By screening a custom library of shRNAs directed toward known drug targets in a genetically defined Myc-driven hepatocellular carcinoma model, Huang et al. identified Cdk9 as required for disease maintenance. Pharmacological or shRNA-mediated CDK9 inhibition led to robust anti-tumor effects that correlated with MYC expression levels and depended on the role that both CDK9 and MYC exert in transcription elongation. This study proposes CDK9 inhibition as a therapeutic strategy for MYC-overexpressing liver tumors and highlights the relevance of transcription elongation in the addiction of cancer cells to MYC.
Keywords: RNAi screen, CDK9, MYC, oncogene addiction, transcription elongation
Abstract
One-year survival rates for newly diagnosed hepatocellular carcinoma (HCC) are <50%, and unresectable HCC carries a dismal prognosis owing to its aggressiveness and the undruggable nature of its main genetic drivers. By screening a custom library of shRNAs directed toward known drug targets in a genetically defined Myc-driven HCC model, we identified cyclin-dependent kinase 9 (Cdk9) as required for disease maintenance. Pharmacological or shRNA-mediated CDK9 inhibition led to robust anti-tumor effects that correlated with MYC expression levels and depended on the role that both CDK9 and MYC exert in transcription elongation. Our results establish CDK9 inhibition as a therapeutic strategy for MYC-overexpressing liver tumors and highlight the relevance of transcription elongation in the addiction of cancer cells to MYC.
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related mortality worldwide (Villanueva et al. 2013). A major risk factor for HCC, the most common type of primary liver cancer, is cirrhosis, frequently caused by chronic viral hepatitis, alcohol abuse, and nonalcoholic fatty liver disease (Farazi and DePinho 2006). Although treatment of HCC has greatly improved over the last decades, most HCC patients diagnosed at advanced stages are ineligible for curative ablative therapies such as liver resection or transplantation (Villanueva et al. 2013). The use of the multikinase inhibitor sorafenib in patients with advanced HCC suggests that targeted therapies could be beneficial in this cancer; however, this regimen only extends life expectancy from 8 to 11 mo (Llovet et al. 2008), highlighting the urgent need for new therapeutic approaches.
Recent developments in gene expression profiling technologies have enabled the molecular classification of HCCs into defined subclasses, creating a solid foundation on which to build more informative clinical trials (Hoshida et al. 2009). Furthermore, exhaustive genomic studies have identified MYC genomic amplifications, β-catenin mutations, and tumor suppressor TP53 inactivation as frequent events in HCC (Laurent-Puig et al. 2001; Okamoto et al. 2003). However, unlike other tumor types, which present genetic drivers that can be therapeutically exploited, such as EGFR mutations in lung cancer and BRAF mutations in melanoma (Lynch et al. 2004; Flaherty et al. 2010), HCC is genetically heterogeneous and lacks clearly targetable mutant drivers (Villanueva et al. 2013). Thus, it seems likely that more insights into the function of currently “undruggable” genetic lesions will be necessary to develop rational therapies for this disease.
The MYC oncoprotein is an example of a well-validated but currently undruggable driver in HCC. MYC overexpression induces aberrant proliferation by affecting different biological processes, including gene transcription, protein translation, and DNA replication (Zhang et al. 2009; Conacci-Sorrell et al. 2014). Sustained MYC activation in mice creates a state of oncogene addiction, while MYC withdrawal in established tumors, including liver carcinomas, leads to tumor involution (Shachaf et al. 2004; Soucek et al. 2008). Additionally, owing to its role in mediating oncogenic signals, MYC is required for the maintenance of some tumors in which it is not amplified, including murine lung adenomas driven by KRAS and leukemia driven by MLL-AF9 (Zuber et al. 2011b; Soucek et al. 2013). In principle, the identification of critical molecules and processes required for MYC action in cancer provides an alternative strategy for targeting MYC-driven tumors (Dawson et al. 2011; Delmore et al. 2011; Zuber et al. 2011c).
RNAi technology enables a systematic interrogation of genes whose loss of function affects cell proliferation and viability (Ashworth and Bernards 2010; Kessler et al. 2012; Kumar et al. 2012). While a powerful method for identifying novel therapeutic targets, genome-wide RNAi screens can be laborious and expensive, requiring substantial infrastructure and specialized expertise for their execution. For these reasons, we favor focused shRNA libraries targeting a manageable set of genes with biological properties predicted to be important for the desired phenotype. Accordingly, we generated a customized shRNA library capable of suppressing proteins for which small molecule inhibitors are available; consequently, any validated hit in the screen should have a chemical probe to explore the underlying biology and serve as a basis for developing pharmacological approaches for modulating the phenotype. By screening the “drug target” library in a murine HCC model driven by Myc overexpression and p53 loss, we identified cyclin-dependent kinase 9 (Cdk9), a key component of the positive transcription elongation factor b (P-TEFb) complex, as required for the aberrant proliferation of MYC-overexpressing tumors. Our studies establish CDK9 as a target for a subset of HCC tumors and document a critical role for transcription elongation in sustaining the proliferation of MYC-overexpressing cancers.
Results
RNAi screen for genes encoding known drug targets
To systematically probe candidate drug targets required for HCC maintenance, we developed a screening platform and a focused shRNA library to facilitate the identification of cancer dependencies in a defined genetic context. For our screening system, we established a murine HCC model driven by Myc overexpression and p53 loss, which mimics two of the most common genetic drivers in human HCC (Supplemental Fig. 1A,B; Beroukhim et al. 2010; Shibata and Aburatani 2014). These cells also expressed a reverse tetracycline transactivator (rtTA3) that enabled efficient induction of tetracycline-responsive transgenes introduced by retroviral-mediated gene transfer (Supplemental Fig. 1C,D; for details, see the Supplemental Material). We envisioned that the use of a murine model produced by defined genetic drivers would avoid some of the confounding effects created by the unknown and heterogeneous dependencies occurring in human cancer cell lines.
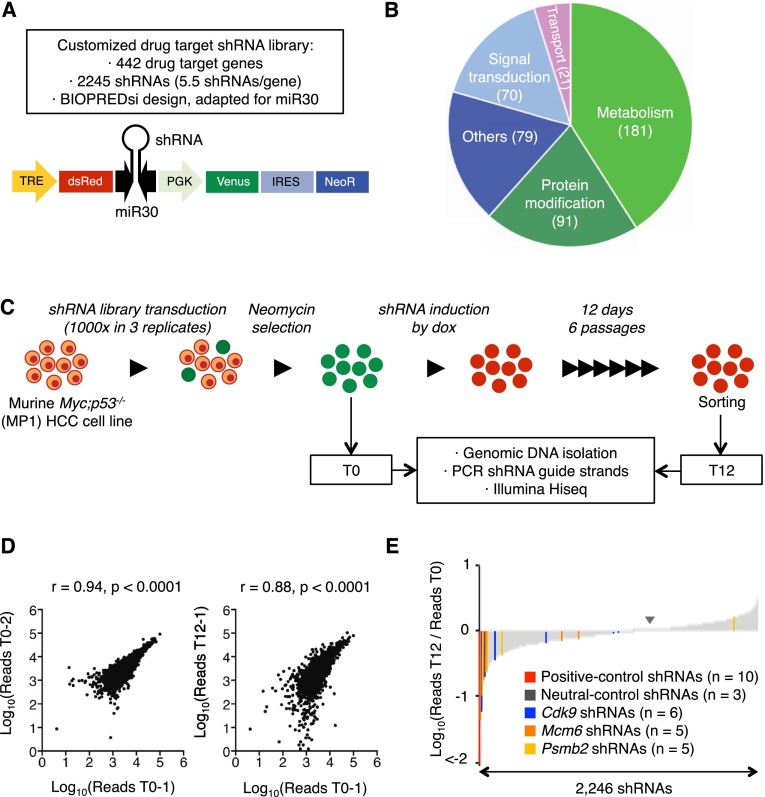
To identify genes whose protein products can be targeted by established agents, we built a custom shRNA library against 442 genes encoding known drug targets (about six shRNAs per gene) (Fig. 1A; Supplemental Table 1). This target list consisted of genes involved in metabolism, protein modifications, signal transduction, and macromolecular transport (Fig. 1B) and was biased for receptors and kinases (Supplemental Fig. 1E). The shRNAs were cloned downstream from a tetracycline-responsive promoter in TRMPV-neo (Fig. 1A), an inducible expression vector that was previously optimized for negative selection RNAi screens (Zuber et al. 2011a,c).
Figure 1.
RNAi screen for genes encoding known drug targets. (A) Library features and schematic of the TRMPV-neo vector. (B) Pathway categories of “drug target” genes included in the library. Numbers indicate the number of genes in each category. All shRNA sequences are provided in Supplemental Table 1. (C) RNAi screening strategy. (D) Representative scatter plots illustrating the correlation of normalized reads per shRNA between replicates at the beginning of the experiment (left) and replicates at different time points (right) (related to Supplemental Table 2). (E) Pooled negative selection screening results in MP1 (Myc;p53−/− clone 1 murine HCC cells) murine HCC cells. After eliminating underrepresented shRNAs at T0 (beginning of the experiment), shRNA abundance ratios of 2246 shRNAs were calculated as the number of normalized reads after 12 d of culture on dox (T12, end) divided by the number of normalized reads before dox treatment (T0) and are plotted as the mean of three replicates in ascending order.
The library was transduced as one pool in triplicate into murine Myc;p53−/− HCC cells (hereafter MP1 cells) at low multiplicity of infection (MOI < 1). Transduced cells were cultured such that, in theory, each shRNA was represented in at least 1000 cells throughout the experiment (Fig. 1C). After G418 selection, shRNAs were induced by addition of doxycycline (dox), and changes in shRNA representation after 12 d of culture were quantified using deep sequencing of shRNA guide strands amplified from genomic DNA of sorted shRNA-expressing cells (Fig. 1C). The correlation of normalized shRNA reads present in the replicates at T0 was close to 1 but substantially decreased when comparing T0 and T12 within the same replicate, suggesting changes in library representation associated with shRNA depletion (Fig. 1D; Supplemental Table 2).
