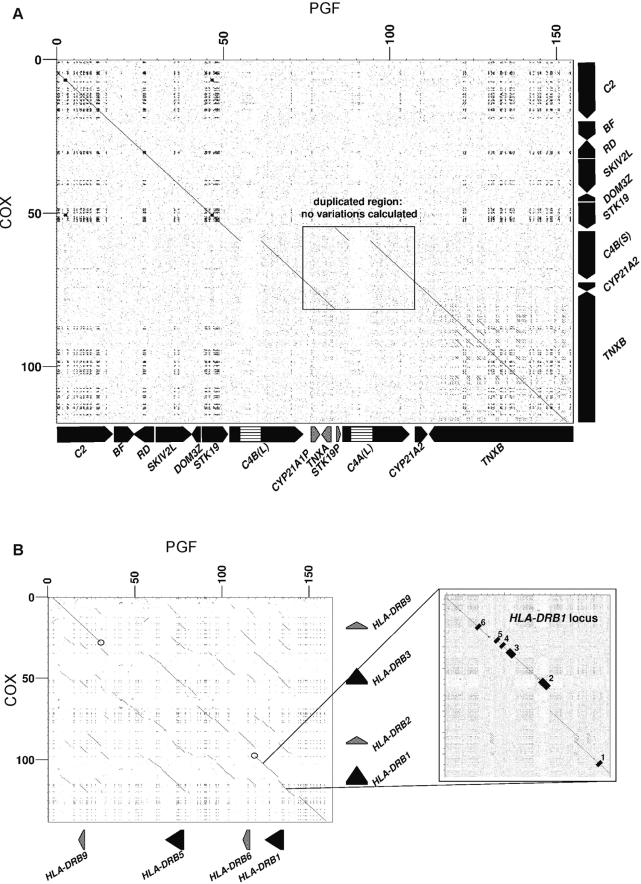
Two genomic regions with repetitive structures were counted as single indels and required exclusion from automated analysis of variation. The RCCX module and HLA-DRB loci are known to be extremely polymorphic owing to insertion and deletion of large fragments of genomic sequence (Dunham et al. 1989). Figure 3 shows the dot-matrix (Sonnhammer and Durbin 1995) comparisons of these regions between the PGF and COX haplotypes, illustrating the complexity.
Figure 3.
Dot-matrix comparison of the PGF and COX sequences spanning the RCCX (A) and HLA-DRB (B) regions. The X-axes represent the PGF contig and encoded genes and the Y-axes display those of COX. Axis numbering is in kilobases. Coding genes are labeled in black and pseudogenes in gray. (A) No variations were calculated from comparison of the duplicated region (boxed region of homology) of PGF with COX. Striped boxes represent endogenous retrovirus sequences that identify long (L) C4 genes. The dot-matrix analysis was performed using sequences taken for PGF from contig position 26,001 to 180,559 of AL645922, and for COX from 12,001 of AL662849 to 55,541 of AL662828. (B) No PGF/COX variations were calculated between sequences bordered by open circles through which the gene content differs between haplotypes. A more detailed comparison of the HLA-DRB1 loci with exon locations is shown. The dot-matrix analysis was performed using sequences taken for PGF from contig position 128,102 of AL662796 to 27,557 of AL662789, and for COX from 130,001 of AL670296 to 108,819 of AL662842.
Individual MHC haplotypes can contain up to four copies of the gene for complement component C4. The genetic basis for this polymorphism is tandem duplication of genomic DNA termed the RCCX module, which includes part of the STK19 (RP) gene, C4A/B, CYP21, and part of TNXB. Additional copies of the RCCX module contain copies of the C4A/B gene and the pseudogenes CYP21A1P, TNXA, and STK19P (Shen et al. 1994; Chung et al. 2002). The structure in the COX haplotype is monomodular with single copies of STK19, C4B, CYP21A2, and TNXB, whereas PGF is bimodular, including not only these genes but also C4A and the above pseudogenes (Fig. 3A). The duplicated region in the PGF sequence has no equivalent in COX and, therefore, was counted as a single insertion (see Methods).
HLA-DRB loci are encoded in a region that has HLA-DRB1 at the centromeric end and the pseudogene HLA-DRB9 at the telomeric end. Between these two loci, different haplotypes have either no other loci or alternative arrangements of other HLA-DRB genes and pseudogenes (Marsh et al. 2000). The dot-matrix comparison of the regions from the PGF and COX haplotypes illustrates the modular repetition of HLA-DRB loci and discontinuous homology between the two haplotypes (Fig. 3B).
