Centromere identity and its epigenetic maintenance require the incorporation of the histone H3 variant CENP-A at centromeres. CENP-A mislocalization may disrupt chromatin-based processes and chromosome segregation. Here, Deyter and Biggins identify a role for the conserved chromatin-modifying complex FACT in preventing CENP-ACse4 mislocalization to euchromatin by mediating its proteolysis. The budding yeast Spt16 subunit of the FACT complex binds to Psh1, an E3 ubiquitin ligase that targets CENP-ACse4 for degradation. A Psh1 mutant that cannot associate with FACT has a reduced interaction with CENP-ACse4 in vivo.
Keywords: CENP-A, FACT, Psh1, proteolysis, ubiquitylation, centromere
Abstract
Centromere identity and its epigenetic maintenance require the incorporation of a histone H3 variant called CENP-A at centromeres. CENP-A mislocalization to ectopic sites may disrupt chromatin-based processes and chromosome segregation, so it is important to uncover the mechanisms by which this variant is exclusively localized to centromeres. Here, we identify a role for the conserved chromatin-modifying complex FACT (facilitates chromatin transcription/transactions) in preventing budding yeast CENP-ACse4 mislocalization to euchromatin by mediating its proteolysis. The Spt16 subunit of the FACT complex binds to Psh1 (Pob3/Spt16/histone), an E3 ubiquitin ligase that targets CENP-ACse4 for degradation. The interaction between Psh1 and Spt16 is critical for both CENP-ACse4 ubiquitylation and its exclusion from euchromatin. We found that Psh1 cannot efficiently ubiquitylate CENP-ACse4 nucleosomes in vitro, suggesting that additional factors must facilitate CENP-ACse4 removal from chromatin in vivo. Consistent with this, a Psh1 mutant that cannot associate with FACT has a reduced interaction with CENP-ACse4 in vivo. Together, our data identify a previously unknown mechanism to maintain centromere identity and genomic stability through the FACT-mediated degradation of ectopically localized CENP-ACse4.
Chromosome segregation is an orchestrated process by which the replicated genome is distributed to successive cellular generations. Critical to chromosome segregation is the kinetochore, a megadalton-sized DNA/protein complex that dynamically associates with spindle microtubules to mediate chromosome movement (Biggins 2013). Kinetochores assemble at a unique chromosomal locus known as the centromere, which is epigenetically maintained at least in part by a histone H3 variant called CENP-A (also called CenH3) that replaces canonical H3 in nucleosomes (Westhorpe and Straight 2013). CENP-A nucleosomes have a number of unique physical properties that promote kinetochore assembly and the maintenance of centromeres (Black et al. 2004; Foltz et al. 2006; Carroll et al. 2010; Panchenko et al. 2011; Tachiwana et al. 2011; Kato et al. 2013). CENP-A must be exclusively maintained at centromeres to avoid the formation of dicentric chromosomes, which are associated with chromosome instability and have been postulated to drive tumorigenesis (Tomonaga et al. 2003; Amato et al. 2009; Gascoigne and Cheeseman 2013; Lacoste et al. 2014).
Cells use multiple mechanisms to inhibit CENP-A localization to ectopic sites. The presence of H3 nucleosomes appears to provide a barrier for CENP-A incorporation into euchromatin. The budding yeast CENP-ACse4 localizes to ectopic sites in the absence of the H3 chaperones Caf1 and Hir1 or in cells that have decreased H3 expression (Au et al. 2008; Lopes da Rosa et al. 2011). This mislocalization is likely due to reduced H3 incorporation into nucleosomes, which allows CENP-ACse4 to populate genomic regions normally occupied by H3. Chromatin remodeling enzymes have also been ascribed a role in regulating CENP-ACse4 incorporation. For example, the Swi/SNF ATP-dependent chromatin remodeling complex has been shown to restrain CENP-ACse4 localization to centromeres by evicting CENP-ACse4 from noncentromeric loci (Gkikopoulos et al. 2011).
Another important mechanism that ensures the exclusive localization of centromeres is the control of CENP-A protein levels via ubiquitin-mediated proteolysis (Collins et al. 2004; Hewawasam et al. 2010; Ranjitkar et al. 2010; Moreno-Moreno et al. 2011; Au et al. 2013). In budding yeast, the Psh1 E3 ligase is a critical regulator of CENP-ACse4 stability (Hewawasam et al. 2010; Ranjitkar et al. 2010). CENP-ACse4 degradation is impaired in psh1∆ cells, leading to the accumulation of CENP-ACse4 in euchromatin. Overexpression of CENP-ACse4 in psh1∆ cells is lethal, possibly due to perturbation of euchromatin-based processes. Psh1 recognizes the CATD (CENP-A targeting domain) of CENP-A, which includes loop 1 and helix 2 of the histone fold domain, to mediate CENP-A degradation, a property that is conserved with the Drosophila Ppa1 F-box protein that mediates fly CENP-ACID ubiquitylation (Ranjitkar et al. 2010; Moreno-Moreno et al. 2011). However, structural analysis of CENP-A nucleosomes has revealed that a substantial portion of the CATD is buried within the nucleosome, so it is unclear whether Psh1 can act on CENP-ACse4 nucleosomes or instead targets a soluble pool (Tachiwana et al. 2011). Indeed, nucleosome structure hinders DNA-based processes, so cells use nucleosome remodeling, histone eviction, and histone tail post-translational modifications to alleviate the repressive nature of chromatin. It is not known whether Psh1 requires any of these processes to promote CENP-ACse4 degradation.
One important factor that influences chromatin structure is the FACT (facilitates chromatin transcription/transactions) complex (Orphanides et al. 1998; Reinberg and Sims 2006). FACT facilitates many chromatin-based processes, including transcription initiation, transcription elongation, and DNA replication and repair (Belotserkovskaya et al. 2003; Biswas et al. 2005; VanDemark et al. 2006; Abe et al. 2011; Dinant et al. 2013). To promote these functions, FACT generates a more open chromatin configuration via multiple contacts with nucleosomes to allow polymerases to move through chromatin (Belotserkovskaya et al. 2003; Rhoades et al. 2004; Xin et al. 2009; Winkler et al. 2011). FACT also re-establishes chromatin structure in the wake of polymerase passage, thereby maintaining epigenetic states of chromatin (Kaplan et al. 2003; Jamai et al. 2009). Recently, FACT and other transcription elongation factors have been shown to have an indirect role in excluding fission yeast CENP-ACnp1 from euchromatin by reassembling H3-containing nucleosomes during transcription elongation (Choi et al. 2012).
Here, we uncover a previously unknown role for the FACT complex in mediating proteolysis of the centromeric H3 variant. We found that the incorporation of CENP-ACse4 into nucleosomes occludes access of CENP-ACse4 to Psh1-mediated ubiquitylation in vitro, suggesting that additional factors must facilitate its degradation in vivo. Consistent with this, we identified a domain in the E3 ligase Psh1 that directly interacts with the Spt16 FACT subunit and mediates CENP-ACse4 degradation to prevent its accumulation in euchromatin. Together, these data suggest that FACT destabilizes CENP-ACse4 nucleosomes to facilitate Psh1-mediated degradation of CENP-ACse4, revealing a critical role for the conserved FACT complex in maintaining genomic integrity by ensuring that CENP-ACse4 localizes exclusively to centromeres.