Using the scoring criterion of more than fivefold average depletion in three independent replicates, 43 shRNAs were strongly depleted (Fig. 1E; Supplemental Table 3); these included all positive control shRNAs targeting essential genes (Rpa1, n = 1; Rpa3, n = 5; and Pcna, n = 1) as well as three shRNAs targeting Myc, the driving oncogene in our model. For a “hit” to undergo further analysis, we required that at least two independent shRNAs targeting a particular gene were identified in the primary screen. Genes fulfilling these criteria included the proteasome component Psmb2 (proteasome subunit β type 2), the replication factor Mcm6 (minichromosome maintenance complex component 6), and the transcription elongation factor Cdk9.
CDK9 is required for the proliferation of some HCC cell lines
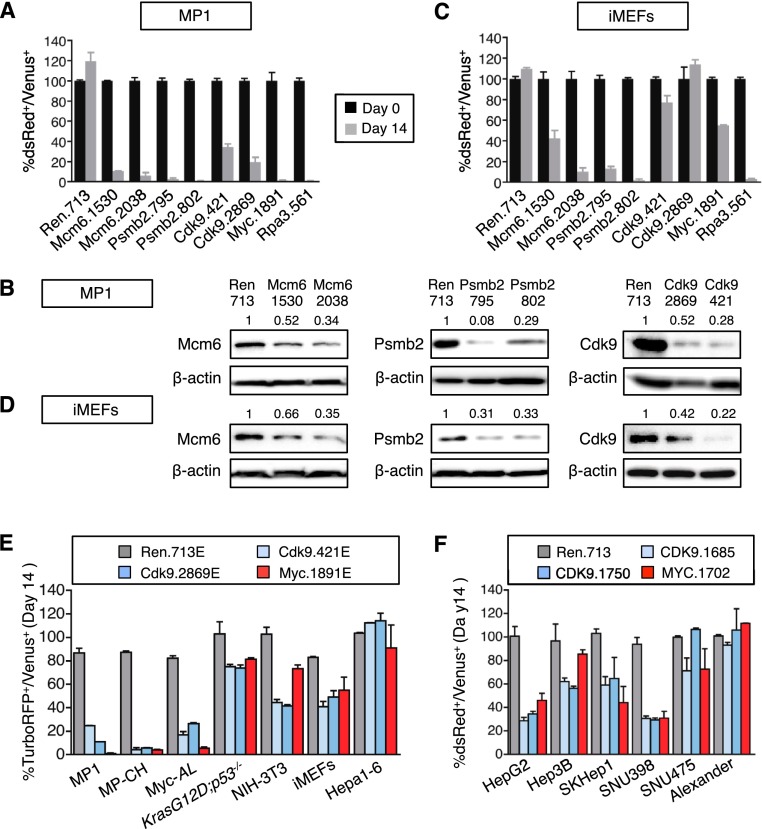
As our intent was to identify targets whose inhibition showed selective anti-proliferative effects in cancer cells, we validated the positive hits above in the MP1 cells used in the screen and then filtered out those that showed a similar effect on nontransformed immortalized mouse embryonic fibroblasts (iMEFs). shRNAs targeting Rpa3, an essential gene, and Myc, the oncogenic driver in the screened MP1 cell line, were used as positive controls; a Renilla luciferase shRNA was used as negative control. All selected shRNAs produced a competitive disadvantage in MP1 cells (Fig. 2A), confirming that the screen performance and the selection criteria were sufficient to remove false positive events. Moreover, each validated shRNA showed substantial knockdown of the intended protein, further indicating that the observed phenotypes were due to on-target effects (Fig. 2B).
Figure 2.
CDK9 is required for the proliferation of some HCC cell lines. (A) Competitive proliferation assay. G418-selected Venus+ cells were mixed with untransduced cells at 1:1 ratio and subsequently cultured in the presence of dox. The percentage of Venus+dsRed+ (shRNA-expressing) cells was determined at different time points (results at day 0 and day 14 are shown and are relative to day 0). Changes were used as readout of growth inhibitory effects. Values are mean + SD of three independent experiments. The graphs show the validation of the candidate shRNAs as well as control shRNAs (Ren.713, Myc.1891, and Rpa3.561) in MP1 murine HCC cells. (B) Immunoblots showing the knockdown induced by shRNAs expressed from TRMPV-neo in MP1 murine HCC cells. β-Actin was used as loading control. The numbers indicate protein levels relative to β-actin. (C) Competitive proliferation assay of control and candidate shRNAs expressed from TRMPV-neo in iMEFs, as described in A. Color code is as in A. (D) Immunoblots showing the knockdown induced by shRNAs expressed from TRMPV-neo in iMEFs. β-Actin was used as loading control. The numbers indicate protein levels relative to β-actin. (E,F) Competitive proliferation assay of control (Renilla and MYC) and CDK9 shRNAs expressed from TRMPV-neo-miR-E (E) or TRMPV-neo (F) in different murine (E) and human (F) cell lines, as described in A. The percentage of shRNA-expressing cells at day 14 relative to day 0 is shown. Values are mean + SD from two independent experiments.
shRNAs targeting Mcm6 and Psmb2 inhibited iMEFs and MP1 cells to a similar extent, suggesting that inhibition of their target proteins was generally lethal, much like the control Rpa3 shRNA (Fig. 2A,C). In contrast, Cdk9 shRNAs showed a reduced ability to inhibit proliferation in iMEFs as compared with MP1 cells (Fig. 2A,C; Supplemental Fig. 2A), an effect that was similar to the Myc shRNA and could not be accounted for by differences in proliferation rates of iMEFs and MP1 cells or in shRNA knockdown (Fig. 2B,D). Owing to this apparent specificity, we selected Cdk9 as a candidate for more detailed analysis.
To confirm that the impact of CDK9 inhibition on cancer cell proliferation was not limited to one experimental HCC line, we studied additional murine and human cell lines engineered to express rtTA3 to allow dox-dependent shRNA induction. Control (Renilla and Myc) and Cdk9 shRNAs were subcloned into miR-E, an optimized miR-30-based backbone that increases knockdown efficiency, particularly for those shRNAs with intermediate potency (Supplemental Fig. 2B,C; Fellmann et al. 2013). All three experimental murine HCC cell lines that overexpressed Myc (MP1, MP-CH, and Myc-AL) were highly sensitive to Cdk9 inhibition, while murine HCC cells expressing mutant KrasG12D, Hepa1-6 hepatoma cells, NIH-3T3 fibroblasts, and iMEFs showed modest to no sensitivity (Fig. 2E). Similarly, human HCC cell lines showed a range of responses to human CDK9 shRNAs, with some being sensitive and others being more resistant (Fig. 2F; Supplemental Fig. 2D). Again, these differential responses were independent of the proliferation rates of various cell lines (Supplemental Fig. 2E,F) and the extent of CDK9 knockdown (Supplemental Fig. 2B,G). In contrast to certain cell types in which CDK9 inhibition triggers cell death, CDK9 knockdown did not lead to apoptosis in HCC cells (data not shown) but primarily affected proliferation (Supplemental Fig. 2A,D). Together, these results indicate that CDK9 is a critical requirement for the ongoing proliferation of certain cancer cell lines.
Pharmacological inhibition of CDK9 in HCC cell lines
The overarching rationale for our screening setup was that validated hits would also be targets of existing small molecule inhibitors. CDK9 is a member of the CDK family of serine–threonine kinases that forms the catalytic core of P-TEFb and, in the presence of cyclin T, stimulates transcription elongation by RNA polymerase II (RNA Pol II) (Peterlin and Price 2006). Many small molecule drugs initially designed to target other CDKs also target CDK9, and it is now recognized that CDK9 inhibition contributes to their anti-proliferative effects (Wang and Fischer 2008). However, molecular determinants that dictate sensitivity to CDK9 inhibition are not well established, and the potential toxicities of various nonspecific CDK inhibitors have produced uncertainty as to whether CDK9 is a cancer-specific therapeutic target. Nonetheless, our studies using RNAi argue that selective inhibition of CDK9 can have potent and specific anti-cancer effects.