Indels
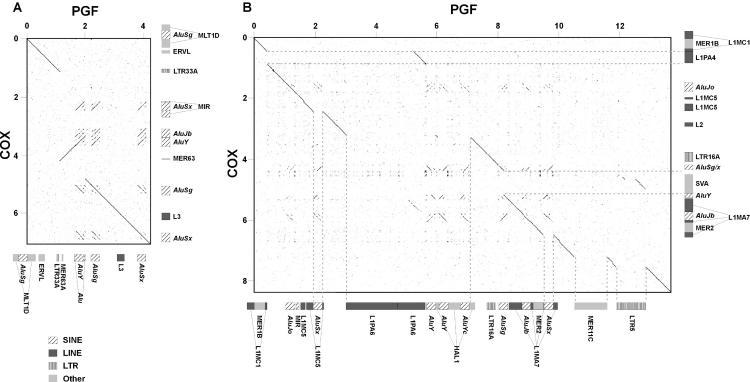
In addition to the above gene polymorphisms, 38 large indels were observed when comparing the PGF and COX haplotypes (Table 3). A selection of these indels have been previously identified (e.g., Dangel et al. 1994; Horton et al. 1998; Gaudieri et al. 1999; Dunn et al. 2002, 2003). Most large indels observed were due to the presence/absence of Alu sequences, of which the majority were of the younger AluY type, and in particular those of the AluYb8 and AluYa5 types, which emerged after the divergence of humans and African apes (Carroll et al. 2001). However, other, older AluS sequences and repeats of the LINE, HERV, LTR, MER, and SVA families also differ between the two haplotypes. The majority of these indels are located in the HLA-D region, with a few in close proximity to classical HLA class I genes, consistent with ancient divergence of the haplotypes in these regions. In addition to indels composed of single copies of repetitive elements, for example, SINEs and LINEs, some complex indels were observed. The 4-kb-long indel ∼600 bp centromeric of RFP (Table 3, labeled C) comprises non-repeat sequence along with SINE and MER sequences, and has probably arisen by nonhomologous recombination between the two flanking AluS sequences. Figure 4A shows a comparison of the PGF and COX sequences over the indel labeled A in Table 3. With respect to PGF, part of the COX haplotype is inverted, with two insertions on either side. Figure 4B shows the complex series of indels (labeled B in Table 3) distributed between HLA-DQA1 and HLA-DQB1.
Table 3.
Additional Major Indels Between the PGF and COX Haplotypes
| Indel family | Indel type | Additional DNA present in | Context |
|---|---|---|---|
| Simple repeat | (CTTT)n(CCTT)n(CTTT)n | PGF | Satellite repeat of CC/TTT at position where COX has six copies of an ATTT repeat. Positioned ∼5 kb telomeric of 5′ C6orf101 |
| LINE | L1PA2 | PGF | Clean indel positioned ∼35 kb centromeric of 5′ HLA-DRB1 and ∼12 kb telomeric of 5′ HLA-DQA1 |
| LIM2 | COX | Clean indel positioned ∼37 kb centromeric of 5′ HLA-DRB1 and ∼11 kb telomeric of 5′ HLA-DQA1 | |
| BL1PA4 | COX | Centromeric of HLA-DQA1 and telomeric of HLA-DQB1 | |
| SINE | AluYa5/8 | PGF | Clean indel. Positioned ∼450 bp telomeric of 5′ OR12D2 |
| AluYb8 | COX | Within a MER65-int element. Between exon 2 and exon 3 of a variant of C6orf12 (HTEX4.3) | |
| AluSg | COX | Within a LTR10E element. Positioned ∼57 kb centromeric of 5′ HLA-C and ∼25 kb telomeric of 3′ HLA-B | |
| AluYb8 | PGF | Within L2 element. Within C6orf10 (TSBP like) intron 8, ∼4 kb from centromeric exon 8 and ∼3 kb from telomeric exon 9 | |
| AlYa5 | PGF | Within an LTR12 element. Within HLA-DRB1 intron 5, ∼100 bp from centromeric exon 5 and ∼700 bp from telomeric exon 6 | |
| AluY | PGF | Clean insertion positioned ∼14 kb centromeric of 5′ HLA-DRB1 and ∼33 kb telomeric of 5′ HLA-DQA1 | |
| AluY | PGF | Clean indel positioned ∼36 kb centromeric of 5′ HLA-DRB1 and ∼11 kb telomeric of 5′ HLA-DQA1 | |
| BAluSx | PGF | Centromeric of HLA-DQA1 and telomeric of HLA-DQB1 | |
| BAluSx | PGF | Centromeric of HLA-DQA1 and telomeric of HLA-DQB1 | |
| AluY | COX | Within HLA-DQB1 intron 2 ∼2 kb centromeric of exon 3 and ∼1.2 kb telomeric of exon 2 | |
| AluYa5 | COX | Positioned ∼8.5 kb centromeric of HLA-DQB3 and ∼1.2 kb telomeric of 5′ HLA-DQA2 | |
| AluY | PGF | Positioned ∼18 kb centromeric of 5′ HLA-DQB2 and ∼30 kb telomeric of 3′ HLA-DOB | |
| AluYb8 | PGF | Clean indel within intron 2 of HLA-DPB2 pseudogene, positioned ∼8 kb centromeric of exon 2 and ∼2 kb telomeric of exon 3 | |
| HERV | HERVK9 | PGF | Within a MER9 element. Positioned ∼24 kb telomeric of MICF and ∼6 kb centromeric of 5′ end HLA-H |
| HERVCK4 | PGF | Within C4B intron 9, ∼300 bp from telomeric exon 9 and ∼130 bp from centromeric exon 10 | |
| LTR | BLTR5 | PGF | Centromeric of HLA-DQA1 and telomeric of HLA-DQB1 |
| MER | BMER11C | PGF | Centromeric of HLA-DQA1 and telomeric of HLA-DQB1 |
| SVA | SVA | PGF | Clean indel. Positioned ∼8 kb telomeric of HLA-A |
| SVA | PGF | Positioned ∼52 kb centromeric of POU5F1 and ∼23 kb telomeric of HLA-C | |
| SVA | COX | Within an HERV1 element. Positioned ∼57 kb centromeric of 5′ HLA-C and ∼25 kb telomeric of 3′ HLA-B | |
| SVA within which there are two copies of a 38-mer | PGF | Within an L14MC element. Positioned ∼6.5 kb telomeric of 5′ HCP5, between MICA and MICB | |
| Combination indels | SVA and (TCTCCC)×38 | PGF | Clean indel between Charlie9 repeat and MLT1E3 repeat. Positioned ∼3 kb telomeric of 5′ HLA-F |
| AluSp and L1PA13 | PGF | Positioned ∼52 kb centromeric of 5′ HLA-C and ∼28 kb telomeric of 3′ HLA-B | |
| AluSq and AluY | COX | Within an L2 element. Within HLA-DRB1 intron 1, ∼2.5 kb from centromeric exon 1 and ∼3.5 kb from telomeric exon 2 | |
| SVA and other sequence | PGF | Positioned ∼4.5 kb centromeric of 5′ HLA-DRB1 and ∼41 kb telomeric of HLA-DQA1 | |
| L1PA4 and (A)n stretch | COX | Positioned ∼2 kb centromeric of 3′ HLA-DQA1 and ∼14 kb telomeric of 3′ HLA-DQB1 | |
| BL1PA6 and AluY | PGF | Centromeric of HLA-DQA1 and telomeric of HLA-DQB1 | |
| BT-rich repeats and SVA | COX | Centromeric of HLA-DQA1 and telomeric of HLA-DQB1 | |
