DNA replication remains unfinished in many Drosophila polyploid cells, which harbor disproportionately fewer copies of late-replicating chromosomal regions. Using NextGen sequencing of DNA from giant polytene cells of the larval salivary gland, Yarosh and Spradling show that sporadic, incomplete replication during the endocycle S phase alters the Drosophila genome at thousands of sites that differ in every cell; similar events occur in the ovary. The authors propose that the extensive somatic DNA instability described here underlies position effect variegation and molds the structure of polytene chromosomes.
Keywords: polyploid, polytene, underreplication, genome instability, DNA replication
Abstract
DNA replication remains unfinished in many Drosophila polyploid cells, which harbor disproportionately fewer copies of late-replicating chromosomal regions. By analyzing paired-end high-throughput sequence data from polytene larval salivary gland cells, we define 112 underreplicated (UR) euchromatic regions 60–480 kb in size. To determine the effects of underreplication on genome integrity, we analyzed anomalous read pairs and breakpoint reads throughout the euchromatic genome. Each UR euchromatic region contains many different deletions 10–500 kb in size, while very few deletions are present in fully replicated chromosome regions or UR zones from embryo DNA. Thus, during endocycles, stalled forks within UR regions break and undergo local repair instead of remaining stable and generating nested forks. As a result, each salivary gland cell contains hundreds of unique deletions that account for their copy number reductions. Similar UR regions and deletions were observed in ovarian DNA, suggesting that incomplete replication, fork breakage, and repair occur widely in polytene cells. UR regions are enriched in genes encoding immunoglobulin superfamily proteins and contain many neurally expressed and homeotic genes. We suggest that the extensive somatic DNA instability described here underlies position effect variegation, molds the structure of polytene chromosomes, and should be investigated for possible functions.
The idea that metazoan tissue cells contain identical genomes has long served as a convenient fiction appropriately termed “the dogma of DNA constancy” (Gilbert 2013). In reality, despite highly faithful polymerases and repair systems, all organisms begin to sporadically accumulate DNA sequence alterations at low levels beginning with the first embryonic divisions (Kazazian 2011; Reizel et al. 2012; Grandi and An 2013). If replication is stressed (Lambert and Carr 2012) or the cell cycle is altered (Fox et al. 2010), greater levels of DNA changes may occur. Although well documented, these genome alterations have no known functional importance and are thought to be neutral or deleterious. At the other end of the spectrum, in relatively few organisms and cells, somatically programmed genomic changes generate useful differences. Eggshell genes are specifically amplified (Calvi and Spradling 1999), antibody genes are productively rearranged (Alt et al. 2013), and whole-ciliate genomes are re-engineered (Chalker and Yao 2011).
The polytene cells of Dipterans such as Drosophila represent an intermediate case. During the growth of such cells via as many as 10 consecutive endocycles (cell cycles without cytokinesis), most euchromatic chromosome regions are fully replicated, but pericentromeric genomic regions rich in satellite DNA sequences are not (Gall et al. 1971). In the best-studied system, the Drosophila larval salivary gland (Fig. 1A), late-replicating euchromatic regions (“intercalary heterochromatin”) also underreplicate to varying degrees (for review, see Spradling and Orr-Weaver 1987; Belyaeva et al. 2008). Thirty to 52 underreplicated (UR) regions 90–570 kb in length have been precisely mapped using DNA arrays (Belyakin et al. 2005; Nordman et al. 2011; Sher et al. 2012). These UR zones correspond closely to regions of repressive chromatin, sparse replication origins, and mostly silent genes (Belyakin et al. 2005; Pindyurin et al. 2007; Filion et al. 2010; Nordman et al. 2011; Belyaeva et al. 2012; Sher et al. 2012; Maksimov et al. 2013). The repressive chromatin state and late replication timing of UR regions are thought to be responsible for their susceptibility to incomplete replication.
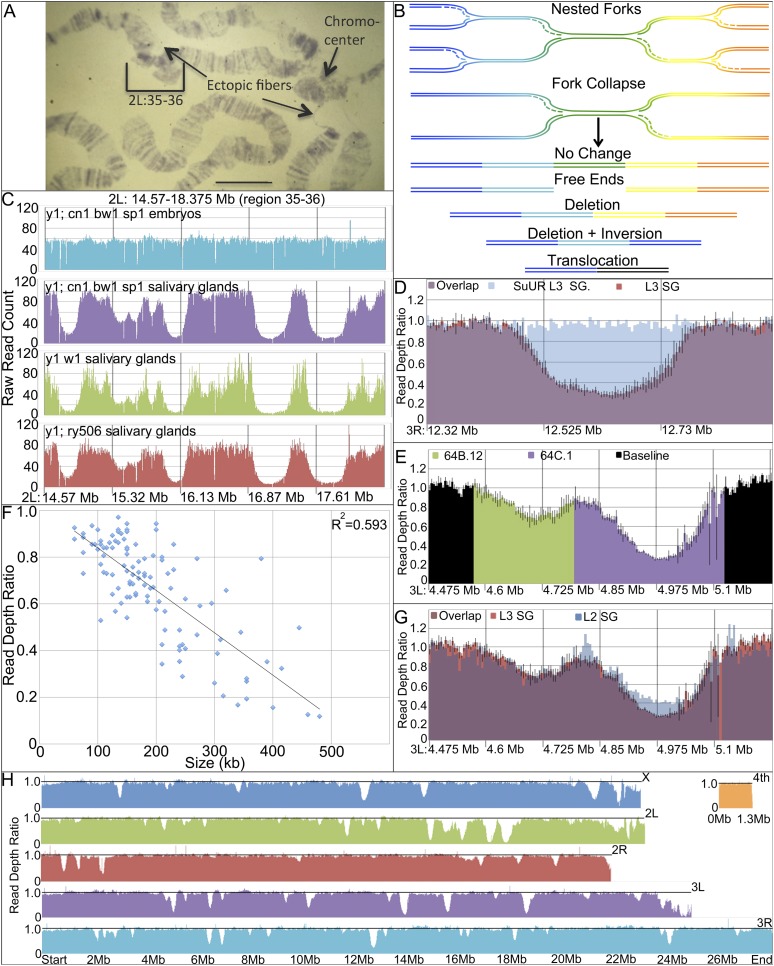
Figure 1.
Mapping underreplication in L3 salivary gland DNA by sequencing. (A) Drosophila polytene salivary gland chromosomes showing banded euchromatic arms (bracket; regions 2L: 35–36 = 14.5–18.0 Mb), ectopic fibers (arrows), and chromocenter (shorter arrow). (B) Models of underreplication. (Top) Stalled forks may be stable, forming inverse “nested forks”. Alternatively, stalled forks may collapse and undergo repair (arrow), leading to novel DNA junctions and genomic alterations. (C) Read counts in 2L: 14.5–18.0 Mb, the same region shown in brackets in A. The read depth is uniform in embryo DNA, whereas multiple UR regions are seen in salivary gland DNA from three strains. (D) UR89E.1 is underreplicated in wild-type L3 salivary gland (SG); average SG/embryo read ratio (orange and purple) ± SD (black bars; N = 3). UR89E.1 is fully replicated in SuUR mutant SG. SG/embryo read ratio (blue and purple). (E) Partitioning example: adjacent UR regions 64B.12 (green) and 64C.1 (purple). Average SG/embryo read ratio ± SD (black bars; N = 3). (F) Underreplication (read depth ratio) is proportional to UR size (in kilobases). (G) Underreplication in UR64B.12 and UR 64C.1 in L2 (blue and purple) and L3 salivary glands (orange and purple). Underreplication increases between L2 and L3 in UR64C.1 but not in UR64B.12. (H) Average SG/embryo read ratio is plotted throughout the Drosophila genome to summarize distribution of UR regions.
The biological significance of underreplication has remained unclear. Most UR regions are the same in polytene fat body, midgut, and salivary gland tissues, but a few show tissue specificity suggestive of a developmental function; moreover, genes in UR regions in fat body are also more frequently expressed (Nordman et al. 2011). Genetic evidence suggests that any such function is nonessential, however. A specific gene, suppressor of underreplication (SuUR), encoding a novel protein, is required for differential replication in euchromatin, but mutants are viable (Belyaeva et al. 1998). SuUR is found within many UR regions (Pindyurin et al. 2007; Nordman et al. 2011; Sher et al. 2012) but is also distributed widely elsewhere in Drosophila chromatin (Filion et al. 2010; Maksimov et al. 2013). SuUR is proposed to slow the progress of replication forks preventing S-phase completion in susceptible regions (Sher et al. 2012; Kolesnikova et al. 2013). Failure to complete replication might cause a mitotically proliferating cell to undergo apoptosis; however, at least some endocycling cells down-regulate the normal apoptotic response to unrepaired DNA damage (Mehrotra et al. 2008).