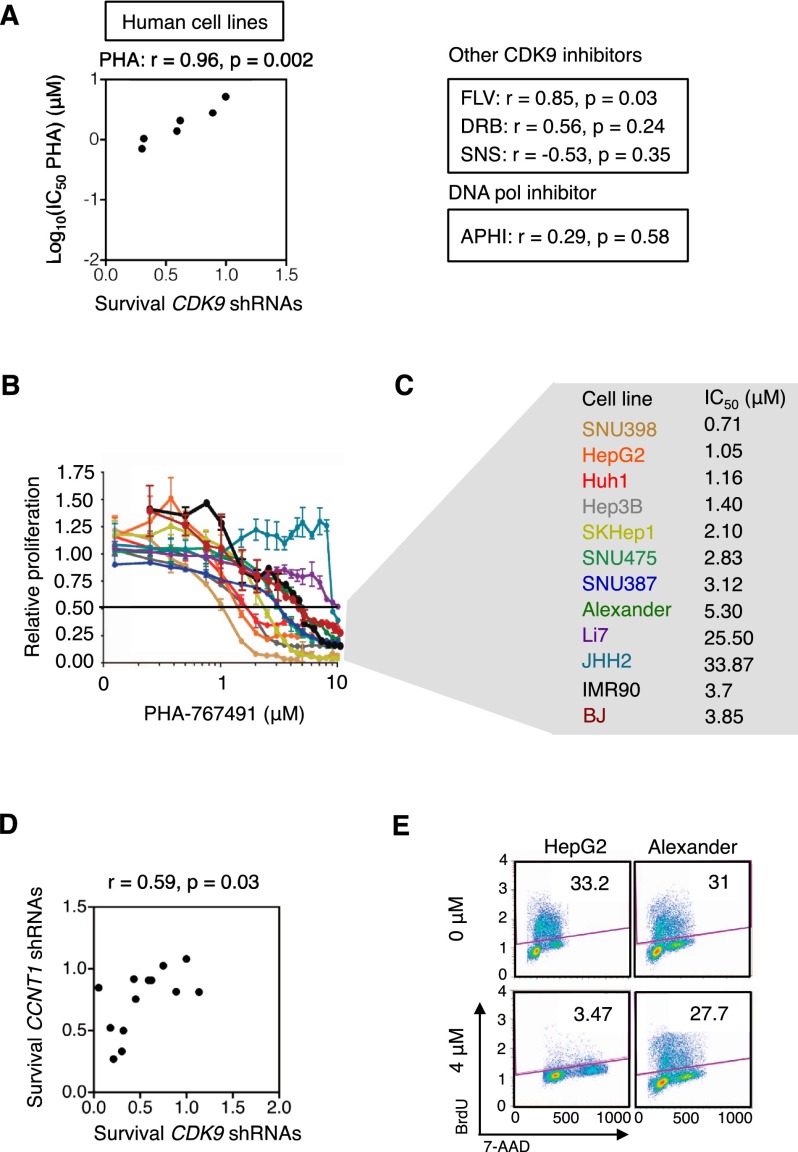
Although all currently available CDK9 inhibitors exhibit multiple off-target effects, we hypothesized that those that are most specific in inhibiting CDK9 would be the most likely to show a spectrum of activity similar to CDK9 shRNAs. We therefore examined the growth inhibitory effects of several CDK9 inhibitors and compared the IC50 of each drug with the anti-proliferative effects of CDK9 shRNAs in competitive proliferation assays (as in Fig. 2E,F). After testing four different CDK9 inhibitors (Supplemental Fig. 3A; Wang and Fischer 2008), we found that PHA-767491, a dual CDK9 and CDC7 (cell division cycle 7) kinase inhibitor (Montagnoli et al. 2008), most closely recapitulated shRNA-mediated CDK9 inhibition in both human and murine cell lines (Fig. 3A–C; Supplemental Fig. 3B–D). Notably, CDC7 in complexes with its allosteric regulator, DBF4, is required for initiation of DNA replication (Jiang et al. 1999). However, exposure of cells to aphidicolin, an inhibitor of DNA polymerase, did not mimic the effects of CDK9 shRNAs (Fig. 3A; Supplemental Fig. 3B). In contrast, shRNAs directed to Cyclin T1 (CCNT1), an obligate allosteric regulator of the CDK9 kinase, recapitulated the results obtained with CDK9 shRNAs (Fig. 3D), thereby highlighting the specificity of our results.
Figure 3.
Pharmacological inhibition of CDK9 in HCC cell lines. (A) Scatter plot illustrating the correlation between anti-proliferative effects of CDK9 shRNAs and the IC50 of PHA-767491 in six human cell lines (from Fig. 2F). The correlation and P-values of three additional CDK9 inhibitors and one DNA replication inhibitor (aphidicolin) are also shown in the right panel. The survival is defined as the average of the survival ratio of two shRNAs in competitive proliferation assay. (FLV) Flavopiridol; (PHA) PHA-767491; (SNS) SNS-032; (APHI) aphidicolin. (B) Proliferation rates of PHA-767491-treated human cells, calculated by measuring the change in viable cell number after 72 h in culture and fitting data to an exponential growth curve. Results were normalized to the proliferation rate of vehicle (H2O)-treated cells, set to 1. Values are mean ± SD of three independent replicates. (C) Summary of PHA-767491 IC50 values of human cell lines in B. (D) Scatter plot illustrating the correlation between survival with CDK9 shRNAs and survival with CCNT1 shRNAs in murine and human cell lines (from Fig. 2E,F). (E) Representative flow cytometry plots showing cell cycle analysis (BrdU+7-AAD+ double staining) of cells after 48 h of PHA-767491 treatment. The experiment was performed twice, and values indicate the mean ± SD.
We evaluated the effect of PHA-767491 in additional human HCC cell lines (Fig. 3B,C) and observed broad anti-proliferative activity in the most sensitive cell lines (IC50 < 2 μM) (Fig. 3E; Supplemental Fig. 3E). PHA-767491 treatment triggered cell cycle arrest, similar to the effects of CDK9 shRNAs (Fig. 3E; Supplemental Figs. 2D, 3E); however, the anti-proliferative effects mediated by PHA-767491 were more pronounced. Of note, BJ, IMR90, and AML12 (α mouse liver 12), three nontransformed cell lines, were consistently less sensitive to PHA-767491 treatment (Fig. 3B,C; Supplemental Fig. 3C,D), further supporting increased sensitivity of certain cancer cells to CDK9 inhibition. While some of these phenotypes may be due to CDC7 inhibition (Supplemental Fig. 3A), the significant correlation between the IC50 of PHA-767491 and the anti-proliferative effects of CDK9 shRNAs implies that a major component of PHA-767491 activity is through CDK9 inhibition. These data illustrate how using RNAi and small molecule inhibitors as orthogonal approaches can assist target validation and indicate that CDK9 can be required for HCC proliferation.
MYC expression predicts response to CDK9 inhibition
Since our screening system was driven by p53 loss and Myc overexpression, we next asked whether alterations in either gene could dictate sensitivity to CDK9 inhibition. We cross-referenced our data on the relative sensitivity of HCC cell lines to CDK9 inhibition (measured as the growth inhibitory effects of CDK9 shRNAs in competitive proliferation assays or the IC50 of PHA-767491) to mutational and gene expression profiles of the same cell lines available from the Cancer Cell Line Encyclopedia (CCLE) (Barretina et al. 2012). We found no correlation between the anti-proliferative response to CDK9 inhibition and p53 expression or mutational status (Supplemental Fig. 4A,B), consistent with previous findings indicating that PHA-767491 inhibits cancer cell proliferation through p53-independent mechanisms (Montagnoli et al. 2008).
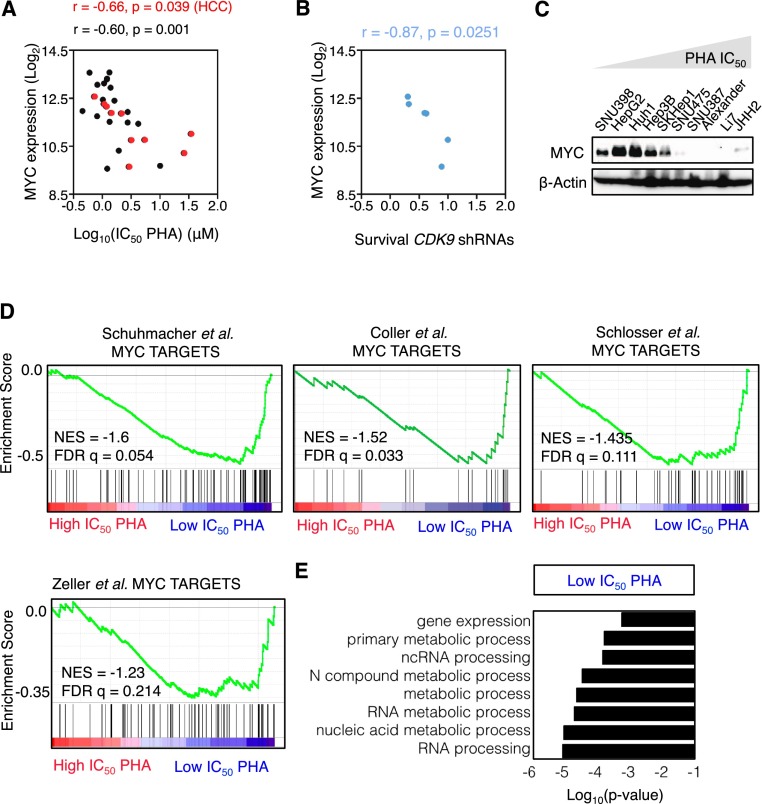
In contrast, there was a highly significant correlation between the response to CDK9 inhibition and MYC mRNA expression (Fig. 4A,B; Supplemental Table 4), an effect confirmed by immunoblotting for MYC protein (Fig. 4C; Supplemental Fig. 4C). A similar trend was noted in murine cell lines, further reinforcing these observations (Supplemental Fig. 4D). This significant correlation was not limited to HCC cell lines but was also observed in a panel of lung and hematopoietic cancer cell lines available in our laboratory (Fig. 4A; Supplemental Fig. 4E). In all instances, no correlation was observed between sensitivity to CDK9 inhibition and the relative proliferative rates of individual cell lines (Supplemental Fig. 4F). Furthermore, no correlation was observed between sensitivity to CDK9 and sorafenib, a multikinase inhibitor that is the only drug that confers a significant, albeit limited, survival benefit to HCC patients (Llovet et al. 2008), or JQ1, a bromodomain inhibitor known to inhibit MYC in hematological malignancies (Supplemental Fig. 4G; Delmore et al. 2011; Zuber et al. 2011c). Accordingly, the sensitivity of HCC cells to sorafenib and JQ1 did not correlate with MYC expression levels (Supplemental Fig. 4H).
Figure 4.