| L1MC5 with LTR42 in middle | PGF | Positioned ∼47 kb centromeric of 5′ HLA-DQB2 and ∼700 bp telomeric of 3′ HLA-DOB | |
| Complex indels | CSome non-repeat sequence, MIR, MER41B, MER115, AluSx, Flam_C, AluSg, AluY, AluSx and L2 with MER38 in middle | PGF | Probable recombination between two nearby Alus: PGF has AluSc bordering the telomeric end of the indel and an AluSx bordering the centromeric end. Positioned ∼600 bp centromeric of 5′ of RFP, and ∼15 kb telomeric of C6orf100 |
| ∼4 kb indel containing a possible CpG island through the centromeric 2 kb. EST matches are observed around the CpG island. One match is observed to a small ∼260-bp fragment of the KIAA1545 cDNA | COX | Positioned ∼16 kb centromeric of 3′ HLA-G and ∼5.5 kb telomeric of MICF pseudogene | |
| ACompared with PGF, COX has part of the sequence inverted and two inserted sequences either side of the inversion | COX | Positioned ∼50 kb centromeric of C6orf205 | |
| L2, Tigger4, and MER20 each separated by non-repeat element sequence | COX | Located within an MER53 element in HLA-DRB1 intron 1, ∼5 kb from centromeric exon 1 and ∼900 bp from telomeric exon 2 | |
| BHAL1 with an AluY and an AluYc in it | PGF | Centromeric of HLA-DQA1 and telomeric of HLA-DQB1 |
SVA repeats contain several Alu sequences and a fragment of LTR5. Dot-matrix comparisons of PGF and COX sequences over indels marked with superscript A and B are shown in Figures 3A and 3B, respectively. See Supplemental Table 4 for genomic locations of each indel.
Figure 4.
Dot-matrix comparisons of PGF and COX sequences spanning selected indels. (A) Complex inversion centromeric of C6orf205 and (B) multiple indels located between the HLA-DQA1 and HLA-DQB1 loci. PGF is represented on the X-axis and COX on the Y-axis. Scales are in kilobases. Repetitive elements identified by RepeatMasker are shown. Sequence is taken from (A) AL669830 134,100 to 138,400 and AL663093 82,300 to 89,300 and (B) AL662789 57,100 to 70,800 and AL731683 8,700 to 17,100.
Intrahaplotype Variation
As a quality control check for the accuracy of sequence data and to ensure that BAC clones were, indeed, derived from a single haplotype, we compared the sequences of overlapping BACs derived from the same consanguineous cell line. Whereas finished BAC sequences are only submitted with 2 kb of overlap between them, unfinished sequence derived from the shotgun sequencing strategy can extend >100 kb into finished sequence of a neighboring BAC clone. Certain PGF but not COX overlaps showed discrepancies inconsistent with the assumed origin of the BAC clones from a single haplotype. Of 45 overlaps examined between PGF-derived BAC clones, 31 showed no variations between BAC sequences. Another 12 overlaps had minor discrepancies, most frequently indels of single or dinucleotide repeat sequences, consistent with a single haplotype origin and minor variations arising during propagation of the PGF cell line or derivative BAC clones. However, in two comparisons between PGF-derived BAC clones, a great degree of variation was observed, inconsistent with their derivation from a single haplotype. Comparisons of AL662791 with AL645937, and AL662860 with AL645936, gave a total of 75 variations through ∼166 kb of overlapping sequence. Both of these overlaps occur between BACs that represent the extended MHC class I region. These regions are all telomeric of the HLA genes tested during both our tissue-typing and that used to define the homozygous nature of the cell line (i.e., telomeric of HLA-A). There are two possible explanations for the differences observed. The PGF cell line may not be truly consanguineous. Alternatively, a recombination event could have occurred telomeric of HLA-A at some time following the bifurcation of the common ancestor of the PGF consanguineous chromosomes. This finding of haplotype divergence telomeric of classical HLA loci has additionally been made for several other consanguineous HLA homozygous cell lines (Ehlers et al. 2000). Nevertheless, these cell lines remain invaluable in obtaining homozygous DNA from the classical MHC.
DISCUSSION
We have sequenced two complete MHC haplotypes that are strongly associated with common diseases, including type 1 diabetes and multiple sclerosis (Hall and Bowness 1996; Price et al. 1999; Warrens and Lechler 1999). The SNP content and variation between them has been fully described. A total of 18,414 variations including 16,013 SNPs have been identified by comparing the high-quality finished genomic sequence data from these haplotypes. These data and those from the other six haplotypes in progress (Allcock et al. 2002) provide an essential resource in the search for MHC-encoded disease alleles. More than 28,000 SNPs from the MHC Haplotype Project have been submitted to dbSNP to date, of which >21,000 have not previously been reported. These SNPs and those from other projects (Geraghty et al. 2002; T. Shiina and H. Inoko, pers. comm.) will contribute to the ongoing construction of a dense SNP map (Walsh et al. 2003), which will allow the selection of informative haplotype tag SNPs (htSNPs). htSNP typing reduces the cost of genotyping by at least 50% (Johnson et al. 2001) in the large disease samples that will require collection and analysis to map the disease-susceptibility loci in the MHC.