Results
FACT has a role in CENP-ACse4 degradation
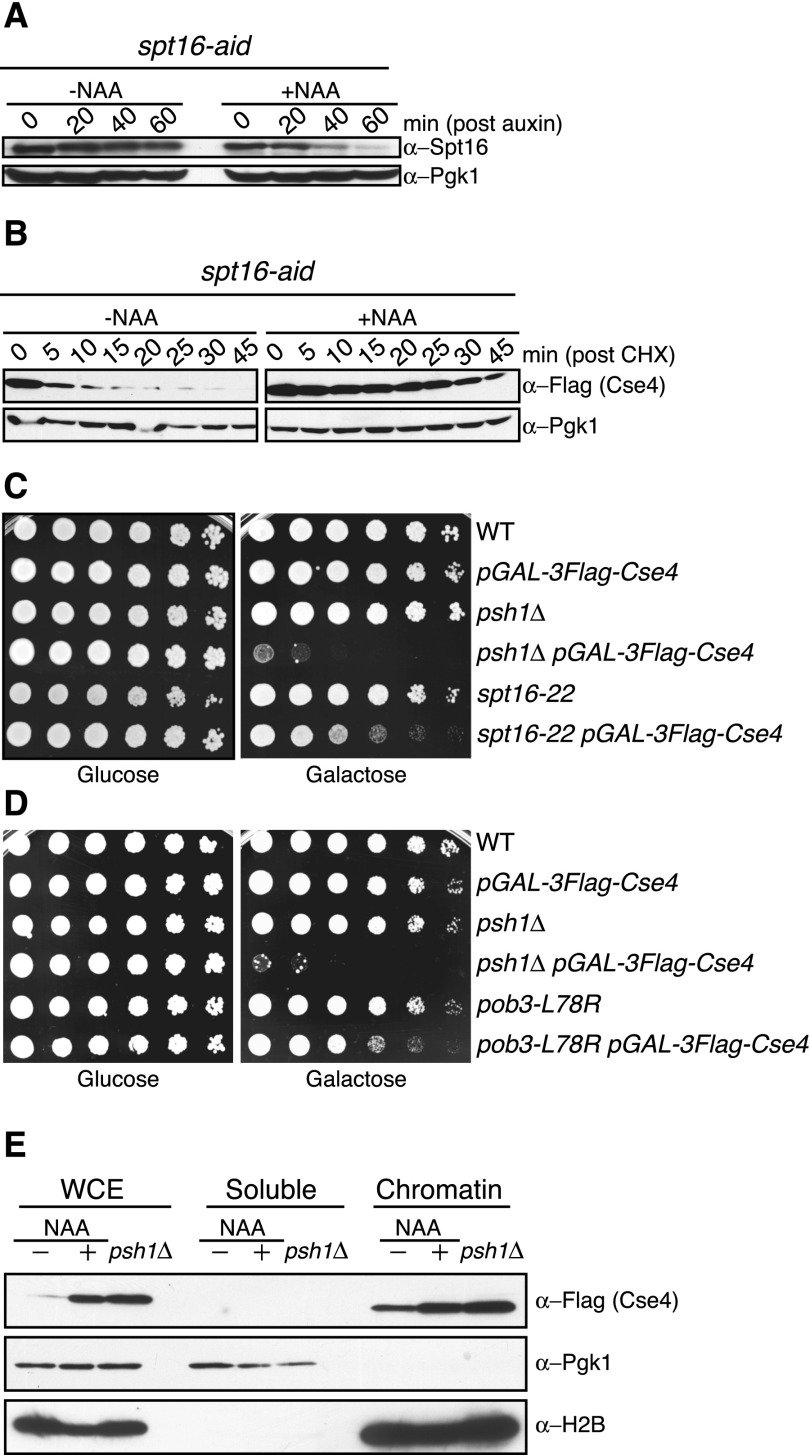
The budding yeast FACT complex consists of a highly stable and abundant heterodimer of Spt16 and Pob3 (Brewster et al. 1998). Psh1 (Pob3/Spt16/histone) was identified in a coprecipitation experiment with the Pob3 and Spt16 proteins (Krogan et al. 2002), so we tested whether FACT affects CENP-ACse4 stability. Because SPT16 and POB3 are essential genes, we generated a conditional allele of SPT16 by fusing the C terminus of the endogenous copy to an auxin-inducible degradation signal (AID) (Nishimura et al. 2009). spt16-aid cells grew normally, but their growth was inhibited in the presence of the synthetic auxin hormone 1-naphthaleneacetic acid (NAA) (data not shown). We analyzed the levels of the Spt16-AID protein in the presence and absence of NAA and found that the bulk of Spt16-AID was degraded 60 min after NAA addition (Fig. 1A).
Figure 1.
The FACT complex facilitates CENP-ACse4 degradation in vivo. (A) spt16-aid (SBY11265) cells were treated with vehicle (95% ethanol [−NAA]) or auxin (+NAA) at time 0, and lysates were analyzed for Spt16-AID levels using α-Spt16 antibodies at the indicated time points. Pgk1 is shown as a loading control. (B) spt16-aid (SBY11268) cells containing pGAL-3Flag-CSE4 were treated with vehicle (−NAA) or auxin (+NAA) to degrade Spt16 for 1 h, followed by a 1-h galactose induction of 3Flag-Cse4. Protein synthesis was inhibited by cycloheximide (CHX) addition at time 0, and lysates were monitored for 3Flag-Cse4 levels with α-Flag antibodies at the indicated time points. Pgk1 is a loading control. Quantification of the stability is in Supplemental Figure S1. (C) Fivefold serial dilutions of the indicated strains (SBY3, SBY8851, SBY8336, SBY8903, SBY6006, and SBY11186) were plated on glucose or galactose medium at 30°C. (D) Fivefold dilutions of the indicated strains (SBY3, SBY8851, SBY8336, SBY8903, SBY11295, and SBY11296) were plated on glucose or galactose medium at 23°C. (E) pGAL-3Flag-CSE4 was overexpressed with galactose in either spt16-aid (SBY11268) cells treated with (+NAA) or without (−NAA) auxin as in B or psh1∆ (SBY8903) cells. This was followed by cycloheximide addition for 30 min, and then whole-cell extracts (WCE) were fractionated into soluble and chromatin fractions. Cse4 levels were monitored in each fraction with α-Flag antibodies. Pgk1 and H2B are markers of the soluble and chromatin fractions, respectively.
To determine whether FACT has a role in CENP-ACse4 stability, we transiently overexpressed CENP-ACse4 after depleting Spt16, inhibited translation with cycloheximide, and monitored CENP-ACse4 levels. Although CENP-ACse4 was quickly degraded in control cells, it was stabilized twofold in Spt16-depleted cells (Figs. 1B; Supplemental Fig. S1A). We were unable to generate a viable pob3-aid strain, so the contribution of Pob3 to CENP-ACse4 degradation could not be analyzed. The FACT complex requires Nhp6, a high-mobility group (HMG) domain-containing protein that binds DNA in a sequence-independent manner, to localize to chromatin (Formosa et al. 2001). We therefore tested whether it also affects CENP-ACse4 stability. Nhp6 is encoded by a gene duplication, and nhp6a∆ nhp6b∆ cells (hereafter called nhp6∆) are viable but exhibit a slow growth phenotype. Surprisingly, CENP-ACse4 levels are not stabilized in nhp6∆ mutants (Supplemental Fig. S1B), indicating that the canonical method for FACT recruitment to nucleosomes is not involved in CENP-ACse4 degradation.
We previously found that CENP-ACse4 overexpression in psh1∆ cells is lethal (Ranjitkar et al. 2010), so we asked whether CENP-ACse4 overproduction affects the viability of FACT mutants. We analyzed the spt16-22 and pob3-L78R point mutants that are thermosensitive, hydroxyurea-sensitive, and have an spt− phenotype (Schlesinger and Formosa 2000; Formosa et al. 2002). Indeed, spt16-22 and pob3-L78R cells exhibited partial sensitivity to CENP-ACse4 overexpression (Fig. 1C,D), similar to findings in fission yeast (Choi et al. 2012). However, we were unable to detect a significant change in CENP-ACse4 stability in these mutants (data not shown), so additional mechanisms must contribute to the underlying synthetic sickness. In contrast, nhp6∆ cells were insensitive to CENP-ACse4 overexpression (Supplemental Fig. S1C), consistent with the canonical FACT recruitment mechanism not being required for CENP-ACse4 degradation.
Psh1-mediated degradation of CENP-ACse4 prevents its accumulation in euchromatin (Hewawasam et al. 2010; Ranjitkar et al. 2010). To determine whether this is also true in cells depleted of Spt16, we overexpressed CENP-ACse4 in spt16-aid cells and analyzed its levels in the soluble and chromatin fractions after inhibiting translation with cycloheximide. CENP-ACse4 incorporation into the chromatin fraction was increased in cells depleted of Spt16, similar to the levels that occur in psh1∆ cells (Figs. 1E; Supplemental Fig. S1D). Because budding yeast CENP-ACse4 is constitutively localized to the 125-base-pair (bp) point centromere, all additional CENP-ACse4 in the chromatin pool must be mislocalized to euchromatin (Biggins 2013). Soluble CENP-ACse4 levels also showed a modest increase in psh1∆ cells (Ranjitkar et al. 2010), and darker exposures of the immunoblots revealed a similar increase in Spt16-depleted cells (data not shown). Together, these data reveal the existence of a novel FACT mechanism independent of Nhp6 that regulates CENP-ACse4 stability and inhibits its euchromatic localization.