MYC expression predicts response to CDK9 inhibition. (A) Scatter plot illustrating the correlation between PHA-767491 (PHA) IC50 values and MYC expression levels in human HCC (red; from Fig. 3B,C), leukemia, lymphoma, and lung cancer cell lines (n = 28). (B) Scatter plot illustrating the correlation between survival with CDK9 shRNAs and MYC expression levels in a panel of different human HCC cell lines (from Fig. 2F). The survival is defined as the average of the survival ratio of two shRNAs in competitive proliferation assay. (C) Immunoblots showing MYC protein levels in 10 human HCC cell lines. β-Actin was used as loading control. (D) GSEA plot evaluating the association between low IC50 of PHA-767491 and MYC targets. (NES) Normalized enrichment score; (FDR) false discovery rate. (E) GO term analysis of the genes that are significantly associated with sensitivity to PHA-767491.
The transcriptional profiles of the 10 human HCC cell lines with defined sensitivities to PHA-767491 were also subjected to gene set enrichment analysis (GSEA) and gene ontology (GO) analysis to point toward genes and processes that might underlie their differential sensitivity to CDK9 inhibition. This analysis revealed a significant correlation between low IC50 (sensitivity to CDK9 inhibition) and four canonical transcriptional signatures of MYC-dependent genes (Fig. 4D; Supplemental Table 5; Coller et al. 2000; Schuhmacher et al. 2001; Zeller et al. 2003; Schlosser et al. 2005). MYC is a global regulator of gene expression that affects overall transcription, ribosomal biogenesis, protein translation, and cellular metabolism (Dang 2012; Conacci-Sorrell et al. 2014), and, interestingly, transcripts overrepresented in sensitive compared with resistant cells were linked to “gene expression,” “RNA metabolic process,” and “RNA processing” (Fig. 4E; Supplemental Table 6). Taken together, these results indicate that CDK9 might be crucial for the maintenance of MYC-overexpressing tumors and identify a potential patient population whose tumors might be responsive to CDK9 inhibition.
CDK9 mediates transcription elongation of MYC targets in MYC-overexpressing cancer cells
After transcription initiation, RNA Pol II is trapped near the promoter of many genes, a process known as proximal promoter pausing (Core et al. 2008; Zhou et al. 2012; Lee and Young 2013). For productive transcription, P-TEFb is recruited, and CDK9 phosphorylates Ser2 in the C-terminal domain (CTD) of RNA Pol II, inducing pause release and subsequent transcription elongation (Zhou et al. 2012; Lee and Young 2013). MYC has previously been shown to participate in transcription elongation by regulating this pause release mechanism (Eberhardy and Farnham 2001; Kanazawa et al. 2003; Rahl et al. 2010). Hence, in MYC-overexpressing tumor cells, MYC accumulates in the promoter region of many transcriptionally active genes, recruiting the P-TEFb complex and amplifying transcription of MYC target genes (Lin et al. 2012b; Nie et al. 2012).
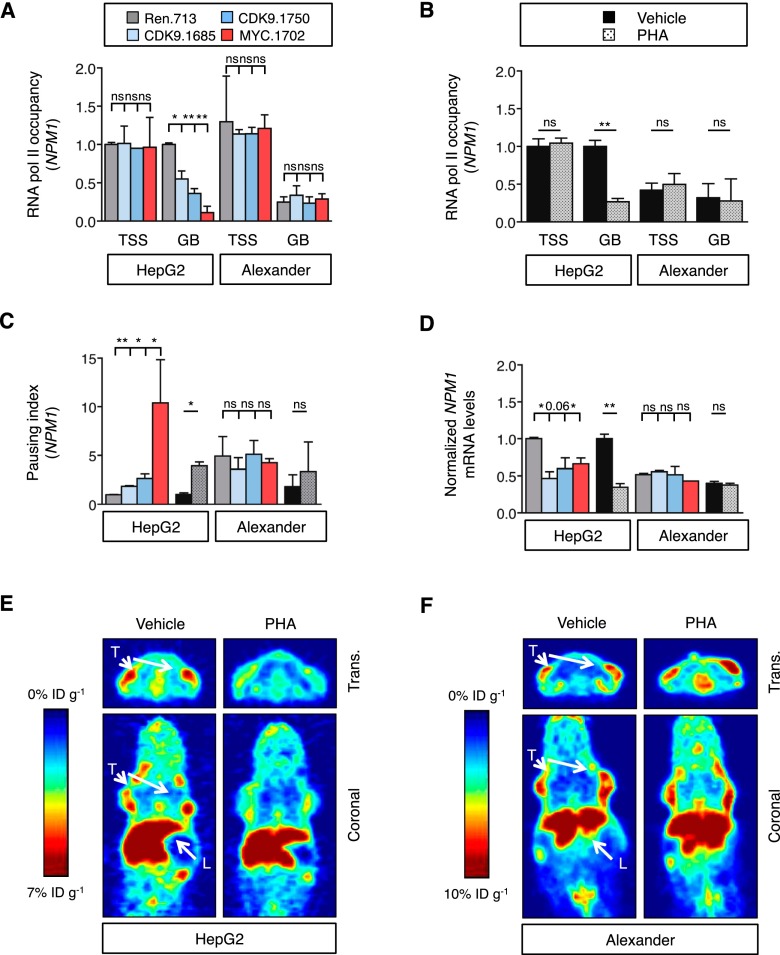
To explore the role of CDK9 in MYC-mediated transcription elongation in HCC, we first investigated the effects of CDK9 inhibition in two human HCC cell lines, HepG2 and Alexander, which express different levels of MYC (Fig. 4C; Supplemental Table 4). As expected, pharmacological or RNAi-mediated suppression of CDK9 led to a decrease in Ser2 phosphorylation in both cell lines (Supplemental Fig. 5A). In order to investigate whether these changes correlated with changes in transcription elongation, we calculated the pausing index or traveling ratio, in which the ratio of RNA Pol II-binding density in the proximal promoter region is compared with that in the gene body (Rahl et al. 2010). In HepG2 cells expressing high levels of MYC, inhibition of CDK9 (using small molecules or shRNAs) or MYC caused a significant repression of transcription elongation of NPM1 and MCM4, two select MYC targets, and an increase in their pausing index (Fig. 5A–C; Supplemental Fig. 5B). However, no changes were observed for BRG1, whose transcription is not impacted by MYC (Supplemental Fig. 5C; Rahl et al. 2010). In contrast, in Alexander cells expressing much lower MYC levels, the pausing index at the NPM1 and MCM4 genes was already high in untreated cells and did not change substantially following CDK9 or MYC inhibition (Fig. 5C; Supplemental Fig. 5B). Accordingly, CDK9 or MYC inhibition reduced mRNA levels of MYC target genes in HepG2 cells but not Alexander cells (Fig. 5D; Supplemental Fig. 5D).
Figure 5.
CDK9 mediates transcription elongation of MYC targets in MYC-overexpressing cancer cells. (A,B) Chromatin immunoprecipitation combined with quantitative PCR (ChIP-qPCR) performed in human HCC cells expressing CDK9 and MYC shRNAs (A) or treated with PHA-767491 (PHA; 6 h at 4.5 μM) (B), with RNA Pol II antibody and primers located in either the transcription start site (TSS) or the gene body (GB) of NPM1. RNA Pol II occupancy relative to control condition in HepG2 cells is shown. Values are mean + SD from two independent experiments. (C) Pausing index of NPM1 in human HCC cell lines. The pausing index, also known as traveling ratio, was calculated as the ratio between the RNA Pol II bound to the TSS and the RNA Pol II bound to the GB. Values are mean + SD from two independent experiments. Color code and statistics are as in A and B. (D) Quantitative RT–PCR of NPM1 in human HCC cell lines treated with PHA-767491 (16 h at 4.5 μM) or with CDK9 shRNAs. Data are relative to expression in the untreated cells or Renilla-shRNA in HepG2 cells, normalized to the average expression of the housekeeping gene GAPDH. Values are mean + SD from two independent experiments. Color code and statistics are as in A and B. (E,F) PET imaging with 89Zr-transferrin of HepG2 (E) or Alexander (F) tumors with or without 3-d treatment with PHA-767491. (L) Liver; (T) tumor; (trans.) transverse. The color scale for all PET image data shows radiotracer uptake in units of injected dose per gram (%ID/g), with red corresponding to the highest activity, and blue corresponding to the lowest activity.
MYC is known to stimulate transcription of transferrin receptor 1 (TFRC) (Supplemental Fig. 5E,F; O’Donnell et al. 2006), leading to an increase in steady-state protein levels. By using 89Zr-desferrioxamine-labeled transferrin (89Zr-transferrin), a new positron emission tomography (PET) radiotracer that binds TFRC (Holland et al. 2012), it is possible to indirectly assess MYC activity in vivo. HepG2 and Alexander cell lines were implanted subcutaneously, as the TFRC levels in the liver are already high via MYC-independent mechanisms (Sciot et al. 1990). Upon tumor formation, mice were treated with PHA-767491 during 3 d, and PET was performed 2 d after. PHA-767491 treatment had no effect on the strong PET signal in the liver and did not change the signal in low-MYC-expressing Alexander-derived tumors (Fig. 5E,F; Supplemental Fig. 5G,H). In contrast, PHA-767491 treatment caused a marked decrease in PET signal in the MYC-overexpressing HepG2 line (Fig. 5E,F; Supplemental Fig. 5G,H). Together, these results indicate that CDK9 regulates the transcription elongation of several MYC targets in the context of high MYC expression and that CDK9 inhibition can reverse these effects.