A better understanding of the molecular consequences of underreplication would likely help reveal its significance. If stalled replication forks remain stable, UR domains would contain nested replication forks directed toward their centers (see Fig. 1B; Laird 1980; Sher et al. 2012). In contrast, if forks undergo breakage, then free ends would be produced, which, if repaired, would cause deletions and DNA rearrangements (Spradling 1993; Leach et al. 2000; Andreyeva et al. 2008). Previous searches failed to detect accumulated replication forks in an UR region (Glaser et al. 1992). Moreover, novel DNA bands were observed in Southern blots of DNA from polytene tissues, consistent with DNA breakage (Glaser et al. 1992, 1997; Spradling 1993; Leach et al. 2000). Distinctive features of polytene chromosome structure, including the mesh-like chromocenter and ectopic fibers such as those consistently observed in polytene region 35–36 (Fig. 1A), might also be explained by high levels of breakage and repair. The genetic phenomenon of position effect variegation (PEV) has also been ascribed to DNA alterations (Karpen and Spradling 1990). However, most investigators have rejected the idea of somatic DNA instability (Ahmad and Golic 1996).
Here we analyze polytene DNA using high-throughput sequencing and show that DNA alterations are generated at many sites throughout the genomes of salivary gland and ovarian cells. DNA deletions 10–500 kb in size are found throughout 112 UR zones, comprising 19% of salivary gland euchromatin, but are rare within fully replicated regions or the corresponding regions of early embryonic diploid cells. Thus, during polytenization, unfinished replication forks break and efficiently rejoin to nearby free ends. An even higher level of underreplication and deletion formation likely takes place in heterochromatic regions whose repetitive sequences prevented detailed analysis using our methods. Our results show that somatic DNA instability is a widespread feature of polyploid Drosophila cells. The significance of somatic DNA alterations for chromosome structure, PEV, and developmental function deserves further study.
Results
Deep sequence analysis of underreplication
DNA from early Drosophila embryos, whose cells are predominantly diploid, and third instar larval salivary glands, which are composed mostly of highly polyploid cells, was prepared in order to study underreplication by deep sequencing (Supplemental Table S1). The y; cn bw sp reference strain, used to determine the Drosophila genome sequence, was employed in order to minimize alignment ambiguities; the y w and y; ry[506] strains were also used. Each DNA preparation was sheared, and libraries were prepared and subjected to paired-end sequencing with 100-base-pair (bp) reads on an Ilumina HiSeq2000. Sequences were aligned to the Drosophila genome R5.33 using ELAND and BWA software (see the Materials and Methods).
We tested the utility of sequencing for analyzing changes in genomic copy number by examining the behavior of heterochromatic sequences. The severe underreplication of heterochromatin in salivary gland DNA was evident from the fraction of reads that aligned to heterochromatic versus euchromatic zones of the genome. In embryo DNA, 35.6% of read pairs mapped to heterochromatic sequence contigs, whereas only 2.2% of pairs from the salivary gland aligned to heterochromatin (see Supplemental Table S2). Raw sequencing reads were queried to estimate sequence underreplication “digitally” based on read frequencies and were compared with previous “analog” assessments based on nucleic acid hybridization (Supplemental Table S3). Generally, 0.15%–1.3% of reads from embryo DNA but only 0.005%–0.05% of reads from salivary gland DNA were homologous to individual satellite DNAs. Thus, satellite DNAs appear to be UR ∼30-fold (about five rounds of replication) during salivary gland development, a somewhat smaller degree of underreplication than previously reported in Drosophila melanogaster (Rudkin 1969) or Drosophila virilis (Gall et al. 1971). The read frequency of rDNA sequences in the salivary gland averaged 21% ± 1.0% that of the embryo (N = 3) (Supplemental Table S3), consistent with the fourfold underrepresentation previously reported (Spear and Gall 1973). 5S rDNA, which is encoded in a separate locus, replicated fully as expected (Hammond and Laird 1985).
More than 100 chromosome regions underreplicate in larval salivary glands
Plotting the frequency of reads along the euchromatic Drosophila genome sequence potentially provides a highly sensitive measure of replication uniformity. When DNA was sheared but not narrowly sized prior to library construction, read frequency from embryo DNA was highly uniform along the five major chromosome arms (Fig. 1C; see the Materials and Methods). In contrast, plotting the read frequency in salivary gland DNA revealed many chromosome regions in which read frequency declines smoothly over a distance of 50–100 kb and then increases back to the genome average in a strain-independent manner (Fig. 1C). The most strongly affected zones correspond to the major UR regions mapped previously (Belyakin et al. 2005; Sher et al. 2012), such as the UR zones in chromosome 2L regions 35 and 36 (Fig. 1C).
UR regions were characterized more accurately by averaging reads in 5-kb windows across the genome and normalizing salivary gland reads to embryo reads in each window to minimize perturbations caused by the presence of repetitive DNA. Normalized 5-kb read values were calculated based on three separate experiments, each involving one preparation of embryo (RefEmb1-3) and salivary gland (RefSG1-3) DNA from the reference strain that was analyzed separately. The average ratios and standard deviations were plotted to determine the replication profiles (Fig. 1D,E). This approach revealed that more euchromatic UR regions exist than described previously but that most of them show low levels of underreplication. Overall, we defined 112 euchromatic regions 60–480 kb in size that were consistently UR in salivary gland DNA (Table 1). Twenty-five regions were between twofold and 8.46-fold UR, while the remaining 87 zones were reduced 1.06-fold to 1.99-fold. The level of underreplication correlated with the size of the region (Fig. 1F). Together, the UR domains account for 21.76 Mb/115.7 Mb = 19% of the euchromatic genome.
Table 1.
UR region properties
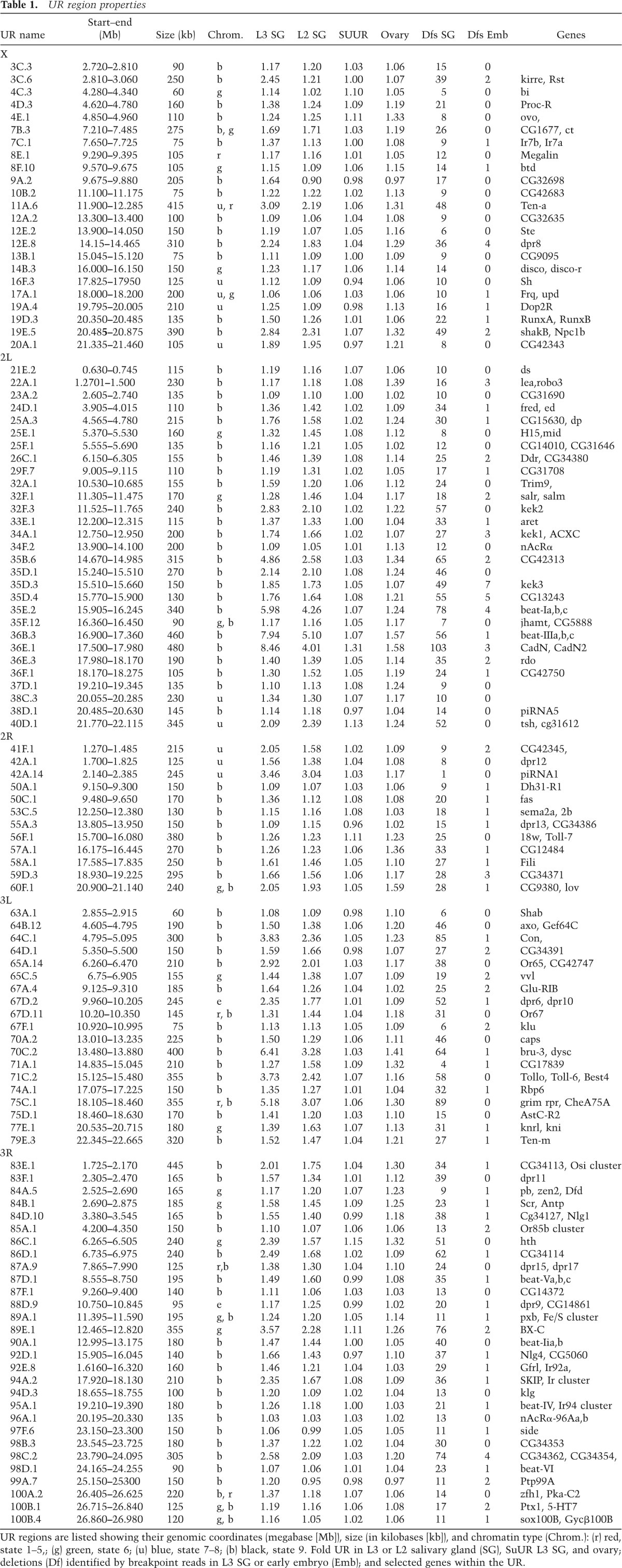
Previous studies have shown that euchromatic underreplication is greatly reduced or absent in SuUR mutant salivary glands (Belyaeva et al. 1998; Sher et al. 2012). When salivary gland and embryo DNA from SuUR−/− animals was examined, nearly all of the UR regions, including those with low UR values, were greatly attenuated, further supporting their validity (Fig. 1D; Table 1). We also found (Table 1; Supplemental Fig. S1), as previously reported for strong UR regions (Sher et al. 2012), that virtually all of the UR regions corresponded closely to domains of repressive chromatin as defined in genomic studies (Karchenko et al. 2012). To investigate when underreplication occurs during development, we analyzed second instar larval (L2) salivary glands, which have completed about seven endocycles. Underreplication in most regions was nearly complete by the second instar (Fig. 1G; Table 1). However, the most strongly UR regions were exceptional; in the second instar, these regions showed less underreplication than in mature third instar (L3) glands. Rather than supporting developmental regulation, these are the results expected if replication failure has a constant probability characteristic of each UR region during each endocycle.