The Spt16 subunit of the FACT complex interacts with Psh1
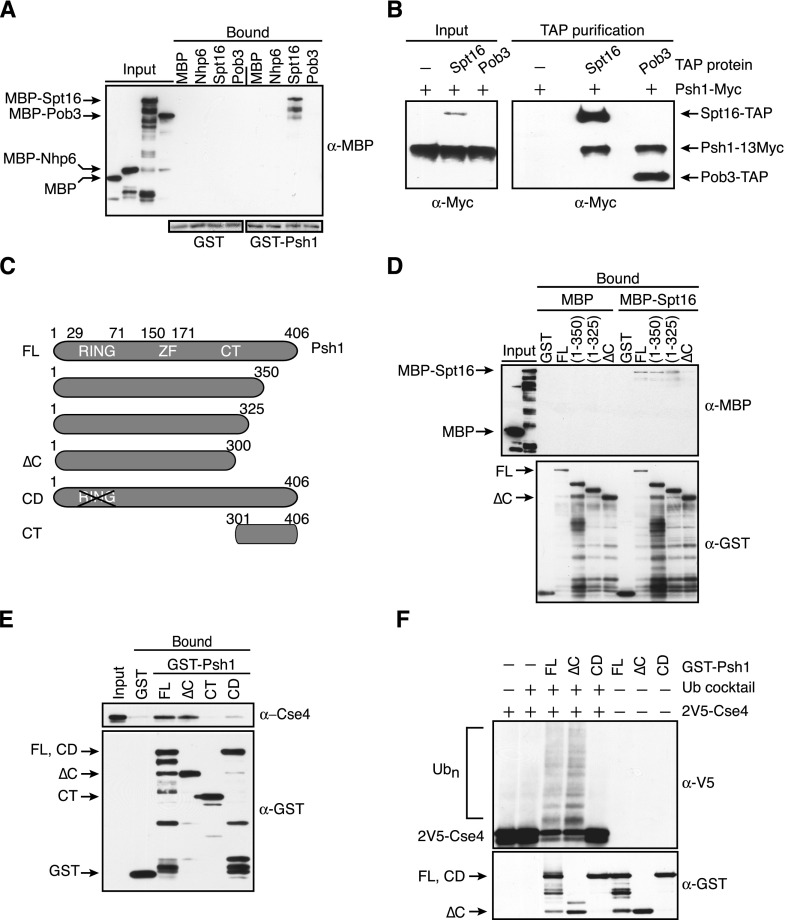
Although FACT could exclude CENP-ACse4 from euchromatin by promoting H3 incorporation (Choi et al. 2012), the association of FACT with Psh1 in coimmunoprecipitation experiments suggested a more direct role in CENP-ACse4 degradation (Krogan et al. 2002; Ranjitkar et al. 2010). We therefore asked whether Psh1 directly interacts with FACT in vitro. We expressed and purified recombinant FACT subunits (Nhp6, Pob3, and Spt16) that were fused to the maltose-binding protein (MBP) and assayed for binding to immobilized glutathione-S-transferase (GST) or GST-Psh1. We detected an interaction between recombinant GST-Psh1 and MBP-Spt16 (Fig. 2A). To narrow down the domain in Spt16 that interacts with Psh1, we tested Spt16 fragments in the binding assays. Prior work defined four structured domains in Spt16: an N-terminal domain that binds weakly to H3/H4, a Pob3 dimerization domain, an M domain that also contributes to H3/H4 binding, and a C-terminal domain that binds histone H2A/H2B and can actively displace nucleosomal DNA (Supplemental Fig. S2A; O’Donnell et al. 2004; VanDemark et al. 2006; Stuwe et al. 2008; Kemble et al. 2013). An N-terminal Pob3 fragment has been reported to bind to the dimerization domain (VanDemark et al. 2006), while another study showed that full-length Pob3 binds with highest affinity to a Spt16 construct that contained the dimerization domain and an N-terminal region of the M domain (O’Donnell et al. 2004). In our assays, we found that Pob3 and Psh1 both interact with a large fragment containing the dimerization and M domain (residues 485–804) (Supplemental Fig. S2B; data not shown). Because this fragment may contain an N-terminal unstructured region that could mediate nonspecific interactions (O’Donnell et al. 2004), we tested a smaller fragment lacking this region (residues 643–804) and found that both Psh1 and Pob3 associate with this smaller region in the M domain (Supplemental Fig. S2B).
Figure 2.
Psh1 interacts with the Spt16 subunit of the FACT complex. (A) Glutathione–Sepharose resin bound to GST or GST-Psh1 was incubated with MBP or the indicated MBP fusion proteins. The Sepharose was washed and eluted, and the presence of the MBP-tagged proteins was analyzed with α-MBP antibodies. (Bottom) The blot was stained with Ponceau S to reveal GST and GST-Psh1 levels. (B) IgG Sepharose beads were used to affinity-purify Spt16-TAP (SBY9977) or Pob3-TAP (SBY9978) from cells expressing a 13Myc epitope fusion to Psh1. Cells expressing full-length Psh1-13Myc but no TAP-tagged protein (SBY6423) served as a control. The samples were subsequently analyzed by immunoblotting with α-Myc antibodies. The TAP tag contained a calmodulin-binding peptide, a TEV cleavage site, and two IgG-binding domains of protein A, the latter of which interacts with the α-Myc antibodies, allowing detection of the TAP-tagged proteins along with Psh1-13Myc. (Note: Pob3-TAP is detected in the input on longer exposures that are not shown here.) (C) Schematic of Psh1 and the fragments used in D–F. (FL) Full-length; (∆C) C-terminal deletion; (CD) catalytically dead; (CT) C-terminal region. (D) Immobilized GST alone, GST-fused full-length Psh1 [FL (1–406)], or the indicated Psh1 fragments were incubated with MBP or MBP-Spt16. The resin was washed and eluted, and the levels of the MBP and GST proteins were analyzed with α-MBP and α-GST antibodies. (E) 2V5-Cse4 octamers were added to immobilized GST alone or GST-fused full-length Psh1 (1–406), Psh1 (1–300), Psh1 (301–406), and a Psh1 RING domain mutant (C45S, C50S), followed by washing and elution from the resin. The levels of the GST proteins were assessed with α-GST antibodies, and the presence of Cse4 was assessed with α-Cse4 antibodies. (F) The indicated GST-Psh1 proteins and 2V5-Cse4 octamers were added to reactions in the presence (+) or absence (−) of a ubiquitin (Ub) cocktail. The top immunoblot shows Cse4 ubiquitylated species, while Psh1 levels are shown in the bottom immunoblot. The blots were not probed with α−ubiquitin antibodies, since we cannot distinguish Cse4 ubiquitylation from Psh1 autoubiquitylation. Furthermore, Psh1 autoubiquitylation makes the comparison of Psh1 levels in the reactions difficult, so parallel reactions were performed in the absence of the ubiquitin cocktail and 2V5-Cse4 octamers (shown in the three right lanes).
We were not able to identify distinct binding domains for Pob3 and Psh1 within the M domain of Spt16 (data not shown), so we tested whether Pob3 and Psh1 binding to Spt16 might be mutually exclusive. To do this, we performed tandem affinity purification (TAP) tag isolations of Spt16 and Pob3 and tested whether Psh1 copurifies. Spt16 is not known to form homodimers, so the presence of Psh1 in Pob3-TAP purifications could not be a consequence of dimerization of Spt16/Pob3 with Spt16/Psh1. We found that Psh1 is present in both Pob3 and Spt16 TAP tag purifications, suggesting that it likely associates with the FACT complex (Fig. 2B). However, because quantitative immunoblotting revealed that Spt16 is at least 30-fold more abundant than Psh1 (Supplemental Fig. S2C; data not shown), Psh1 is bound to only a subset of FACT complexes.
We next identified a region of Psh1 required for Spt16 association. Psh1 contains a catalytic RING domain that mediates ubiquitylation of CENP-ACse4 in the N terminus (residues 29–71), a zinc finger motif (residues 150–171), and a C-terminal region lacking obvious motifs (residues 171–406) (Fig. 2C). The only region of Psh1 that directly interacted with recombinant Spt16 was the C-terminal region (data not shown). Consistent with this, a truncation of the C-terminal 106 amino acids of GST-Psh1 (residues 301–406) eliminated the interaction between Psh1 and MBP-Spt16 in vitro (Fig. 2D). Together, these data indicate that the C terminus of Psh1 is required for the Psh1 interaction with Spt16.
The identification of a region in Psh1 required for Spt16 binding allowed us to ask whether this domain is required for Psh1 activity in vitro. First, we tested whether a Psh1 mutant lacking this domain (hereafter called Psh1ΔC) affects CENP-ACse4 binding. Bacterially expressed and purified CENP-ACse4 octamers were used in these assays, since CENP-ACse4 is an unstable protein in bacteria when produced in the absence of the other histones (data not shown). In contrast to Spt16, CENP-ACse4 associated normally with GST-Psh1∆C in vitro (Fig. 2E). Consistent with this, we did not detect an interaction between the C terminus (CT) of Psh1 and CENP-ACse4. However, mutations in the catalytic domain (CD) affected CENP-ACse4 binding, as previously shown (Hewawasam et al. 2010). We next tested the catalytic activity of GST-Psh1∆C toward CENP-ACse4 in ubiquitylation assays. CENP-ACse4 octamers were incubated in the presence of the requisite E1 and E2 proteins, ubiquitin, ATP, and either GST-Psh1, GST-Psh1∆C, or a catalytically dead RING mutant, GST-Psh1(CD). When analyzed by immunoblotting, the unmodified CENP-ACse4 band predominates in the absence of Psh1, as previously described (Ranjitkar et al. 2010). However, the addition of either GST-Psh1 or GST-Psh1∆C produced similar levels of ubiquitin-conjugated CENP-ACse4 species and a corresponding decrease in unmodified CENP-ACse4 (Fig. 2F). Therefore, the GST-Psh1∆C mutant protein binds to and ubiquitylates CENP-ACse4 in vitro as efficiently as full-length GST-Psh1. Together, these data identify a specific region of Psh1 required for association with the Spt16 subunit of FACT that is separable from the domains required for CENP-ACse4 binding and ubiquitylation in vitro.