Transcription elongation is required to maintain proliferation in MYC-overexpressing HCC
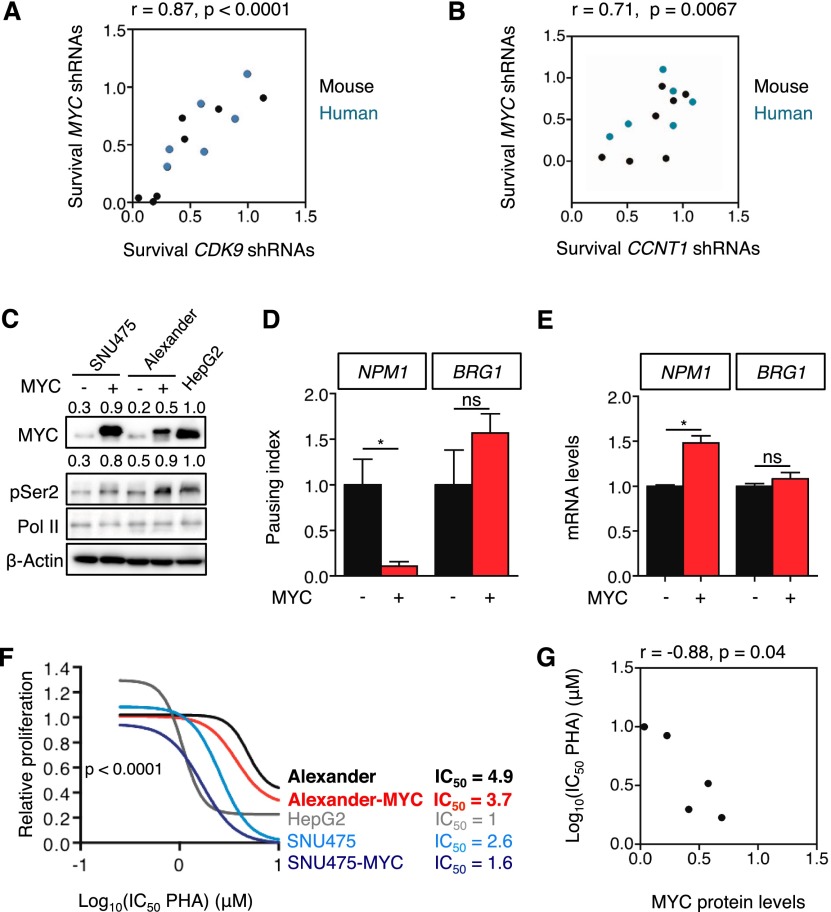
MYC has been implicated in DNA replication, transcriptional activation, transcription elongation, and other processes (Dang 2012; Conacci-Sorrell et al. 2014), although the relative contribution of each process to tumor initiation and maintenance is not well understood. Despite the diversity of factors involved in these processes, we noticed that CDK9 and MYC shRNAs displayed similar depletion patterns across multiple lines (Fig. 2E,F), and, in fact, there was a significant correlation between the anti-proliferative effects of CDK9 and MYC shRNAs in competitive proliferation assays (Fig. 6A). A similar correlation existed between the anti-proliferative effects of Cyclin T1 and Myc shRNAs (Fig. 6B; Supplemental Fig. 6A–C). However, no correlation was observed between these shRNAs and the control Renilla shRNA, confirming the specificity of these results (Supplemental Table 7).
Figure 6.
Transcription elongation is required to maintain proliferation in MYC-overexpressing HCC. (A,B) Scatter plot illustrating the correlation between survival with MYC shRNAs and survival with CDK9 shRNAs (A) or CCNT1 shRNAs (B) in mouse (black) and human (blue) cell lines (from Fig. 2E,F). The survival is defined as the ratio of surviving cells in the competitive proliferation assays. In the case of CDK9 and CCNT1, the average of the survival of two different shRNAs was used. (C) Immunoblots showing the effect of MYC overexpression on Ser2 phosphorylation of RNA Pol II in low-MYC-expressing SNU475 and Alexander cells. β-Actin was used as loading control. Values indicate normalized protein levels, normalized with β-actin or RNA Pol II and relative to the levels in HepG2 cells. (D) Pausing index of NPM1 and BRG1 in Alexander cells overexpressing MYC. Pausing index, also known as traveling ratio, was calculated as the ratio between the RNA Pol II bound to the TSS and the RNA Pol II bound to the gene body. Values are mean + SD from two independent experiments. (E) Quantitative RT–PCR of NPM1 and BRG1 in Alexander cells overexpressing MYC. Data are relative to expression in the cells expressing an empty vector, normalized to the average expression of the housekeeping gene GAPDH. Values are mean + SD from two independent experiments. (F) Proliferation rates of PHA-767491 (PHA)-treated cells in C, calculated by measuring the increase in viable cell number after 72 h in culture and fitting data to an exponential growth curve. Results are normalized to the proliferation rate of vehicle (H2O)-treated cells, set to 1. Values are mean ± SD of two independent replicates. The IC50 values are included at the right in micromolar (μM). (G) Scatter plot illustrating the correlation between MYC protein levels and the IC50 of PHA-767491 on the different cell lines (related to C and F).
To directly determine whether modulation of MYC activity could influence cellular dependence on CDK9, we tested the ability of enforced MYC expression to influence sensitivity to CDK9 inhibition. Ectopic MYC expression in the low-MYC Alexander cells produced MYC levels ∼50% of those measured in sensitive HepG2 cells and an increase in MYC-dependent transcription elongation as measured by a reduced pausing index at the NPM1 and MCM4 but not BRG1 genes (Fig. 6C,D; Supplemental Fig. 6D). This effect on transcription elongation was accompanied by significant increases in NPM1 and MCM4 mRNAs (Fig. 6E; Supplemental Fig. 6E), while levels of BRG1 mRNA did not change (Fig. 6E). Concordantly, MYC-overexpressing Alexander cells became more sensitive to CDK9 inhibition by PHA-767491 or CDK9 shRNAs (Fig. 6F; Supplemental Fig. 6F). While enforced MYC expression in these cells did not produce the same sensitivity of high-MYC-expressing HepG2 cells, enforced MYC expression in SNU-475 cells achieved MYC levels equivalent to those observed in HepG2 cells (Fig. 6C) and produced a similar sensitivity to PHA-767491 (Fig. 6F,G), underscoring that high MYC expression levels are required to establish sensitivity to CDK9 inhibition. Conversely, MYC suppression in HepG2 and SNU398 cells, but not Alexander cells, decreased their sensitivity to PHA-767491; however, as the MYC shRNAs alone reduce their proliferation rate, we cannot rule out the possibility that this reduced drug sensitivity is an indirect effect (data not shown). Taken together, the phenotypic similarities between MYC inhibition and CDK9/CCNT1 inhibition point to a key role for transcription elongation in mediating MYC function in tumor maintenance.
CDK9 is required for initiation and maintenance of MYC-overexpressing liver tumors
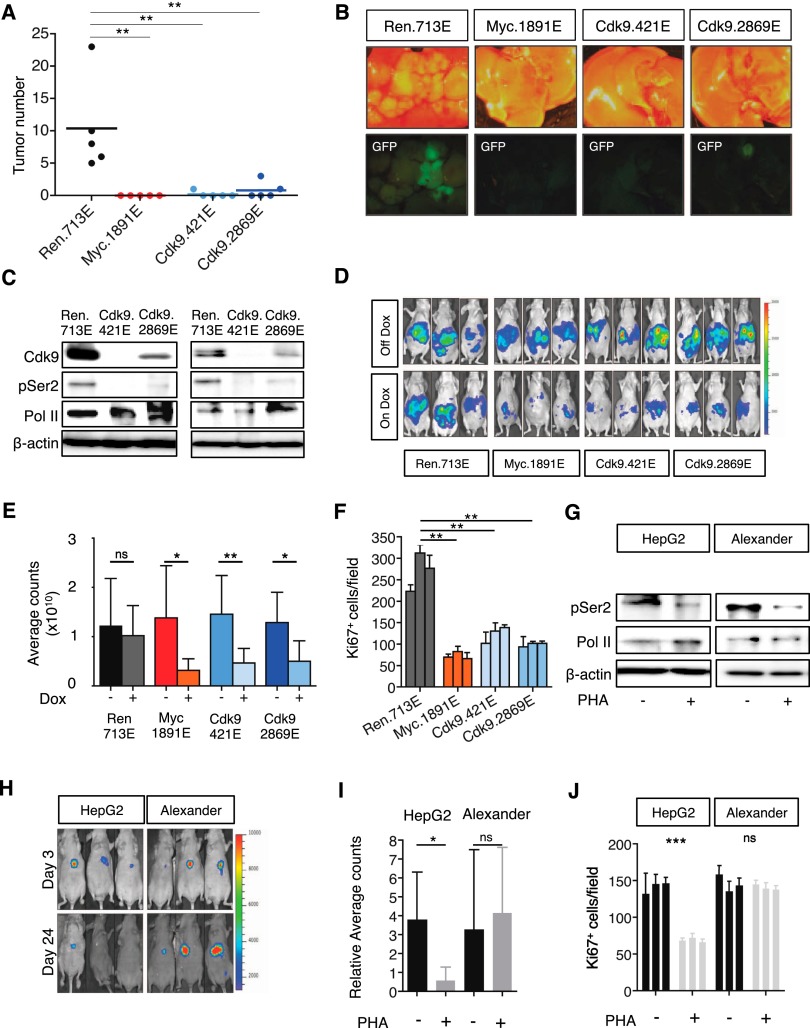
We also investigated the relevance of Cdk9 on HCC initiation and progression in vivo. To test the role of Cdk9 on Myc-driven tumorigenesis, we performed hydrodynamic tail vein injections of transposons expressing Myc and different shRNAs, which are integrated into the liver following transient expression of transposase from a recombinant transposon vector (Supplemental Fig. 7A; Yant et al. 2000). Seven weeks after injection, numerous tumors were observed in the livers expressing control shRNA; however, Cdk9 or Myc silencing completely abrogated tumor formation (Fig. 7A,B), indicating that Cdk9 is required for Myc-driven tumorigenesis.
Figure 7.