In addition to their relevance for genetic studies, many of the variations identified have the potential to affect gene expression or function. In all, 194 nonsynonymous coding substitutions were observed, of which 67 are within 42 genes other than those for classical MHC molecules. Many SNPs and indels were also found in UTRs and possible regulatory regions and hence have the potential to affect gene expression.
The data presented here form the foundation for a refined knowledge of variation and its haplotype structure throughout the extremely polymorphic and gene-rich MHC. These haplotype-specific sequences can now be used as reference sequences for other haplotypes and sequences. This knowledge is an essential resource to precisely define the so-far-elusive disease-associated polymorphisms that are encoded in this genomic region. The completeness of the polymorphism map from SNPs to large indels involving highly repetitive sequences vindicates our choice of experimental strategy: cloning and shotgun sequencing, instead of sequencing of PCR products. In the latter approach, not only are many sequences difficult to PCR, particularly the GC-rich 5′-exons of genes, but also the reliable and accurate calling of heterozygous bases is still not a fully automated process. This consideration, although obviated by the use of homozygous cell lines, suggests that using clone-based sequences for the assembly of complete polymorphism maps is a vital and valid strategy.
METHODS
Cell Lines
Two HLA-homozygous typing, consanguineous cell lines, PGF (DR15, Caucasoid, England) and COX (DR3, Caucasoid, South Africa) lines were selected from the 10th International Histocompatibility Workshop panel (Dupont and Ceppellini 1989). PGF types as A*0301, Cw*07, B*0702, DRB1*15011, DRB5*01011, DRB6*0201, DQA1*01021, DQB1*0602, DPA1*01, and DPB1*0401. COX types as A*0101, Cw*0701, B*0801, G*01012, DRA*0102, DRB1*0301, DRB3*0101, DQA1*05011, DQB1*0201, DPA1*01, and DPB1*0301 (http://www.ebi.ac.uk/imgt/hla). Following cultivation, DNA from these cell lines was typed to confirm identity and singularity as PGF: A3, B7, Bw6, C*070, DR15(2), DRw51, DQ6(1), DQB1*0602/0611, and COX: A1, B8, Bw6 (Tissue Typing Laboratory, Addenbrooke's NHS Trust, Cambridge, UK).
BAC Clone Library Construction
The PGF and COX libraries were made to ∼10 times genome representation as follows. Cells were embedded in agarose (0.5%, InCert agarose: FMC) solidified in disposable plug molds (BioRad). The plugs were treated with cell lysis solution (2 mg/mL Proteinase K, Roche; 2% N-lauroyl sarcosine; and 0.4 M EDTA) and washed with PMSF solution (0.1 mM PMSF, 10 mM Tris-HCl at pH 8.0, 50 mM EDTA), as described in detail previously (Osoegawa et al. 1999). High-molecular-weight DNA was partially digested with a combination of EcoRI and EcoRI Methylase and size-fractionated by pulse-field electrophoresis (CHEF system; BioRad). Size-fractionated DNA was electroeluted and ligated between the EcoRI sites of the pTARBAC2.1 vector (http://bacpac.chori.org/ptarbac21.htm) and transformed into Escherichia coli DH10B cells (Invitrogen). The libraries derived from PGF (CHORI-501) and COX cell lines (CHORI-502) were arrayed into 672 and 576 384-well microtiter dishes, respectively. For hybridization screening, the BACs were gridded onto 26 (14 and 12) distinct nylon high-density colony filters of 22 × 22 cm, respectively. Each hybridization membrane represents the colonies from 48 dishes through duplicate colonies (∼18,000 BAC clones per sheet). Further details about the two BAC libraries can be found at http://bacpac.chori.org/mhc501.htm and http://bacpac.chori.org/mhc502.htm.
Mapping and Sequencing
Genomic sequences (∼200 bases) were selected at an interval of 30–40 kb from the 4.75-Mb MHC region (HSA 6p21.3). A total of 157 pairs of 24-nt oligonucleotides with eight bases 3′ overlap “overgo pairs” (Ross et al. 1999) were designed using the overgo script (http://genome.wustl.edu/tools/?overgo=1). The resulting 40 base sequences were searched using BLAST to confirm the uniqueness of the sequence in the human genome. A full listing of probe sequences is provided in Supplemental Table 2. The overgo partners were combined, annealed by heating for 5 min at 80°C, and subsequently incubating for 10 min at 37°C. The annealed oligonucleotides were extended with Klenow polymerase using a mixture of dGTP, dTTP, [α-32P]dCTP, and [α-32P]dATP at room temperature (McPherson et al. 2001). The probes were purified by spin-dialysis through Sephadex G-50 columns in 96-well plates. The labeled probes were pooled into three mixtures of 52–53 each, then denatured and hybridized to the high-density filters in hybridization buffer (0.5 M sodium phosphate at pH 7.2, 7% SDS, and 1 mM EDTA) overnight at 60°C. The filters were washed sequentially at 60°C with 1.5× SSC, 0.1% SDS and 0.5× SSC, 0.1% SDS. The filters were exposed overnight to a phosphor screen for subsequent scanning in a PhosphorImager (STORM860; Amersham). Hybridization positive signals were automatically scored with ArrayVision Ver6.0 (Imaging Research Inc), although some editing of the results was required. All candidate MHC BACs (1150 clones from the CHORI-501 library, 1523 clones from the CHORI-502 library) were manually rear-rayed into 12 and 16 96-well dishes, respectively, and reprobed with individual probes. Clones that remained positive were subsequently restriction (HindIII) fingerprinted, the gel images processed using IMAGE (http://www.sanger.ac.uk/Software/Image/; Sulston et al. 1989), and the fingerprints assembled into contigs using FPC (http://www.genome.arizona.edu/fpc/; Soderlund et al. 2000). This process was repeated until a single contig was obtained from which a minimal tiling path was selected for sequencing, comprising 50–60 BAC clones per haplotype.