Replication appeared to be uniform across the genome, except within the 112 UR regions (Fig. 1H). The measured copy number values indicate that even within UR regions, complete replication is the norm. In only a few regions, such as UR36B.3, UR36E.1, and UR70C.2, is the number of copies reduced more than twofold over the course of 10 doublings. Thus, copy number changes during salivary gland development arise stochastically in multiple regions by relatively low absolute levels of incomplete replication.
Ovary DNA also contains UR regions
Many larval and adult Drosophila tissues in addition to the salivary gland are made up predominantly of polyploid cells that contain polytene chromosomes and underreplicate heterochromatin (Ashburner 1970; Spradling and Orr-Weaver 1987). Genomic analyses have shown that at least two such tissues, larval fat body and larval midgut, underreplicate most of the same euchromatic regions as the salivary gland (Nordman et al. 2011). We sequenced DNA prepared from ovaries, which derives predominantly from polytene nurse and follicle cells, to examine the euchromatic regions that underreplicate in adult polytene cells. Most UR regions that underreplicate strongly in larval salivary glands duplicated incompletely during ovary development, but the level of reduction in ovary DNA was much less, in part due to the presence of diploid ovarian cells (Fig. 2A; Table 1). Thus, the UR regions defined for the salivary gland are likely to be similar in a wide range of polyploid cells, including ovarian polyploid cells.
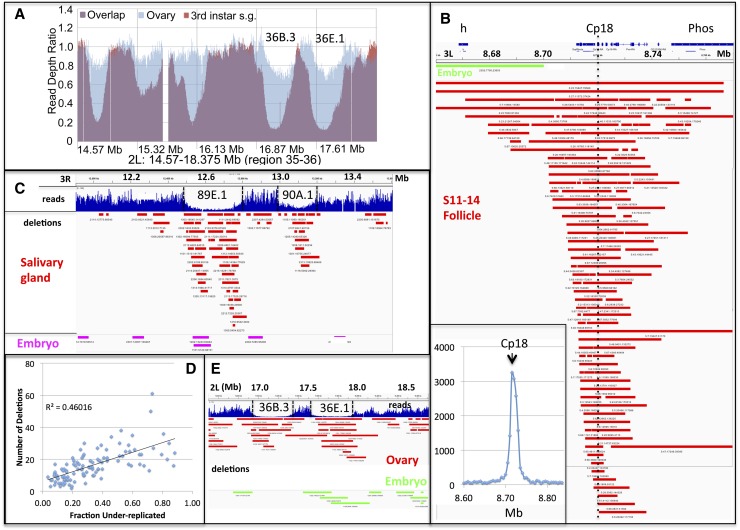
Figure 2.
Replication fork instability leads to DNA deletions detected using anomalous pairs. (A) Ovary DNA (purple and blue) contains the same major UR regions as L3 salivary gland DNA (purple and orange). (Ovary or salivary gland)/embryo read ratio is plotted in region 35–36, and UR36B.3 and UR36E.1 are indicated. (B) Replication forks break and generate deletions (bars) during chorion gene amplification. Deletions defined by anomalous pairs are plotted in the vicinity of the 66D chorion gene cluster at 3L: 8.66–8.76 Mb and center around the major amplification origins located near Cp18 (dashed line). (Red bars) Follicle cell deletions,; (green bars) embryo deletions; (blue bars at top) genes. (Inset) Reads in the 66D region are plotted in 10-kb windows to reveal 50-fold amplification of this region. (C) Deletions 10–100 kb in length defined by anomalous pairs are more abundant in salivary gland DNA (red) than in embryo DNA (purple) and cluster at UR regions. Reads (blue) are plotted in 3R: 11.9–13.6 Mb, and URs 89E.1 and 90A.1 are indicated (dashed lines). (D) The number of deletions within 111 UR regions is proportional to the degree of underreplication. (E) Deletions (10–500 kb) defined by anomalous pairs are more abundant in ovary DNA (red) than in embryo (green) DNA and cluster in some UR regions. Reads in 2L: 16.5–18.7 Mb are shown, and URs 36B.3 and 36E.1 are indicated.
Replication forks are unstable during chorion gene amplification in ovarian follicle cells
In order to distinguish whether polytene DNA contains stalled forks or has undergone breakage/repair, methods are needed that can identify rare molecules with novel junctions. To assess the ability of paired-end sequencing to detect rare products of replication fork instability, we initially investigated dense zones of replication forks that are generated during chorion gene amplification. Amplification generates a high density of replication forks because multiple rounds of replication initiate at just a few genomic locations during a final S-like phase in stage 10B of oogenesis (Calvi and Spradling 1999). Subsequently, during stages 11–14, these forks continue to elongate, moving away from the initiation region on each side (Claycomb et al. 2002). If replication forks break and are repaired in vivo, novel junctions will be generated that could be detected by paired-end sequencing.
We sequenced DNA from stage 11–14 follicles and looked for anomalous read pairs; i.e., those whose component reads align at sites incompatible with normal sheared DNA. The read profile revealed the dramatic nature of amplification. For example, in the largest amplified domain on chromosome 3L, comprising ∼100 kb centered on Cp18 and three other chorion genes, the read profile increased in exactly the location expected from previous studies, with a peak value 47 times higher than the average in unamplified regions (Fig. 2B, inset). More than 70 anomalous read pairs were identified in the amplified region, all indicative of deletions mostly centered around Cp18, whereas only one deletion was found in embryo DNA (Fig. 2B). Similar deletions (but less enriched) were observed around Cp18 using the ovary DNA sample (Supplemental Fig. S2). We calculated from these results that ∼2% of the amplified strands break and undergo repair to form deletions during amplification (Materials and Methods) and conclude that these rare events can be detected by paired-end sequencing. The major origins used during amplification are located near Cp18, so the presence of small deletions flanking this region suggests that some forks stall shortly after initiation, break, and are ligated to other broken ends. These DNA alterations were not observed in earlier studies of chorion gene amplification, which illustrates the difficulty of detecting rare DNA derivatives that differ from each other.
Anomalous read pairs identify a class of deletions enriched in salivary gland DNA
With this encouragement, we took the same approach to look for DNA alterations generated during underreplication in salivary glands. Following alignment of paired-end sequences from salivary gland DNA to the Drosophila genome, we first analyzed anomalous pairs. Ideally, these pairs come from reads in which the unsequenced center of the fragment contains a deletion breakpoint. However, a background of misleading anomalous read pairs will also be generated when reads are misaligned to the genome due to the presence of local repeats such as transposons or duplicated genes. Additionally, hybrid DNAs generated by the ligation of unrelated fragments during library preparation will also produce misleading anomalous pairs. Random ligation will generate “translocations” and large “deletions” preferentially, since the chance that two randomly joined fragments come from nearly the same chromosome region is relatively low. The most important test of whether predicted DNA alterations truly result from DNA underreplication rather than methodological artifacts is that they should be enriched in salivary gland DNA compared with embryo DNA and in UR regions compared with normal regions. We initially focused on deletions in the size range from 10 to 500 kb, since 10 kb is large enough to exclude transposon polymorphisms, and 500 kb is the upper limit of the measured UR size.
We identified all anomalous read pairs predicting 10- to 500-kb deletions among all salivary gland and embryo read pairs from the three replicate experiments using the reference strain. Pairs in which the sequence quality of one of the reads was questionable were excluded. We also removed read pairs with identical reads, since they are a product of PCR amplification, and read pairs aligned within heterochromatin, including repetitive, unmapped portions of the genome (chrU and chrUextra). It was important to align against these sequences initially, however, to prevent matching reads from being force-aligned elsewhere in the genome.
The remaining read pairs were examined to determine whether they might be related to underreplication. Salivary gland DNA contained more than three times as many deletion read pairs as embryo DNA within euchromatin as a whole (Table 2). However, when the location of the salivary gland deletions was plotted, they showed only a slight specificity for UR regions (data not shown). Because large deletions are more likely to be caused by random ligation, we tried plotting only those deletions with predicted sizes between 10 kb and 100 kb. Anomalous pairs in this size regime were fourfold enriched in salivary gland versus embryo DNA (Table 2), and the deletions that they specify strongly clustered within UR regions (Fig. 2C). Significantly, the number of excess deletions in salivary gland DNA compared with embryo DNA in the UR regions correlated with the degree of underreplication (Fig. 2D). We concluded that 10- to 100-kb deletions are generated by DNA underreplication.
