The exon junction complex (EJC) plays a central role in controlling RNA fate and aids faithful splicing of pre-mRNAs containing large introns via an unknown mechanism. Brennecke and colleagues show that the core EJC plus the accessory factors RnpS1 and Acinus aid in the definition and efficient splicing of neighboring introns. Interestingly, the most highly affected intron belongs to the piwi locus, which explains the reported transposon desilencing in EJC-depleted Drosophila ovaries. Based on transcriptome-wide analysis, the authors propose that the dependency of splicing on the EJC is exploited to control the temporal order of splicing events.
Keywords: Piwi–piRNA pathway, SR proteins Acinus and RnpS1, exon junction complex, intron definition, splicing, transposon silencing
Abstract
Splicing of pre-mRNAs results in the deposition of the exon junction complex (EJC) upstream of exon–exon boundaries. The EJC plays crucial post-splicing roles in export, translation, localization, and nonsense-mediated decay of mRNAs. It also aids faithful splicing of pre-mRNAs containing large introns, albeit via an unknown mechanism. Here, we show that the core EJC plus the accessory factors RnpS1 and Acinus aid in definition and efficient splicing of neighboring introns. This requires prior deposition of the EJC in close proximity to either an upstream or downstream splicing event. If present in isolation, EJC-dependent introns are splicing-defective also in wild-type cells. Interestingly, the most affected intron belongs to the piwi locus, which explains the reported transposon desilencing in EJC-depleted Drosophila ovaries. Based on a transcriptome-wide analysis, we propose that the dependency of splicing on the EJC is exploited as a means to control the temporal order of splicing events.
Expression of protein-coding genes involves a series of interlinked and interdependent molecular events, including nuclear steps such as pre-mRNA transcription, intron removal via splicing, 5′ and 3′ modifications, and export, followed by cytoplasmic events such as mRNA translation and degradation. A central factor connecting nuclear pre-mRNA maturation to mRNA fate is the exon junction complex (EJC), a multisubunit protein complex that is deposited ∼24 nucleotides (nt) upstream of individual exon–exon boundaries after splicing (Le Hir et al. 2000a).
Assembly of the EJC on an RNA depends on tight RNA binding of a trimeric nuclear complex of the sequence-independent DEAD-box RNA clamp eIF4AIII and a heterodimer of Mago and Y14/Tsunagi (Tsu) (Shibuya et al. 2004; Gehring et al. 2009). Upon nuclear mRNA export to the cytoplasm, these three factors recruit a fourth EJC core component, Barentsz (Btz)/MLN51 (Degot et al. 2004).
The EJC is a binding platform for several peripheral factors, which provide mechanistic links to various processes in mRNA fate control (Tange et al. 2004; Bono and Gehring 2011). Among these are the nuclear mRNA export factors UAP56, REF/Aly, and TAP/NFX1:p15 (Kataoka et al. 2000; Luo et al. 2001); the nonsense-mediated mRNA decay factor Upf3 (Gehring et al. 2003); and the ASAP complex (Acinus [Acn], RnpS1, and SAP18), whose subunits have been individually implicated in mRNA quality control, pre-mRNA splicing, and transcriptional regulation (Zhang et al. 1997; Mayeda et al. 1999; Schwerk et al. 2003; Sakashita et al. 2004).
Most molecular processes that depend on the EJC are post-splicing events. However, the EJC core also interacts with proteins implicated in splicing (e.g., the splicing coactivators/alternative splicing factors SRm160 and Pinin) (Le Hir et al. 2000b; Li et al. 2003), suggesting that the EJC plays a direct role in splicing itself. Indeed, the nuclear EJC core factors eIF4AIII, Mago, and Tsu, together with RnpS1, are required for the faithful splicing of several Drosophila pre-mRNAs harboring large introns, often encoded at heterochromatic gene loci (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010). Loss of the EJC results in frequent exon skipping and reduced mRNA levels, the most prominent example being the ERK-encoding rolled transcript.
It is unclear how the EJC facilitates splicing mechanistically—in particular, whether the EJC aids splicing of neighboring introns after it has been deposited at flanking spliced junctions or whether individual EJC subunits directly facilitate splicing independently of their assembly into an EJC.
Here, we demonstrate that the nuclear EJC core factors Mago, Tsu, and eIF4AIII and the peripheral EJC components RnpS1 and Acn are essential for the accurate transcript splicing of piwi, which encodes an essential component of the ovarian defense pathway for transposable element (TE) silencing in Drosophila. We show that the nuclear EJC is required for splicing of intron4 of piwi and that expression of piwi transcripts lacking this intron is EJC-independent. In the absence of the EJC recruited to a flanking exon–exon junction, intron4 is inefficiently spliced, even in wild-type cells. A transcriptome-wide analysis reveals that several other mRNAs also exhibit an intron retention phenotype upon EJC depletion, indicating that the EJC facilitates the removal of adjacent/nearby introns with suboptimal splicing characteristics.
