KDM2A, a histone lysine demethylase, specifically targets lower methylation states of H3K36. Here, Cheng et al.’s structural studies reveal that H3K36 specificity for KDM2A is mediated by the U-shaped threading of the H3K36 peptide through a catalytic groove within KDM2A. Key residues contributing to K36me specificity on histone H3 were identified. Structure-guided substitutions of residues in the KDM2A catalytic pocket abrogate KDM2A-mediated functions important for suppression of cancer cell phenotypes. These results provide insights into the molecular basis underlying KDM2A regulation of the methylated H3K36 mark.
Keywords: histone demethylase, KDM2A, mechanism, substrate specificity
Abstract
The dynamic reversible methylation of lysine residues on histone proteins is central to chromatin biology. Key components are demethylase enzymes, which remove methyl moieties from lysine residues. KDM2A, a member of the Jumonji C domain-containing histone lysine demethylase family, specifically targets lower methylation states of H3K36. Here, structural studies reveal that H3K36 specificity for KDM2A is mediated by the U-shaped threading of the H3K36 peptide through a catalytic groove within KDM2A. The side chain of methylated K36 inserts into the catalytic pocket occupied by Ni2+ and cofactor, where it is positioned and oriented for demethylation. Key residues contributing to K36me specificity on histone H3 are G33 and G34 (positioned within a narrow channel), P38 (a turn residue), and Y41 (inserts into its own pocket). Given that KDM2A was found to also bind the H3K36me3 peptide, we postulate that steric constraints could prevent α-ketoglutarate from undergoing an “off-line”-to-“in-line” transition necessary for the demethylation reaction. Furthermore, structure-guided substitutions of residues in the KDM2A catalytic pocket abrogate KDM2A-mediated functions important for suppression of cancer cell phenotypes. Together, our results deduce insights into the molecular basis underlying KDM2A regulation of the biologically important methylated H3K36 mark.
Histone lysine methylation is an important component of the histone code (Jenuwein and Allis 2001), with methylation marks on Lys4 (H3K4) and Lys36 (H3K36) of histone 3 being linked to gene activation. In contrast, the corresponding marks on Lys9 (H3K9) and Lys27 (H3K27) are linked with gene repression. Therefore, these marks underpin a multilayer regulation of chromatin structure and gene expression (Martin and Zhang 2005). Lysine can be mono-, di-, or trimethylated, with these marks installed by specific “writer” modules (Cheng et al. 2005; Qian and Zhou 2006) and read by specific “reader” modules (Taverna et al. 2007; Musselman et al. 2012; Patel and Wang 2013).
Lysine methylation was regarded as a stable modification until the discovery of the first histone lysine demethylase, LSD1 (Shi et al. 2004). LSD1 and its enzymatic homologs are flavin-dependent amine oxidases that remove mono- and dimethylation but not trimethylation lysine marks due to the requirement for protonation of the methylated ɛ-ammonium group. Soon afterward, a superfamily of Jumonji C (JmjC) domain-containing histone lysine demethylases was discovered, which are α-ketoglutarate (αKG) and Fe2+-dependent oxygenases belonging to the cupin superfamily (Cloos et al. 2006; Tsukada et al. 2006; Whetstine et al. 2006; Yamane et al. 2006). By employing a different demethylation mechanism from that employed by LSD1, the JmjC-containing oxygenases are able to remove not only mono- and dimethylated lysine marks but also their trimethylated counterparts from histone tails (Klose et al. 2006; Anand and Marmorstein 2007; Cheng and Zhang 2007; Mosammaparast and Shi 2010; Kooistra and Helin 2012). How JmjC-containing proteins recognize the sequence context and methylation state in their capacity as “erasers” remains a subject of continuing investigation (Hou and Yu 2010; McDonough et al. 2010; Upadhyay et al. 2011).
KDM2A (previously named JHDM1A), a modular protein containing JmjC, CXXC, and PHD finger domains (Fig. 1A), was the first JmjC-containing histone lysine demethylase identified in the literature (Tsukada et al. 2006) and exhibits specificity for removal of methyl groups from lower methylation states (Kme1 and Kme2) of H3K36. The CXXC domain interacts with nonmethylated CpG DNA, thereby tethering KDM2A to nonmethylated CpG islands and depleting H3K36me1/me2 marks (Blackledge et al. 2010). Subsequently, KDM4A (previously named JMJD2A)—composed of JmjN and JmjC domains, a zinc finger motif, and a C-terminal domain—was shown to exhibit specificity for removal of methyl groups from higher methylation states (Kme2 and Kme3) of K9 and K36 (Whetstine et al. 2006). The structure of KDM4A in the free state (Chen et al. 2006) and bound to methylated H3K9 and H3K36 peptides, αKG cofactor, and metal clarified aspects of the catalytic mechanism, with substrate recognition achieved by a network of hydrogen bonds and van de Waals contacts between KDM4A and its methylated substrate peptides (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007).
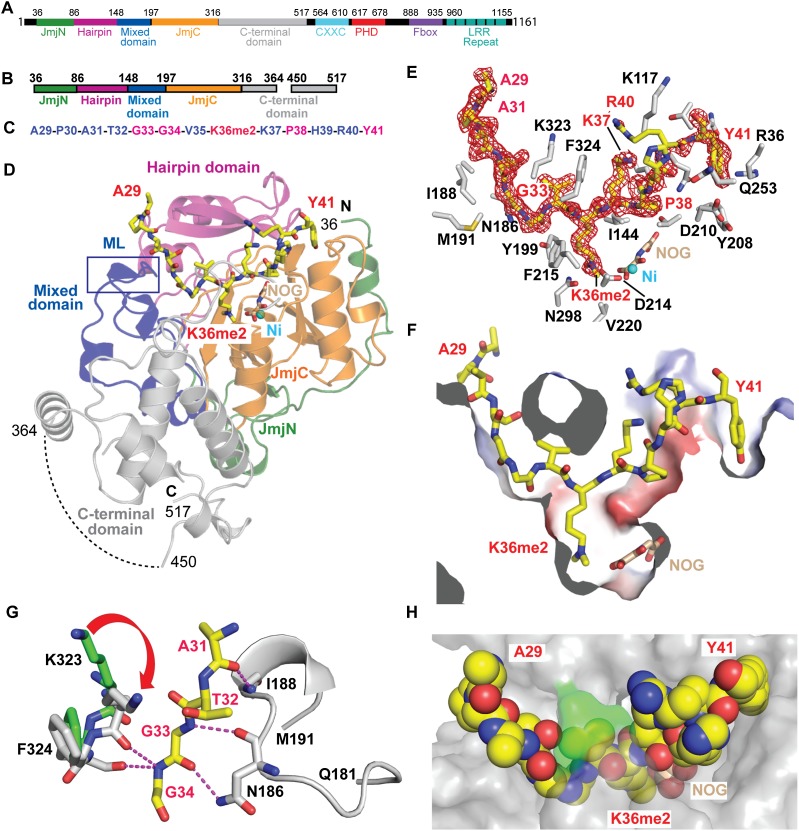
Figure 1.
Structure of the H3(A29–Y41)K36me2–KDM2A complex and conformational transitions on complex formation. (A) The domain architecture of full-length KDM2A. The domains are labeled and color-coded. (B) The truncated KDM2A construct (36–517) used for growing crystals of the complex lacked 1–35 and 365–449 peptide segments. The various domains are color-coded. (C) The sequence of the K36me2-containing H3(A29–Y41) peptide. Residues highlighted in red play key intermolecular recognition roles in complex formation. (D) The structure of the H3K36me2–KDM2A complex in the presence of NOG and Ni2+. The color-coding of the various domains of KDM2A in a ribbon representation are as shown in A. The bound H3K36me2 peptide in yellow and NOG in gold are shown in a stick representation, while Ni2+ is represented by a cyan ball. The internal loop (365–449) deletion position is shown by a dashed line. (E) The Fo − Fc omit map for the bound peptide (3σ) positioned in the peptide-binding channel with K36me2 inserted into the catalytic pocket of KDM2A in the structure of the complex. The peptide residue labels are colored in red, and the protein residue labels are colored in black, with both in a stick representation. (F) A surface cutaway view highlighting the bound H3K36me2 peptide (colored yellow in a stick representation) positioned in the peptide-binding channel, with K36me2 inserted into the catalytic pocket of KDM2A in the structure of the complex. NOG and Ni2+ are color-coded as in C. Surface is colored by electrostatic potential. (G) Conformational transitions on complex formation. The Q181–M191 peptide segment of the mixed domain is disordered in the free state of KDM2A but becomes ordered on complex formation with H3K36me2 peptide. The K323–F324 segment undergoes a conformational transition on proceeding from the free state (in green) to the bound state (in silver) on complex formation. (H) The K323–F324 segment that undergoes a conformational change on complex formation forms a flap (in green) over the bound peptide (drawn in space-filling representation) in the structure of the complex.