CDK9 is required for initiation and maintenance of MYC-overexpressing liver tumors. (A) Dot plot representation of the number of liver tumors after the hydrodynamic injection of Myc oncogene and the corresponding shRNAs. Bars represent the mean ± SD of five independent mice. (B) Representative bright-field and fluorescent images of the livers in A. Tumors are positive for GFP. (C) Immunoblots showing the knockdown induced by Cdk9 shRNAs in two representative tumors. Cdk9 inhibition leads to a decrease in the levels of phosphorylation of Ser2 of RNA Pol II (pSer2) and mild changes in total RNA Pol II levels (Pol II). β-Actin was used as loading control. (D) Bioluminescent imaging of representative mice orthotopically transplanted with MP1 (Myc;p53−/− murine HCC clone #1) cells harboring the indicated TRMPV-Neo-miR-E shRNAs 8 d after dox treatment. Dox was administered upon disease onset, 7 d after transplant. (E) Quantification of bioluminescent imaging responses with or without dox treatment. Values are mean + SD of six independent tumors. (F) Quantification of the number of Ki67-positive cells per field after analyzing three fields per animal and three animals per condition. Values are mean + SD. (G) Immunoblot showing the effects caused by PHA-767491 in two representative tumors. PHA-767491 treatment leads to a decrease in the levels of phosphorylation of Ser2 of RNA Pol II (pSer2) and mild changes in total RNA Pol II levels (Pol II). β-Actin was used as loading control. (H) Bioluminescent imaging of representative mice orthotopically transplanted with either HepG2 or Alexander HCC cells. PHA-767491 was administered upon disease onset (considered as day 0), 28 d after transplant. Days 3 and 24 of treatment are shown. (I) Quantification of bioluminescent imaging responses with or without PHA-767491 treatment. Values are mean + SD of seven or eight independent tumors. (J) Quantification of the number of Ki67-positive cells per field after analyzing three fields per animal and three animals per condition. Values are mean + SD.
To suppress Cdk9 in established tumors, MP1 murine HCC cells were transduced with Luciferase and dox-inducible TRMPV-Neo-miR-E constructs containing Cdk9 shRNAs or control shRNAs (Renilla and Myc) and were transplanted into the livers of recipient mice by subcapsular injection (Supplemental Fig. 7B). Upon detection of a luminescent signal, the animals were randomized and treated with dox to induce shRNA expression. Tumors bearing Cdk9 shRNAs exhibited a prominent decrease in the levels of Ser2 phosphorylation of RNA Pol II, implying that transcription elongation was efficiently repressed (Fig. 7C). By day 8, bioluminescent imaging revealed that mice from the untreated group (no dox) displayed large hepatic tumors; however, knockdown of Cdk9 or Myc led to a comparable and significant delay in tumor growth (Fig. 7D,E; Supplemental Fig. 7C). Ki67 staining of histological sections revealed that tumors expressing shRNAs for Cdk9 or Myc showed less proliferation compared with tumors expressing the control Renilla shRNA (Fig. 7F; Supplemental Fig. 7D). Therefore, RNAi-mediated suppression of Cdk9 approximates the effect of Myc inhibition in eliciting anti-tumor effects in HCC in vivo.
We also examined growth inhibitory effects of pharmacological CDK9 inhibition in a series of human HCC xenografts expressing luciferase. Treatment with PHA-767491 (50 mg/kg, twice per day, 5 d per week) or vehicle was initiated upon detection of bioluminescence. Consistent with previous findings (Montagnoli et al. 2008), PHA-767491 treatment was well tolerated in mice (Supplemental Fig. 7E) and triggered a decrease in Pol II Ser2 phosphorylation in the emerging tumors (Fig. 7G). In high-MYC-expressing HepG2 cells, PHA-767491 also decreased tumor cell proliferation, which was associated with substantial inhibition of tumor growth and even some tumor regressions (Fig. 7H–J; Supplemental Fig. 7F–H). In marked contrast, the same treatment of low-MYC-expressing Alexander cells produced little if any effect (Fig. 7H–J; Supplemental Fig. 7F–H) despite a detectable decrease in Pol II Ser2 phosphorylation (Fig. 7G). Thus, CDK9 is required for the maintenance of MYC-overexpressing HCC, implicating transcription elongation as important for MYC dependency in vivo.
Discussion
HCC is among the most lethal cancers owing to its aggressive nature and frequently late detection (Villanueva et al. 2013). While genome characterization efforts have identified druggable cancer drivers for many malignancies, these studies have yet to provide focus for future HCC therapies, and it seems likely that more functional insights into the mechanism of action of high-frequency yet not obviously druggable drivers will be needed to develop rational therapies for this disease. In this regard, the MYC oncogene is a well-established driver whose overexpression is required for the maintenance of murine and human HCC (Shachaf et al. 2004), although, as a transcription factor, it has not proven amenable to small molecule inhibition. Here, we show that MYC sustains malignancy in part through CDK9, thereby supporting the importance of transcription elongation for MYC action in HCC and implicating CDK9 and the processes it controls as rational targets for therapies to treat MYC-overexpressing cancers.
Our study used experimental RNAi to identify genes that are selectively required for the proliferation or survival of cancer cells (Ashworth and Bernards 2010; Kessler et al. 2012; Kumar et al. 2012). We designed a focused shRNA library to identify small molecules that could be repurposed for new applications such as treatment of HCC. Both RNAi and small molecule screens have limitations, mainly related to distinct types of off-target effects, but the combination of these orthogonal approaches enabled us to identify and validate CDK9 as a promising HCC target despite the use of imperfect reagents. Additionally, the establishment of a genetically defined murine HCC model appeared to bias the screen toward identifying dependencies produced by one of the drivers without the need to extensively screen many cell lines. Underscoring the utility of this approach, we were able to validate the screening results in human cancer cell lines with complex genetic alterations. Similarly focused RNAi screens can be combined with an existing drug to identify novel therapeutic combinations or performed in vivo, where hundreds of genes or drug targets can be evaluated simultaneously in a single animal (Schramek et al. 2014).
CDK9 has previously been proposed as a potential therapeutic target for cancer and other diseases (Cho et al. 2010; Romano 2013). CDK9 inhibitors such as flavopiridol, SNS-032, or roscovitine have proven particularly efficient for multiple myeloma or chronic lymphocytic leukemia through the down-regulation of anti-apoptotic genes XIAP and Mcl-1 (Gojo et al. 2002; Hahntow et al. 2004; Chen et al. 2009). Interestingly, these cancer types are often MYC-driven, although whether this explains their drug sensitivity is not well established. Nonetheless, further drug development into novel therapies has been hampered by lack of specificity of drugs and uncertainty about which patient population would most likely benefit from such agents. By performing experiments using RNAi triggers and small molecules in cells from two different species, we identified CDK9 as a selective therapeutic target for a subset of HCCs. Because CDK9 is not evidently mutated in HCC (Barretina et al. 2012), the sensitivity of liver cancer cells to CDK9 inhibition could not have been revealed simply through genomic studies alone.
In HCC cells, CDK9 inhibition mainly inhibits proliferation, although tumor regression is also observed in vivo. While further studies will be required to understand the precise mechanism of action, our work clearly identifies MYC overexpression as a key molecular determinant that dictates sensitivity to CDK9 inhibition. Therefore, we predict that HCCs with high MYC levels will be most likely to respond to improved CDK9 inhibitors. Thus, our study identifies a drug target (CDK9), tool compounds (e.g., PHA-767491), a patient selection criteria (high MYC), and a pharmacodynamic marker (Ser2 phosphorylation on the CTD of Pol II) that will greatly facilitate preclinical and clinical development of this strategy in both liquid and solid cancers.
CDK9 constitutes the catalytic core of P-TEFb and stimulates transcription elongation of most protein-coding genes (Peterlin and Price 2006). CDK9 kinase activity is stimulated to phosphorylate the CTD of RNA Pol II through its association with cyclin T and, consistent with a requirement of CDK9 activity for transcription elongation, cyclin T1 shRNAs phenocopied the effects of CDK9 inhibitors at impairing RNA Pol II Ser2 phosphorylation, transcription elongation of MYC target genes, and cancer cell proliferation. In agreement with a role for MYC in these effects, both MYC and CDK9 shRNAs had similar effects in all of the above assays. Importantly, these effects correlated with basal MYC levels; hence, tumor cells with high MYC expression show more transcription elongation on MYC target genes than those with low MYC levels, and enforced MYC expression in low-MYC cancer cell lines increases elongation and sensitivity to the anti-proliferative effects of CDK9 inhibition. Our data support previous work showing that MYC recruits the CDK9/P-TEFb complex to specific promoters (Eberhardy and Farnham 2001; Kanazawa et al. 2003; Rahl et al. 2010), thereby enhancing transcriptional output by stimulating transcript elongation via pause release (Lin et al. 2012b; Nie et al. 2012). As such, the P-TEFb complex is required for MYC to promote transcription elongation, and, as shown here, MYC-overexpressing tumor cells appear to depend on this activity. Interestingly, CDK9 inhibitors seem to primarily affect short-lived mRNAs, such as MYC, suggesting that MYC deregulation itself may contribute to the anti-tumor effects by CDK9 inhibition (Lam et al. 2001).
