To develop stem cell therapy for small intestinal (SI) diseases, it is essential to determine whether SI stem cells in culture retain their tissue regeneration capabilities. Fukuda et al. demonstrate that cultured murine SI epithelial organoids are able to reconstitute self-renewing epithelia in the colon. The SI-derived grafts maintain many features unique only to the SI. This study provides evidence that cultured adult SI stem cells could be a source for cell therapy of intestinal diseases, maintaining their identity along the gastrointestinal tract through an epithelium-intrinsic mechanism.
Keywords: small intestinal stem cell, transplantation, stem cell niche, Paneth cell, regenerative medicine
Abstract
To develop stem cell therapy for small intestinal (SI) diseases, it is essential to determine whether SI stem cells in culture retain their tissue regeneration capabilities. By using a heterotopic transplantation approach, we show that cultured murine SI epithelial organoids are able to reconstitute self-renewing epithelia in the colon. When stably integrated, the SI-derived grafts show many features unique only to the SI but distinct from the colonic epithelium. Our study provides evidence that cultured adult SI stem cells could be a source for cell therapy of intestinal diseases, maintaining their identity along the gastrointestinal tract through an epithelium-intrinsic mechanism.
Recent advances have enabled long-term culture and expansion of epithelial stem cells of the small intestine (SI) and colon in vitro (Ootani et al. 2009; Sato et al. 2009, 2011a; Jung et al. 2011). With the advent of such technologies, there has emerged a growing interest in the use of these cultured adult stem cells in replacement therapy for intestinal epithelial injuries. We demonstrated previously that Lgr5+ colonic stem cells are able to regenerate normal epithelia when expanded in culture and transplanted onto a damaged colon in mice (Yui et al. 2012). To extend such an approach to SI diseases, it is imperative to test whether SI stem cells in culture retain their tissue regeneration capabilities.
Despite common features, epithelia of the SI and colon show many differences. The former builds protruding villus structures that are connected to glandular structures called crypts, whereas the latter only forms crypts (Sancho et al. 2004). They also differ in their cellular components, as represented by Paneth cells that reside only in the SI and play a role in innate immunity (Clevers and Bevins 2013). Such heterogeneity may result from organ-specific, developmentally acquired programs in respective epithelium and environmental cues from the surrounding nonepithelial components in respective tissues. However, how epithelium-intrinsic and epithelium-extrinsic factors contribute to the establishment and maintenance of intestinal identity remains unclear.
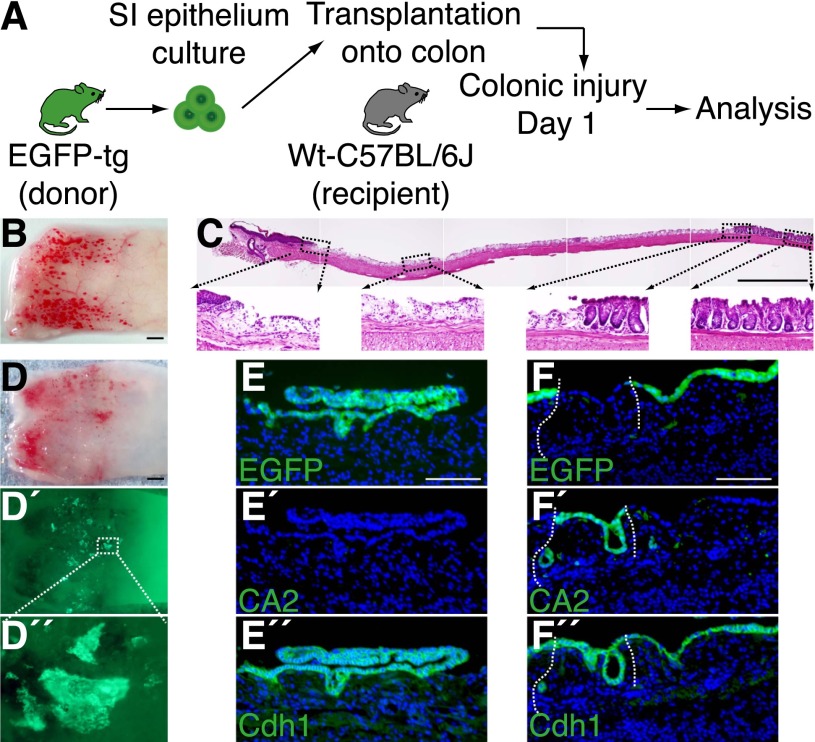
In this study, by employing the cell transplantation technique developed in our laboratory, we investigated whether cultured SI stem cells are capable of regenerating epithelia in vivo and how adult SI epithelial cells behave when heterotopically transplanted onto the colon (Fig. 1A).