The Psh1 mutant that cannot bind FACT is defective in CENP-ACse4 degradation in vivo
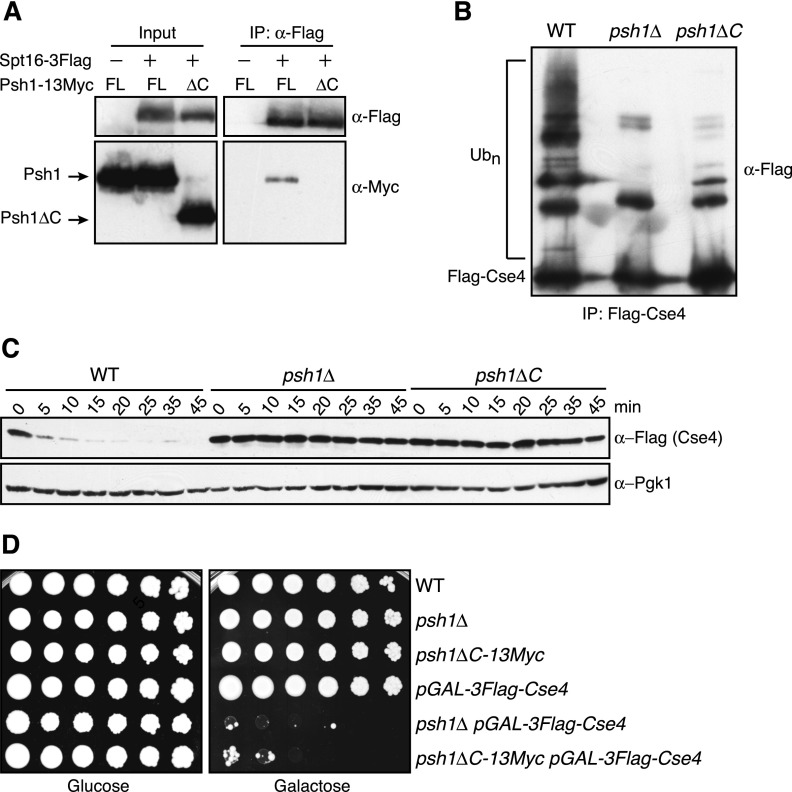
We next analyzed the function of the interaction between Spt16 and Psh1 in vivo. We incorporated 13Myc epitope tags after residue 300 of Psh1 (hereafter psh1∆C-13Myc), to simultaneously truncate the domain required for Spt16 binding and to epitope-tag Psh1. Consistent with our in vitro binding results, Spt16-3Flag coimmunoprecipitated full-length Psh1-13Myc from yeast extracts but failed to associate with Psh1∆C-13Myc (Fig. 3A). To determine whether FACT association with Psh1 influences CENP-ACse4 ubiquitylation in vivo, we analyzed the ubiquitin conjugates of CENP-ACse4 isolated from wild-type, psh1∆, and psh1∆C-13myc cells. Multiple slower-migrating CENP-ACse4 species that correspond to ubiquitin conjugates were detected when CENP-ACse4 was overexpressed and isolated from wild-type cells (Figs. 3B; Supplemental Fig. S3; Ranjitkar et al. 2010). As expected, these species were substantially decreased but not eliminated in psh1∆ cells, consistent with additional Psh1-independent pathways contributing to CENP-ACse4 ubiquitylation (Collins et al. 2004; Au et al. 2013). Strikingly, CENP-ACse4 purified from psh1∆C-13Myc cells also displayed a significant decrease in CENP-ACse4 ubiquitylation (Figs. 3B; Supplemental Fig. S3). To confirm that the decreased CENP-ACse4 ubiquitylation in psh1∆C-13Myc cells affects CENP-ACse4 degradation, CENP-ACse4 protein levels were monitored after transient overexpression followed by translational repression with cycloheximide. CENP-ACse4 was stabilized in psh1∆C-13Myc cells to an extent similar to psh1∆ cells (Fig. 3C), indicating that the Psh1 C terminus that is required for Spt16 association is also required for CENP-ACse4 degradation.
Figure 3.
The Psh1 domain required for Spt16 binding facilitates CENP-ACse4 degradation. (A) α-Flag-conjugated beads were used to immunoprecipitate Spt16-3Flag from cells expressing a 13Myc epitope fusion to either full-length (FL) Psh1 (SBY6439) or a Psh1 mutant (SBY10917) truncated at its C terminus (∆C). Cells expressing full-length Psh1-13Myc but not Spt16-3Flag (SBY6423) served as a control. The samples were subsequently analyzed by immunoblotting with α-Flag and α-Myc antibodies. (B) Wild-type (WT) (SBY8851), psh1∆ (SBY8903), and psh1∆C-13Myc (SBY10921) cells expressing pGAL-3Flag-CSE4 were grown in galactose, and Cse4 was immunoprecipitated with α-Flag antibodies. Cse4 and its ubiquitin conjugates (Ubn) were detected on immunoblots by probing with α-Flag antibodies. (C) Wild-type (SBY8851), psh1∆ (SBY8903), and psh1∆C-13Myc (SBY10921) cells expressing pGAL-3Flag-CSE4 were grown in galactose, protein synthesis was inhibited by cycloheximide addition at time 0, and lysates were monitored for Cse4 levels at the indicated time points with α-Flag antibodies. Pgk1 served as a loading control. (D) Fivefold serial dilutions of the indicated strains (SBY3, SBY8336, SBY10919, SBY8851, SBY8903, and SBY10921) were plated on glucose or galactose medium and incubated at 23°C.
Because CENP-ACse4 overexpression is lethal in a psh1∆ mutant, we tested how overexpression affects the viability of psh1∆C cells. PSH1 is a nonessential gene, and psh1∆C cells grew similarly to both wild-type and psh1∆ cells (Fig. 3D). However, CENP-ACse4 overexpression strongly inhibited the growth of psh1∆C-13Myc cells. Because the Psh1∆C mutant has normal activity toward CENP-ACse4 in vitro (Fig. 2), these data reveal a critical role for the C terminus of Psh1 to mediate CENP-ACse4 degradation in vivo. Although the Psh1∆C mutant may affect interactions with other factors, the similar phenotypes between this mutant that is defective in Spt16 association with cells that are depleted of Spt16 strongly suggest that FACT binding to Psh1 plays a crucial role in CENP-ACse4 degradation.
Psh1 requires the FACT association domain to inhibit CENP-ACse4 euchromatin localization
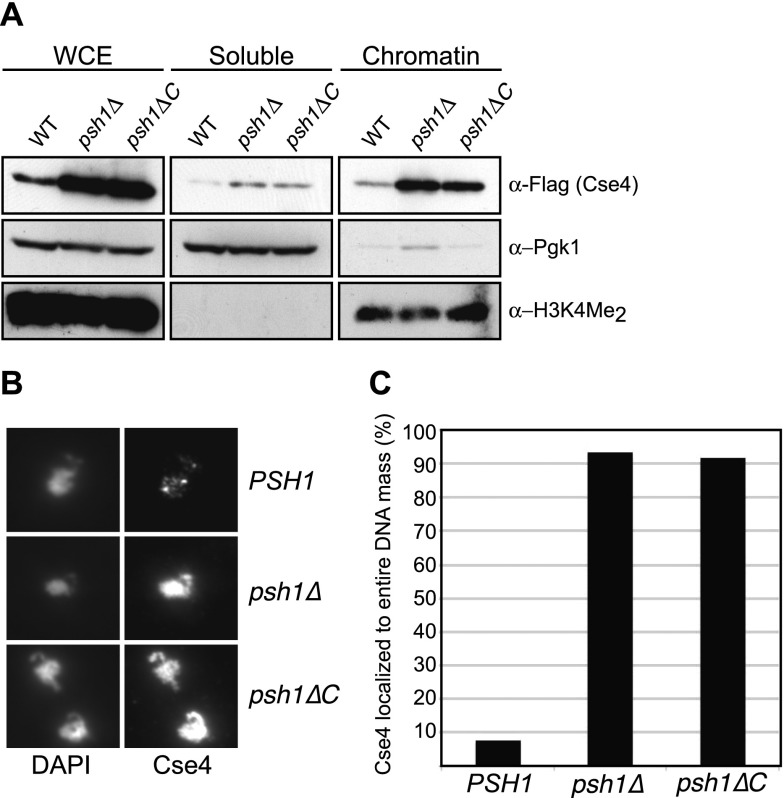
To determine the consequences of the increased levels of CENP-ACse4 in psh1ΔC cells, we analyzed CENP-ACse4 localization. We fractionated wild-type, psh1∆, and psh1∆C-13Myc yeast lysates after CENP-ACse4 overexpression. There was a slight increase in the soluble pool of CENP-ACse4 and a substantial increase in the amount incorporated into the chromatin in both psh1∆ and psh1∆C-13Myc cells (Fig. 4A). As an independent assessment of euchromatin localization of CENP-ACse4 in psh1∆C-13Myc cells, we also performed chromosome spreads. We overexpressed CENP-ACse4 in wild-type cells and found that the majority of the protein localized to discrete foci previously shown to correspond to budding yeast kinetochore clusters (Fig. 4B; Ranjitkar et al. 2010). In contrast, overexpressed CENP-ACse4 was not limited to centromeres in psh1∆ or psh1∆C-13Myc cells. Instead, CENP-ACse4 localized with the bulk of chromatin in >90% of chromosome spreads prepared from the mutants (Fig. 4C), suggesting that FACT binding to Psh1 prevents the ectopic localization of CENP-ACse4. These data are consistent with a crucial role for the Psh1/FACT interaction in CENP-ACse4 ubiquitylation and degradation, which ultimately prevents it from stably mislocalizing to euchromatin.