Table 2.
Identification of deletions by paired-end sequencing
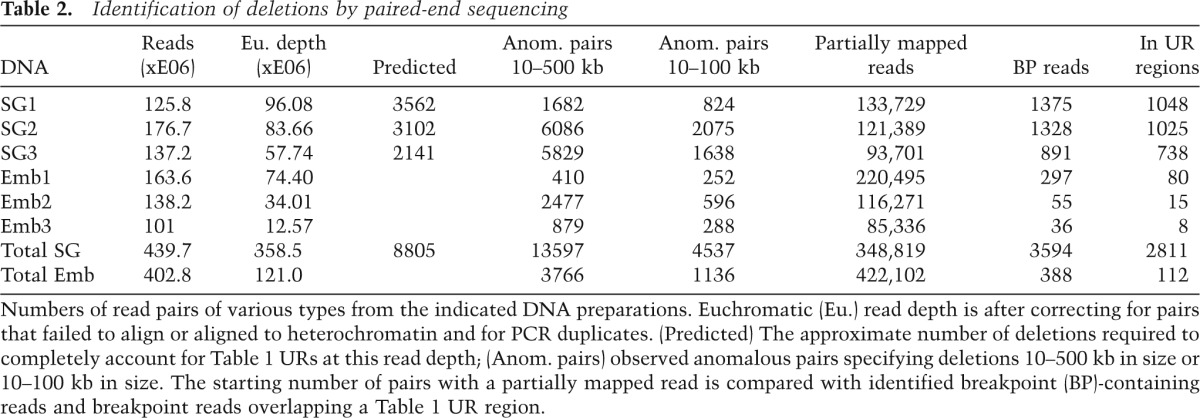
Anomalous pairs from ovary DNA also predicted an excess of deletions (1629 vs. 410) compared with those seen in embryo DNA (Emb1). However, the distribution of ovarian deletions was less specific for UR regions than in the case of the salivary gland. Strong UR regions such as 36B.3 and 36E.1 that underreplicate in the ovary (Table 1) contained more deletions in ovary DNA than in embryo DNA (Fig. 2E). However, the overall level of enrichment in ovary URs compared with non-UR regions was only twofold. It would be worthwhile to analyze nurse cells separately to see whether they underreplicate in a manner different from that of somatic cells. As expected, a peak of deletions was seen around Cp18 (Supplemental Fig. S2), documenting that biologically significant deletions were being observed.
These results provide strong evidence that underreplication generates somatic deletions in UR regions due to fork breakage and repair. However, the deletions defined by anomalous read pairs did not appear to reveal the entire distribution of rearrangements associated with underreplication. Large deletions showed little specificity, and the number of such deletions varied significantly between experiments, suggesting that many arose during library construction (Table 2). Consequently, while enough predicted smaller deletions were present in salivary gland DNA sequences to establish a correlation with UR regions, we sought to identify a more representative and highly enriched collection of deletions associated with underreplication.
Analyzing rearrangement breakpoints
Identifying junction reads—i.e., individual 100-bp reads that transition across a sequence gap—appeared to be a way to increase specificity. DNAs generated by random ligation should be larger, on average, than individual DNAs, and end sequence reads might only rarely be long enough to cross artificial junctions. Another reason for analyzing junction sequences is that they potentially provide information on the mechanism of break repair. However, most alignment programs such as Eland and BWA do not efficiently identify junction reads. Such reads end up as unaligned or partially aligned depending on the location of the junction and the parameters of the alignment algorithm.
Empirically, we found that carrying out BLAT or BLAST searches with unaligned or partially aligned reads frequently revealed new alignment information, including the identity of junction reads. Consequently, we scrutinized all high-quality unmapped reads from read pairs with only one aligned read as well as all reads in which at least 15 bp were unmatched in euchromatic genomic regions (Table 2). By realigning these 770,921 reads to the genome using BLAT, we identified 3594 reads from salivary gland DNA that spanned the breakpoint of a deletion 10–500 kb in size with 99%–100% sequence matches on both ends. Identical treatment of embryo DNA reads yielded only 388 potential breakpoint reads.
If the salivary gland deletions defined by junction reads are generated by incomplete replication and represent the molecular mechanism of sequence underrepresentation, then they should be preferentially located within the UR regions. Plotting their position showed that this was indeed the case (Fig. 3A,B). For example, in UR-rich regions 35 and 36 on chromosome 2L, the distribution of deletions precisely mimics the location of UR zones (Fig. 3A). Most deletions are located within a single UR, but in the case of nearby regions, such as UR36B.3 and UR36E.1, at least six deletions span the two regions (Fig. 3A). Equally, precise localization is observed in the Ubx region (UR89E.1) and nearby UR89F.2 (Fig. 3B). In contrast, the deletions identified from embryo DNA were not enriched in UR regions. Those appearing in small clusters were likely to be alignment artifacts associated with regions containing short tandem repeats separated by 10–500 kb, and these deletions were seen in similar numbers in the salivary gland as well.
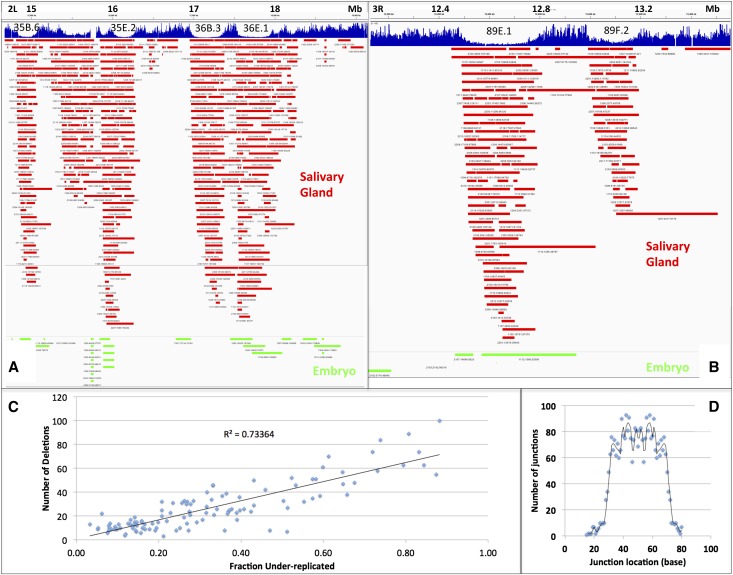
Figure 3.
Salivary gland-specific deletions detected from breakpoint reads recapitulate UR regions. (A) Deletions (10–500 kb) defined by breakpoint reads ([red] salivary gland; [green] embryo) are shown in region 35–36: 2L: 14.6–19.1 Mb. Salivary gland-specific deletions are highly localized to UR regions or span adjacent URs; major URs 35B.6, 35E.2, 36B.3, and 36E.1 are indicated. (B) Same as A but showing region 3R: 11.9–13.6 Mb. The major URs 89E.1, containing the Ultrabithorax complex, and 89F.2, containing beat-IIa,b genes, are shown. (C) The number of deletions within 111 UR regions is highly proportional to their fractional degree of underreplication. (D) Breakpoint location within 3659 salivary gland junction reads. Nearly all breakpoints fall between nucleotides 30 and 70 in the 100-nt reads. Since orientation is arbitrary, both fragments are counted, generating a symmetrical plot.
If we are detecting the deletions that give rise to salivary gland sequence underrepresentation, then every UR should contain an excess of deletions compared with the embryo controls or normally replicated regions. We found this to be the case when we calculated the number of deletions from salivary gland or embryo DNA that lie wholly or partially within each UR region (Table 1). The only exception was UR42B containing the major Drosophila piRNA locus, a highly repetitive region. Moreover, the number of salivary gland deletions in a UR region was strongly related to its level of sequence reduction. For example, in strong UR regions, such a UR89E.1 containing the bithorax complex, 61 deletions in salivary gland DNA recapitulate the read profile, but only one deletion was found in embryo DNA, and it overlapped only partially. The 350 kb just prior to Ubx, which does not underreplicate, contains no salivary gland deletions (Fig. 3B). Similarly, in region 36 of chr2L, there were 55 deletions with UR36B.5 and 65 deletions in UR36C.10 (compared with two in embryo DNA), while only 10 deletions were present in a much larger region, chr2L: 1–1,000,000, which lacks URs. The 33 weakest URs that could be identified based on copy number reduction, which had underreplication values <1.2, each still contained an average of 11.7 deletions in the salivary gland but only 0.63 deletions in the embryo. Decisive evidence that the deletions cause the copy number reductions seen in UR regions was provided by the strong correlation that we observed between deletion number and UR value (Fig. 3C). Overall, 2811 of the 3594 salivary gland deletions (78%) were located within the mapped UR regions. However, many of the remaining salivary gland deletions mapping outside the URs in Table 1 were present in clusters of four to eight deletions within zones of repressive chromatin and probably correspond to additional real UR regions that were too weak to document by copy number reduction.