Our studies suggest that CDK9 has a “synthetic lethal” relationship with MYC in that its requirement for cancer cell proliferation is revealed in the presence of MYC overexpression (Luo et al. 2009). Analogous cofactors have also been identified, and some are being developed as therapeutic targets (Goga et al. 2007; Zuber et al. 2011c; Kessler et al. 2012; Lin et al. 2012a; Toyoshima et al. 2012). For example, BRD4 was identified as a therapeutic target for acute myeloid leukemia and other blood cancers by virtue of its ability to sustain MYC transcription, and small molecule BRD4 inhibitors show potent anti-leukemic effects (Dawson et al. 2011; Delmore et al. 2011; Zuber et al. 2011c). Interestingly, BRD4 can either act to sustain MYC transcription by binding superenhancers in the MYC promoter or cooperate with MYC to stimulate transcription elongation through its ability to recruit P-TEFb (Bisgrove et al. 2007; Loven et al. 2013). Nonetheless, what determines sensitivity to BRD4 inhibition is not understood, and it seems likely that BRD4 is not required to regulate MYC transcription in all cell types (Lockwood et al. 2012). Accordingly, Brd4 shRNAs did not score in a parallel screen using the screened HCC MP1 cell line (A Lujambio, C-H Huang, and SW Lowe, unpubl.), and the impact of BRD4 shRNAs and JQ1, a BRD4 inhibitor, on proliferation were not related to MYC expression in the human HCC lines studied here. Still, while CDK9 inhibition broadens the possibilities for “anti-MYC” therapy, its impact is not as great as MYC inhibition. Perhaps combinatorial therapies using the multikinase inhibitor sorafenib, a recently developed covalent CDK7 inhibitor that targets transcription regulation (Kwiatkowski et al. 2014), or those targeting other MYC activities will synergize with CDK9 inhibition in producing strong anti-tumor effects.
As one of the first oncogenes identified to actively contribute to human cancer (Dalla-Favera et al. 1982; Vennstrom et al. 1982), there has been substantial interest in understanding how MYC drives tumorigenesis and the consequences of MYC inhibition on tumor growth (Felsher 2010). Collectively, these studies indicate that MYC can influence a wide range of cellular processes and that tumor cells often become addicted to MYC activity such that MYC withdrawal or inhibition leads to marked tumor regression (Shachaf et al. 2004; Soucek et al. 2008). Interestingly, since MYC can mediate the oncogenic potential of other driving oncogenes (Zuber et al. 2011b; Soucek et al. 2013), MYC need not be a genetically altered driver to produce cancer-specific dependencies. By establishing an epistatic relationship between MYC and CDK9 in cancer maintenance, we demonstrate that CDK9 is a key effector of MYC tumor-promoting activities. While our results do not exclude the possibility that other MYC functions also play a role (Sabo et al. 2014; Walz et al. 2014), they imply that transcription elongation mediated by MYC is essential for oncogene addiction and provide a strategy for therapeutically targeting this addiction in cancer patients.
Materials and methods
Pooled negative selection RNAi screening
A custom shRNA library focused on 442 drug target genes was designed using miR30-adapted BIOPREDsi predictions (six shRNAs per gene) and constructed by PCR-cloning a pool of oligonucleotides synthesized on 55k customized arrays (Agilent Technologies) as previously described (Zuber et al. 2011a,c). The list of genes was obtained from DrugBank (version 2.5; http://www.drugbank.ca) and was manually curated, excluding ambiguous or redundant targets. After sequence verification, 2245 shRNAs (five to six per gene) were combined with several positive and neutral control shRNAs (n = 20) at equal concentrations in one pool.
The library was cloned into TRMPV-Neo and transduced into Tet-on murine HCC MP1 cells using conditions that predominantly lead to a single retroviral integration and represent each shRNA in a calculated number of at least 1000 cells. Transduced cells were selected for 5 d using 1 mg/mL G418 (Invitrogen); at each passage, >20 million cells were maintained to preserve library representation throughout the experiment. After drug selection, T0 samples were obtained (20 million cells per replicate) and sorted for Venus+ cells. After 12 d (six passages, T12), ∼20 million shRNA-expressing (dsRed+Venus+) cells were sorted for each replicate using a FACSAriaII (BD Biosciences). Genomic DNA from T0 and T12 samples was isolated by two rounds of phenol extraction using PhaseLock tubes (5 Prime) followed by isopropanol precipitation.
Animal studies
All mouse experiments were approved by the Memorial Sloan-Kettering Cancer Center (MSKCC) Animal Care and Use Committee (protocol no. 11-06-011). Mice were maintained under specific pathogen-free conditions, and food and water were provided ad libitum.
Plasmids
For conditional RNAi experiments, shRNAs were expressed from the TRMPV-Neo vector from either miR-30 or miR-E backbones, which have been described previously (and are available from Addgene, catalog no. 27990, or on request) (Zuber et al. 2011a; Fellmann et al. 2013). To produce Tet-on murine HCC MP1 cells, liver progenitor cells from p53Loxp/Loxp mice were transduced with CreER and Myc-IRES-rtTA3. For MYC rescue experiments, the wild-type human MYC cDNA was subcloned into MSCV-PGK-Puro-IRES-GFP (MSCV-PIG) (Hemann et al. 2003). For the in vivo experiments, cancer cells were infected with Luciferase-hygro. Human cell lines were infected with MSCV-RIEP (MSCV-rtTA3-IRES-EcoR-PGK-Puro) (Zuber et al. 2011b). Knockdown efficiency or overexpression was evaluated by immunoblotting. shRNA sequences are available in Supplemental Tables 1 and 8. The pT3 transposon and pT3-EF1a-Myc vectors were a kind gift of Dr. Xin Chen, University of California at San Francisco. To generate the constitutive expression vector pT3-EF1a (Tschaharganeh et al. 2014), the CpG-free EF1a promoter from pCpGfree-vitroBmcs (InvivoGen) was inserted into pT3, and a GFP-miR-E fragment was cloned following the EF1a promoter.
Immunoblotting
Liver tissues and cell pellets were lysed in Laemmli buffer or protein lysis buffer (200 mM NaCl, 0.2% NP40, 50 mM Tris at pH 7.5, 1% Tween20, protease and phosphatases inhibitors) using a tissue homogenizer. Equal amounts of protein were separated on 12% SDS–polyacrylamide gels and transferred to PVDF membranes. The abundance of β-actin was monitored to ensure equal loading. Images were analyzed using the AlphaView software (ProteinSimple). Detection in immunoblots was performed using antibodies for CDK9 (Santa Cruz Biotechnology), MCM6 (Santa Cruz Biotechnology), PSMB2 (Santa Cruz Biotechnology), β-Actin (AC-15, Sigma), p53 (Leica Biosystems), MYC (Abcam), CCNT1 (Santa Cruz Biotechnology), phopho-Ser2 RNA Pol II (Cell Signaling), and RNA Pol II (Santa Cruz Biotechnology). For Ki67 staining (VP-K451, Vector Laboratories), organ samples were fixed in fresh 4% paraformaldehyde overnight at 4°C and further subjected to routine histological procedures for embedding in paraffin. Images were taken on a Zeiss Axio Imager Z2 system.
Proliferation assays
Competitive proliferation assays using shRNAs in TRMPV-Neo vector (with miR-30 or miR-E backbone) were performed as described previously (Zuber et al. 2011c). Proliferation assays for PHA-767491 were performed in vitro by counting the viable cell numbers over 72 h in the presence of different PHA-767491 concentrations. Dead cells were excluded using propidium iodide (PI) staining to score cells with sub-2N DNA content. Measurements of cell concentration were performed on a Guava Easycyte (Millipore), gating only viable cells (FSC/SSC/PI−). Proliferation rates were calculated by dividing cell concentration at 72 h and cell concentration at 0 h, divided by 72. Relative proliferation rates were calculated by normalizing to the rate of vehicle-treated cells. Population doublings were calculated by calculating log2 (cells at end point/cells at initial point) divided by time in days.
shRNA experiments in human HCC cell lines
HepG2, Hep3B, SKHep1, SNU398, SNU475, and Alexander cells were modified to express the ecotropic receptor and rtTA3 by transducing MSCV-RIEP (MSCV-rtTA3-IRES-EcoR-PGK-Puro) followed by drug selection (1 μg/mL puromycin for 1 wk). The resulting cell lines were transduced with ecotropically packaged TRMPV-Neo-shRNA retroviruses with either miR-30 or miR-E backbone, selected with 1 mg/mL G418 for 1 wk, and treated with 1 μg/mL dox to induce shRNA expression. The relative change in Venus+dsRed+ (shRNA+) cells was monitored on a Guava Easycyte (Millipore) by performing proliferation competitive assays.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as previously described (Bracken et al. 2006). Briefly, cross-linking was performed with 1% formaldehyde, and cells were lysed in SDS buffer. DNA was fragmented by sonication (Bioruptor). ChIP for RNA Pol II was performed using a specific antibody (N-20, Santa Cruz Biotechnology). DNA enrichment was measured by quantitative PCR performed using SYBR Green (ABI) on a ViiA 7 (Life Technologies). Each reaction was done in triplicate using gene-specific primers. Each immunoprecipitate signal was referenced to an input standard curve dilution series (immunoprecipitate/input) to normalize for differences in starting cell number and for primer amplification efficiency. Pausing index, also known as traveling ratio, was calculated as the ratio between the RNA Pol II bound to the transcription start site and the RNA Pol II bound to the gene body. The list of ChIP primers is included in Supplemental Table 9.
Small animal PET imaging
For in vivo assays, human holo-Transferrin was labeled with 89Zr. The 89Zr was prepared as previously described (Holland et al. 2009). Female athymic nude mice were inoculated with 5 × 106 to 10 × 106 cells reconstituted in a 1:1 mixture of medium and Matrigel in the shoulder. After tumors reached ∼500 mm3, mice were treated with PHA-767491 for 3 d and, 2 d after, injected with 275–300 mCi of 89Zr-holoTransferrin via the tail vein. At various time points following injection (24–48 h), mice were scanned using a MicroPET Focus 120 Scanner (Concorde Microsystems). Approximately 5 min before recording PET images, mice were anesthetized by inhalation of 1%–2% isoflurane (Baxter Healthcare) in an oxygen gas mixture and placed on the scanner bed. Image reconstruction and processing details have been reported elsewhere (Holland et al. 2010). For biodistribution studies, mice (n = 5; 10 tumors) were euthanized by CO2 at 48 h post-injection, and organs were harvested immediately. The radioactivity in each organ was counted alongside a known amount of 89Zr, the counts were correlated to activity, and decay was corrected. The organs were then weighed, and the percentage of the injected dose per gram (%ID/g) in each tissue was calculated.