For the shotgun phase (Bankier et al. 1987), the selected tile path BACs were subcloned into pUC plasmids and sequenced from both ends using the dideoxy chain terminator method (Sanger et al. 1977) with different versions of big dye terminator chemistry (Rosenblum et al. 1997). The resulting sequencing reactions were analyzed on various models of ABI sequencing machines, and the generated data were processed by a suite of inhouse programs (http://www.sanger.ac.uk/Software/sequencing/) prior to assembly with the PHRED (Ewing and Green 1998; Ewing et al. 1998) and PHRAP (http://www.phrap.org/) algorithms. For the finishing phase, we used the GAP4 program (Bonfield et al. 1995) to help assess, edit, and select reactions to eliminate ambiguities and close sequence gaps. Sequence gaps were closed by a combination of primer walking, PCR, short/long insert sublibraries (McMurray et al. 1998), oligonucleotide probe screens of such sublibraries, and transposon sublibraries. Unless annotated otherwise, each clone has been finished according to the agreed international finishing standard (http://genome.wustl.edu/gsc/Overview/finrules/hgfinrules.html).
Gene Annotation
The finished genomic sequence was analyzed using an automatic Ensembl pipeline (Hubbard et al. 2002) with modifications as described in Mungall et al. (2003). Interspersed repeats were identified using RepeatMasker (http://repeatmasker.genome.washington.edu/; A.F.A. Smit and P. Green, unpubl.); and simple repeats were detected by Tandem Repeat Finder (Benson 1999). Sequence with repeats masked was searched against vertebrate cDNAs and ESTs using WU-BLASTN and EST_GENOME, and against a nonredundant SWISS-PROT/TrEMBL database using WU-BLASTX. Genes were annotated according to human annotation workshop (HAWK) guidelines (http://www.sanger.ac.uk/HGP/havana/hawk.shtml). Owing to their highly restricted expression, some olfactory receptor genes were annotated based on protein homology to olfactory receptors with known expression.
Variation Analysis
Before analyzing the PGF and COX haplotypes for sequence differences, we compared different methods of detection. The “cross_match” program eventually used was compared with ssahaSNP (Ning et al. 2001). At low SNP densities, good agreement could be found between results generated by each program. However, at higher densities it proved impossible for ssahaSNP to replicate cross_match results. A comparison of cross_match and the latest version of ssahaSNP (Version 1.08; J.C. Mullikin, pers. comm.) between regions with low and high SNP counts illustrates this finding. Consistent, low SNP counts were given by both programs when comparing BACs AL662799 and AL662827 over 36 kb at the centromeric end of the haplotypes. Both cross_match and ssahaSNP reported 34 SNPs (0.94 SNP/kb), although the latter also reported three further SNPs that cross_ match identified as insertions or deletions. These were positioned within repetitive sequence, which probably accounted for different interpretation by the two programs. At high SNP density, however, such as in the 47-kb comparison between AL662789 and AL662842 within the highly variable region just centromeric of HLA-DRB1, cross_match reported 1741 SNPs (37.4 SNPs/kb), whereas ssahaSNP found only 911. Independent assessment of sequence alignments revealed that cross_match was reporting genuine sequence differences. Although the maximum allowed variation rate within ssahaSNP was set in this case at the relatively high value of 45 SNPs/kb, there were several segments of the sequence that were excluded by ssahaSNP because of greater SNP counts. In our hands, cross_match reported variations more thoroughly than ssahaSNP, making it more amenable for our purposes and the program of choice for this study.
Pairs of overlapping BAC sequences, one each from the two haplotypes PGF and COX, were compared using the cross_match program (P. Green, unpubl.), an implementation of the Smith-Waterman sequence alignment algorithm (Smith and Waterman 1981). Lists of variations in the form of substitutions, insertions, and deletions were generated using the program's “discrepancy list” option.
The cross_match program (version 0.990319) was used with option values set in such a way as to enable fast running of the program and suppression of many spurious matches between repeat elements (in general minmatch: 127, maxmatch: 127, minscore: 30, bandwidth: 14, indexwordsize: 10, vector_bound: 0, word_raw: 0, masklevel: 80; minmatch was reduced in some cases for the analysis of short matches).
Discrepancy lists were examined and edited to remove data beyond the range of overlapping BAC sequences. Where microsatellites were present there was a tendency for cross_match to report discrepancies twice such that a base in one sequence could be counted as having two SNPs at different positions in the other. Such overlaps were removed by manual editing.