The breakpoint reads also identified candidate translocation junctions; i.e., reads in which the sequence switched between two different chromosomes. However, most of these joints probably do not correspond to true translocations generated during salivary gland underreplication in vivo. There was no enrichment for such translocations, since a total of 13,379 junctions was identified within euchromatin in the three Ref strain salivary gland experiments, while 14,822 candidates were identified in the corresponding Ref embryo DNAs. Moreover the chromosomal location of the putative translocation pairs appeared to be random (data not shown). The great majority are probably caused by ligation during library preparation. A low level of real translocations generated by the repair of broken forks in UR regions on different chromosomes would have been hidden by this background.
Properties of UR-associated deletions
Analysis of the salivary gland deletion junctions defined by breakpoint analysis revealed additional information. The location of the deletion junction within the 100-bp sequence fell overwhelmingly between positions 30 and 70 (Fig. 3D). Recovery of more asymmetric deletion junction reads using our methods must be much less efficient.
About half of the junctions occur at sites with no nucleotide overlap, while the remaining joints show very limited homology of 1–6 bp and no evidence of a consensus. Only 6% of joints showed homology at the site of joining that was >6 bp. One-thousand-seven-hundred-six of the deletion reads involved no change of chromosome orientation across the breakage site, and these tended to be larger and to span the edges of the UR (Fig. 4A). Such molecules may be generated when two approaching forks on a single DNA molecule stall near each edge of a UR and undergo breakage, and the two free ends join to each other. In contrast, 1887 deletion chromosomes reversed their orientation across the break. Presumably, when more than one stalled fork is present on the same edge of a UR region, a situation expected during later endocycles, forks may break and resolve by ligating to a free end originating from a different fork on the same side, thereby generating a giant inverted repeat or isochromosome (Fig. 4B). Sequences within the UR would be lost as a consequence of such events in addition to those between the breakpoints of this class of deletion. The net result would be to generate strands within a UR as shown (Fig. 4C).
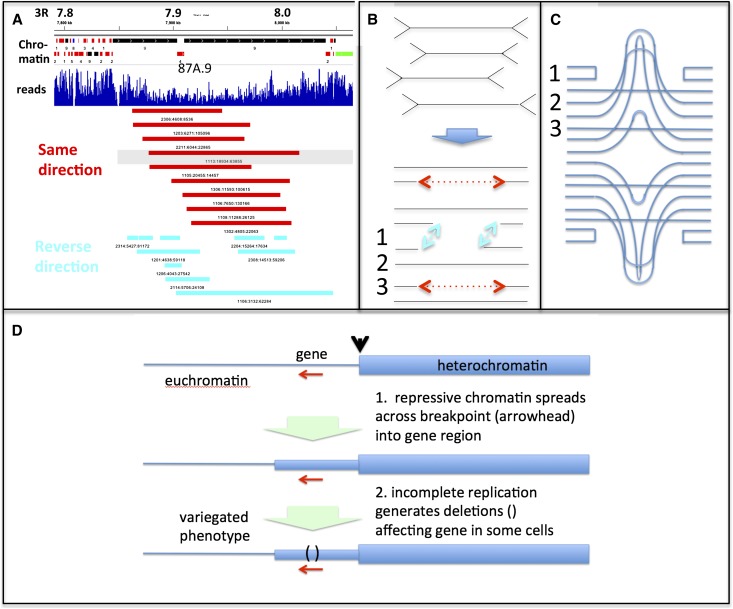
Figure 4.
Structure and significance of polyploid cell somatic DNA instability. (A) Two classes of deletion chromosomes are generated by underreplication. Chromosomes with no orientation change at the deletion (red; “same direction”) tend to span URs, while deletion chromosomes that “reverse direction” (blue) cluster at UR edges. Region 3R: 7.8–8.1 Mb with UR87A.9; chromatin types (see Supplemental Fig. S1) are shown above. (B) Model for two types of deletion-bearing chromosome. Four DNAs with eight stalled forks are shown; following breakage (arrow), they generate four complete and four gapped DNAs. Repair on the same UR side (1; blue arrows) generates an inversion chromosome (see A, blue); repair across the UR (3; red arrows) generates unaltered polarity (see A, red); unbroken strands remain unaltered (2). (C) Model of the DNAs from B in a hypothetical UR: (1) DNAs religated to adjacent copies generating inversion chromosomes, (2) unchanged DNAs, and (3) DNAs religated across the UR to DNA in the same orientation. (D) Model of PEV at a euchromatin–heterochromatin junction. Heterochromatin spreads into adjacent euchromatin, creating a new UR containing a gene (red arrow) and affecting its expression. Variegated expression would result from the different somatic deletions generated in this region during development within individual cells.
UR zones are enriched in immunoglobulin superfamily genes and genes involved in the nervous system
The more complete census of salivary gland UR sites made it possible to search the genome more thoroughly than previously possible for classes of genes that are associated with these regions (Table 1). As previously noted, one subset of URs corresponds to the major Drosophila Hox gene clusters, and we observed ∼10 additional URs containing Polycomb-rich chromatin. Interestingly, we found that URs are also enriched in a class of genes not previously associated with repressive chromatin domains; namely, genes encoding IgG superfamily (IgSF) proteins (Özkan et al. 2013). Among the four classes of IgSF proteins characterized by Özkan et al. (2013), 13 of 14 Beat class, seven of eight Side class, five of 11 DIP class, and nine of 20 Dpr class IgSF genes are in UR regions (Table 1). Since UR regions constitute only 19% of euchromatin, finding 34 of 53 IgSF genes in UR regions is unlikely to occur by chance (P < 10−13; binomial distribution). Many other genes involved in cellular adhesion are also located in UR regions, including proteins involved in IgSF and LRR interactions such as Robo, Robo3, Lea, and Caps as well as CadN, CadN2, Connectin, Fred, Ed, Rst, Kirre, and Snap25. Many genes in UR regions—including many of the Hox, adhesion, and IgSF genes—are unusually large due to the presence of large introns. Among the large genes found in UR regions were many others that, like many IgSF genes, are expressed during neural development (Table 1).
Discussion
Polytene salivary gland cells undergo extensive genomic alterations during development
Our results show that larval salivary gland cells covalently alter their somatic genome structure at hundreds of sites within 112 dispersed euchromatic domains (Table 1). The affected zones, known as UR regions, were first identified because they display a reduced copy number relative to the ploidy of the genome as a whole (Spradling and Orr-Weaver 1987; Belyakin et al. 2005; Pindyurin et al. 2007; Sher et al. 2012). By deep sequencing, we report a more complete census of UR regions, revealing that they encompass 19% of euchromatin and house a substantial fraction of Drosophila genes. In striking contrast to the assumption that UR regions contain only stalled, nested replication forks (Sher et al. 2012), we found that all but one of the 112 UR regions contain a diverse array of DNA deletions at levels sufficient to entirely explain the copy number changes. Similar deletions are seen only at much lower levels, if at all, in UR region DNA from early embryos or at non-UR euchromatic sites within the salivary gland.
The sufficiency of the observed deletions to explain the copy number reductions is based on simple calculations. From three salivary gland sequencing experiments with a total read depth of 237, we observed 3659 deletions defined by junction reads, of which 2811 overlapped defined UR regions. However, the distribution of breakpoints within these reads (Fig. 3D) showed that only breaks within the central 40% of the read had been efficiently recovered. So, a better estimate of the total number of deletions in UR regions would be 2811/0.4 = 7027. Any UR-associated deletions that fall outside the 10- to 500-kb window were also not counted. For comparison, if all copy number variation in the UR regions results from somatic deletions, then 8805 deletions should have been observed (see the Materials and Methods). The close agreement of these numbers indicates that the great majority of sequence underrepresentation results from deletions rather than nested forks or free DNA ends (neither of which will generate novel junctions upon sequencing).
Stalled replication and repair generates UR-associated deletions
A great deal of evidence had accumulated previously that polytene underreplication is caused in some way by replication fork stalling. The most strongly affected regions replicate late in S phase (Belyakin et al. 2005; Pindyurin et al. 2007; Belyaeva et al. 2012), mostly from external origins (Sher et al. 2012), which renders them susceptible to incompletion. Furthermore, UR zones are characterized by repressive chromatin (Belyakin et al. 2005; Pindyurin et al. 2007; Sher et al. 2012; Maksimov et al. 2013), a property that we confirmed for all but one of the 112 UR regions (Table 1). Replication forks probably have difficulty elongating through these regions in polytene cells (Sher et al. 2012), and SuUR binds to late-replicating regions (Makunin et al. 2002; Filion et al. 2010) and may directly contribute to fork slowing (Kolesnikova et al. 2013). We observed that the domain boundaries of many strong UR regions corresponds closely to the junctions of dozens of deletions, suggesting that replication forks frequently stall almost immediately after encountering a UR domain. UR regions are also known to be sites of elevated DNA repair. Salivary gland chromosomes contain elevated amounts of phosphorylated His2Av, and sites enriched in phosphorylated His2Av correspond to many UR regions (Andreyeva et al. 2008). However, earlier studies could not determine whether the repair activity maintained nested fork structures, generated broken DNA ends, or led to novel DNA junctions.