Figure 4.
CENP-ACse4 localizes to the euchromatin in psh1∆C cells. (A) Whole-cell extracts (WCE) from wild-type (WT) (SBY8851), psh1∆ (SBY8903), and psh1∆C-13Myc (SBY10921) cells expressing pGAL-3Flag-CSE4 were fractionated into soluble and chromatin fractions. Cse4 levels were monitored in each fraction with α-Flag antibodies. Pgk1 and dimethylation of histone H3 at Lys4 (H3K4Me2) are markers of the soluble and chromatin fractions, respectively. (B) 3Flag-Cse4 was transiently overexpressed for 1 h in wild-type (SBY8851), psh1∆ (SBY8903), and psh1∆C-13Myc (SBY10921) cells, and its localization to chromatin was analyzed by immunofluorescence on chromosome spreads. DAPI staining was used to visualize DNA, while α-Flag staining revealed Cse4 localization. (C) Quantification of the percentage of chromatin masses in which Cse4 staining was coincident with total DNA staining instead of with kinetochores that contain discrete Cse4 foci. Wild type (SBY8851), n = 132; psh1∆ (SBY8903), n = 137; psh1∆C-13Myc (SBY10921), n = 130.
Nucleosome structure is a barrier to CENP-ACse4 ubiquitylation by Psh1
Because our data indicated a role for FACT in mediating CENP-ACse4 degradation, we considered the possibility that its role in nucleosome disassembly might facilitate Psh1 degradation by destabilizing CENP-ACse4 from chromatin. Consistent with this, Psh1 interacts with CENP-ACse4 via CATD residues that exist in the histone fold domain, and much of the CATD is occluded within the nucleosome (Tachiwana et al. 2011). Our in vitro ubiquitylation assays were performed with CENP-ACse4 octamers, which presumably behave like H3 octamers and dissociate into a CENP-ACse4/H4 tetramer and two H2A/H2B dimers in the low-salt buffers used for the ubiquitylation reaction (Eickbush and Moudrianakis 1978). We therefore tested whether CENP-ACse4 nucleosomes, which have a more tightly associated histone core that is stabilized by nucleosome-wrapped DNA, can serve as a substrate for Psh1.
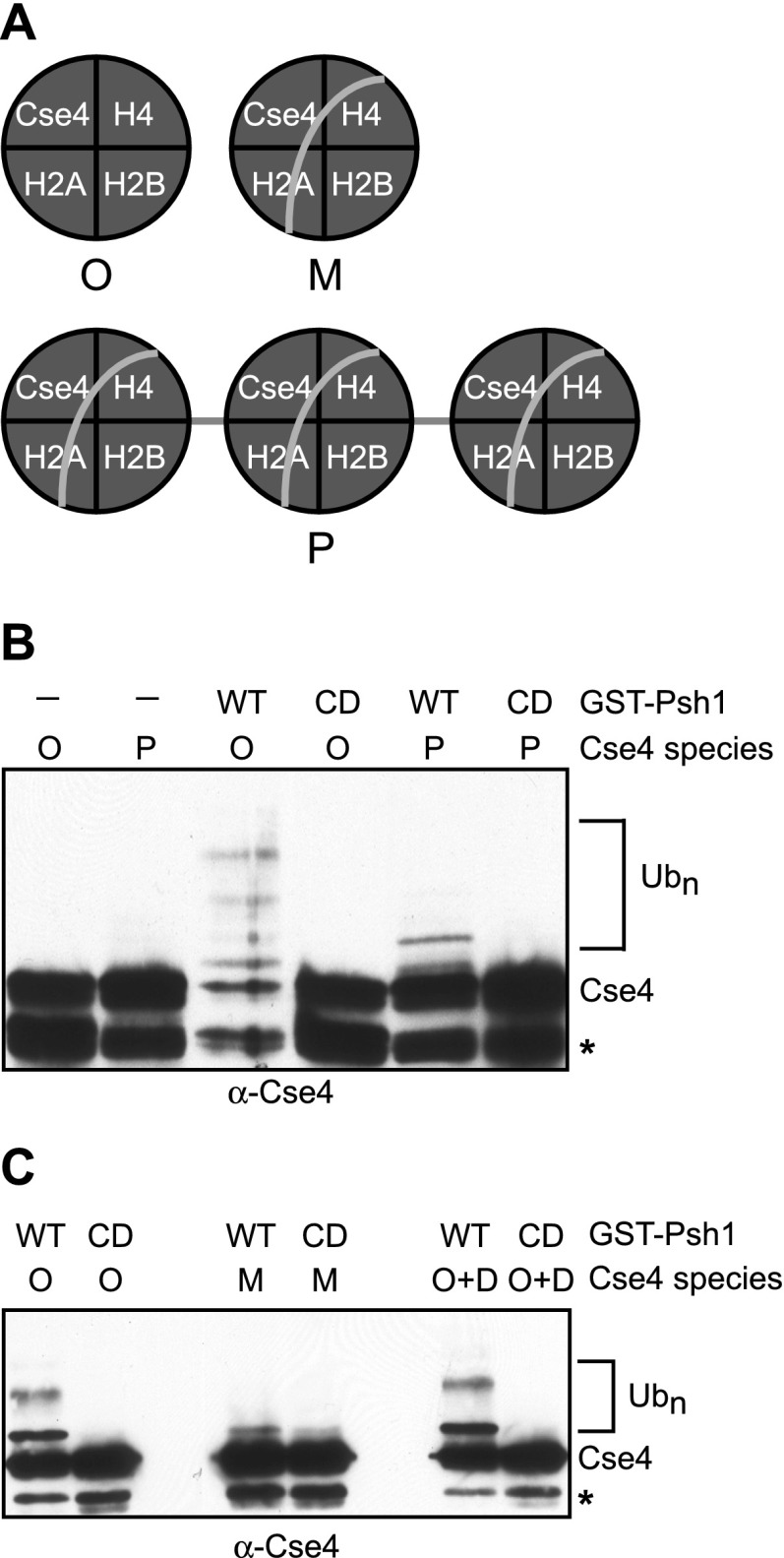
We generated two species of CENP-ACse4 nucleosomes in vitro (Fig. 5A). Mononucleosomes were assembled by the traditional step-salt gradient dialysis of recombinant histones and a 146-bp DNA sequence containing a strong nucleosome-positioning sequence as described (Kingston et al. 2011). In addition, polynucleosomes were made to test whether linker DNA or multiple CENP-ACse4 nucleosomes on the same template would influence Psh1 activity. The production of CENP-ACse4 mononucleosomes was confirmed by nondenaturing PAGE (Supplemental Fig. S4), and the stoichiometry of the histones from the polynucleosomal preparations was assessed on silver stain gels as described (Gelbart et al. 2001; data not shown). There was a substantial decrease in CENP-ACse4 ubiquitylation by Psh1 when either CENP-ACse4 polynucleosomes (Fig. 5B) or mononucleosomes (Fig. 5C) were used as the substrate instead of octamers in ubiquitylation assays in vitro. We speculate that the slight ubiquitylation of CENP-ACse4 nucleosomes by Psh1 reflects ubiquitylation of lysines in the N-terminal tail of CENP-ACse4 (Au et al. 2013). To ensure that the decrease in activity was not due to direct inhibition of Psh1 by DNA, we added the DNA used to assemble the mononucleosomes to the ubiquitylation assays containing CENP-ACse4 octamers. The ubiquitylation of CENP-ACse4 was unaltered, indicating that Psh1 ubiquitin ligase activity is unaffected by the presence of DNA (Fig. 5C). Together, these results reveal that nucleosome structure is a barrier to CENP-ACse4 ubiquitylation by Psh1 in vitro.