Structure–function studies have also been reported on additional families of JmjC domain-containing KDMs. PHF8 and KIAA1718 have been identified to be H3K9-specific and H3K27-specific (embedded in a common A-R-K-S motif) lysine demethylases, with structural studies establishing that their specificity is regulated by the N-terminal PHD finger located outside of the JmjC domain (Horton et al. 2010). The KDM6 subfamily, which includes JmjD3 and UTX, interacts specifically with higher methylation marks of H3K27 (Sengoku and Yokoyama 2011; Kruidenier et al. 2012). Of special interest was the recent identification of selective small molecule inhibitors of the KDM6 subfamily and their modulation of the proinflammatory macrophage response (Kruidenier et al. 2012).
The present study focuses on KDM2A, given that this subset of lysine demethylases (KDM2A/2B) has key regulators of vitamin C-induced reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) (Wang et al. 2011). In addition, KDM2B accelerates cell cycle progression and suppresses cell senescence by repressing the ink4/arf locus (He et al. 2008) while repressing the transcription of ribosomal RNA through binding the promoter region of rDNA and reducing H3K36me1/me2 levels (Frescas et al. 2007; Tanaka et al. 2010). To better understand the mechanism underlying the lysine demethylase activity of KDM2A, we successfully crystallized and solved the structure of a series of methylated H3K36-truncated KDM2A complexes in the presence of Ni2+ (KDM2A was purified on Ni-NTA columns, whereby Ni2+ replaced endogenous Fe2+) and bound to either cofactor αKG or the nonreactive analog N-oxalylglycine (NOG). The structural studies on truncated KDM2A bound to methylated H3K36 peptides were complemented by enzymatic studies on structure-guided mutants of both the peptide and the lysine demethylase so as to provide insights into the enzymatic mechanism of KDM2A and the molecular basis underlying its sequence and methylation state-specific demethylase activity. We also found that full-length KDM2A functions to maintain genomic stability and inhibits the ability of cells to grow under anchorage-independent conditions and be transformed by the Ras oncogene. These cellular functions of KDM2A are abolished by structure-based mutations that target the catalytic pocket of KDM2A. The state of methylation at H3K36 is linked to dramatically different oncogenic consequences (Dalgliesh et al. 2010; Kuo et al. 2011; Black et al. 2013). Thus, our study provides insights into aspects of the molecular basis by which KDM2A influences methylation dynamics at the critical H3K36 site.
Results
The domain architecture of KDM2A is shown in Figure 1A. In an effort to facilitate crystallization, an earlier structural study focused on a truncated KDM2A construct (36–517), including deletion of a flexible loop segment (365–449) within this construct, as shown in Figure 1B. The structure of this KDM2A construct was solved in the apo state in the presence of αKG and Ni2+ (Han et al. 2007).
Crystal structure of the H3K36me2 peptide–KDM2A complex
We extended this earlier study by generating a complex of the same truncated KDM2A construct (Fig. 1B) bound to a H3(A29-Y41)K36me2 peptide (Fig. 1C) and grew crystals that diffracted to 1.75 Å resolution in the presence NOG and Ni2+ (X-ray statistics in Supplemental Table S1). The overall structure of the complex is shown in Figure 1D. The Fo − Fc omit map (3σ level) of the bound H3K36me2 peptide in the complex is shown in Figure 1E and readily allows tracing of the backbone and side chains of the bound peptide, including insertion of the K36me2 into the catalytic pocket. A cutaway view of the H3K36me2 peptide positioned within the binding channel of KDM2A is shown in Figure 1F.
Conformational change in KDM2A on complex formation with the H3K36me2 peptide
Comparison of the structures of the KDM2A construct in the free (Protein Data Bank [PDB] ID: 2YU1) and H3K36me2 peptide-bound states identified conformational transitions associated with complex formation. Specifically, a loop segment spanning residues Q181 to M191, which is disordered in the free state (Han et al. 2007), forms a 310 helix in the H3K36me2 peptide-bound state whereby it interacts with and stabilizes the A31 to G34 segment of the bound peptide (Fig. 1G). In addition, a conformational transition is observed in K323–F324 upon binding of the H3K36me2 peptide (Fig. 1G), resulting in formation of a flap that covers the bound peptide (Fig. 1H).
Key molecular interactions within the KDM2A–H3K36me2 peptide complex
The bound H3(A29–Y41)K36me2 peptide forms intermolecular contacts with multiple domains of KDM2A, including the hairpin, mixed, JmjC, and C-terminal domains, with the identified intermolecular contacts shown schematically in Figure 2A. The U-shaped architecture of the bound peptide is facilitated by changes in chain direction at residues G34 and P38. Complex formation results in the burial of ∼1031 Å2 (58.6%) of the bound peptide within KDM2A.
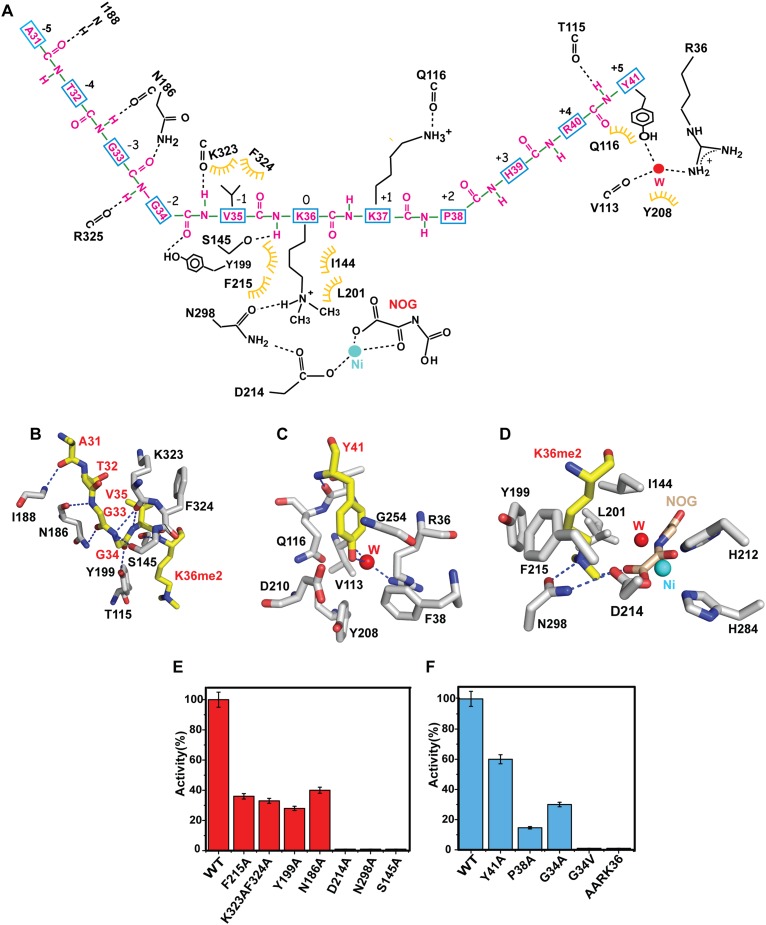
Figure 2.
Details of peptide–protein interactions in the H3K36me2–KDM2A complex and kinetics of demethylation of H3K36me2 by wild-type KDM2A and mutants of peptide and protein residues involved in intermolecular recognition. (A) Schematic outlining details of intermolecular hydrogen-bonding and hydrophobic contacts between the backbone and side chains of amino acids (labeled in red) of the H3(A29–Y41)K9me2 peptide and the backbone and side chains of amino acids (labeled in black) of the KDM2A in the complex. The A29–P30 segment was involved in crystal packing but not in intermolecular contacts. (B) Intermolecular hydrogen-bonding and hydrophobic contacts associated with the H3(A31–K36me2) peptide segment positioned within the binding channel of KDM2A. The peptide segment is in yellow, and amino acids of KDM2A are in silver. (C) Intermolecular hydrogen-bonding and hydrophobic contacts associated with insertion of Y41 into its binding pocket in the complex. Water molecules are represented by red spheres. (D) Intermolecular hydrogen-bonding and hydrophobic contacts associated with insertion of K36me2 into the catalytic pocket in the complex. NOG is colored in gold, and Ni2+ and water are colored as cyan and red spheres, respectively. (E) The impact of KDM2A mutants involved in intermolecular recognition on H3K36me2 demethylation enzymatic activity. (F) The impact of H3K36me2 peptide mutants involved in intermolecular recognition on H3K36me2 demethylation enzymatic activity by wild-type KDM2A.