Our experiments demonstrate that DNA replication forks are unstable in polytene cells and can be efficiently repaired. We observed high levels of fork instability during chorion gene amplification that is followed by repair to form deletions of heterogeneous size. Many of the deletions begin close to the site of amplification initiation near Cp18. Overall, ∼2% of all amplified DNA strands contain a deletion in our experiments. In UR regions, damage repair by end joining occurs predominantly within the same UR.
Many studies prior to the advent of high-throughput sequencing support the idea that incomplete replication forks are processed to deletions in UR regions. Nested replication forks from amplified chorion genes were readily observed by electron microscopy of ovarian follicle DNA (Osheim and Miller 1983) but were never detected in salivary gland DNA, either using electron microscopy or by two-dimensional gel analysis of a UR region (Glaser et al. 1992). In UR regions, small restriction fragments undergo detectable changes in abundance but not in size (Spierer and Spierer 1984; Karpen and Spradling 1990). However, by including a UR region on a minichromosome or analyzing large restriction fragments within heterochromatic regions, consistent DNA changes in polytene DNA were observed, and the sizes of the novel fragments were heterogeneous (Spradling 1993; Leach et al. 2000; Andreyeva et al. 2008), as predicted by the results reported here.
DNA alterations probably occur in many polyploid Drosophila cells
While patterns of DNA replication are developmentally regulated (Nordman and Orr-Weaver 2012), some regions of the Drosophila genome in both cultured cells and salivary glands unexpectedly share similarities in replication origins (Belyakin et al. 2005; Nordman et al. 2011; Sher et al. 2012), late replication (Eaton et al. 2011), and heterochromatin (Karchenko et al. 2012). Mostly the same regions underreplicate in polytene larval midgut, fat body, and salivary gland cells (Nordman et al. 2011; Sher et al. 2012), although the level of copy number reduction can vary. Our experiments show that all UR regions identified by Nordman et al. (2011) in three tissues underreplicate at some level in the salivary gland. The UR regions that could be mapped in ovary DNA again matched the strongest of these same regions. This suggests that the program of late replication and underreplication may be less flexible than other aspects of replication programming; for example, early replication that correlates with transcription and differs between cell types. However, early nurse cells do differ from other polyploid cells by fully replicating their heterochromatin (Dej and Spradling 1999).
Stochasticity of underreplication enhances genetic diversity within individual cells
The similarity in underreplication within both the second and third instar salivary glands, except in the most strongly UR regions, suggests that underreplication occurs extensively before the second instar and probably takes place throughout all of the salivary gland endocycles. Indeed, a failure to undergo a full doubling of DNA content during the first endocycle has been documented in many types of polyploid cells (for review, see Spradling and Orr-Weaver 1987). This suggests that each UR has a low intrinsic probability of incomplete replication during every endocycle and that UR values simply represent the effects of this propensity averaged over multiple cells and endocycles. This situation will result in a wide range of deletion abundances within individual polyploid cells and might act to enhance their significance.
Consider a UR in which forks stall and generate a deletion, on average, once every 10 replications. Among the ∼100 main cells of the salivary gland, there will be ∼20 (since each contains two chromosomes) that will generate deletions during the first endocycle. Each new variant will thereafter constitute 25% of the alleles at the locus. Even more cells will generate different deletions during the second endocycle (since there are more replicating normal strands), and each of these will occupy 12.5% of the final chromosomes. A still larger fraction of cells will generate deletions on the third endocycle. Thereafter, with 16 or more strands replicating the region in question, essentially every cell will generate novel derivatives on at least one of their strands, and these new changes will continue to replicate. However, a probability of fork stalling of 0.1 only corresponds to an underreplication value of 2 (Materials and Methods). Even regions with much lower UR values will still generate substantial genetic diversity, and some of the novel products will still be present at a high copy number in a few cells. Thus, for a tissue like the salivary gland that undergoes 10 endocycles, essentially all cells will contain many different rearrangements at widely different ploidy levels in every one of the 122 UR regions. For each region, a variable but significant subset of tissue cells will harbor alterations in a very high copy number.
Previously, regions with underreplication values less than ∼2 were thought to be unimportant, because the only effects of underreplication were assumed to be on gene dosage. Now that we know that underreplication generates genomic novelty and that stochastic replication spreads that novelty among subpopulations of tissue cells at high levels, the true potential of underreplication to generate significant somatic variation can be better appreciated. This predicted cell-to-cell variation has already been observed. The level of underreplication of particular classes of rDNA repeat varied stochastically from cell to cell in the salivary gland (Belikoff and Beckingham 1985). The copy number of the yellow gene when located in a UR region varied widely between individual salivary gland cells when assayed by in situ hybridization, and yellow expression variegated in polytene bristle cells (Karpen and Spradling 1990).
Significance for polytene chromosome structure
Our finding that hundreds or thousands of new junctions are produced in each polytene cell strongly supports the idea that somatic DNA modifications contribute to the characteristics of polytene chromosomes (Ashburner et al. 1970; Spradling et al.1992; Leach et al. 2000; Andreyeva et al. 2008). Cytogenetic regions 35 and 36 on chr2L contain the most frequent and deepest UR regions, and we identified many deletions within and between nearby URs. These polytene regions are most frequently subject to structural disruption and ectopic fiber formation in salivary gland chromosomes (Fig. 1A), suggesting that ectopic fibers result from the mispairing caused by strands of very different length and sequence content. We expect that replication fork stalling, breakage, and repair are also responsible for the copy number reductions in heterochromatic regions. Further improvement in methods, such as long sequence reads, might allow such changes to be mapped, including those that probably underlie chromocenter formation.
Deletion formation may cause variegation
When euchromatic genes are rearranged near centromeric heterochromatin, they usually display the phenomenon of PEV. In recent decades, only epigenetic explanations for PEV have been considered (Ahmad and Golic 1996; for review, see Elgin and Reuter 2013). However, previous evidence suggested that somatic genetic changes related to underreplication contribute to some examples of PEV; for example, when the affected gene is located in a new UR generated by rearrangements such as In(1)sc8 (Karpen and Spradling 1990; Spradling 1993; Glaser et al. 1997; Leach et al. 2000).
We propose a model for PEV that incorporates both heterochromatin spreading and the somatic mutation process described here (Fig. 4D). Following chromosome rearrangements juxtaposing heterochromatin and euchromatin, heterochromatin would invade adjacent euchromatin, as currently envisaged. However, we propose that gene expression variegates within the affected euchromatic region because the new domain of repressive chromatin arrests replication forks, leading to somatic DNA mosaicism for deletions or other rearrangements that alter gene expression. Consistent with this proposal, SuUR mutation suppresses both the PEV and underreplication of rearranged genes in In(1)sc8 and In(1)wm4 (Belyaeva et al. 2003).
Somatic DNA instability may be functional
The nonrandom location of IgSF and other genes in UR zones raises the question of whether underreplication has a biological function. IgSF genes are expressed on cell surfaces and govern cellular interactions that are important during multiple developmental processes, especially in the nervous system (Özkan et al. 2013). Beat proteins are expressed in different subgroups of neural cells (Pipes et al. 2001), and some individual family members have been shown to guide motor axons (Fambrough and Goodman 1996). Combinations of IgSF molecules may affect synaptic adhesion (Yamagata et al. 2003). Additional diversity within Ig proteins is generated by DNA alterations in the vertebrate immune system (Alt et al. 2013), while in Drosophila, the Dscam family of IgSF proteins is extensively diversified by differential splicing (Wojtowicz et al. 2007). Consequently, the location of IgSF genes within UR regions might generate somatic diversity of IgSF copy number, expression, and gene structure by virtue of DNA rearrangements even though our data showed no evidence of sequence- or gene-related specificity. Even immunoglobulin gene rearrangement is highly error-prone, and many rearranged genes are nonfunctional. The generation of useful diversity even by an error-prone process and at a relatively low level might be significant, especially if a cell expressing such a surface protein was subject to selection.
The gene structure of IgSF genes and other adhesion protein genes in UR regions might lend itself to such a purpose. Many of these genes as well as other genes located in URs, such as HOX genes, are characterized by long transcription units with multiple long introns. Large genes would provide a greater cross-section for regionally localized rearrangements to generate fusion genes with altered expression patterns and/or coding capacity. Gene organization, chromatin structure, and replication timing may have been optimized by evolution to generate potentially useful diversity using a semirandom mechanism. Because the late replication program appears to be similar between many cell types, genes might undergo similar rearrangements in multiple tissues, including many in which they are inactive; activity in only one critical tissue and developmental stage might be sufficient for the system to have selective value.