Figure 5.
Nucleosome structure inhibits CENP-ACse4 ubiquitylation by Psh1. (A) Schematic of the Cse4 species used in the assays. (O) Octamer; (M) mononucleosome; (P) polynucleosomes. Gray lines indicate DNA. (B) Cse4 polynucleosomes (P) or octamers (O) were incubated in a Ub cocktail. (Lanes 1,2) The absence (−) of GST-Psh1 reveals the level of unmodified Cse4 present in the reactions. Where indicated, full-length wild-type (WT) GST-Psh1 or a catalytically dead (CD) RING mutant with C45S and C50S mutations was added to the reactions. Immunoblots were probed with α-Cse4 antibodies to analyze Cse4 ubiquitylation (Ubn). (*) Cse4 degradation product. (C) Cse4 mononucleosomes (M), octamers (O), or octamers with exogenously added DNA (O+D) were incubated with the Ub cocktail and either wild-type GST-Psh1 (WT) or the RING mutant (CD). Immunoblots were probed with α-Cse4 antibodies to analyze Cse4 ubiquitylation (Ubn). (*) Cse4 degradation band.
FACT facilitates the interaction between Psh1 and CENP-ACse4
Because FACT is known to facilitate chromatin-based processes by destabilizing nucleosomes, we speculated that it might help expose CENP-ACse4 nucleosomes to Psh1 to promote ubiquitylation. We therefore tested whether the addition of FACT enhanced the ubiquitylation of CENP-ACse4 in vitro. However, FACT inhibited the in vitro ubiquitylation reactions containing either Psh1 or Psh1∆C (Supplemental Fig. S5A). Because we do not know the underlying reason for the inhibition, we could not further test whether FACT facilitates the ubiquitylation of CENP-ACse4 incorporated into nucleosomes.
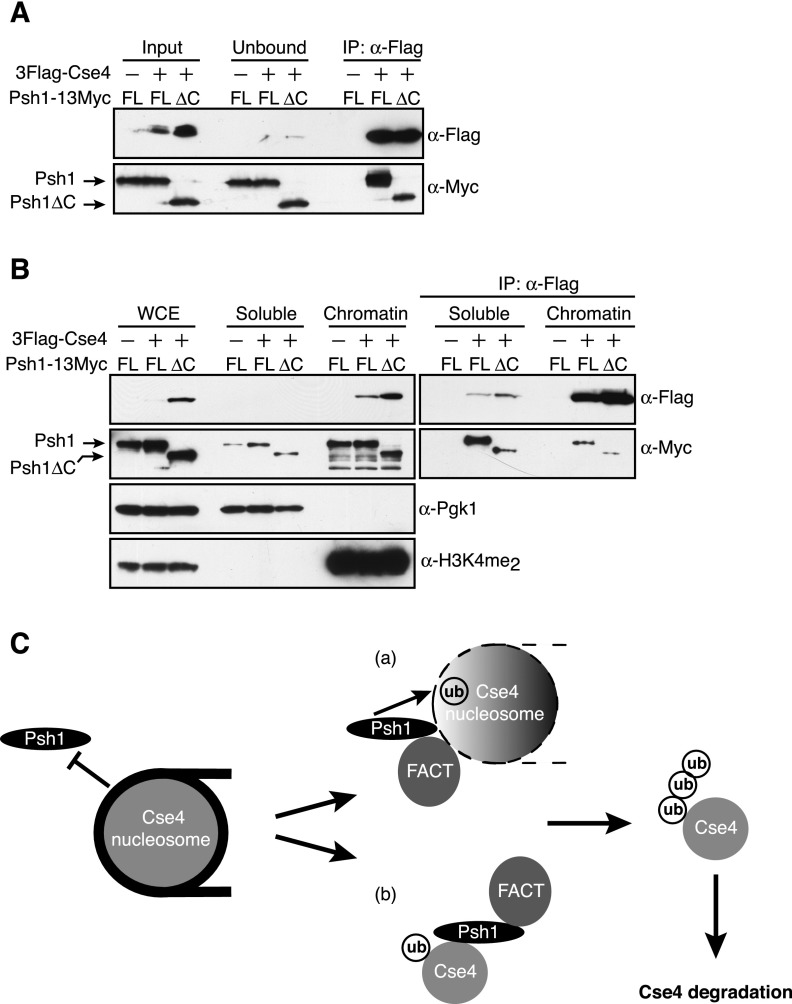
If FACT releases CENP-ACse4 from nucleosomes to promote an interaction with Psh1, we reasoned that FACT might enhance the association of Psh1 with CENP-ACse4 in vivo. To test this, 3Flag-CENP-ACse4 was immunoprecipitated from whole-cell extracts, and the amount of coprecipitating full-length Psh1-13Myc or Psh1∆C-13Myc was analyzed. Indeed, the interaction of CENP-ACse4 with Psh1∆C-13Myc was strongly reduced compared with full-length Psh1 (Fig. 6A). To determine whether a specific pool of CENP-ACse4 was defective in binding to Psh1∆C-13Myc, we immunoprecipitated overexpressed 3Flag-CENP-ACse4 from the soluble and chromatin fractions in cells expressing full-length Psh1-13Myc or Psh1∆C-13Myc. Although the Psh1∆C-13Myc protein localized to chromatin normally, it had impaired binding to CENP-ACse4 in both the soluble and chromatin fractions (Fig. 6B). These data are consistent with the idea that FACT promotes an interaction between Psh1 and CENP-ACse4 that facilitates CENP-ACse4 degradation.
Figure 6.
The Psh1 mutant that is defective in binding to FACT has reduced association with CENP-ACse4 in vivo. (A) α-Flag-conjugated beads were used to immunoprecipitate 3Flag-Cse4 from cells expressing 13Myc-tagged full-length (FL) (SBY11102) or the C-terminal mutant (∆C) Psh1-13Myc (SBY11104). Cells expressing full-length Psh1-13Myc but not 3Flag-Cse4 (SBY6423) served as a control. The immunoblots were probed with α-Flag and α-Myc antibodies to detect the levels of Cse4 and Psh1, respectively. (B) pGAL-3Flag-CSE4 was overexpressed with galactose in cells expressing 13Myc-tagged full-length (FL) or the C-terminal mutant (∆C) Psh1-13Myc (SBY11090 and SBY10920). Cells expressing full-length Psh1-13Myc but not 3Flag-Cse4 (SBY6423) served as a control. Whole-cell extracts (WCE) were fractionated into soluble and chromatin fractions. α-Flag-conjugated beads were used to immunoprecipitate 3Flag-Cse4 from the soluble and chromatin fractions. The immunoblots were probed with α-Flag and α-Myc antibodies to detect the levels of Cse4 and Psh1, respectively. Pgk1 and H3K4me2 are markers of the soluble and chromatin fractions, respectively. (C) In this model, Cse4 mislocalizes to the euchromatin, where it is protected from Psh1-dependent ubiquitylation due to the repressive nature of chromatin structure on trans-acting factors. In order to override this accessibility barrier, Cse4 nucleosomes must be restructured so that Psh1 can bind and ubiquitylate Cse4. The FACT complex plays a crucial role in this process by increasing the association of Psh1 with Cse4. In part a, FACT reorganizes nucleosome structure (denoted by the black and white gradient) to facilitate the interaction and subsequent ubiquitylation of Cse4 by Psh1. (Part b) Additionally, FACT activity might release Cse4 from nucleosomes to create a soluble pool of Cse4 that is competent in interacting with Psh1. In either scenario, FACT exposes critical Cse4 residues to which Psh1 binds, ultimately leading to Cse4 ubiquitylation, degradation, and elimination from euchromatin.
Discussion
Here we identify a direct physical interaction between Psh1, an E3 ubiquitin ligase that targets CENP-ACse4 for degradation, and the Spt16 subunit of the FACT complex. Cells depleted of Spt16 stabilize CENP-ACse4, which leads to CENP-ACse4 accumulation in the euchromatin. Similarly, a psh1 mutant that has perturbed Spt16 association cannot efficiently degrade CENP-ACse4 or prevent its localization to euchromatin in vivo despite its efficient activity against CENP-ACse4 in vitro. We found that chromatin structure inhibits CENP-ACse4 ubiquitylation by Psh1 in vitro, and FACT potentiates the interaction between Psh1 and CENP-ACse4 in vivo. We therefore propose that the nucleosome-reorganizing activity of FACT is a key factor that promotes the Psh1-mediated ubiquitylation of CENP-ACse4 in vivo at least in part by increasing the association of Psh1 with CENP-ACse4.