Hydrodynamic tail vein injection
A sterile 0.9% NaCl solution/plasmid mix was prepared containing 5 µg of DNA of pT3-EF1a-Myc and 20 µg of DNA of pT3-EF1a-GFP-miRe Transposon vector together with CMV-SB13 Transposase (1:5 ratio) for each injection. FVBN mice from JAX were injected with the 0.9% NaCl solution/plasmid mix into the lateral tail vein with a total volume corresponding to 10% of body weight in 5–7 sec.
Statistics
Statistical significance was calculated by two-tailed Student’s t-test. Correlation was calculated by Pearson test. Prism 5 software was used to calculate the IC50 values. Significance values are P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
Acknowledgments
We gratefully thank B. Ma, S. Muller, A. Shroff, J. Ahn, E. Manchado, D. Grace, J. Simon, M. Taylor, K.E. Carnazza, H. Zhao, C. Ruland, and C.-W. Chen for excellent technical assistance; O. Ouerfelli and G. Yang for PHA-767491 production and useful discussion; D. Quail and J.P. Morris IV for carefully editing the manuscript; J. Massagué, A. Ventura, and R. White for useful comments on the project; and J. Pelletier for helping to design the shRNA library. We also thank members of the Lowe laboratory for stimulating discussions. We thank MSKCC Animal Facilities, MSKCC Molecular Cytology Core, MSKCC Genomics Core, and MSKCC Organic Synthesis Core. C.-H.H. is funded by the Ministry of Education of Taiwan; A.L is funded by an EMBO Long-Term fellowship; J.Z. was supported by a research fellowship from the German Research Foundation (DFG) and by the Andrew Seligson Memorial Clinical Fellowship; T.K. and D.F.T. are funded by the German Research Foundation (DFG); and S.W.L., C.J.S., and C.L.S. are investigators of the Howard Hughes Medical Institute. This work was supported in part by grants CA008748 (E.d.S.), CA176671 (J.S.L. and M.J.E.), 5U01 CA168409-02 (S.W.L.), and CA013106 (S.W.L.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.244368.114.
References
- Ashworth A, Bernards R. 2010. Using functional genetics to understand breast cancer biology. Cold Spring Harb Perspect Biol 2: a003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. 2007. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci 104: 13690–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Wierda WG, Chubb S, Hawtin RE, Fox JA, Keating MJ, Gandhi V, Plunkett W. 2009. Mechanism of action of SNS-032, a novel cyclin-dependent kinase inhibitor, in chronic lymphocytic leukemia. Blood 113: 4637–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SJ, Lee SS, Kim YJ, Park BD, Choi JS, Liu L, Ham YM, Moon Kim B, Lee SK. 2010. Xylocydine, a novel Cdk inhibitor, is an effective inducer of apoptosis in hepatocellular carcinoma cells in vitro and in vivo. Cancer Lett 287: 196–206 [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci 97: 3260–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conacci-Sorrell M, McFerrin L, Eisenman RN. 2014. An overview of MYC and its interactome. Cold Spring Harb Perspect Med 4: a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. 1982. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci 79: 7824–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 2012. MYC on the path to cancer. Cell 149: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J Biol Chem 276: 48562–48571 [DOI] [PubMed] [Google Scholar]
- Farazi PA, DePinho RA. 2006. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 6: 674–687 [DOI] [PubMed] [Google Scholar]
- Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, et al. 2013. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 5: 1704–1713 [DOI] [PubMed] [Google Scholar]
- Felsher DW 2010. MYC inactivation elicits oncogene addiction through both tumor cell-intrinsic and host-dependent mechanisms. Genes Cancer 1: 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O’Dwyer PJ, Lee RJ, Grippo JF, Nolop K, et al. 2010. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. 2007. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med 13: 820–827 [DOI] [PubMed] [Google Scholar]
- Gojo I, Zhang B, Fenton RG. 2002. The cyclin-dependent kinase inhibitor flavopiridol induces apoptosis in multiple myeloma cells through transcriptional repression and down-regulation of Mcl-1. Clin Cancer Res 8: 3527–3538 [PubMed] [Google Scholar]
- Hahntow IN, Schneller F, Oelsner M, Weick K, Ringshausen I, Fend F, Peschel C, Decker T. 2004. Cyclin-dependent kinase inhibitor Roscovitine induces apoptosis in chronic lymphocytic leukemia cells. Leukemia 18: 747–755 [DOI] [PubMed] [Google Scholar]
- Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C, Hannon GJ, Lowe SW. 2003. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet 33: 396–400 [DOI] [PubMed] [Google Scholar]
- Holland JP, Sheh Y, Lewis JS. 2009. Standardized methods for the production of high specific-activity zirconium-89. Nucl Med Biol 36: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JP, Divilov V, Bander NH, Smith-Jones PM, Larson SM, Lewis JS. 2010. 89Zr–DFO–J591 for immunoPET of prostate-specific membrane antigen expression in vivo. J Nucl Med 51: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JP, Evans MJ, Rice SL, Wongvipat J, Sawyers CL, Lewis JS. 2012. Annotating MYC status with 89Zr-transferrin imaging. Nat Med 18: 1586–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, et al. 2009. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 69: 7385–7392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, McDonald D, Hope TJ, Hunter T. 1999. Mammalian Cdc7–Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J 18: 5703–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM. 2003. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene 22: 5707–5711 [DOI] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. 2012. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 335: 348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, Armenteros-Monterroso E, Lassailly F, Matthews N, Nye E, et al. 2012. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell 149: 642–655 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, Dastur A, Amzallag A, Ramaswamy S, Tesar B, et al. 2014. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature doi: 10.1038/nature13393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, Averett LM, Zhao H, Davis RE, Sathyamoorthy Met al. 2001. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol 2: research0041–research0041.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Puig P, Legoix P, Bluteau O, Belghiti J, Franco D, Binot F, Monges G, Thomas G, Bioulac-Sage P, Zucman-Rossi J. 2001. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology 120: 1763–1773 [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. 2013. Transcriptional regulation and its misregulation in disease. Cell 152: 1237–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Nasr Z, Premsrirut PK, Porco JA Jr, Hippo Y, Lowe SW, Pelletier J. 2012a. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Reports 1: 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. 2012b. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, et al. 2008. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 359: 378–390 [DOI] [PubMed] [Google Scholar]
- Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. 2012. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci 109: 19408–19413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. 2009. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, Tibolla M, Tenca P, Brotherton D, Albanese C, et al. 2008. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol 4: 357–365 [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Yu D, Zeller KI, Kim JW, Racke F, Thomas-Tikhonenko A, Dang CV. 2006. Activation of transferrin receptor 1 by c-Myc enhances cellular proliferation and tumorigenesis. Mol Cell Biol 26: 2373–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H, Yasui K, Zhao C, Arii S, Inazawa J. 2003. PTK2 and EIF3S3 genes may be amplification targets at 8q23-q24 and are associated with large hepatocellular carcinomas. Hepatology 38: 1242–1249 [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305 [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano G 2013. Deregulations in the cyclin-dependent kinase-9-related pathway in cancer: implications for drug discovery and development. ISRN Oncol 2013: 305371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, et al. 2014. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511: 488–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser I, Holzel M, Hoffmann R, Burtscher H, Kohlhuber F, Schuhmacher M, Chapman R, Weidle UH, Eick D. 2005. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene 24: 520–524 [DOI] [PubMed] [Google Scholar]
- Schramek D, Sendoel A, Segal JP, Beronja S, Heller E, Oristian D, Reva B, Fuchs E. 2014. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 343: 309–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al. 2001. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res 29: 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciot R, Verhoeven G, Van Eyken P, Cailleau J, Desmet VJ. 1990. Transferrin receptor expression in rat liver: immunohistochemical and biochemical analysis of the effect of age and iron storage. Hepatology 11: 416–427 [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. 2004. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 431: 1112–1117 [DOI] [PubMed] [Google Scholar]
- Shibata T, Aburatani H. 2014. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol 11: 340–349 [DOI] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. 2008. Modelling Myc inhibition as a cancer therapy. Nature 455: 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB, Evan GI. 2013. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev 27: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN, Frazier J, Chau BN, Loboda A, Linsley PS, et al. 2012. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci 109: 9545–9550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaharganeh DF, Xue W, Calvisi DF, Evert M, Michurina TV, Dow LE, Banito A, Katz SF, Kastenhuber ER, Weissmueller S, et al. 2014. p53 dependent Nestin regulation links tumor suppression to cellular plasticity in liver cancer. Cell doi: 10.1016/j.cell.2014.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennstrom B, Sheiness D, Zabielski J, Bishop JM. 1982. Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J Virol 3: 773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A, Hernandez-Gea V, Llovet JM. 2013. Medical therapies for hepatocellular carcinoma: a critical view of the evidence. Nat Rev Gastroenterol Hepatol 10: 34–42 [DOI] [PubMed] [Google Scholar]
- Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M, et al. 2014. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511: 483–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Fischer PM. 2008. Cyclin-dependent kinase 9: a key transcriptional regulator and potential drug target in oncology, virology and cardiology. Trends Pharmacol Sci 29: 302–313 [DOI] [PubMed] [Google Scholar]
- Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. 2000. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet 25: 35–41 [DOI] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol 4: R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Edwards JP, Mosser DM. 2009. The expression of exogenous genes in macrophages: obstacles and opportunities. Methods Mol Biol 531: 123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu Rev Biochem 81: 119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. 2011a. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol 29: 79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, Shi J, Weissmueller S, Fellmann C, Taylor MJ, et al. 2011b. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev 25: 1628–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011c. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]