Underreplication may be a widespread feature of polyploid cells
After the present study was submitted for publication, Hannibal et al. (2014) reported that polytene giant trophoblast cells from the mouse placenta contain at least 47 UR regions dispersed throughout the genome. Mouse UR domains are individually much larger than Drosophila URs but share many other key features. These include late replication, an association with repressive chromatin, low gene density, and low gene expression. Like the majority of the domains characterized here, the levels of underreplication are less than twofold. However, somatic deletions were not detected, although the methods employed may not have been as sensitive as those used here. Interestingly, like Drosophila URs, mouse URs are enriched for genes involved in adhesion and neurogenesis.
Significance for somatic mutation in diploid cells
Replication fork arrest, breakage, and repair are not confined to polyploid cells but are basic aspects of cell cycle physiology that normally occur in all cells. Recently, evidence has been found that replication timing, including late replication, strongly affects the accumulation of mutations in diploid cells (for review, see Lambert and Carr 2012). In cancer cells, some of which lack P53 function like the Drosophila salivary gland (Mehrotra et al. 2008), large-scale structural variations are greatly increased in late-replicating genomic regions and preferentially join regions with proximity in the nucleus (De and Michor 2011). The diverse DNA break sites in some cancer rearrangements (Ross et al. 2013) resemble the distributed breakpoints observed at the junctions of strong UR regions in the salivary gland. Consequently, diploid cells may sometimes acquire somatic mutations by replication fork stalling, breakage, and repair, like underreplicating Drosophila cells. Thus, studying insect polytene chromosomes promises to shed new light on general processes important for genome evolution and carcinogenesis.
Materials and methods
Drosophila strains
Strains iso-1: y[1]; Gr22b[1] Gr22d[1] cn[1] CG33964[R4.2] bw[1] sp[1]; LysC[1] MstProx[1] GstD5[1] Rh6[1], y w: y[1]w[1], y ry: y[1]; ry[506], and SuUR−: In(1)scV2, scV2; SuURES were obtained from the Bloomington stock center. Stocks were maintained on standard fly food, which was supplemented with additional yeast beginning 1 wk before tissue collection.
DNA isolation
All samples were prepared independently. Embryos were collected using grape juice agar caps during a 0.5- to 2.5-h collection window and were dechorionated using bleach. Salivary glands were dissected from 300–400 larvae per preparation (late third instar except as noted) in cold Grace’s insect medium (Life Technologies). Adult females were anesthetized using CO2, and ovaries were dissected and late stage follicle were separated using jeweler’s forceps. Tissues were flash-frozen in liquid nitrogen immediately after collection. DNA was purified using a QIAamp DNA minikit (Qiagen) and RNase A (Qiagen) according to the manufacturer’s protocol.
DNA and library preparation
Libraries for paired-end sequencing were prepared using Illumina’s TruSeqDNA sample prep kit LT using the LS and gel-free options. Fragmentation was carried out with a Diagenode Bioruptor sonicator, with the power setting “low” for 15 min (intervals of 30 sec of sonication, 30 sec without sonication over 30 min) at 4°C. One-hundred nanograms of sonicated DNA was used as input. The amplification reaction was altered (while still using kit reagents and cycling conditions) to the following: 3 μL of ligated DNA, 25 μL of PCR master mix, 5 μL of PCR primer cocktail, and 17 μL of resuspension buffer. The size of the resulting DNA was 300–500 bp, including 120 bp of linkers. Most libraries were prepared on different days, but RefSG2, RefSG3, RefEmb2, and RefEmb3 were prepared in the same batch.
Chromatin domain analysis
The chromatin state of UR and surrounding regions was classified based on the nine-state model described in Karchenko et al. (2012) and applied to the S2 cell line. For further simplicity in viewing, similar states were grouped as follows: red (“active:” states 1–5), green (“Polycomb-regulated:” state 6), blue (“heterochromatic:” states 7–8), and black (state 9). Note that state 9 corresponds to the “black” chromatin described by Filion et al. (2010). When downloaded chromatin data were displayed relative to salivary gland read data, there was a striking correspondence to chromatin domains (Supplemental Fig. S1). The predominant chromatin state associated with each UR is listed in Table 1.
Alignment and identification of UR regions
Sequencing was carried out on an Illumina HiSeq2000, and reads were aligned to the Drosophila genome version R5.33 using ELAND or BWA software and visualized using IGV. UR regions were automatically flagged if the salivary gland/embryo read average fell more than two to three standard deviations from the mean for five or more consecutive 5-kb windows. Most such regions have a characteristic shape profile consisting of a monotonic decrease to a minimum value near the center followed by a similar rise back to the baseline (Fig. 1E–G). In addition, <10 of the weakest UR regions were added or had a boundary adjusted because the shape of their deviation from baseline over 70–200 kb appeared very similar to those of stronger UR regions. Overlapping URs were separated at the local maximum point (Fig. 1E). The depth of each UR region was determined as the ratio of its low point and baseline. Underreplication was the inverse of this value. Further analyses were carried out using SAMtools, IGV tools, FileMaker Pro, Microsoft Excel, Python, C++, and the Unix command line (see below).
Calculations and formulas
Fraction of strands with deletions
For a UR region that is 2× UR, half of the DNA strands must contain a deletion (or other rearrangement). Hence, the number of predicted deletion reads would be 0.5 times the read depth (additional smaller deletions off center are ignored). For an underreplication value of UR, the general formula for the fraction of strands with deletions is f = 1 − 1/UR.
Predicted total deletions from UR values
We used the underreplication values in Table 1 to calculate f for each UR and multiplied the sum of these values by the total single copy read depth (after correction for PCR duplicates), 237.5, to yield the predicted number of unique deletions in the RefSG salivary gland DNA sequences (Table 2).
Fraction of mutated strands in amplified chorion DNA
Seventy-two read pairs specifying deletions in the 66D chorion region/total number of reads in the region (3250) = 2.2% (see Fig. 2B, inset).
Probability of fork arrest vs. UR value
UR value is total strands (2n)/unmutated strands [2n(1 − p)] = 2np, where n is the number of endocycles and p is the probability of fork arrest per endocycle. Values of p calculated from the L3 UR values in Table 1 accurately predicted the differences between L2 and L3 UR values in most regions, with n = 3 as the number of intervening endocycles (round 7 to round 10). The formula yields UR = 2 for p = 0.1, as discussed in the text.
Analysis of deletions defined by anomalous reads
Read pairs aligned but separated by 10–500 kb were sorted by AWK and loaded into a custom FileMaker Pro database with all associated samfile data. Reads pairs aligned to heterochromatic regions or mitochondrial DNA were removed, as were reads with quality fields containing more than four #′s and duplicate read pairs. Both reads matched the genome 100% in >95% of these pairs. Scripts were used to output display files. Each deletion is denoted by the shortened name of the read pair from which it is derived (i.e., HWI-ST375:119:D0A2LACXX:6:2205:1397:93872 is shown as 2205:1397:93872.)
Identification of breakpoint reads
For each experiment, all reads from pairs mapped within euchromatin that were unaligned and all mapped euchromatic reads in which a run of 15–80 bases was not aligned (“soft-clipped”) were collected using AWK, and the 100-bp sequences were aligned to the Drosophila 5.33 genome locally using BLAT. The BLAT output was parsed at six matches per sequence and loaded along with the corresponding samfile data into a FileMaker Pro database. Deletions identified by BLAT were accepted if Tgapbases was between 10,000 and 500,000 and if one segment of the deletion-containing read was on the same arm and at a proper distance from the mate. Additional deletions were identified if BLAT aligned the sequence on two separate lines corresponding to genome regions on the same arm separated by between 10 and 500 kb and together matched at least 97% of the input sequence. One of the matching segments was required to be on the same arm and within a proper distance from the mate. More than 95% of the deletions matched 100% of the input sequence and 100% of the target, with the deletion as the only gap. However, candidate deletions involving repetitive sequences with more than four BLAT matches or involving input sequences with long homopolymeric tracts were excluded.
Data archive
The data from this project have been submitted to the National Institutes of Health Short Read Archive: SubmissionID, SUB495929; BioProject ID, PRJNA244953; and title, D. melanogaster polytene cell sequencing.
Acknowledgments
We are grateful to Allison Pinder for expert assistance with library construction and DNA sequencing. We thank Fred Tan and Nick Ingolia for valuable advice on sequence analysis. Steve DeLuca provided valuable assistance with chromatin analysis. A.C.S is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.245811.114.