The residues that comprise the N-terminal segment of the bound peptide prior to the K36me2 position (A29-P-A-T-G-G-V35) enter into a narrow channel within KDM2A, which, due to steric constraints, can only accommodate Gly residues at adjacent positions 33 and 34 in the peptide sequence (Figs. 1F, 2B). This segment is characterized by intermolecular hydrogen bonds involving the backbone of the bound peptide (G33–G34 and G34–V35 steps) with the backbone and side chains of KDM2A (Fig. 2A,B). In contrast, the C-terminal segment of the bound peptide spanning residues after the K36me2 position (K37-P-H-R-Y41), which contains larger side chains, is readily accommodated in a much wider segment of the channel (Fig. 1F).
Indeed, low average B factors (<20 Å2) are observed for the A31 to K36me2 segment of the peptide in the H3K36me2–KDM2A complex, while the B factors increase for the flanking A29 to P30 and K37 to R40 segments (Supplemental Fig. S1A). The B-factor profile of the bound peptide is consistent with our structural observation that A31–K36 is constrained in a narrow channel, while K37–Y41 fits in a spacious groove (Fig. 1F).
The aromatic side chain of Y41, the C-terminal residue of the bound H3K36me2 peptide, is inserted into its own pocket in KDM2A, with its hydroxyl group forming water-mediated intermolecular hydrogen bonds (Fig. 2C).
The side chain of H3K36me2 inserts into the catalytic pocket of KDM2A. The methylene side chain of K36me2 forms van der Waals contacts with aromatic (F215 and Y199) and hydrophobic (L201 and I144) side chains of KDM2A, while the dimethylammonium group is directed toward the charged (D214), polar (N298) KDM2A residues and the carboxylate end of the NOG cofactor (Fig. 2A,D).
The insertion of K36me2 into the catalytic pocket and anchoring of Y41 in its own pocket together contribute to the positioning of the peptide in a defined register within the U-shaped binding channel of KDM2A.
Impact of KDM2A protein mutations involved in intermolecular recognition
We measured the impact on activity of mutations of KDM2A residues involved in intermolecular recognition using a fluorescence-based assay. Mutations positioned at the base of the catalytic pocket (D214A and N298A) inactivate the enzyme, while those that line the walls (Y199A and F215A) reduce activity to 30%–40% of that of the wild type (Fig. 2E). In addition, we also monitored the impact of mutations from the mixed loop (N186A) and the segment that forms a flap over the bound substrate (K323A/F324A), which leads to an approximately threefold reduction in activity (Fig. 2E).
Impact of H3K36me2 peptide mutations involved in intermolecular recognition
We monitored the impact of mutations G34A and P38A (Fig. 2F), given that the bound U-shaped H3K36 peptide significantly changes chain direction at G34 and P38 in the H3(A29–Y41)K36me2–KDM2A complex, leading to an approximately threefold and sevenfold reduction in activity, respectively (Fig. 2F). In addition, we also monitored the impact of the G34V pediatric glioblastoma-linked mutation (Fig. 2F), which leads to inactive enzyme. The contribution of Y41 in retaining the bound peptide in a particular register in the complex was tested through modification of Y41 (Y41A), leading to a less than twofold impact on activity (Fig. 2F). We also monitored the impact on replacement of the G33–G34–V35–K36me2 segment in the H3K36 peptide by A33–A34–R35–K36me2 to mimic the equivalent H3K27 sequence and observed no activity toward this peptide (Fig. 2F).
Complex formation as a function of K36 methylation with bound NOG
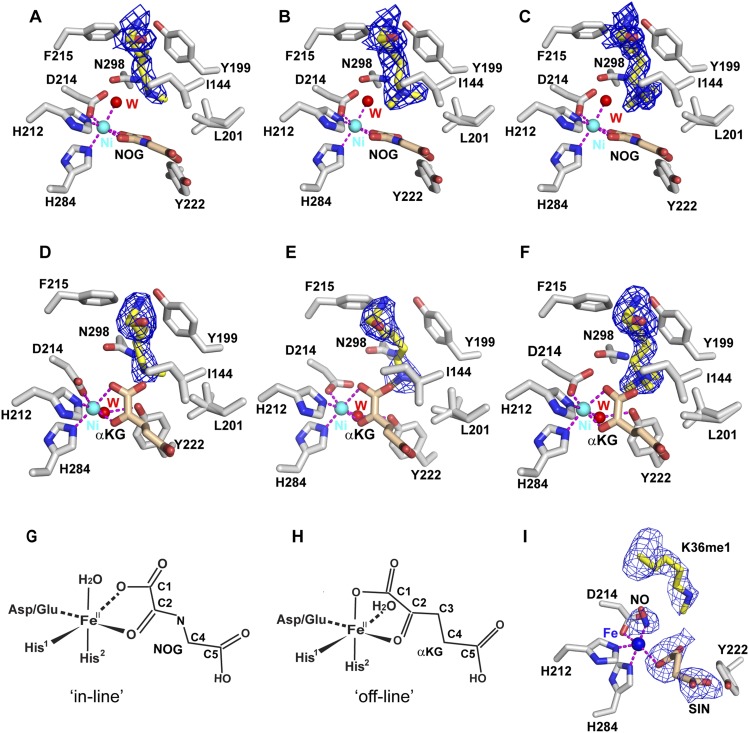
Most αKG/Fe2+-dependent enzymes contain a His1-X-Asp/Glu-Xn-His2 sequence motif required for chelation of the Fe2+ atom. Based on the conformations of the bound cofactor, the active site structures have been classified into two categories, representing “in-line” and “off-line” modes (Hausinger 2004). We observed both of these modes when using NOG and αKG as cofactors (Fig. 3). In order to interrogate the relationship of the methylation state and the ligand conformation, we first crystallized and solved the structures of truncated KDM2A bound to H3K36me-containing peptides in the presence of NOG and Ni2+ as a function of methylation state (for X-ray statistics, see Supplemental Table S2). The positioning of the K36me1, K36me2, and K36me3 of the bound peptides in the catalytic pockets of the complexes, together with the coordination geometries of the Ni2+, are shown in Figure 3, A–C, respectively. All three structures of the complexes are very similar to each other from the perspective of both insertion of the methylated lysine side chains into the catalytic pockets and the coordination geometries of the Ni2+. The Ni2+ is six-coordinate, and very similar to the Fe2+ six-coordinate geometry observed in other αKG/Fe2+-dependent enzymes (Valegard et al. 2004). As such, the Fe2+ coordination geometry in the presence of NOG can be inferred from the Ni2+-containing methylated H3K36–KDM2A complexes (Fig. 3A–C) and is shown schematically in Figure 3G. The bound NOG adopts an “in-line” mode (Hausinger 2004) in which the C1 carboxylate oxygen of NOG is positioned opposite His1 (H212 of KDM2A) in an approximate square planar arrangement with D214 and the C2 carbonyl of NOG, while a water molecule and His2 (H284 of KDM2A) occupy apical positions (Fig. 3G). The side chain of Y222 is directed away from the metal site in all three “in-line” mode complexes independently of the methylation state of H3K36 (Fig. 3A–C). These data revealed that the NOG conformation remains the same regardless of the methylation state of the substrate in the pocket and is representative of the “in-line” mode. In this mode, oxygen displaces the apical water, and when activated to ferryl iron, this is correctly positioned to perform the chemistry.
Figure 3.
Details of positioning of K36me, Ni2+, and either NOG (“in-line” mode) or αKG (“off-line” mode) in the catalytic pocket in the complex with KDM2A as a function of K36 methylation state. (A) The H3K36me1–KDM2A complex in the presence of NOG and Ni2+. (B) The H3K36me2–KDM2A complex in the presence of NOG and Ni2+. (C) The H3K36me3–KDM2A complex in the presence of NOG and Ni2+. (D) The H3K36me1–KDM2A complex in the presence of αKG and Ni2+. (E) The H3K36me2–KDM2A complex in the presence of αKG and Ni2+. (F) The H3K36me3–KDM2A complex in the presence of αKG and Ni2+. Omit maps (3σ) for methylated K36 are shown in A–F. (G) Schematic postulating the coordination geometry of the Fe2+ together with the role of NOG associated with coordination in the complex for the “in-line” mode. (H) Schematic postulating the coordination geometry of the Fe2+ together with the role of αKG associated with coordination in the complex for the “off-line” mode. (I) The coordination geometry of Fe2+ in the complex with succinate (SIN) together with the orientation of Y222 in the NO-treated H3K36me1–KDM2A complex. The observed electron density involved in direct coordination to Fe2+ has been tentatively assigned as NO.