Figure 1.
Cultured SI epithelial cells repopulate onto injured colons upon transplantation. (A) Experimental protocol. (B) Colonic injury model used in the present study. Stereoscopic image of the distal colon immediately after generation of injury. (C) H&E staining of the longitudinally sectioned colon shown in B. The anal side is shown on the left, and the oral side is on the right. High-power views of areas in dotted boxes are shown below. (D) Recipient colon at 1 d after transplantation. Stereoscopic image (D) and its fluorescent (EGFP) view (D′). High-power view of the dotted rectangle is shown in D″. (E,F) Histological analysis of the graft shown in D. EGFP fluorescence (E,F), immunohistochemistry for CA2 (E′,F′) and Cdh1 (E″,F″) are shown. The top two images (E,E′,F,F′) are the results sequentially obtained from the same section, and the bottom images (E″,F″) are their adjacent sections. Note that the antigen retrieval procedure before immunostaining completely clears endogenous EGFP signals. Fluorescent images are presented with DAPI staining. White dotted lines show borders between EGFP+ and EGFP− epithelia. Bars: B–D, 1 mm; E,F, 100 μm.
Results and Discussion
To obtain denuded colonic regions that would potentially allow engraftment of transplanted SI cells, we developed a new mouse recipient model of colonic mucosal injury. The distal colonic lumen was exposed to a chelating agent, and then mechanical epithelial abrasion was performed. This procedure yielded mucosal injuries along the whole circumference of the distal colonic segment (Fig. 1B). Histology showed that the epithelial layer was largely stripped, with small, discrete epithelial elements left behind, whereas the underlying tissue remained almost intact (Fig. 1C). Shortly thereafter, epithelial repair occurred rapidly and was nearly completed within 4 wk (Supplemental Fig. S1).
SI epithelial cells were isolated from adult EGFP transgenic mice (Okabe et al. 1997) and cultured for 7 d as stem cell-containing organoids (Sato et al. 2009). They were then collected and instilled into the colonic lumen of syngeneic, immunocompetent wild-type recipients in which colonic epithelial injury was generated beforehand (day 1). One day after transplantation (day 2), we found that EGFP+ cells showed a scattered distribution on the recipient colon (Fig. 1D–D″). The EGFP+ areas were mostly composed of a cluster of cells, implying that the transplanted cells adhered to the tissue still partly preserving their organoid structure (Fig. 1D′,D″). Histological analysis revealed a variety of appearances in those areas. Some displayed a convoluted, continuous lining of EGFP+ cells, a part of which directly contacted with the denuded tissue (Fig. 1E). In other areas, the EGFP+ cells covered the luminal surface as a flat lining of single-layered cells (Fig. 1F). The grafted cells were shown to remain as epithelial cells, since they were all positive for Cdh1 (Fig. 1E″,F″). Fordham et al. (2013) recently reported that SI epithelial progenitors of fetal origin are able to grow as fetal enterospheres (FEnSs) in vitro. When transplanted onto the adult colon, FEnS-derived cells showed plasticity in regard to their cell fate by expressing CA2, a marker protein of colonic epithelium (Fordham et al. 2013). Interestingly, the EGFP+ areas derived from adult SI cells in this study did not show obvious expression of CA2 (Fig. 1E′). This was clearly visible in contrast to its exclusive expression in the EGFP− epithelium of recipient origin, which survived the damage and intervened in two separate EGFP+ areas of donor origin (Fig. 1F′). It is thus shown that adult SI epithelial cells in culture are able to repopulate onto the colon in a manner different from that of fetal SI progenitor cells.