References
- Ahmad K, Golic K. 1996. Somatic reversion of chromosomal position effects in Drosophila melanogaster. Genetics 144: 657–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. 2013. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 152: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyeva EN, Kolesnikova TD, Belyaeva ES, Glaser RL, Zhimulev IF. 2008. Local DNA underreplication correlates with accumulation of phosphorylated H2Av in the Drosophila melanogaster polytene chromosomes. Chromosome Res 16: 851–862 [DOI] [PubMed] [Google Scholar]
- Ashburner M 1970. Function and structure of polytene chromosomes during insect development. Adv. Insect Physiol. 7: 1–95 [Google Scholar]
- Belikoff EJ, Beckingham K. 1985. A stochastic mechanism controls the relative replication of equally competent ribosomal RNA gene sets in individual dipteran polyploid nuclei. Proc Natl Acad Sci 82: 5045–5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva ES, Zhimulev IF, Volkova EI, Alekseyenko AA, Moshkin YM, Koryakov DE. 1998. Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc Natl Acad Sci 95: 7532–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva ES, Boldyreva LV, Volkova EI, Nanayev RA, Alekseyenko AA, Zhimulev IF. 2003. Effect of the Suppressor of Underreplication (SuUR) gene on position-effect variegaion silencing in Drosophila melanogaster. Genetics 165: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva ES, Andreyeva EN, Belyakin SN, Volkova EI, Zhimulev IF. 2008. Intercalary heterochromatin in polytene chromosomes of Drosophila melanogaster. Chromosoma 117: 411–418 [DOI] [PubMed] [Google Scholar]
- Belyaeva ES, Goncharov FP, Demakova OV, Kolesnikova TD, Boldyreva LV, Semeshin VF, Zhimulev IF. 2012. Late replication domains in polytene and non-polytene cells of Drosophila melanogaster. PLoS ONE 7: e30035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyakin SN, Christophides GK, Alekseyenko AA, Kriventseva EV, Belyaeva ES, Nanayev RA, Makunin IV, Kafatos FC, Zhimulev IF. 2005. Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc Natl Acad Sci 102: 8269–8274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi BR, Spradling AC. 1999. Chorion gene amplification in Drosophila: a model for metazoan origins of DNA replication and S-phase control. Methods 18: 407–417 [DOI] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. 2011. DNA elimination in ciliates: transposon domestication and genome surveillance. Annu Rev Genet 45: 227–246 [DOI] [PubMed] [Google Scholar]
- Claycomb JM, MacAlpine DM, Evans JG, Bell SP, Orr-Weaver TL. 2002. Visualization of replication initiation and elongation in Drosophila. J Cell Biol 159: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Michor F. 2011. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat Biotechnol 29: 1103–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dej K, Spradling AC. 1999. The endocycle controls nurse cell polytene chromosomes during Drosophila oogenesis. Develop. 126: 293–303 [DOI] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM. 2011. Chromatin signatures of the Drosophila replication program. Genome Res 21: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SC, Reuter G. 2013. Position-effect variegation, heterochromatin formation and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 5: a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrough DM, Goodman CS. 1996. The Drosophila beaten path gene encodes a novel secreted protein that regulates defasciculation at motor axon choice points. Cell 87: 1049–1058 [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D, Gall JG, Spradling AC. 2010. Error-prone polyploid mitosis during normal Drosophila development. Genes Dev 24: 2294–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Cohen EH, Polan ML. 1971. Repetitive DNA sequences in Drosophila. Chromosoma 33: 319–344 [DOI] [PubMed] [Google Scholar]
- Gilbert S. 2013. Developmental biology, 10th ed. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Glaser RL, Karpen GH, Spradling AC. 1992. Replication forks are not found in a Drosophila mini-chromosome demonstrating a gradient of polytenization. Chromosoma 102: 15–19 [DOI] [PubMed] [Google Scholar]
- Glaser RL, Leach TJ, Ostrowski SE. 1997. The structure of heterochromatic DNA is altered in polyploid cells. Mol Cell Biol 17: 1254–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi FC, An W. 2013. Non-LTR retrotransposons and microsatellitels: partners in genomic variation. Mob Genet Elements 3: e25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond MP, Laird CD. 1985. Chromosome structure and DNA replication in nurse cells and follicle cells of Drosophila melanogaster. Chromosoma 91: 267–278 [DOI] [PubMed] [Google Scholar]
- Hannibal RL, Chuong EB, Rivera-Mulia JC, Gilbert DM, Valouev A, Baker JC. 2014. Copy number variation is a fundamental aspect of the placental genome. PLoS Genet 10: e1004290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchenko PV, Aleksehyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2012. Comphrensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Spradling AC. 1990. Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila. Cell 63: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH 2011. Mobile DNA transposition in somatic cells. BMC Biol 9: 62 doi: 10.1186/1741-7007-9-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova TD, Posukh OV, Andreyeva EN, Bebyakina DS, Ivankin AV, Zhimulev IF. 2013. Drosophila SUUR protein associates with PCNA and binds chromatin in a cell cycle-dependent manner. Chromosoma 122: 55–66 [DOI] [PubMed] [Google Scholar]
- Laird CD 1980. Structural paradox of polytene chromosomes. Cell 22: 869–874 [DOI] [PubMed] [Google Scholar]
- Lambert S, Carr AM. 2012. Replication stess and genome rearrangements: lessons from yeast models. Curr Opin Genet Dev 23: 132–139 [DOI] [PubMed] [Google Scholar]
- Leach TJ, Chotkowski HL, Wotring MG, Dilwith RL, Glaser RL. 2000. Replication of heterochromatin and structure of polytene chromosomes. Mol Cell Biol 20: 6308–6316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimov DA, Koryakov DE, Belyakin SN. 2013. Developmental variation of the SUUR protein binding correlates with gene regulation and specific chromatin types in D. melanogaster. Chromosoma. doi: 10.1007/s00412-013-0445-6 [DOI] [PubMed] [Google Scholar]
- Makunin IV, Volkova EI, Belyaeva ES, Nabirochkina EN, Pirrotta V, Zhimulev IF. 2002. The Drosophila suppressor of underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics 160: 1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, Calvi BR. 2008. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev 22: 3158–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J, Orr-Weaver TL. 2012. Regulation of DNA replication during development. Development 139: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J, Li S, Eng T, Macalpine D, Orr-Weaver TL. 2011. Developmental control of the DNA replication and transcription programs. Genome Res 21: 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, Miller OL Jr. 1983. Novel amplification and transcriptional activity of chorion genes in Drosophila melanogaster follicle cells. Cell 33: 543–553 [DOI] [PubMed] [Google Scholar]
- Özkan E, Carrillo RA, Eastman CL, Weiszmann R, Waghray D, Johnson KG, Zinn K, Celniker SE, Garcia KC. 2013. An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell 154: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pindyurin AV, Moorman C, de Wit E, Belyakin SN, Belyaevab ES, Christophides GK, Kafatos FC, van Steensel B, Zhimulev IF. 2007. SUUR joins separate subsets of PcG, HP1 and B-type lamin targets in Drosophila. J Cell Sci 120: 2344–2351 [DOI] [PubMed] [Google Scholar]
- Pipes GC, Lin Q, Riley SE, Goodman CS. 2001. The Beat generation: a multigene family encoding IgSF proteins related to the Beat axon guidance molecule in Drosophila. Development 128: 4545–4552 [DOI] [PubMed] [Google Scholar]
- Reizel Y, Itzkovitz S, Adar R, Elbaz J, Jinich A, Chapal-Ilani N, Maruvka YE, Nevo N, Marx Z, Horovitz I, et al. 2012. Cell lineage analysis of the mammalian female germline. PLoS Genet 8: e1002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DM, O’Hely M, Bartley PA, Dang P, Score J, Goyne JM, Sobrinho-Simoes M, Cross NC, Melo JV, Speed TP, et al. 2013. Distribution of genomic breakpoints in chronic myeloid leukemia: analysis of 308 patients. Leukemia 27: 2105–2107 [DOI] [PubMed] [Google Scholar]
- Rudkin, G 1969. Non-replicating DNA in Drosophila. Genetics 61: 227-238 [PubMed] [Google Scholar]
- Sher N, Bell GW, Li S, Nordman J, Eng T, Eaton ML, Macalpine DM, Orr-Weaver TL. 2012. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res 22: 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear BB, Gall JG. 1973. In dependent control of ribosomal gene replication in polytene chromosomes of Drosophila melanogaster. Proc Natl Acad Sci 70: 1359–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spierer A, Spierer P. 1984. Similar level of polyteny in bands and interbands of Drosophila giant chromosomes. Nature 307: 176–178 [DOI] [PubMed] [Google Scholar]
- Spradling AC 1993. Position-effect variegation and genomic instability. Cold Spring Harb Symp Quant Biol 58: 585–596 [DOI] [PubMed] [Google Scholar]
- Spradling AC, Orr-Weaver T. 1987. Regulation of DNA replication during Drosophila development. Annu Rev Genet 21: 373–403 [DOI] [PubMed] [Google Scholar]
- Spradling AC, Karpen GH, Glaser R, Zhang P. 1992. Evolutionary conservation of developmental mechanisms: DNA elimination in Drosophila. Symp Soc Dev Biol 50: 39–48 [Google Scholar]
- Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. 2007. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell 130: 1134–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR, Weiner JA. 2003. Synaptic adhesion molecules. Curr Opin Cell Biol 15: 621–632 [DOI] [PubMed] [Google Scholar]