Complex formation as a function of K36 methylation with bound αKG
We also crystallized and solved the structures of truncated KDM2A bound to H3K36me-containing peptides in the presence of αKG and Ni2+ as a function of methylation state (for X-ray statistics, see Supplemental Table S2). The structures of the complexes with K36me1-containing (Fig. 3D; Supplemental Fig. S1B), K36me2-containing (Fig. 3E; Supplemental Fig. S1C), and K36me3-containing (Fig. 3F; Supplemental Fig. S1D) peptides are very similar to each other from the perspective of both insertion of the methylated lysine side chains into the catalytic pockets and the coordination geometries of the Ni2+. The metal (represented by Fe2+) coordination geometry of the H3K36me1–KDM2A complex in the presence of αKG is shown schematically in Figure 3H. The bound αKG adopts an “off-line” mode (Hausinger 2004) in which the C2 carbonyl oxygen of αKG is positioned opposite a carboxylate oxygen of Asp (D214 in KDM2A) in an approximate square planar arrangement with His1 (H212 of KDM2A) and a water, while the C1 carboxylate oxygen of αKG and His2 (H284 in KDM2A) occupy apical positions (Fig. 3H). Here the water is equatorial, and one of the carboxylate oxygens of αKG occupies the axial position. The side chain of Y222 adopts two alternate positions, with the major conformer directed toward the Ni2+ (Fig. 3D–F), and the minor conformer (observed only in the H3K36me1 and H3K36me2 complexes and similar to its alignment for the “in-line” mode) directed away from the Ni2+. The αKG conformation is the same regardless of the methylation state of the substrate in the pocket and is representative of the “off-line” mode.
Complex formation in the presence of Fe2+, succinate, and NO
Since we observed the “off-line” mode for αKG and the “in-line” mode for NOG, we then asked whether having the cognate metal (Fe2+) would favor an “in-line” binding state and, if not, whether O2 could induce this change. To determine this, we replaced the Ni2+ with Fe2+ and used NO as an oxygen analog to mimic the Fe2+–O2 intermediate (Yukl et al. 2011). Anaerobic crystallization of the Fe2+–H3K36me1–KDM2A complex was unsuccessful, and so crystals of the apo–H3K36me1–KDM2A complex in the presence of αKG were grown aerobically and moved into an anaerobic glove box. These were reconstituted anaerobically with Fe2+ and then exposed to NO. Fe2+ reconstituted crystals were in a different space group and were of inferior diffraction quality (for X-ray statistics, see Supplemental Table S3). Despite attempts to make the aerobically grown crystals anaerobic prior to Fe2+ reconstitution, αKG had oxidized to succinate (although K36 remained methylated), with additional density (tentatively assigned to NO rather than water because of the diatomic shape of the density) observed bound to Fe2+ at the active center (Fig. 3I). In the proposed NO-bound structure of the complex, NO in an apical position is positioned opposite H1 (H284), while one oxygen atom of succinate is positioned opposite D214 in a monodentate-binding mode. The aromatic side chain of Y222 is directed away from the active center, with the OH group of Y222 forming a hydrogen bond with a carboxylic oxygen from the succinate molecule (Fig. 3I). Thus, the alignment of residues coordinated to the Fe2+ in the proposed NO-bound structure of the H3K36me1–KDM2A complex in the presence of succinate (Fig. 3I) resembles the alignment of the complex in the presence of NOG (Fig. 3A–C, “in-line” mode) rather than in the presence of αKG (Fig. 3D–F, “off-line” mode). This observation for the complex in the presence of NO implies that when O2 enters into the active site of the complex, the cofactor needs to undergo a conformational change from “off-line” to the “in-line” alignment mode.
Residues involved in recognition of H3K36 peptide are critical for KDM2A cellular functions
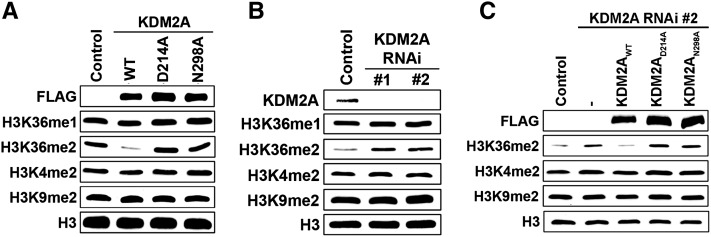
Alanine substitutions of two key residues positioned at the base of the catalytic pocket, D214A and N298A, abrogate KDM2A catalytic activity in vitro (Fig. 2E). We tested the role of these residues on KDM2A activity in cells. As shown in Figure 4A, overexpression of wild-type KDM2A, but not the structure-guided KDM2AD214A and KDM2AN298A mutants, resulted in depletion of bulk H3K36me2 signal without affecting H3K4me2 and H3K9me2 levels. Next, a KDM2A complementation system was used to investigate the biological significance of the structure-guided mutants. First, KDM2A expression was stably knocked down in the fibrosarcoma HT1080 cell line by RNAi using two independent shRNA constructs targeting the 3′ untranslated region of KDM2A (Fig. 4B). As expected, depletion of endogenous KDM2A by both shRNAs resulted in an increase of total H3K36me2 relative to control knockdown cells, without alterations in H3K4me2 and H3K9me2 signal (Fig. 4B). Complementation of KDM2A knockdown cells with RNAi-resistant wild-type KDM2A, but not RNAi-resistant KDM2AD214A and KDM2AN298A, restored H3K36me2 levels to those observed in the control cells (Fig. 4C). These results establish a system for structure–function investigation of KDM2A catalytic activity.
Figure 4.
Structure-guided mutations on KDM2A abolish its H3K36me2 demethylation activity in cells. (A) Overexpression of wild-type KDM2A, but not KDM2A carrying D214A or N298A mutations, reduces global H3K36me2 level in cells. Western analysis with the indicated antibodies of whole-cell extract (WCE) from 293T cells transfected with control vector, wild-type KDM2A, or catalytically inactive mutants. Total H3 is shown as loading control. (B) Depletion of KDM2A by RNAi increases global H3K36me2 level in cells. Western analysis as in A of whole-cell extract from HT1080 cells carrying control vector or stably expressing two independent RNAi targeting KDM2A transcript. (C) Complementation of KDM2A-depleted HT1080 cells with wild-type KDM2A, but not KDM2AD214A or KDM2AN298A, restores global H3K36me2 to the control level. Western analysis as in B of whole-cell extract from HT1080 control cells and KDM2A-depleted cells stably expressing KDM2AWT or catalytically inactive KDM2A.
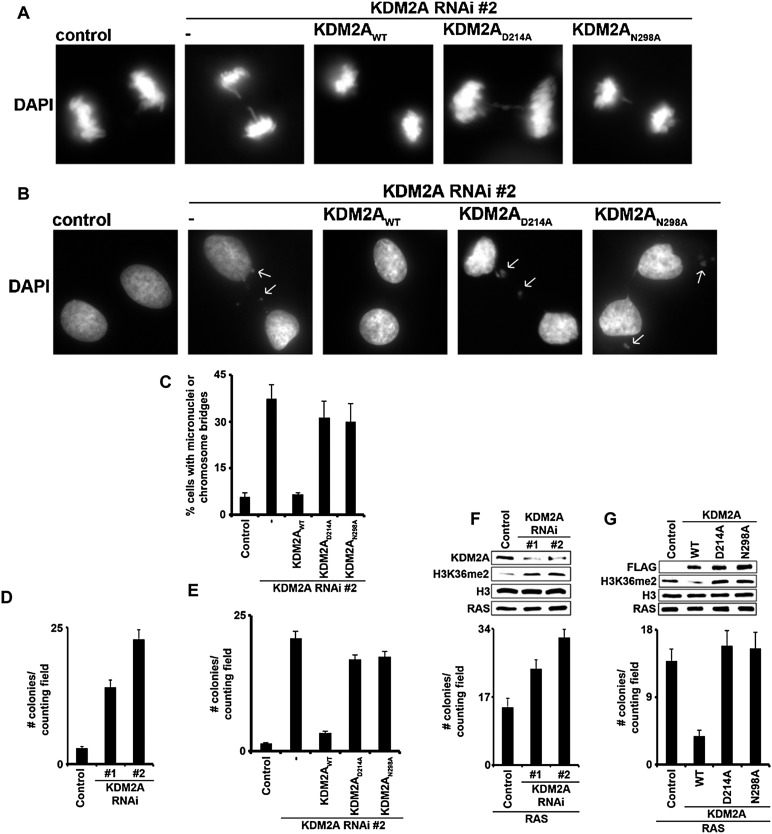
KDM2A has been implicated in regulating genomic stability through stabilization of centromere chromatin during mitosis, with knockdown of KDM2A triggering formation of chromosome bridges (Frescas et al. 2008). Indeed, we observed the formation of chromosome bridges in mitotic HT1080 cells depleted of KDM2A, which could be rescued by expression of RNAi-resistant KDM2A but not the catalytically dead KDM2AD214A and KDM2AN298A mutants (Fig. 5A). As chromosome bridges often result in micronuclei formation, we determined the relationship between KDM2A enzymatic activity and the frequency of these two forms of genomic instability using cycling HT1080 cells and the KDM2A complementation system. Knockdown of KDM2A results in a several-fold increase in the percentage of cells showing micronuclei or chromosome bridges relative to control cells, which can be restored to the levels observed in control cells by complementation with wild-type KDM2A (Fig. 5B,C). In contrast, complementation with the catalytically dead KDM2AD214A and KDM2AN298A mutants failed to rescue the genomic instability induced by KDM2A depletion (Fig. 5B,C), implicating a role for H3K36me2 demethylation in maintenance of genomic stability.