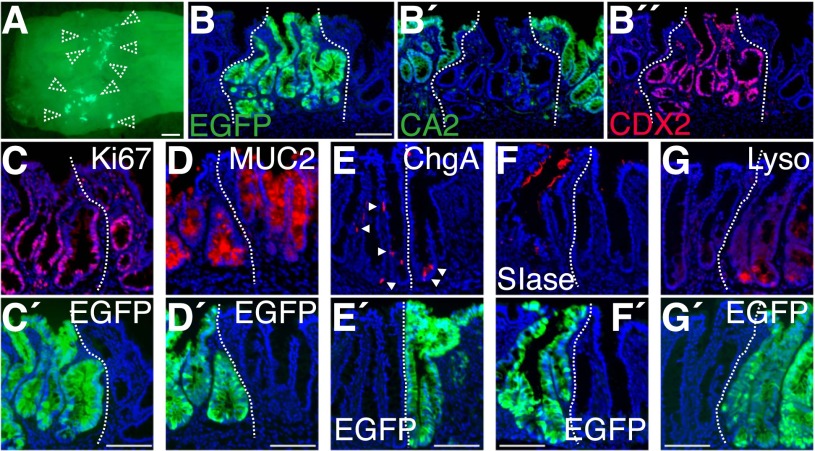
At 2 wk post-transplantation (Fig. 2A), EGFP+ cells displayed intricate structures containing many invaginations extending downward (Fig. 2B). They were still devoid of CA2 expression (Fig. 2B′). We found that expression of CDX2, an intestine-specific transcription factor that plays important roles in regional maintenance of gastrointestinal epithelial cells (Silberg et al. 2000; Gao et al. 2009), was clearly demarcated by the borders between EGFP− and EGFP+ epithelia (Fig. 2B″). This recapitulated its high expression in the SI and low abundance in the distal colon of adult mice (Silberg et al. 2000), suggesting that adult SI-derived donor cells retain their original identity.
Figure 2.
SI epithelial cells reconstitute self-renewing epithelia of SI phenotype in the colon. (A) Recipient colon at 2 wk post-transplantation. Stereoscopic image overlaid with EGFP fluorescence. Dotted arrowheads show EGFP+ grafts. (B) EGFP fluorescence of a section obtained from the tissue shown in A. The same section was double-stained for CA2 (B′) and CDX2 (B″). (C–G) Immunofluorescence for Ki67 (C), MUC2 (D), ChgA (E), SIase (F), and lysozyme (G). Arrowheads point to ChgA+ cells. Images of adjacent sections stained with GFP-specific antibody (C′,G′) or endogenous EGFP fluorescence of the same sections (D′,E′,F′) are shown at the bottom. All fluorescent images are presented with DAPI staining. White dotted lines show borders between EGFP+ and EGFP− epithelia. Bars: A, 1 mm; B–G, 100 μm.
EGFP+ areas contained Ki67+ cells at the lower compartment of invaginations, indicating that SI-derived cells proliferated actively in the colon (Fig. 2C). Differentiated cell types—such as Goblet cells, enteroendocrine cells, and sucrase isomaltase (SIase)-expressing absorptive cells of the SI—were also detected (Fig. 2D–F). In addition, lysozyme+ Paneth-like cells were detected exclusively in the EGFP+ epithelium (Fig. 2G). These results suggested that the transplanted cells are able to reconstitute self-renewing epithelia of SI phenotype in vivo even after being heterotopically engrafted. The retention of SI phenotype is unlikely to be a feature acquired through the culture process, as we obtained similar results when freshly isolated SI crypts were immediately transplanted and then recipients were analyzed at 2 wk (Supplemental Fig. S2).
At 4 wk, the EGFP+ epithelia were found to be stably incorporated in the colon (Fig. 3A). Within some but not all areas of grafts, there emerged structures reminiscent of typical architecture of the SI epithelium. Cells constituting finger-like projections were directly linked to flask-shaped, crypt-like invaginations (Fig. 3A″). The tissue lying beneath the EGFP+ epithelium contained a number of cellular components of recipient origin, such as immune cells (CD3+ and F4/80+) and CD31+ blood vessel endothelial cells (Supplemental Fig. S3), indicating that epithelial and nonepithelial elements of different origins cooperatively assembled into the crypt/villus structure. A recent report demonstrated that the villus formation occurs as a result of stepwise differentiation of the outer smooth muscle layers, which restricts the planar expansion of growing epithelium/mesenchyme and thereby generates protrusions into the lumen (Shyer et al. 2013). This, however, described the process of villi specification in fetal stages and may not account for our data in which epithelial reorganization occurred in fully mature adult colons. Other reports showed that the epithelial–mesenchymal cross-talk regulates the patterning of villi through a mechanism involving PDGFRα-expressing mesenchymal cells (Madison et al. 2005; Walton et al. 2012). In our attempts, however, the clusters of PDGFRα+ cells, which were reported to emerge at the site of villus formation (Walton et al. 2012), were not detectable in subepithelia of any grafts (data not shown). This suggests that some additional mechanisms might be involved in morphogenesis of the villus-like structures in this artificial situation in which epithelial and nonepithelial components of different origins coexist.
Figure 3.