The discrepancy lists were subsequently parsed for reading into an ACeDB database (Durbin and Thierry-Mieg 1994) containing the gene annotation data. All data were subsequently written from the database in General Feature Format (gff), enabling the distribution of variation relative to sequence contig position and gene locus to be determined. In the case of two complex polymorphic regions, the RCCX module and HLA-DRB region, the procedure was modified. As the duplicated RCCX region in the PGF sequence has no equivalent in that of COX, no variations could be calculated. This region was defined as bases 101,638 to 134,373 of BAC AL645922. This represents an insertion with respect to COX after base 85,169 of BAC AL662849. Determination of variations between HLA-DRB loci was restricted to the region extending from base 105,201 to the end of AL713966 and base 68,683 to the end of AL662842 corresponding to the HLA-DRB1 locus, and was accomplished by increasing the sensitivity of cross_match. Analysis of these complex polymorphic regions and major insertions/deletions was performed using the dotter dot-matrix analysis program (Sonnhammer and Durbin 1995).
Resources
All sequences presented in this paper have been submitted to the EMBL/Genbank/DBB database and allocated accession numbers (see Supplemental Table 3). For purposes of clarity, all BAC clones are referred to using their accession numbers. The annotation of each haplotype has been entered in the Vertebrate Genome Annotation (VEGA) database and is accessible through its browser (http://vega.sanger.ac.uk). All variations from the study were submitted to dbSNP (http://www.ncbi.nlm.nih.gov/SNP) using the submitter handle SI_MHC_SNP. BAC clones from the CHORI-501 (PGF) and CHORI-502 (COX) libraries can be requested from BACPAC resources (http://www.chori.org/bacpac). A regularly updated Web site for the MHC Haplotype Project is found at http://www.sanger.ac.uk/HGP/Chr6/MHC/.
Acknowledgments
We thank J.C. Mullikin for critical reading of this manuscript, J.G.R. Gilbert and S.J. Keenan for assistance with the VEGA database, all staff of the DNA Sequencing Division at the Wellcome Trust Sanger Institute (www.sanger.ac.uk), and, in particular, the following finishers: J.P. Almeida, J.Y. Brown, C. Griffiths, J.L. Harley, M.D. Humphries, C.M. Johnson, D.A. Leongamornlert, M. Mashreghi-Mohammadi, A.I. Peck, S. Squares, N. Sycamore, A. Tracey, J.M.D. Wood, and J. Wyatt. This work was supported by a joint grant (048880) from the Wellcome Trust to S.B., S.S., J.A.T., and J.T.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2188104. Article published online before print in May 2004.
Footnotes
[Supplemental material is available online at www.genome.org. All sequences presented in this paper have been submitted to EMBL and allocated accession numbers (see Supplemental material). All variations from the study were submitted to dbSNP (http://www.ncbi.nlm.nih.gov/SNP) using the submitter handle SI_MHC_SNP.]
References
- Allcock, R.J., Atrazhev, A.M., Beck, S., de Jong, P.J., Elliott, J.F., Forbes, S., Halls, K., Horton, R., Osoegawa, K., Rogers, J., et al. 2002. The MHC haplotype project: a resource for HLA-linked association studies. Tissue Antigens 59: 520–521. [DOI] [PubMed] [Google Scholar]
- Altshuler, D., Pollara, V.J., Cowles, C.R., Van Etten, W.J., Baldwin, J., Linton, L., and Lander, E.S. 2000. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407: 513–516. [DOI] [PubMed] [Google Scholar]
- Ashurst, J.L. and Collins, J.E. 2003. Gene annotation: Prediction and testing. Annu. Rev. Genomics Hum. Genet. 4: 69–88. [DOI] [PubMed] [Google Scholar]
- Balendran, N., Clough, R.L., Arguello, J.R., Barber, R., Veal, C., Jones, A.B., Rosbotham, J.L., Little, A.M., Madrigal, A., Barker, J.N., et al. 1999. Characterization of the major susceptibility region for psoriasis at chromosome 6p21.3. J. Invest. Dermatol. 113: 322–328. [DOI] [PubMed] [Google Scholar]
- Bankier, A.T., Weston, K.M., and Barrell, B.G. 1987. Random cloning and sequencing by the M13/dideoxynucleotide chain termination method. Methods Enzymol. 155: 51–93. [DOI] [PubMed] [Google Scholar]
- Benson, G. 1999. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfield, J.K., Smith, K., and Staden, R. 1995. A new DNA sequence assembly program. Nucleic Acids Res. 23: 4992–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill, M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Shaw, N., Lane, C.R., Lim, E.P., Kalyanaraman, N., et al. 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231–238. [DOI] [PubMed] [Google Scholar]
- Carroll, M.L., Roy-Engel, A.M., Nguyen, S.V., Salem, A.H., Vogel, E., Vincent, B., Myers, J., Ahmad, Z., Nguyen, L., Sammarco, M., et al. 2001. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J. Mol. Biol. 311: 17–40. [DOI] [PubMed] [Google Scholar]
- Chung, E.K., Yang, Y., Rennebohm, R.M., Lokki, M.L., Higgins, G.C., Jones, K.N., Zhou, B., Blanchong, C.A., and Yu, C.Y. 2002. Genetic sophistication of human complement components C4A and C4B and RP-C4-CYP21-TNX (RCCX) modules in the major histocompatibility complex. Am. J. Hum. Genet. 71: 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman, I., Eaves, I.A., Kosoy, R., Morrison, V.A., Heward, J., Gough, S.C., Allahabadia, A., Franklyn, J.A., Tuomilehto, J., Tuomilehto-Wolf, E., et al. 2002. Parameters for reliable results in genetic association studies in common disease. Nat. Genet. 30: 149–150. [DOI] [PubMed] [Google Scholar]
- Dangel, A.W., Mendoza, A.R., Baker, B.J., Daniel, C.M., Carroll, M.C., Wu, L.C., and Yu, C.Y. 1994. The dichotomous size variation of human complement C4 genes is mediated by a novel family of endogenous retroviruses, which also establishes species-specific genomic patterns among Old World primates. Immunogenetics 40: 425–436. [DOI] [PubMed] [Google Scholar]
- Dunham, I., Sargent, C.A., Dawkins, R.L., and Campbell, R.D. 1989. An analysis of variation in the long-range genomic organization of the human major histocompatibility complex class II region by pulsed-field gel electrophoresis. Genomics 5: 787–796. [DOI] [PubMed] [Google Scholar]
- Dunn, D.S., Naruse, T., Inoko, H., and Kulski, J.K. 2002. The association between HLA-A alleles and young Alu dimorphisms near the HLA-J, -H, and -F genes in workshop cell lines and Japanese and Australian populations. J. Mol. Evol. 55: 718–726. [DOI] [PubMed] [Google Scholar]
- Dunn, D.S., Inoko, H., and Kulski, J.K. 2003. Dimorphic Alu element located between the TFIIH and CDSN genes within the major histocompatibility complex. Electrophoresis 24: 2740–2748. [DOI] [PubMed] [Google Scholar]
- Dupont, B. and Ceppellini, R. 1989. Immunobiology of HLA. Springer-Verlag, New York.