FACT may have multiple functions in regulating CENP-A chromatin. Although our data identify a role for FACT in preventing ectopic localization of CENP-ACse4 via an association with the CENP-ACse4 degradation machinery, FACT has also been implicated in CENP-A deposition at centromeres. Human FACT copurifies with inner kinetochore proteins that associate with centromeres and has been postulated to be involved in CENP-A deposition (Walfridsson et al. 2005; Foltz et al. 2006; Okada et al. 2009). However, the conserved histone chaperone HJURP/Scm3 appears to be the key deposition factor both in vivo and in vitro (Camahort et al. 2007; Foltz et al. 2009). FACT might facilitate CENP-A deposition and kinetochore assembly indirectly through its role in forming pericentromeric heterochromatin in organisms with regional centromeres (Lejeune et al. 2007). FACT has also been implicated in recruiting CHD1, an ATP-dependent chromatin remodeler that influences histone H4 deacetylation, to centromeres to promote CENP-A deposition (Walfridsson et al. 2005; Okada et al. 2009). However, Chd1 is absent from budding yeast centromeres (Zentner et al. 2013), and it is unknown whether FACT influences CENP-ACse4 deposition in this organism.
Our work identifies a previously unknown role for FACT in facilitating the degradation of the centromeric H3 variant. Although most of our experiments focused on the Spt16 protein, a pob3 mutant was also sensitive to increased CENP-ACse4 dosage and Psh1 coprecipitated with Pob3. While it is formally possible that Psh1 interacts with a pool of Spt16 that does not include Pob3, the simplest interpretation is that the FACT complex mediates CENP-ACse4 degradation. FACT defects were previously associated with CENP-ACnp1 misincorporation into euchromatin (Choi et al. 2012), consistent with evidence that maintaining the integrity of H3 chromatin via FACT and other elongation factors provides a crucial barrier to CENP-A misincorporation. Here, we found that FACT also has a more direct role in controlling CENP-A chromatin association by promoting its proteolysis to prevent ectopic localization.
The role of FACT in CENP-ACse4 degradation may be dependent on events that occur during transcription. Transcription elongation factors, including Spt16 and Spt6, have been shown to inhibit CENP-ACnp1 incorporation into euchromatin (Choi et al. 2012), and CENP-ACse4 stability is partially dependent on RNA polymerase II (Pol II) transcription (Lopes da Rosa et al. 2011). An attractive possibility is that FACT’s role in re-establishing H3-containing chromatin during transcription is coupled to its role in CENP-ACse4 degradation to ensure the exclusion of CENP-ACse4 from euchromatin. However, FACT may also have a role in mediating CENP-ACse4 degradation independent of transcription, since FACT can regulate chromatin disassembly at promoter regions outside of transcriptional activation of the downstream gene (Takahata et al. 2009). Moreover, CENP-A mislocalizes to promoter regions that have high histone turnover (Krassovsky et al. 2012; Lacoste et al. 2014), and FACT can evict histones at promoters (Ransom et al. 2009; Takahata et al. 2009). In the future, it will be important to determine whether polymerases or other factors exist in the Psh1-associated FACT complex to couple CENP-ACse4 degradation to chromatin-based processes.
We found that the mechanism by which FACT facilitates CENP-ACse4 degradation is independent of Nhp6, the protein known to mediate FACT association with canonical nucleosomes (Formosa et al. 2001). Nhp6 constitutes an initial step in FACT-dependent nucleosome reorganization that involves DNA bending and altered histone–DNA contacts to allow FACT to substantially change nucleosome structure (Xin et al. 2009). Recent data suggest that DNA topology at the nucleosomal entry/exit site might influence FACT activity on nucleosomes (Hondele et al. 2013). Reconstituted CENP-ACse4 nucleosomes have weakened DNA–histone contacts at the DNA entry/exit site compared with H3 nucleosomes (Dechassa et al. 2011; Tachiwana et al. 2011), which may generate an open chromatin configuration that allows FACT to function on CENP-ACse4 nucleosomes independent of Nhp6. Psh1 has a pair of C4-type zinc finger motifs that are found in many DNA-binding proteins, so it may bind to CENP-ACse4 nucleosomes to recruit FACT.
FACT is required for the association of Psh1 with both the soluble and chromatin pools of CENP-ACse4 in vivo, although it is not required for Psh1 and CENP-ACse4 to interact in vitro. We speculate that the altered interaction in the chromatin reflects a requirement for FACT to expose CENP-ACse4 when it is incorporated into nucleosomes to the Psh1 ligase in vivo. The decreased interaction of Psh1 with CENP-ACse4 in the soluble pool may also be a result of the decreased chromatin interaction in the absence of FACT. Alternatively, Psh1 may prevent the soluble FACT complex from binding to CENP-ACse4 to prevent FACT from misincorporating CENP-ACse4 into euchromatin.
Taken together, our data lead to the following model (Fig. 6C). CENP-ACse4 overexpression leads to its misincorporation into euchromatin (Camahort et al. 2009; Lefrancois et al. 2009; Krassovsky et al. 2012; Lacoste et al. 2014). Psh1 must recognize and target the mislocalized CENP-ACse4 for degradation, but we found that CENP-ACse4 nucleosomes are refractory to its activity. The FACT complex destabilizes nucleosomes and interacts with Psh1, so we propose that FACT exposes CENP-ACse4 for Psh1 association and subsequent degradation in vivo. Psh1 can still associate with chromatin when it does not interact with FACT, so Psh1 may help FACT associate with chromatin. CENP-ACse4 localized to the centromere is protected from proteolysis (Collins et al. 2004), and this may be because the histone chaperone Scm3 binds to the CATD of CENP-ACse4 at the centromere (Zhou et al. 2011). However, we also found that FACT inhibited Psh1 activity in vitro, so it may also have a role in protecting CENP-ACse4 from degradation at the centromere. Additional mechanisms might also inhibit Psh1 at the centromere. For example, CENP-ACse4 proline isomerization is required for its interaction with Psh1, so this activity may be inactive at centromeres (Ohkuni et al. 2014). Together, our work identifies a previously unknown function for the conserved FACT complex in maintaining euchromatin integrity through degradation of the CENP-ACse4 histone variant. In the future, it will be critical to further understand the mechanisms that promote FACT activity on CENP-ACse4 nucleosomes to maintain genomic stability.
Materials and methods
Strain construction and microbial techniques
Microbial techniques and media were as described (Sherman et al. 1974; Rose et al. 1990). Yeast strains were constructed using standard genetic techniques. PCR integration was used to truncate the C terminus of PSH1 with 13myc (Longtine et al. 1998). All oligonucleotide sequences are available on request. Specific plasmid construction and yeast strains used in this study are described in the Supplemental Material and Supplemental Tables S1 and S2. For Spt16-AID degradation experiments, NAA (dissolved in 95% ethanol) was added to a final concentration of 500 μM. For galactose induction experiments, cells were grown in YEP + lactic acid medium to mid-log phase, and galactose was added to a final concentration of 2%. For nocodazole arrest, nocodazole was added to a final concentration of 75 μg/mL.
Stability assays
Cells were grown in YEP + lactic acid until mid-log phase, followed by a 1-h 3Flag-Cse4 induction with 2% galactose. Glucose was added to a final concentration of 2% to inhibit further 3Flag-Cse4 transcription, and cycloheximide was added to a final concentration of 50 μg/mL. Time point 0 was taken immediately, followed by sample collection at the indicated time points. Extracts were prepared and analyzed as described in the Supplemental Material.
Ubiquitylation assays
Ubiquitylation assays were performed as described (Ranjitkar et al. 2010) with the following slight modifications. Twenty-microliter reactions containing 150 ng of E1 enzyme (human Uba1; catalog no. E-305, Boston Biochemicals), 115 ng of E2 enzyme (human UbcH5a; catalog no. E2-616, Boston Biochemicals), 100 ng of purified GST-Psh1 (or C45S, C50S RING mutant), ∼250 ng of Cse4 octamers, 2.5 μg of ubiquitin, 2 mM Mg-ATP, 50 mM Tris-HCl (pH 7.5), 2.5 mM MgCl2, and 0.5 mM DTT were incubated for 15–30 min at 23°C. Reactions were stopped by adding sample buffer and boiling for 4 min. Ubiquitin and Mg-ATP were obtained from Boston Biochemicals. Cse4 ubiquitin conjugates were detected by immunoblotting with anti-V5 or anti-Cse4 antibodies (gift from Carl Wu, National Institutes of Health) as indicated.
For ubiquitylation assays with Cse4 nucleosomes, ∼250 ng of nucleosomal template (mononucleosomes or polynucleosomes) conjugated to streptavidin–Dynabeads was washed twice with TE (10 mM Tris-HCl at pH 8.0, 1 mM EDTA), resuspended in TE, and added to the ubiquitylation reactions. For ubiquitylation assays with octamers and exogenously added DNA, an equal amount of biotinylated 146-bp DNA (purified as described in the Supplemental Material) to Cse4 octamer was added to the reactions.