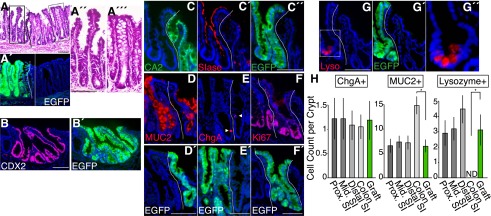
SI phenotypes are preserved in stably integrated SI-derived grafts at 4 wk post-transplantation. (A) Histological analysis of the recipient colon at 4 wk post-transplantation. H&E staining of a section (A) and its EGFP fluorescence (A′). High-power views of areas in dotted boxes in A are shown as A″ and A′″. (B,B′) Immunostaining for CDX2 (B) and an adjacent section stained with GFP-specific antibody (B′). (C–C″) Section double-stained with CA2 (C) and SIase (C′) is presented, with the adjacent section stained with GFP-specific antibody (C″). (D–G) Immunofluorescence for MUC2 (D), ChgA (E), Ki67 (F), and lysozyme (G). Arrowheads point to ChgA+ cells. Images of adjacent sections stained with GFP-specific antibody (F′) or those of endogenous EGFP fluorescence of the same sections (D′,E′,G′) are also shown. High-power view of the area within the dotted box or rectangle in G is shown in G″. (H) Quantification of cellular components. Mean cell counts of ChgA+, MUC2+, or lysozyme+ cells per crypt–villus or crypt unit are presented for the proximal (Prox. SI), middle (Mid. SI), and distal (Distal SI) SI; the colon; and EGFP+ grafts (Graft). Error bars indicate SEM. (*) P < 0.05. All fluorescent images are shown with DAPI staining. Dotted lines indicate borders between EGFP+ and EGFP− epithelia. Bars: A–G, 100 μm.
EGFP+ epithelial cells at 4 wk retained a higher expression of CDX2 than the surrounding recipient colon (Fig. 3B). They were negative for CA2 expression (Fig. 3C), which was in sharp contrast to the inverted expression pattern of SIase (Fig. 3C′). Goblet cells (Fig. 3D), enteroendocrine cells (Fig. 3E), and Ki67+ proliferating cells (Fig. 3F) were present in the EGFP+ epithelium. In addition, lysozyme+ Paneth cells were found at the bottom-most regions of the EGFP+ crypts, as in physiological SI epithelium (Fig. 3G). Even 4 mo after transplantation, multidifferentiation and self-renewal capabilities of the grafted cells remained stable. The EGFP+ epithelia formed distinguishable crypt/villus structures that contained cells of multilineages as well as proliferating cells (Supplemental Fig. S4). When quantitative analysis was performed with the grafts at 4 wk post-transplantation, the numbers of Goblet cells (MUC2+) and Paneth cells (lysozyme+) in EGFP+ areas showed a significant difference from those in normal colonic epithelium. Rather, they were comparable with those in the SI epithelium from which the grafted cells originated (Fig. 3H).
To verify this observation at the molecular level, gene expression profiles of EGFP+ epithelium in the graft (4 wk post-transplantation) and control colonic epithelium were assessed by laser capture microdissection and microarray analysis (Supplemental Fig. S5). Comparative analysis of the data identified 452 probe sets (see the Supplemental Material) that show enrichment in either of the two data sets by more than threefold (Supplemental Table S1). Of note, among those that are highly enriched in the graft are many genes known to be expressed in Paneth cells, such as α-defensins (Amid et al. 2009; Ouellette 2011) and lyz1 (Keshav 2006). Genes involved in transporter activity are also expressed differently in these two tissues. For example, compared with the colonic epithelium, the graft epithelium showed higher expression of genes encoding solute carrier (SLC) family members (slc26a6 and slc7a9) and non-SLC transporters (fabp1 and rbp2) but lower expression of other SLC genes (slc20a1, slc26a2, slc35a1, slc40a1, and slc6a14). Differential expression levels of these transporter genes between these two samples are consistent with their differentially regulated expression patterns in the SI and colon in vivo (Anderle et al. 2005). In addition, the genes enriched in the graft contain pdx1 (Offield et al. 1996) and gata4 (Bosse et al. 2006; Middendorp et al. 2014), both of which encode transcription factors that control region-specific gene expression in the SI. These data suggested that, even in the colonic milieu, the graft epithelium retains their commitment to the SI phenotype with preservation of functional Paneth cells and expression of a particular set of SI-specific genes.