Results
The nuclear EJC is required for transposon silencing in ovaries
TE silencing in Drosophila ovarian tissues in both the germline and associated soma is mediated by the piRNA pathway, a small RNA silencing system. Three Argonaute proteins of the PIWI clade (nuclear Piwi and cytoplasmic Aubergine and AGO3) are at its core. Each is bound to a population of 22- to 30-nt single-stranded piRNAs that act as sequence-specific guides to identify complementary targets for their transcriptional and post-transcriptional silencing (Senti and Brennecke 2010; Siomi et al. 2011).
Recent reverse genetic screens have begun to identify the genetic framework of the Drosophila piRNA pathway (Czech et al. 2013; Handler et al. 2013; Muerdter et al. 2013). Besides factors involved in piRNA biogenesis or piRNA-mediated silencing, several unexpected players were identified. Among these are components of the EJC, whose depletion results in desilencing of piRNA pathway repressed TEs.
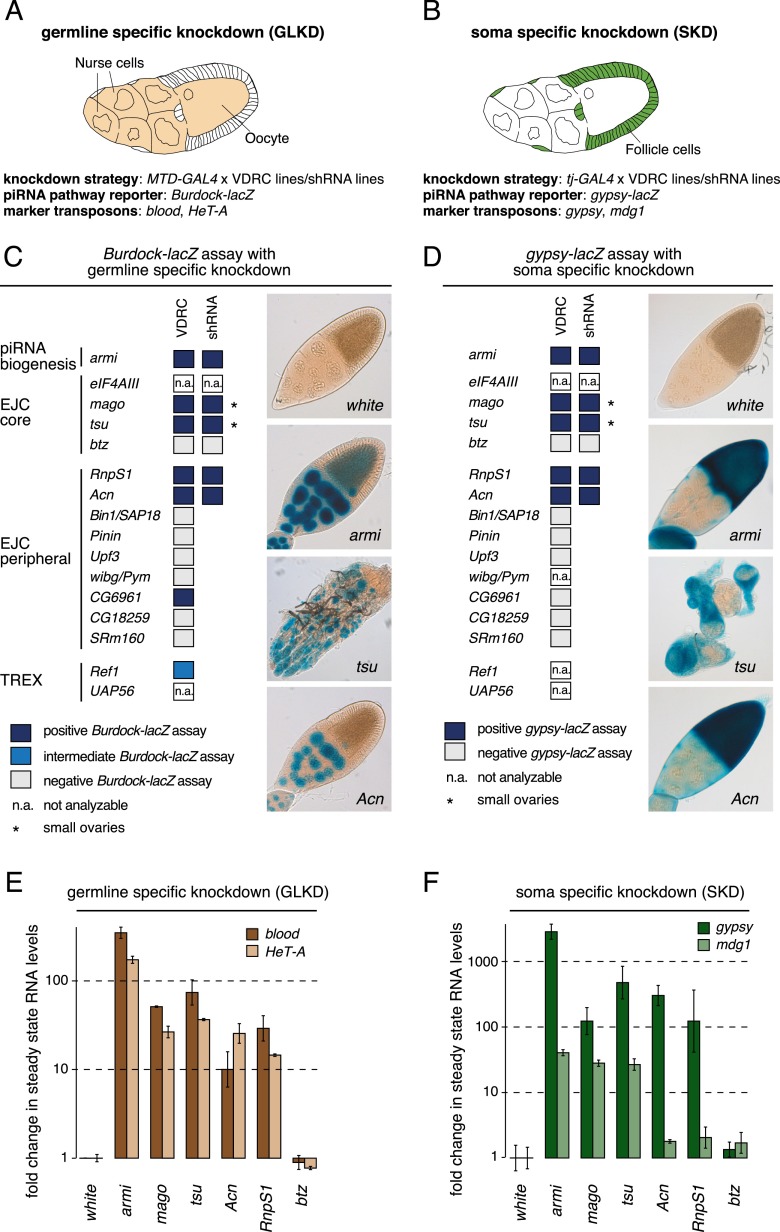
We retested 15 annotated core and peripheral EJC factors for a role in ovarian TE silencing. Using tissue-specific RNAi (Dietzl et al. 2007; Ni et al. 2011), we depleted individual factors in either germline tissue (which comprises accessory nurse cells interconnected with each other and with the oocyte) or the overlying somatic follicular epithelium (Fig. 1A,B; Materials and Methods). piRNA pathway integrity was monitored with lacZ reporter transgenes that are repressed by the pathway in wild-type cells (Fig. 1C,D) and via quantitative RT–PCR (qRT–PCR) to measure steady-state RNA levels of marker TEs (Fig. 1E,F; Materials and Methods). For both assays, depletion of the essential piRNA biogenesis factor Armitage (Armi) served as positive control.
Figure 1.