Figure 5.
Lysine demethylase activity of KDM2A is essential for the maintenance of genomic stability and the suppression of cellular transformation. (A) Loss of KDM2A catalytic activity leads to chromosome missegregation during mitosis. Representative images of 4′,6-diamidino-2-phenylindole (DAPI) staining of mitotic HT1080 cells carrying control vector or stably expressing KDM2AWT, KDM2AD214A, or KDM2AN298A. (B,C) KDM2A catalytic activity is required for the maintenance of genomic stability. (B) Representative images of DAPI staining of the indicated cell lines. Arrows indicate micronuclei or chromosome bridges. (C) Quantification of the abnormal cells based on DAPI staining as in A and B. Error bars indicate SEM from two independent experiments. Total numbers of cells analyzed were as follows for experiment #1/#2,: control, 150/214; RNAi #2, 201/217; KDM2AWT, 254/199; KDM2AD214A, 166/164; and KDM2AN298A, 221/134. (D) Depletion of KDM2A promotes anchorage-independent growth. Soft agar colony formation assay of KDM2A-depleted or control HT1080 cells. The ability of the indicated HT1080 cell line to grow colonies in methylcellulose is shown. Bar graph indicates number of colonies per field 21 d after seeding. Error bars indicate SEM from three independent experiments. (E) Complementation of KDM2A-depleted HT1080 cells with wild-type KDM2A, but not with catalytically inactive mutants, suppresses anchorage-independent growth. Soft agar colony formation assay as in D using the indicated HT1080 cells. Bar graph shows number of colonies per field 21 d after seeding. Error bars indicate SEM from three independent experiments. (F) Loss of KDM2A promotes Ras-induced oncogenic transformation of p19ARF−/− MEFs. (Top) Western analysis of whole-cell extract (WCE) from p19ARF−/− MEFs stably expressing RAS and control vector or two independent RNAi against the untranslated region of KDM2A transcript. (Bottom) Numbers of colonies formed in methylcellulose for each cell line. Error bars indicate SEM from three independent experiments. (G) KDM2AWT, but not the catalytic mutants, suppresses Ras-induced oncogenic transformation of p19ARF−/− MEFs. (Top) Western analysis of whole-cell extract from p19ARF−/− MEFs transduced with Ras and control vector, KDM2AWT, KDM2AD214A, or KDM2AN298A. (Bottom) Soft agar colony formation assay using the indicated cell lines. Bar graph shows number of colonies per field 21 d after seeding. Error bars indicate SEM from three independent experiments.
The role of KDM2A demethylation activity in preventing genomic instability and the observation that KDM2A expression is low in prostate carcinoma suggest that this enzyme may have tumor-suppressive functions (Frescas et al. 2008). In this regard, KDM2A depletion increased the ability of HT1080 cells to grow in soft agar, a measure of anchorage-independent growth and a hallmark cancer cell phenotype (Fig. 5D). Complementation of KDM2A-depleted cells with RNAi-resistant wild-type KDM2A, but not the catalytically dead mutants, inhibited anchorage-independent growth (Fig. 5E). These results suggest that KDM2A catalytic activity is important for suppressing a cancer cell phenotype. We therefore tested the ability of KDM2A to suppress cellular transformation using a primary mouse cell transformation system. In this assay, oncogenic v-Ras expression in p19ARF knockout mouse embryonic fibroblasts (p19ARF−/− MEFs) transforms cells, as measured by colony formation in soft agar assays (Fig. 5F, control). Depletion of KDM2A, which was accompanied by higher H3K36me2 levels, augmented Ras-induced anchorage-independent growth of p19ARF−/− MEFs (Fig. 5F). Wild-type KDM2A, but not the catalytically dead KDM2AD214A and KDM2AN298A mutants, restored both H3K36me2 levels and resistance to anchorage-independent growth in Ras-expressing p19ARF−/− MEFs (Fig. 5G). These data support the structural analysis and argue that KDM2A catalytic activity regulates important cancer cell phenotypes.
Discussion
Peptide-binding channel in the H3K36me2–KDM2A complex
The H3(A31–Y41)K36me2 peptide is positioned in a U-shaped channel upon complex formation with KDM2A (Figs. 1E,F, 2A), with changes in chain direction at positions G34 and P38. The majority of the intermolecular contacts are restricted to the G33–G34–V35–K36me2 segment, together with intermolecular interactions involving the C-terminal Y41 residue (Fig. 2A). Presumably the network of intermolecular hydrogen bonds at the G34–V35 step of the bound peptide, supplemented by hydrophobic interactions involving the side chain of V35 (with the side chain of K323 and the aromatic ring of F234) (Fig. 2A,B), stabilizes the first significant bend in the bound H3K36me2 peptide at G34, while the Pro residue at position 38 facilitates formation of the second direction change within the peptide.
The observed localized conformational transition on proceeding from KDM2A in the free (Han et al. 2007) to the H3K36me2-bound (this study) state, which involves a disordered-to-ordered transition for the Q181–M191 mixed loop segment and a repositioning of the K323–F324 step of KDM2A, not only generates a narrow channel that accommodates the G33–G34 step of the bound histone peptide (Fig. 1G) but also forms a flap that covers the central segment of the bound peptide (Fig. 1H). We monitored the impact on activity (rate of demethylation) of mutating KDM2A residues involved in these two features. The N186A (from the mixed domain loop) and K323A/F324A (forms flap over bound substrate) mutants result in a loss of enzymatic activity to ∼40% of wild type (Fig. 2E) and demonstrate the importance of intermolecular recognition within the narrow channel of the H3K36me2–KDM2A complex (Fig. 2B).
Substrate specificity of the H3K36me2–KDM2A complex
KDM2A specifically targets and demethylates the methylated H3K36 mark and discriminates against methylated H3K4, H3K9, H3K27, H3K79, and H4K20 marks (Tsukada et al. 2006). This sequence specificity can be readily rationalized based on several recognition features that are unique to the structure of the H3K36me2–KDM2A complex. First, the G33–G34 step (positions −3 and −2) relative to K36me2 (position 0) (Fig. 2A) threads through a narrow channel (Fig. 1F) that cannot accommodate side chains at these positions. It should be noted that only the H3K36 sequence context contains Gly residues at adjacent −3 and −2 positions, while all of the other Lys-containing sequence contexts listed above have side chains at these positions (Supplemental Fig. S2A). Indeed, replacement of the adjacent Gly residues (GGVKme2 segment) associated with the H3K36 sequence context by their adjacent Ala counterparts (AARKme2 segment) associated with the H3K27 sequence context resulted in complete loss of KDM2A enzymatic activity (Fig. 2F). Of note, it has been recently shown that G34V and G34R mutants of H3 are driver mutations leading to a lethal form of brain tumor (Schwartzentruber et al. 2012), suggesting a potential link between KDM2A demethylation of H3K36me2 and tumor suppression.
Second, the first direction change in the bound peptide occurs at the G34–V35 step (−2 and −1 positions), while the second occurs at P38 (+2 position) in the complex. Both the G34–V35 step and P38 residue are unique to the H3K36 sequence context (Supplemental Fig. S2A). We observed complete loss in enzymatic activity for the S145A mutant (Fig. 2F) involved in stabilizing the direction-changing segment at G34 of the bound peptide (Fig. 2B). The K37–P38 peptide bond adopts a trans configuration in the H3K36me2–KDM2A complex. It has been previously shown that cis–trans isomerization at H3 P38 regulates lysine methylation and gene expression (Nelson et al. 2006). Indeed, G34V and P38A mutations result in a complete loss in KDM2A demethylase activity (Fig. 2F).