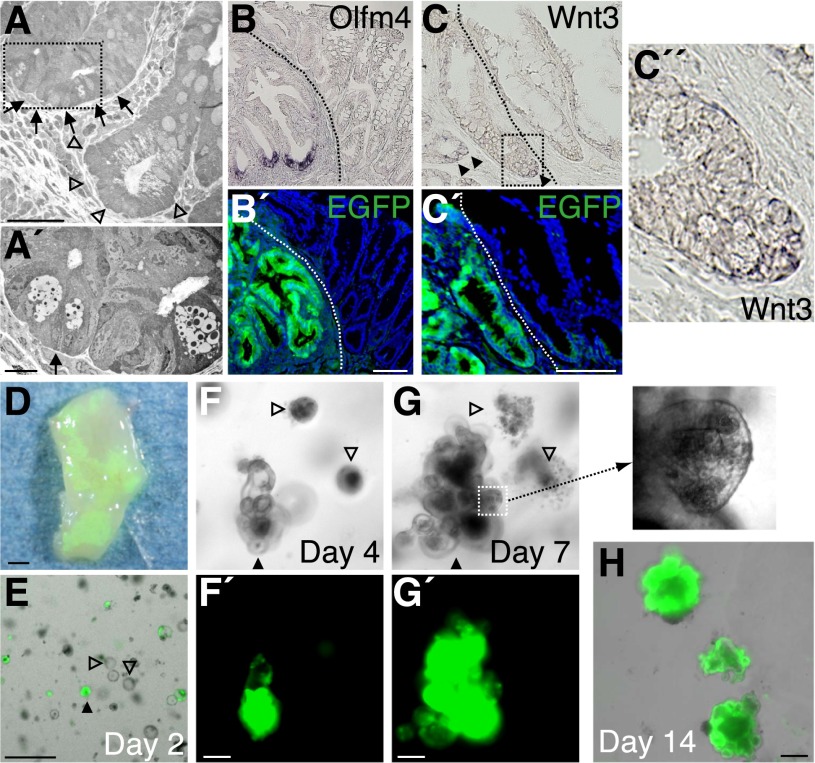
Multipotent, rapidly cycling stem cells known to express Lgr5 reside at the base of crypts in both the SI and colon (Barker et al. 2007). The Lgr5+ SI stem cells are morphologically characterized as crypt base columnar (CBC) cells interspersed with Paneth cells (Sato et al. 2011b). In contrast, the cells of CBC phenotype are not present in the colon, where classical Paneth cells are absent. By transmission electron microscopy, we were able to document that a crypt of SI phenotype, which contained Paneth-like cells with discernible secretory granules, was located immediately adjacent to a “goblet cell-rich” crypt of colonic phenotype on the contiguous mesenchyme (Fig. 4A). When the neighboring section was examined at a higher magnification, a slender columnar cell wedged between two Paneth-like cells was visible, which resembled the CBC cells in the SI (Fig. 4A′). Stem cells in the grafted areas were examined further. Among several genes that label Lgr5+ SI stem cells (van der Flier et al. 2009b; Hao et al. 2012; Koo et al. 2012; Munoz et al. 2012), Olfm4, a gene encoding a secreted protein of the Olfactomedin family, is known to mark SI but not colonic stem cells in mice (van der Flier et al. 2009a,b). When boundary areas were examined by in situ hybridization (ISH), Olfm4 was found to be present at the bottom of EGFP+ crypts but not in EGFP− areas (Fig. 4B). These results suggest that crypt base cells in the graft maintain the structural and functional properties of adult SI stem cells.
Figure 4.
SI-derived grafts contain stem cell compartments of SI phenotype. (A) Transmission electron microscopy analysis for graft at 4 wk post-transplantation. Crypt of SI phenotype (arrows) located immediately adjacent to that of colonic phenotype (arrowheads). High-magnification view of a neighboring section corresponding to the area within the dotted rectangle is shown in A′. The arrow points to a slender columnar cell wedged between Paneth-like cells. (B) In situ hybridization for Olfm4 on tissue 4 wk post-transplantation. (B′) An adjacent section immunostained for EGFP is also shown. (C) In situ hybridization for Wnt3. (C′) An adjacent section immunostained for EGFP is shown. (C″) A high-power view of the area within the dotted box or rectangle is also presented. (D) Portion of recipient colon at 8 wk post-transplantation from which epithelia were reisolated. The epithelium was then cultured in medium supplemented with EGF, Noggin, and R-spondin1 but without Wnt3a. (E) Image of reisolated cells 1 d after the start of culture (day 2). Merged image of phase-contrast and EGFP fluorescence views. (F,G) Images of culture on day 4 (F) and day 7 (G). Phase-contrast images (F,G) and their EGFP signals (F′,G′) are shown. A high-power view of the area in the dotted square is shown at the right. A filled triangle and open triangles in E, F, and G denote the identical structures tracked throughout the experiment. (H) Merged image of phase-contrast and EGFP fluorescence views of day 14 organoids grown after passage. All fluorescent images of fixed tissues are shown with DAPI staining. Dotted lines indicate borders between EGFP+ and EGFP− epithelia. Bars: A, 50 μm; A′, 10 μm; B,C,F–H, 100 μm; E, 500 μm; D, 1 mm.