- Durbin, R., and Thierry-Mieg, J. 1994. The ACeDB Genome Database. In Computational methods in genome research. (ed. S. Suhai), pp. 45–55. Plenum Press, New York.
- Ehlers, A., Beck, S., Forbes, S.A., Trowsdale, J., Volz, A., Younger, R., and Ziegler, A. 2000. MHC-linked olfactory receptor loci exhibit polymorphism and contribute to extended HLA/OR-haplotypes. Genome Res. 10: 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Gaudieri, S., Kulski, J.K., Dawkins, R.L., and Gojobori, T. 1999. Extensive nucleotide variability within a 370 kb sequence from the central region of the major histocompatibility complex. Gene 238: 157–161. [DOI] [PubMed] [Google Scholar]
- Geraghty, D.E., Daza, R., Williams, L.M., Vu, Q., and Ishitani, A. 2002. Genetics of the immune response: Identifying immune variation within the MHC and throughout the genome. Immunol. Rev. 190: 69–85. [DOI] [PubMed] [Google Scholar]
- Hall, F.C. and Bowness, P. 1996. HLA and disease: From molecular function to disease association? In HLA and MHC: Genes, molecules and function (eds. M.J. Browning and A.J. McMichael), pp. 353–381. BIOS Scientific Publishers Ltd, Oxford.
- Halushka, M.K., Fan, J.B., Bentley, K., Hsie, L., Shen, N., Weder, A., Cooper, R., Lipshutz, R., and Chakravarti, A. 1999. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 22: 239–247. [DOI] [PubMed] [Google Scholar]
- Horton, R., Niblett, D., Milne, S., Palmer, S., Tubby, B., Trowsdale, J., and Beck, S. 1998. Large-scale sequence comparisons reveal unusually high levels of variation in the HLA-DQB1 locus in the class II region of the human MHC. J. Mol. Biol. 282: 71–97. [DOI] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The Ensembl genome database project. Nucleic Acids Res. 30: 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A.L. and Nei, M. 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335: 167–170. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L. and Nei, M. 1989. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc. Natl. Acad. Sci. 86: 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A.J., Kauppi, L., and Neumann, R. 2001. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29: 217–222. [DOI] [PubMed] [Google Scholar]
- Johnson, G.C., Esposito, L., Barratt, B.J., Smith, A.N., Heward, J., Di Genova, G., Ueda, H., Cordell, H.J., Eaves, I.A., Dudbridge, F., et al. 2001. Haplotype tagging for the identification of common disease genes. Nat. Genet. 29: 233–237. [DOI] [PubMed] [Google Scholar]
- Kulski, J.K. and Dawkins, R.L. 1999. The P5 multicopy gene family in the MHC is related in sequence to human endogenous retroviruses HERV-L and HERV-16. Immunogenetics 49: 404–412. [DOI] [PubMed] [Google Scholar]
- Li, W.H. and Sadler, L.A. 1991. Low nucleotide diversity in man. Genetics 129: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, S.G.E., Parham, P., and Barber, L.D. 2000. The HLA factsbook. Academic Press, San Diego, CA.