Third, Y41 (+5 position) inserts its aromatic side chain into its own binding pocket and appears to anchor the bound peptide in register, facilitating insertion of the K36me2 side chain into the catalytic pocket (Figs. 1E, 2C). Here again, a Tyr residue at the +5 position is uniquely found within the H3K36 sequence context (Supplemental Fig. S2A). A decrease in enzymatic activity to 60% of wild-type value was observed for the Y41A mutant (Fig. 2E). Previous research has established that H3 Y41 can be phosphorylated by JAK2 (Dawson et al. 2009).
Insertion of methylated K36 into the binding pocket
Insertion of methylated H3K36 into the catalytic pocket is stabilized by hydrophobic interactions between the methylene side chain and KDM2A residues that line the walls of the catalytic pocket and hydrogen bonding and electrostatic interactions between the methylated ammonium group and KDM2A residues that form the base of the pocket as well as the carboxylate group of bound cofactor αKG/NOG (Fig. 2A,D). These amino acids of KDM2A play a key role, as reflected in the impact on mutation of these residues. Thus, mutation of residues Y199 and F215, which line the walls of the catalytic pocket, to alanine results in a loss of enzymatic activity to 35%–40% of the wild-type value (Fig. 2E). In addition, mutation of residues D214 and N298, which line the base of the catalytic pocket, to alanine results in a complete loss of catalytic activity in vitro (Fig. 2E) and in vivo (Fig. 4C). Moreover, as opposed to the wild-type protein, KDM2A harboring either the D214 or N298 mutation is unable to suppress functions that promote cellular transformation (Fig. 5G). Methylation of H3K36 is known to play a central role in the regulation of cancer. SETD2, the enzyme that catalyzes trimethylation of H3K36 (Edmunds et al. 2008; Li et al. 2009; Kuo et al. 2011), is mutated in a number of cancers, and H3.3K36M is a recurrent mutation in chondroblastoma, likely because the substitution renders a dominant-negative activity (Behjati et al. 2013). In contrast to H3K36me3, H3K36me2 is catalyzed by enzymes like NSD2 and is linked to oncogenic programming. Thus, KDM2A removal of H3K36me2 is consistent with this enzyme having a role in holding cellular transformation in check (Fig. 5F). Inspection of somatic mutation databases in cancers reveals mutations in the Jumonji domain of KDM2A. Future work aimed at understanding whether these mutations impact KDM2A enzymatic function and how KDM2A regulation of H3K36me2 impacts and integrates with other H3K36 methylation metabolizing enzymes should reveal important new insights into the role of chromatin in the regulation of cancer pathways.
Structural comparison of methylated lysine recognition by KDM2A and KDM4A
In addition to the structure of methylated H3K36 peptide bound to KDM2A reported in this study, structures have also been reported for methylated H3K9 and H3K36 peptides bound to KDM4A (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007) and for methylated H3K9 and H3K27 peptides bound to PHF8 and KIAA1718, respectively (Horton et al. 2010). The sequence alignments of these and other histone KDMs are listed in Supplemental Figure S3A and shown structurally in Supplemental Figure S3B.
There are distinctly different structural features involved in the recognition of methylated lysines by KDM2A and KDM4A. KDM2A targets and demethylates mono- and dimethylated states of H3K36 (Tsukada et al. 2006), while KDM4A targets and demethylates di- and trimethylated states of both H3K9 and, to a lesser extent, H3K36 (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007). Notably, the KDM2 sequence contains D214 and Y222, while the KDM4/5/6 sequences contain E214 and N222 at equivalent positions (Supplemental Fig. S2B). The KDM2A/B subfamily insertion loop located between α3 and α4 within the mixed domain plays a key role in the specific recognition of methylated lysines in the H3K36 sequence context. In contrast, an insertion loop is located between α5 and α6 within the JmjC domain of other KDMs, such as KDM4, KDM5, and KDM6 (Supplemental Fig. S3B). Interestingly, both KDM2 and KDM4 can bind the H3K36 sequence, but given that the insertion loops are located in different sections in the structure, the bound peptides adopt completely different orientations in the H3K36me3–KDM2A (Supplemental Fig. S4A) and H3K36me3–KDM4A (Supplemental Fig. S4B) complexes. In the case of the H3K36me3–KDM2A complex, the H3K36me3 side chain inserts into a deep cavity sequestered by the K323–F324 flap segment (this study), while in the H3K36me3–KDM4A complex, the peptide lies on the surface of the protein, and the K36me3 inserts into a surface groove (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007). These two distinct types of methylated H3K36-binding pockets in histone KDMs have been classified previously as “cavity insertion” and “surface groove” recognition modes, respectively (Taverna et al. 2007). Thus, histone KDMs can use very different peptide-binding channels and distinct types of methylated lysine recognition pockets to recognize the same sequence context. When overlaid, the active sites of H3K36me3–KDM2A and H3K36me3–KDM4A show several side chains of the KDMs occupying somewhat different positions between the two complexes (Supplemental Fig. S4C).
Structural comparison of methylated lysine recognition by KDM2A and PHF8
The JmjC domain of KDM2A exhibits ∼40% sequence identity with the KDMs of PHF8 and KIAA1718, with the latter enzymes additionally containing a PHD finger that “reads” H3K4me3 marks, while the JmjC domain “erases” H3K9me2 or H3K27me2 marks (Yang et al. 2007; Horton et al. 2010; Yue et al. 2010). The bound peptides adopt similar trajectories but opposite orientations in the H3K36me2–KDM2A (Supplemental Fig. S5A; this study) and H3K9me2–PHF8 (PDB ID: 3KV4) (Supplemental S5B; Horton et al. 2010) complexes. Interestingly, there are common features, despite recognition of different substrates and usage of different segments of their topologies for molecular recognition. PHF8 and KIAA1718 do not have any insertion loops within the JmjC domain, and their substrate specificities are determined by the PHD finger module outside of the JmjC domain (Horton et al. 2010). Thus, while KDM2A uses an insertion loop from the mixed domain that plays an important role in H3K36me2 substrate recognition (Supplemental Fig. S5A), PHF8 and KIAA1718 are promiscuous enzymes whose activity is dependent on the N-terminal PHD finger (reader of H3K4me3 mark) and linker length separating the PHD finger from the JmjC domain (Supplemental Fig. S5B). Thus, for PHF8, where the linker length is short and flexible, PHD domain binding of H3K4me3 enhances its JmjC domain-mediated demethylation of H3K9me2, while the converse holds for KIAA1718, where the linker length is longer but more rigid (Horton et al. 2010). Consistent with their methylation state specificity for lower-methylated lysine states, the active site residues are highly conserved between KDM2A and PHF8 (for stereo view, see Supplemental Fig. S5C), implying that they most likely use the same mechanism to differentiate lower-methylated lysine states from their trimethylated lysine counterparts.
Methylated lysine-binding catalytic pocket in the H3K36me2–KDM2A complex
H3K36me2 inserts into the catalytic pocket, with its methylene side chain accommodated within walls lined by hydrophobic (I144, L201, Y199, and F215) residues and its dimethylammonium group directed toward the base of the pocket lined by polar (D214 and N298) residues. The former hydrophobic residues make van der Waals contacts with the methylene side chain of K36me2, with the Y199A and F215A mutants resulting in a decrease in enzymatic activity to between 30% and 35% of the wild-type value (Fig. 2E). Of the latter polar residues, N298, whose position is buttressed by D214, is directed toward and hydrogen-bonds to the dimethylammonium proton of K36me2 (Fig. 2A,D), with D214A and N298A mutants resulting in complete loss in activity (Fig. 2F).
The bound Ni2+ (and, by inference, Fe2+) is coordinated by two His (H212 and H284) residues, D214, the cofactor (αKG or NOG), and a water molecule (Fig. 3). The coordination geometry of these residues around the Ni2+ is dependent on whether the cofactor analog NOG (Fig. 3G) or αKG (Fig. 3H) is bound. Importantly, the directionality of Y222 is also dependent on the cofactor, with the side chain of this tyrosine directed away from the Ni2+ in the presence of NOG (Fig. 3A–C) and predominantly toward Ni2+ in the presence of αKG (Fig. 3D–F). In the latter orientation, the tyrosyl hydroxyl forms a hydrogen bond with bound water (O•••O distance of 2.7 Å) (Fig. 3D–F).