Both epithelial and nonepithelial cells express Wnts and constitute a niche for Lgr5+ stem cells (Gregorieff et al. 2005; Sato et al. 2011b). In the SI, Paneth cells express many Wnts and thereby function as the major component of “epithelial niche” for adjacent Lgr5+ stem cells (Sato et al. 2011b). Among many Wnts, Wnt3 is considered to be a pivotal niche factor, as its depletion results in the growth impairment of SI epithelial cells in vitro (Farin et al. 2012). Such function of Wnt3 may be predominant in the SI, as it is expressed in Paneth cells but not at a detectable level in the colon (Farin et al. 2012). When examined by ISH, Wnt3 was shown to be expressed only at the base of EGFP+ crypts (Fig. 4C), suggesting that these Wnt3-producing cells—most likely Paneth cells—are functional as a part of the stem cell niche in the grafts of SI origin.
The SI epithelium is known to behave differently from the colonic one in in vitro culture. Since Paneth cells serve as a local source of Wnts, SI cells grow asymmetrically, building many protrusions that resemble in vivo crypt structures (Sato et al. 2009). In contrast, due to the absence of Paneth cells, colonic cells require exogenous Wnts in culture (Jung et al. 2011; Sato et al. 2011a; Yui et al. 2012). To assess more directly the functional properties of graft cells, epithelia were isolated from a recipient colon at 2 mo post-transplantation (Fig. 4D) and cultured in the absence of Wnt3. While EGFP+ graft cells and EGFP− colonic cells both survived as simple spherical cysts for the first few days (Fig. 4E,4F), EGFP− cysts eventually collapsed and degenerated (Fig. 4G), reflecting the absolute dependence of colonic cell culture on exogenous Wnts. On the other hand, EGFP+ cysts matured into organoids of SI phenotype (Fig. 4G) and could be further propagated (Fig. 4H). Notably, the EGFP+ organoids generated many crypt-like budding structures harboring granule-rich Paneth cells (Fig. 4G). These in vitro data clearly show the continued presence of SI stem cells in the graft, which give rise to competent Paneth cells that in turn produce sufficient niche signals for stem cells.
In summary, we show here that cultured adult SI stem cells are capable of reconstituting epithelial tissues of SI phenotype in vivo, with functional Paneth cells that serve as a niche component. In addition to previous studies on adult colon stem cells (Yui et al. 2012) and fetal SI progenitor cells (Fordham et al. 2013), this study adds the adult SI stem cells to the list of transplantable cells for treatment of intestinal diseases. Moreover, our study highlights the epithelium-intrinsic program that allows for maintenance of organ-specific stem cell properties in adult intestines.
Materials and methods
Mice
Six-wk-old to 10-wk-old EGFP transgenic mice of C57BL6 background (Okabe et al. 1997) were used as a source for donor cells. C57BL6 mice of 6–10 wk of age were used as recipients. All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee of Tokyo Medical and Dental University.
Isolation and culture of SI crypts
SI crypts were isolated from EGFP transgenic mice as described previously (Sato et al. 2009). Epithelial reisolation from the recipient colon was performed according to the method previously described for colonic crypt isolation (Yui et al. 2012). A total of 300 crypts was suspended in 30 μL of Matrigel (BD Biosciences) and placed in 24-well plates. After polymerization, 500 μL of advanced DMEM/F12 containing 500 ng/mL mRspo1 (R&D Systems), 20 ng/mL mEGF (Peprotech), and 100 ng/mL mNoggin (R&D Systems) was added to each well. The medium was changed every 3 d until the following use of cultured cells for transplantation experiments.