- Maynard-Smith, J. and Haigh, J. 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- McMurray, A.A., Sulston, J.E., and Quail, M.A. 1998. Short-insert libraries as a method of problem solving in genome sequencing. Genome Res. 8: 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, J.D., Marra, M., Hillier, L., Waterston, R.H., Chinwalla, A., Wallis, J., Sekhon, M., Wylie, K., Mardis, E.R., Wilson, R.K., et al. 2001. A physical map of the human genome. Nature 409: 934–941. [DOI] [PubMed] [Google Scholar]
- The MHC Sequencing Consortium 1999. Complete sequence and gene map of a human major histocompatibility complex. Nature 401: 921–923. [DOI] [PubMed] [Google Scholar]
- Mungall, A.J., Palmer, S.A., Sims, S.K., Edwards, C.A., Ashurst, J.L., Wilming, L., Jones, M.C., Horton, R., Hunt, S.E., Scott, C.E., et al. 2003. The DNA sequence and analysis of human chromosome 6. Nature 425: 805–811. [DOI] [PubMed] [Google Scholar]
- Nair, R.P., Stuart, P., Henseler, T., Jenisch, S., Chia, N.V., Westphal, E., Schork, N.J., Kim, J., Lim, H.W., Christophers, E., et al. 2000. Localization of psoriasis-susceptibility locus PSORS1 to a 60-kb interval telomeric to HLA-C. Am. J. Hum. Genet. 66: 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, Z., Cox, A.J., and Mullikin, J.C. 2001. SSAHA: A fast search method for large DNA databases. Genome Res. 11: 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, A., Tamiya, G., Tomizawa, M., Ota, M., Katsuyama, Y., Makino, S., Shiina, T., Yoshitome, M., Iizuka, M., Sasao, Y., et al. 1999. Association analysis using refined microsatellite markers localizes a susceptibility locus for psoriasis vulgaris within a 111 kb segment telomeric to the HLA-C gene. Hum. Mol. Genet. 8: 2165–2170. [DOI] [PubMed] [Google Scholar]
- Osoegawa, K., de Jong, P.J., Frengen, E., and Ioannou, P.A. 1999. In Current protocols in human genetics (eds. N.C. Dracopoli et al.), pp. 5.15.11–15.15.33. John Wiley, New York. [DOI] [PubMed]
- Price, P., Witt, C., Allcock, R., Sayer, D., Garlepp, M., Kok, C.C., French, M., Mallal, S., and Christiansen, F. 1999. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol. Rev. 167: 257–274. [DOI] [PubMed] [Google Scholar]
- Rosenblum, B.B., Lee, L.G., Spurgeon, S.L., Khan, S.H., Menchen, S.M., Heiner, C.R., and Chen, S.M. 1997. New dye-labeled terminators for improved DNA sequencing patterns. Nucleic Acids Res. 25: 4500–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, M.T., LaBrie, S., McPherson, J.D., and Stanton, V.P. 1999. Screening large-insert libraries by hybridization. In Current protocols in human genetics (eds. N.C. Dracopoli et al.), pp. 5.6.1–5.6.5. Wiley, New York.
- Sachidanandam, R., Weissman, D., Schmidt, S.C., Kakol, J.M., Stein, L.D., Marth, G., Sherry, S., Mullikin, J.C., Mortimore, B.J., Willey, D.L., et al. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928–933. [DOI] [PubMed] [Google Scholar]
- Sanger, F., Nicklen, S., and Coulson, A.R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 74: 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L., Wu, L.C., Sanlioglu, S., Chen, R., Mendoza, A.R., Dangel, A.W., Carroll, M.C., Zipf, W.B., and Yu, C.Y. 1994. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon–intron structure, composite retroposon, and breakpoint of gene duplication. J. Biol. Chem. 269: 8466–8476. [PubMed] [Google Scholar]
- Smith, T.F. and Waterman, M.S. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147: 195–197. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, A., and French, L. 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 10: 1772–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E.L. and Durbin, R. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: GC1–GC10. [DOI] [PubMed] [Google Scholar]
- Sulston, J., Mallett, F., Durbin, R., and Horsnell, T. 1989. Image analysis of restriction enzyme fingerprint autoradiograms. Comput. Appl. Biosci. 5: 101–106. [DOI] [PubMed] [Google Scholar]
- Ueda, H., Howson, J.M., Esposito, L., Heward, J., Snook, H., Chamberlain, G., Rainbow, D.B., Hunter, K.M., Smith, A.N., Di Genova, G., et al. 2003. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423: 506–511. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
- Walsh, E.C., Mather, K.A., Schaffner, S.F., Farwell, L., Daly, M.J., Patterson, N., Cullen, M., Carrington, M., Bugawan, T.L., Erlich, H., et al. 2003. An integrated haplotype map of the human major histocompatibility complex. Am. J. Hum. Genet. 73: 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D.G., Fan, J.B., Siao, C.J., Berno, A., Young, P., Sapolsky, R., Ghandour, G., Perkins, N., Winchester, E., Spencer, J., et al. 1998. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280: 1077–1082. [DOI] [PubMed] [Google Scholar]
- Warrens, A. and Lechler, R. 1999. HLA in health and disease. Academic Press, San Diego, CA.
- Wu, I. and Moses, M.A. 2001. Cloning of a cDNA encoding an isoform of human protein phosphatase inhibitor 2 from vascularized breast tumor. DNA Seq. 11: 515–518. [DOI] [PubMed] [Google Scholar]
- Xie, T., Rowen, L., Aguado, B., Ahearn, M.E., Madan, A., Qin, S., Campbell, R.D., and Hood, L. 2003. Analysis of the gene-dense major histocompatibility complex class III region and its comparison to mouse. Genome Res. 13: 2621–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bacpac.chori.org/; BACPAC resources center home page.
- http://genome.wustl.edu/gsc/Overview/finrules/hgfinrules.html; Human Genome finishing rules at the Genome Sequencing Center, Washington University Medical School.
- http://genome.wustl.edu/tools/?overgo=1; Overgo Maker script at the Genome Sequencing Center, Washington University Medical School.
- http://repeatmasker.genome.washington.edu/; The RepeatMasker server, University of Washington.
- http://vega.sanger.ac.uk/; Vertebrate Genome Annotation (VEGA) database browser.
- http://www.ebi.ac.uk/imgt/hla/; IMGT/HLA Sequence Database.
- http://www.genome.arizona.edu/fpc/; FPC at the Arizona Genomics Institute.
- http://www.ncbi.nlm.nih.gov/SNP/; dbSNP home page.
- http://www.phrap.org/; The Genome Software Development page.
- http://www.sanger.ac.uk/HGP/Chr6/MHC/; The Sanger Institute: The MHC Haplotype Project.
- http://www.sanger.ac.uk/HGP/havana/hawk.shtml; The Sanger Institute Human Annotation Workshops page.
- http://www.sanger.ac.uk/Software/Image/; The Sanger Institute: Informatics software: Image.
- http://www.sanger.ac.uk/Software/sequencing/; The Sanger Institute: Production Software.