Alignment of the methyl groups of methylated H3K36 in the catalytic pocket
Of the two methyl groups of K36me2, one is directed toward Ni2+ and the other is directed toward Y199 in the H3K36me2–KDM2A complex with both NOG (Fig. 3B) and αKG (Fig. 3E). The dimethylammonium proton of K36me2 is directed toward and forms a hydrogen bond with the side chain carbonyl oxygen of Asn298 in the complex with NOG (Fig. 3B) and with the C1 carboxyl oxygen of αKG in the complex with αKG (Fig. 3E). The observed methyl carbon to Ni2+ distance of 4.4 Å (for αKG) to 4.5 Å (for NOG) falls in the range reported for other JmjC-containing KDM complexes involved in lysine demethylation, such as the KDM6 subfamily (separation 4.25–4.3 Å) (Sengoku and Yokoyama 2011; Kruidenier et al. 2012), PHF8 (separation 4.7 Å) (Horton et al. 2010), and the KDM4A subfamily (separation 4.65–5.05 Å) (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007).
The monomethyl group of K36me1 adopts a single conformation directed away from the metal center in H3K36me1–KDM2A complexes with both NOG (Fig. 3A) and αKG (Fig. 3D). This alignment is due to the two monomethylammonium protons hydrogen-bonding to the C1 carboxylate oxygen of the cofactor and the side chain of D214 in the NOG-bound structure of the complex. The hydroxyl group from Y222 forms a hydrogen bond with a monomethylammonium proton of K36me1 in the αKG-bound structure of the complex, thereby contributing to its direction away from the metal center. In contrast, the monomethyl group adopts a single orientation directed toward the Fe2+ center in the H3K36me1–KDM2A complex containing αKG and Fe2+ under NO-producing anaerobic conditions, with a methyl carbon of methylated K36 to Fe2+ distance of 4.1 Å (Fig. 3I).
KDM2A has been shown to demethylate lower methylation states of H3K36 (Tsukada et al. 2006), and hence we were surprised by our ability to crystallize and solve the structures of H3K36me3–KDM2A complexes in the presence of both NOG (Fig. 3C) and αKG (Fig. 3F). The methyl group of H3K36me3 that is directed toward the Ni2+ exhibits carbon–metal distances in the 4.4 Å (for NOG) to 4.55 Å (for αKG) range. Thus, there has to be another explanation for the inability of KDM2A to demethylate the H3K36me3 group, given that the trimethyl group of K36 can be accommodated within the catalytic pocket.
“Off-line”-to-“in-line” transition for KDM2A demethylation of H3K36me2
Hausinger (2004) introduced the concept of “in-line” (Fig. 3G) and “off-line” (Fig. 3H) alignment modes aimed toward an improved structure-based mechanistic understanding of the Fe2+/αKG-dependent oxygenases within the context of the His1-X-Asp/Glu-Xn-His2 sequence motif required for chelation of the Fe2+ atom (for stereo view of superposition of H3K36me3–KDM2A complexes with αKG and NOG, see Supplemental Fig. S1E). In the “in-line” mode, an open metal coordination site (occupied by water) is located in the apical position (opposite His2) and directed toward the dimethylammonium group of K36me2 in the structure of the complex (Fig. 3B,G), thereby providing the optimal alignment for the demethylation reaction. In the “off-line” mode, the open metal coordination site (occupied by water) is located in the equatorial position (opposite His1) and directed away from the substrate in the structure of the complex (Fig. 3E,H). Thus, an “off-line” (observed for the H3K36me2–KDM2A complex in the presence αKG) (Fig. 3E) to “in-line” (observed for the complex in the presence NOG) (Fig. 3B) transition appears to be required for KDM2A-mediated demethylation of H3K36me2.
The KDM4/5/6 demethylases, which are involved in demethylation of higher methylation states of lysine, contain an Asn at the position of Tyr222 in KDM2A (Supplemental Fig. S2B). However, we did not observe H3K36me3-to-H3K36me2 conversion for Y222F and Y222N mutants of KDM2A, as monitored by mass spectrometry (data not shown). Thus, factors in addition to Y222 must contribute to discrimination between trimethylated and low-methylated substrates.
Principles underlying selective erasure of lower methylation states of H3K36 by KDM2A
Earlier studies on substrate binding to the αKG-dependent nonheme iron enzyme clavaminate synthase established that the αKG/Fe2+-dependent oxygenase binds Fe2+, αKG, and substrate in a sequential order (Zhou et al. 1998). A schematic of a proposed model of KDM2A-mediated demethylation as a function of H3K36 methylation state is shown in Supplemental Figure S6. In this proposed model, following Fe2+ binding to KDM2A (Supplemental Fig. S6, panel 1), αKG chelates the metal in an “off-line” mode (Supplemental Fig. S6, panel 2). On addition of substrate, αKG retains its original “off-line” position, with the inserted H3K36me2 (Supplemental Fig. S6, panel 3) and H3K36me3 (Supplemental Fig. S6, panel 4) side chains stabilized through interaction with the inwardly pointing Y222 side chain. Following diffusion of oxygen to the metal center, the αKG is postulated to undergo a rotation from the “off-line” to the “in-line” mode, as shown for the H3K36me2 (Supplemental Fig. S6, panel 5), accompanied by Y222 being directed away from the metal center. Indeed, evidence for metal-centered rearrangements has been reported previously for calvaminate synthase in the presence of Fe2+, αKG, substrate, and NO (Zhang et al. 2002). We also observed a conversion to an “in-line” mode (as reflected in the orientation of Y222), with density assigned to NO in an apical position in the H3K36me1–KDM2A complex in the presence of NO, Fe2+, and succinate (Fig. 3I). Although there are no steric impediments to rotation of αKG from the “off-line” to “in-line” modes for monomethyl- and dimethyl-modified H3K36 in their complexes with KDM2A (Supplemental Fig. S6, panels 3-to-5 step), a potential steric impediment (between the additional methyl group and the C1 carboxyate oxygen) could prevent rotation of αKG to the “in-line” mode in the trimethyl-modified H3K36 in its complex with KDM2A (Supplemental Fig. S6, panels 4-to-6 step). In subsequent steps 7 and 8 in Supplemental Figure S6, the CH3 group is converted to CH2OH, followed by loss of formaldehyde, thereby demethylating K36me2 substrate to K36me1 product.
Conclusion
We solved the crystal structures of KDM2A complexed with H3(29–41) peptides bearing different methylation states at K36 in the presence of Ni2+ and either cofactor αKG or nonreactive analog NOG. The side chain of methylated K36 (n = 0 position) inserts into the catalytic pocket occupied by Ni2+ and cofactor, where it is positioned and oriented for demethylation. The sequence specificity for demethylation of methylated H3K36 by KDM2A arises from a combination of constraints that includes the requirement of a narrow channel that can only be occupied by Gly residues at the n − 2 and n − 3 positions (G33–G34 in H3), the requirement of a turn-facilitating residue at the n + 2 position (P38 in H3), and the requirement of an aromatic residue at the n + 5 position (Y41 in H3) capable of inserting into a separate pocket. We mutated key residues involved in intermolecular contacts on H3K36me2–KDM2A formation, with enzymatic demethylation experiments and cellular assays validating the molecular basis underlying H3K36 sequence specificity of molecular recognition. Moreover, we observed that KDM2A attenuates a number of cellular phenotypes common to cancer cells and in a manner dependent on its catalytic pocket being intact.
Materials and methods
Crystallization and structure determination
Given that the KDM2A was purified on Ni-NTA columns, we checked for the divalent cation positioned within the catalytic pocket. It should be noted that neither Ni2+ nor Fe2+ was added before crystallization. The anomalous map shown in Supplemental Figure S7 indicates that the active center binds a single metal. In an effort to identify the metal type at the active center, we collected anomalous data sets at the K edges for Ni2+ (1.4848 Å) and Fe2+ (1.7389 Å) on the KDM2A complex with H3K36me3 peptide and αKG. The anomalous map of Fe2+ (Supplemental Fig. S7, red) has a much lower signal than that for Ni2+ (Supplemental Fig. S7, green) contoured at 3σ level, consistent with the active site containing predominantly a bound Ni2+ ion (Supplemental Fig. S7).
Crystals of KDM2A (lacking residues 1–35 and 365–449) bound to H3K36 peptide as a function of methylation state in the presence of Ni2+ and either NOG or αKG were grown by the vapor diffusion method at room temperature. The protein concentration was 18 mg/mL. The truncated KDM2A bound to H3K36me as a function of methylation state with either 5mM cofactor αKG or analog NOG and Ni2+ was crystallized in 100 mM Li-citrate and 18%–30% PEG 3350.