Generation of colonic epithelial injury
C57BL6 mice were anesthetized with an intraperitoneal injection of tribromoethanol solution (0.5 mg/g body). Topical exposure of the colonic mucosa to ethylenedinitrilo tetraacetic acid (EDTA) was achieved by the use of a thin catheter (1 mm in diameter) that was equipped with two small balloons (Supplemental Fig. S1). The catheter was inserted 1 cm via a transanal approach, and the balloons were inflated with air to hold them in place. Another thin catheter was inserted, and the luminal space between two balloons was filled with 500 μL of 500 mM EDTA/phosphate-buffered saline (PBS). After 2 min of exposure, the lumen was emptied by aspiration of the EDTA solution. After deflating the balloons, both catheters were removed out of the colon. Epithelial abrasion was performed with an electric brush (Supplemental Fig. S1). The head of the device was inserted 1 cm into the colon, and a rotational movement was given for 1 min to gently scratch the entire circumference of the luminal surface.
Transplantation
Typically, from a single mouse, our isolation procedure liberates as many SI crypts as can be seeded into ∼200 wells on 24-well plates. For transplantation, SI organoids were grown for 7 d and released from Matrigel using cell recovery solution (Corning). Cell suspension was made without further dissociation of the structure, so the organoids equivalent to those harvested from 200 wells were suspended in 2 mL of a diluted Matrigel/PBS (1:10) solution. Two-hundred microliters of cell suspension was instilled into the colonic lumen of each recipient mouse by using a syringe and a thin flexible catheter 4 cm in length and 2 mm in diameter. After infusion, the anal verge was sutured and left for 12 h to prevent luminal contents from being excreted immediately. After the procedure, mice were maintained as usual before they were sacrificed and analyzed.
Stereomicroscopy, histology, and immunohistochemistry
Whole distal colons of recipients and their fluorescence were imaged using a fluorescence stereomicroscope system (MVX10, Olympus). Protocols for histology and immunohistochemistry and the antibodies used in this study are included in the Supplemental Material. Fluorescent images of sections were acquired using a DeltaVision system (Applied Precision). For quantification of cellular components, sections of the proximal, middle, and distal SI, colon, and EGFP+ grafts were subjected to immunohistochemistry for ChgA, MUC2, or lysozyme. Thirty crypt–villus units or crypt units (colon), originating from three independent recipients or normal controls, were analyzed. The numbers of ChgA+, MUC2+, or lysozyme+ cells were counted and presented as mean cell counts per crypt–villus or crypt unit. Statistical significance was determined by Student’s t-test (P < 0.05).
Transmission electron microscopy
Recipient colons were cut at the area of EGFP+ grafts so that the cutting plane would contain EGFP+ epithelium. Pieces of tissues were fixed with 2.5% glutaraldehyde in 0.1 M PBS for 2 h, washed overnight at 4°C in the same buffer, post-fixed with 1% OsO4 buffered with 0.1 M PBS for 2 h, dehydrated in a graded series of ethanol, and embedded in Epon 812. Ultrathin (90-nm) sections were first collected on single-hole meshes in order to acquire low-magnification images that contained crypts of SI and colonic phenotypes located next to each other. Next, the adjacent sections were collected on copper grids to obtain a magnified view of those areas. Sections were double-stained with uranyl acetate and lead citrate and then examined with a transmission electron microscope (H-7100, Hitachi).
ISH
ISH for Olfm4 and Wnt3 was performed essentially as described previously (Gregorieff et al. 2005; Sato et al. 2011b). A pcDNA3 plasmid containing a cDNA fragment of mouse Olfm4 (nucleotides 259–651; GenBank accession no. NM001030294.1) was constructed. A plasmid containing a fragment of mouse Wnt3 (Gregorieff et al. 2005; Sato et al. 2011b) was a kind gift from Professor Hans Clevers (Hubrecht Institute). Single-stranded, digoxigenin-labeled RNA probes were generated by in vitro transcription system (Roche). Frozen sections were rehydrated, treated with HCl, digested in proteinase K solution, post-fixed, treated in acetic anhydride solution, and hybridized overnight at 68°C with probes. After extensive rinsing and washing, sections were then subjected to the immunohistochemistry by using alkaline phosphatase–conjugated anti-digoxigenin antibody (Roche). Sections were then reacted with a nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution for color development. Image were acquired on a microscope (BZ-8000, Keyence).
Comparison of gene expression profiles
Protocols for laser capture microdissection and microarray analysis can be found in the Supplemental Material. The microarray data were deposited into Gene Expression Omnibus (GEO) under the accession number GSE59410.