Immunofluorescence
Cells were grown in YEP + lactic acid until mid-log phase, followed by nocodazole arrest for 3 h. For the last hour of arrest, 3Flag-Cse4 was induced with 2% galactose. Chromosome spreads were performed as described previously (Collins et al. 2004). DAPI (4′,6-diamidino-2-phenylindole; Molecular Probes) was used at a 1 μg/mL final concentration, and Lipsol was obtained from Fisher. Anti-Flag M2 antibodies were used at 1:1000 dilution, and Cy3 secondary antibodies (Jackson Immunoresearch) were used at a 1:1000 dilution.
Chromatin fractionation assays
Chromatin fractionations were performed similar to Keogh et al. (2006) with modifications described in the Supplemental Material.
Protein techniques and Cse4 nucleosome assembly
All protein techniques (purifications, immunoprecipitations, and immunoblotting) as well as Cse4 nucleosome generation are described in the Supplemental Material.
Acknowledgments
We are grateful to T. Formosa, J. Schumacher, R. Singer, M. Singleton, D. Stillman, and C. Wu for reagents. We thank A. Hernandez, T. Furuyama, and T. Tsukiyama for help with CENP-ACse4 octamer purification and advice on generating nucleosomes. We thank members of the Biggins laboratory, T. Tsukiyama, and S. Henikoff for critical reading of the manuscript, as well as insightful discussions with M. Basrai. This work was supported by National Institutes of Health grant GM078069 to S.B., and an American Cancer Society fellowship to G.M.R.D.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.243113.114.
References
- Abe T, Sugimura K, Hosono Y, Takami Y, Akita M, Yoshimura A, Tada S, Nakayama T, Murofushi H, Okumura K, et al. . 2011. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem 286: 30504–30512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A, Schillaci T, Lentini L, Di Leonardo A. 2009. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer 8: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Crisp MJ, DeLuca SZ, Rando OJ, Basrai MA. 2008. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics 179: 263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Dawson AR, Rawson DW, Taylor SB, Baker RE, Basrai MA. 2013. A novel role of the N terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics 194: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093 [DOI] [PubMed] [Google Scholar]
- Biggins S 2013. The composition, functions, and regulation of the budding yeast kinetochore. Genetics 194: 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Yu Y, Prall M, Formosa T, Stillman DJ. 2005. The yeast FACT complex has a role in transcriptional initiation. Mol Cell Biol 25: 5812–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL Jr, Cleveland DW. 2004. Structural determinants for generating centromeric chromatin. Nature 430: 578–582 [DOI] [PubMed] [Google Scholar]
- Brewster NK, Johnston GC, Singer RA. 1998. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J Biol Chem 273: 21972–21979 [DOI] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. 2007. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol Cell 26: 853–865 [DOI] [PubMed] [Google Scholar]
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. 2009. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell 35: 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol 189: 1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi ES, Stralfors A, Catania S, Castillo AG, Svensson JP, Pidoux AL, Ekwall K, Allshire RC. 2012. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A(Cnp1) in fission yeast. PLoS Genet 8: e1002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KA, Furuyama S, Biggins S. 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol 14: 1968–1972 [DOI] [PubMed] [Google Scholar]
- Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. 2011. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun 2: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, Theil AF, van Cappellen WA, Kimura H, Bartek J, et al. . 2013. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell 51: 469–479 [DOI] [PubMed] [Google Scholar]
- Eickbush TH, Moudrianakis EN. 1978. The histone core complex: an octamer assembled by two sets of protein–protein interactions. Biochemistry 17: 4955–4964 [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR 3rd, Cleveland DW. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8: 458–469 [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. 2009. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman DJ. 2001. Spt16–Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J 20: 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades AR, Kaufman PD, Stillman DJ. 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162: 1557–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Cheeseman IM. 2013. Induced dicentric chromosome formation promotes genomic rearrangements and tumorigenesis. Chromosome Res 21: 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol Cell Biol 21: 2098–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikopoulos T, Singh V, Tsui K, Awad S, Renshaw MJ, Scholfield P, Barton GJ, Nislow C, Tanaka TU, Owen-Hughes T. 2011. The SWI/SNF complex acts to constrain distribution of the centromeric histone variant Cse4. EMBO J 30: 1919–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell 40: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondele M, Stuwe T, Hassler M, Halbach F, Bowman A, Zhang ET, Nijmeijer B, Kotthoff C, Rybin V, Amlacher S, et al. . 2013. Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature 499: 111–114 [DOI] [PubMed] [Google Scholar]
- Jamai A, Puglisi A, Strubin M. 2009. Histone chaperone Spt16 promotes redeposition of the original H3–H4 histones evicted by elongating RNA polymerase. Mol Cell 35: 377–383 [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340: 1110–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble DJ, Whitby FG, Robinson H, McCullough LL, Formosa T, Hill CP. 2013. Structure of the Spt16 middle domain reveals functional features of the histone chaperone FACT. J Biol Chem 288: 10188–10194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. . 2006. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 439: 497–501 [DOI] [PubMed] [Google Scholar]
- Kingston IJ, Yung JS, Singleton MR. 2011. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem 286: 4021–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krassovsky K, Henikoff JG, Henikoff S. 2012. Tripartite organization of centromeric chromatin in budding yeast. Proc Natl Acad Sci 109: 243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22: 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, Cantaloube S, Kurumizaka H, Imhof A, Almouzni G. 2014. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell 53: 631–644 [DOI] [PubMed] [Google Scholar]
- Lefrancois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M. 2009. Efficient yeast ChIP-seq using multiplex short-read DNA sequencing. BMC Genomics 10: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune E, Bortfeld M, White SA, Pidoux AL, Ekwall K, Allshire RC, Ladurner AG. 2007. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr Biol 17: 1219–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lopes da Rosa J, Holik J, Green EM, Rando OJ, Kaufman PD. 2011. Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics 187: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Moreno O, Medina-Giro S, Torras-Llort M, Azorin F. 2011. The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3(CID). Curr Biol 21: 1488–1493 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922 [DOI] [PubMed] [Google Scholar]
- O’Donnell AF, Brewster NK, Kurniawan J, Minard LV, Johnston GC, Singer RA. 2004. Domain organization of the yeast histone chaperone FACT: the conserved N-terminal domain of FACT subunit Spt16 mediates recovery from replication stress. Nucleic Acids Res 32: 5894–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K, Abdulle R, Kitagawa K. 2014. Degradation of centromeric Histone H3 variant Cse4 requires the Fpr3 peptidyl–prolyl cis–trans isomerase. Genetics 196: 1041–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Okawa K, Isobe T, Fukagawa T. 2009. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol Biol Cell 20: 3986–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116 [DOI] [PubMed] [Google Scholar]
- Panchenko T, Sorensen TC, Woodcock CL, Kan ZY, Wood S, Resch MG, Luger K, Englander SW, Hansen JC, Black BE. 2011. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc Natl Acad Sci 108: 16588–16593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 40: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M, Williams SK, Dechassa ML, Das C, Linger J, Adkins M, Liu C, Bartholomew B, Tyler JK. 2009. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J Biol Chem 284: 23461–23471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D, Sims RJ 3rd. 2006. de FACTo nucleosome dynamics. J Biol Chem 281: 23297–23301 [DOI] [PubMed] [Google Scholar]
- Rhoades AR, Ruone S, Formosa T. 2004. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol Cell Biol 24: 3907–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Heiter P. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schlesinger MB, Formosa T. 2000. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics 155: 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Lawrence C. 1974. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Stuwe T, Hothorn M, Lejeune E, Rybin V, Bortfeld M, Scheffzek K, Ladurner AG. 2008. The FACT Spt16 ‘peptidase’ domain is a histone H3-H4 binding module. Proc Natl Acad Sci 105: 8884–8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. . 2011. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476: 232–235 [DOI] [PubMed] [Google Scholar]
- Takahata S, Yu Y, Stillman DJ. 2009. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell 34: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. 2003. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res 63: 3511–3516 [PubMed] [Google Scholar]
- VanDemark AP, Blanksma M, Ferris E, Heroux A, Hill CP, Formosa T. 2006. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell 22: 363–374 [DOI] [PubMed] [Google Scholar]
- Walfridsson J, Bjerling P, Thalen M, Yoo EJ, Park SD, Ekwall K. 2005. The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucleic Acids Res 33: 2868–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Straight AF. 2013. Functions of the centromere and kinetochore in chromosome segregation. Curr Opin Cell Biol 25: 334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler DD, Muthurajan UM, Hieb AR, Luger K. 2011. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem 286: 41883–41892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T. 2009. yFACT induces global accessibility of nucleosomal DNA without H2A–H2B displacement. Mol Cell 35: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tsukiyama T, Henikoff S. 2013. ISWI and CHD chromatin remodelers bind promoters but act in gene bodies. PLoS Genet 9: e1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, et al. . 2011. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472: 234–237 [DOI] [PMC free article] [PubMed] [Google Scholar]