For generation of KDM2A–H3K36me1–NO crystals in the presence of αKG and Fe2+, the enzyme KDM2A was first treated by dialysis against 1 mM EDTA in 150 mM NaCl, 20 mM Tris HCl, and 1 mM DTT to remove the Ni2+. Apo-KDM2A was cocrystallized with H3K36me1 peptide in the presence of 5 mM αKG, and the crystal tray was transferred to an anaerobic glove box. The coverslips with the hanging crystal drops were transferred to a new crystal tray containing reservoirs with degassed mother liquor and left overnight to remove residual O2 from the drops. The crystals were reconstituted with Fe2+ by addition of anaerobic ferrous ammonium sulfate to 1.0 mM in the crystal drop mother liquor. After soaking for at least 24 h, crystals were transferred to cryoprotectant solution containing ∼2 mM NO for 2 min and then flash cooled in liquid nitrogen. The NO solution was formed by diluting a concentrated stock solution of the NO donor DEA-NONOate (Sigma) in 10 mM NaOH into an anaerobic cryoprotectant and incubating for 1 h in a sealed container.
The structures were solved by molecular replacement using the peptide-free KDM2A structure (PDB ID: 2YU1) using the program PHASER (McCoy et al. 2007). The peptide structures were determined by difference Fourier method and manually built in Coot (Emsley and Cowtan 2004). All refinements were performed in CNS (Brunger 2007) or phenix.refine in PHENIX (Adams et al. 2002). The final models exhibited good stereochemical quality. Figures representing the crystal structures were prepared with PyMol.
Additional materials and methods are described in detail in the Supplemental Material. These include information listing protein expression and purification and fluorescence-based lysine demethylase assays as well as molecular biology protocols listing reagents, plasmids, and antibodies; cell culture and transfection; RNAi knockdown strategy; immunofluorescence; and soft agar colony formation assay.
Accession codes
Coordinates and structure factors have been deposited with the following accession codes: NOG-bound structures of KDM2A with H3K36me1 (4QXH), H3K36me2 (4QXC), and H3K36me3 (4QXB) peptides; αKG-bound structures of KDM2A with H3K36me1 (4QWN), H3K36me2 (4QX7), and H3K36me3 (4QX8) peptides; and NO-bound structure of KDM2A with H3K36me1 peptide (4TN7).
Acknowledgments
This work was supported in part by grants from the Leukemia and Lymphoma Society and the STARR foundation to D.J.P., and from the National Institutes of Health (NIH) to O.G. (R01 GM079641), C.M.W. (R01 GM066569), and E.T.Y. (F32 GM097779). P.C. was supported by an NIH T32 training fellowship in immunology and rheumatology.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.246561.114.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Anand R, Marmorstein R. 2007. Structure and mechanism of lysine-specific demethylase enzymes. J Biol Chem 282: 35425–35429 [DOI] [PubMed] [Google Scholar]
- Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, et al. 2013. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 45: 1479–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Manning AL, Van Rechem C, Kim J, Ladd B, Cho J, Pineda CM, Murphy N, Daniels DL, Montagna C, et al. 2013. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell 154: 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. 2010. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 38: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT 2007. Version 1.2 of the crystallography and NMR system. Nat Protoc 2: 2728–2733 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, Lei M. 2006. Crystal structure of human histone lysine-specific demethylase 1 (LSD1). Proc Natl Acad Sci 103: 13956–13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zang J, Kappler J, Hong X, Crawford F, Wang Q, Lan F, Jiang C, Whetstine J, Dai S, et al. 2007. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc Natl Acad Sci 104: 10818–10823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Zhang X. 2007. Structural dynamics of protein lysine methylation and demethylation. Mutat Res 618: 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Collins RE, Zhang X. 2005. Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct 34: 267–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. 2006. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442: 307–311 [DOI] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. 2007. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol 14: 689–695 [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. 2010. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463: 360–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. 2009. JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature 461: 819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. 2008. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 27: 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. 2007. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450: 309–313 [DOI] [PubMed] [Google Scholar]
- Frescas D, Guardavaccaro D, Kuchay SM, Kato H, Poleshko A, Basrur V, Elenitoba-Johnson KS, Katz RA, Pagano M. 2008. KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle 7: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Liu P, Gu L, Zhang Y, Li H, Chen S, Chai J. 2007. Structural basis for histone demethylation by JHDM1. Fron Sci 1: 52–67 [Google Scholar]
- Hausinger RP 2004. Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol 39: 21–68 [DOI] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada YI, Zhang Y. 2008. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol 15: 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng XD. 2010. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol 17: 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou HF, Yu HT. 2010. Structural insights into histone lysine demethylation. Curr Opin Struct Biol 20: 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442: 312–316 [DOI] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. 2012. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 13: 297–311 [DOI] [PubMed] [Google Scholar]
- Kruidenier L, Chung CW, Cheng ZJ, Liddle J, Che KH, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, et al. 2012. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488: 404–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, et al. 2011. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell 44: 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ 3rd, Qiao Q, Neubert TA, Xu RM, Gozani O, et al. 2009. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem 284: 34283–34295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. 2005. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough MA, Loenarz C, Chowdhury R, Clifton IJ, Schofield CJ. 2010. Structural studies on human 2-oxoglutarate dependent oxygenases. Curr Opin Struct Biol 20: 659–672 [DOI] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y. 2010. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem 79: 155–179 [DOI] [PubMed] [Google Scholar]
- Musselman CA, Lalonde ME, Cote J, Kutateladze TG. 2012. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 19: 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CJ, Santos-Rosa H, Kouzarides T. 2006. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126: 905–916 [DOI] [PubMed] [Google Scholar]
- Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BMR, Bray JE, Savitsky P, Gileadi O, von Delft F, et al. 2007. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 448: 87–91 [DOI] [PubMed] [Google Scholar]
- Patel DJ, Wang Z. 2013. Readout of epigenetic modifications. Annu Rev Biochem 79: 81–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Zhou MM. 2006. SET domain protein lysine methyltransferases: structure, specificity and catalysis. Cell Mol Life Sci 63: 2755–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231 [DOI] [PubMed] [Google Scholar]
- Sengoku T, Yokoyama S. 2011. Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev 25: 2266–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Okamoto K, Teye K, Umata T, Yamagiwa N, Suto Y, Zhang Y, Tsuneoka M. 2010. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J 29: 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Upadhyay AK, Horton JR, Zhang X, Cheng XD. 2011. Coordinated methyl-lysine erasure: structural and functional linkage of a Jumonji demethylase domain and a reader domain. Curr Opin Struct Biol 21: 750–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegard K, Terwisscha van Scheltinga AC, Dubus A, Ranghino G, Oster LM, Hajdu J, Andersson I. 2004. The structural basis of cephalosporin formation in a mononuclear ferrous enzyme. Nat Struct Mol Biol 11: 95–101 [DOI] [PubMed] [Google Scholar]
- Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G, et al. 2011. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 9: 575–587 [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen ZZ, Spooner E, Li E, Zhang GY, Colaiacovo M, et al. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125: 467–481 [DOI] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125: 483–495 [DOI] [PubMed] [Google Scholar]
- Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, Machius M, Cole PA, Yu H. 2007. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat Struct Mol Biol 14: 535–539 [DOI] [PubMed] [Google Scholar]
- Yue WW, Hozjan V, Ge W, Loenarz C, Cooper CD, Schofield CJ, Kavanagh KL, Oppermann U, McDonough MA. 2010. Crystal structure of the PHF8 Jumonji domain, an Nε-methyl lysine demethylase. FEBS Lett 584: 825–830 [DOI] [PubMed] [Google Scholar]
- Yukl ET, Goblirsch BR, Davidson VL, Wilmot CM. 2011. Crystal structures of CO and NO adducts of MauG in complex with pre-methylamine dehydrogenase: implications for the mechanism of dioxygen activation. Biochemistry 50: 2931–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ren J, Harlos K, McKinnon CH, Clifton IJ, Schofield CJ. 2002. Crystal structure of a clavaminate synthase–Fe(II)-2–oxoglutarate–substrate–NO complex: evidence for metal centered rearrangements. FEBS Lett 517: 7–12 [DOI] [PubMed] [Google Scholar]
- Zhou J, Gunsior M, Bachmann BO, Townsend CA, Solomon EI. 1998. Substrate binding to the α-ketoglutarate-dependent non-heme iron enzyme clavaminate synthase 2: coupling mechanism of oxidative decarboxylation and hydroxylation. J Am Chem Soc 120: 13539–13540 [Google Scholar]