Acknowledgments
We thank Professor Hans Clevers and Dr. Henner Farin (Hubrecht Institute) for technical assistance, and Lesa Thompson for comments on the manuscript. This study was supported by Ministry of Education, Culture, Sports, Science, and Technology (MEXT) KAKENHI (grant no. 24112508), Japan Society for the Promotion of Science (JSPS) KAKENHI (25670106, 24390186, and 22229005), the Regenerative Medicine Realization Base Network Program from the Japan Science and Technology Agency, and Health and Labour Sciences Research Grants for research on Intractable Diseases from the Ministry of Health, Labor, and Welfare of Japan.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.245233.114.
References
- Amid C, Rehaume LM, Brown KL, Gilbert JG, Dougan G, Hancock RE, Harrow JL. 2009. Manual annotation and analysis of the defensin gene cluster in the C57BL/6J mouse reference genome. BMC Genomics 10: 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderle P, Sengstag T, Mutch DM, Rumbo M, Praz V, Mansourian R, Delorenzi M, Williamson G, Roberts MA. 2005. Changes in the transcriptional profile of transporters in the intestine along the anterior-posterior and crypt–villus axes. BMC Genomics 6: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Bosse T, Piaseckyj CM, Burghard E, Fialkovich JJ, Rajagopal S, Pu WT, Krasinski SD. 2006. Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol Cell Biol 26: 9060–9070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers HC, Bevins CL. 2013. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol 75: 289–311 [DOI] [PubMed] [Google Scholar]
- Farin, H.F., Van Es, J.H., and Clevers, H. 2012. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143: 1518–1529.e7 [DOI] [PubMed] [Google Scholar]
- Fordham RP, Yui S, Hannan NR, Soendergaard C, Madgwick A, Schweiger PJ, Nielsen OH, Vallier L, Pedersen RA, Nakamura T, et al. 2013. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell 13: 734–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, White P, Kaestner KH. 2009. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell 16: 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Pinto D, Begthel H, Destree O, Kielman M, Clevers H. 2005. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology 129: 626–638 [DOI] [PubMed] [Google Scholar]
- Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, Lei H, Mickanin C, Liu D, Ruffner H, et al. 2012. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485: 195–200 [DOI] [PubMed] [Google Scholar]
- Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, et al. 2011. Isolation and in vitro expansion of human colonic stem cells. Nat Med 17: 1225–1227 [DOI] [PubMed] [Google Scholar]
- Keshav S 2006. Paneth cells: leukocyte-like mediators of innate immunity in the intestine. J Leukoc Biol 80: 500–508 [DOI] [PubMed] [Google Scholar]
- Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM, et al. 2012. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488: 665–669 [DOI] [PubMed] [Google Scholar]
- Madison BB, Braunstein K, Kuizon E, Portman K, Qiao XT, Gumucio DL. 2005. Epithelial hedgehog signals pattern the intestinal crypt–villus axis. Development 132: 279–289 [DOI] [PubMed] [Google Scholar]
- Middendorp S, Schneeberger K, Wiegerinck CL, Mokry M, Akkerman RD, van Wijngaarden S, Clevers H, Nieuwenhuis EE. 2014. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 32: 1083–1091 [DOI] [PubMed] [Google Scholar]
- Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, et al. 2012. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J 31: 3079–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. 1996. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122: 983–995 [DOI] [PubMed] [Google Scholar]
- Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. 1997. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett 407: 313–319 [DOI] [PubMed] [Google Scholar]
- Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, et al. 2009. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med 15: 701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette AJ 2011. Paneth cell α-defensins in enteric innate immunity. Cell Mol Life Sci 68: 2215–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho E, Batlle E, Clevers H. 2004. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol 20: 695–723 [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. 2009. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459: 262–265 [DOI] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, et al. 2011a. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141: 1762–1772 [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. 2011b. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyer AE, Tallinen T, Nerurkar NL, Wei Z, Gil ES, Kaplan DL, Tabin CJ, Mahadevan L. 2013. Villification: how the gut gets its villi. Science 342: 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg DG, Swain GP, Suh ER, Traber PG. 2000. Cdx1 and cdx2 expression during intestinal development. Gastroenterology 119: 961–971 [DOI] [PubMed] [Google Scholar]
- van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. 2009a. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 137: 15–17 [DOI] [PubMed] [Google Scholar]
- van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, et al. 2009b. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136: 903–912 [DOI] [PubMed] [Google Scholar]
- Walton KD, Kolterud A, Czerwinski MJ, Bell MJ, Prakash A, Kushwaha J, Grosse AS, Schnell S, Gumucio DL. 2012. Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc Natl Acad Sci 109: 15817–15822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K, et al. 2012. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat Med 18: 618–623 [DOI] [PubMed] [Google Scholar]