The EJC is required for transposon silencing in ovarian soma and the germline. (A,B) Cartoons of Drosophila egg chambers to illustrate the tissue-specific gene depletions in germline (A) or somatic (B) cells. The affected cell types (colored), knockdown strategies, TE reporters, and marker TEs expressed in the respective tissues are indicated. (C,D) The results of the TE reporter assays upon germline (C) or soma (D) knockdown of EJC factors. Defective TE silencing is reflected by lacZ expression (blue). The depletion of EJC core factors mago and tsu, but not btz, as well as the depletion of peripheral factors RnpS1 or Acn consistently leads to lacZ expression in both soma and the germline. Stained egg chambers of representative phenotypes are shown: white for no staining, tsu for strong staining but abnormal morphology, and Acn for strong staining and no morphological defects. Knockdown of piRNA biogenesis factor armi served as control. (E,F) Fold changes in steady-state RNA levels of endogenous TEs in ovaries upon depletion of the indicated genes in the germline (E) or soma (F). RNA levels are normalized to rp49 levels, and values indicate averages of three biological replicates relative to control knockdowns; error bars indicate standard deviation. The depletion of EJC core factors (mago and tsu) and peripheral factors (RnpS1 and Acn) causes strong TE derepression almost comparable with armi depletions.
Only the EJC core factors Mago and Tsu and the peripheral factors RnpS1 and Acn give strong, consistent derepression of the reporters and endogenous TEs in both the ovarian germline (blood and HeT-A) (Fig. 1C,E) and somatic cells (gypsy and mdg1) (Fig. 1D,F). Depletion of the core EJC factor eIF4AIII prevents ovarian development entirely and could not be evaluated. Importantly, depletion of the cytoplasmic core EJC factor Btz does not result in any observable TE derepression.
Similar results were obtained in cultured ovarian somatic stem cells (OSCs), which are also subject to piRNA-mediated silencing of TEs (Niki et al. 2006; Saito et al. 2009). RNAi-mediated depletion of Mago, Tsu, RnpS1, and Acn, but not of Btz, causes derepression of gypsy (Supplemental Fig. S1). Together, our results suggest an involvement of the nuclear EJC plus the factors RnpS1 and Acn in the ovarian piRNA pathway.
The EJC is required for a post-transcriptional step in piwi expression
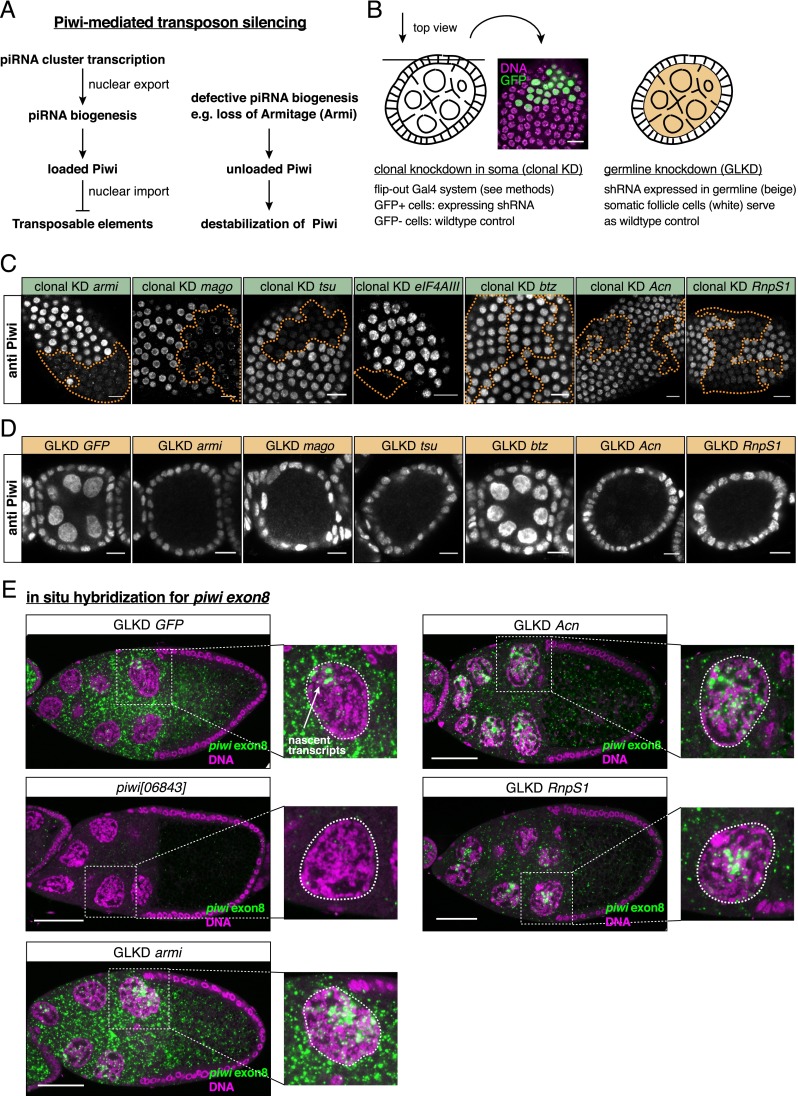
To place the EJC into the hierarchy of the piRNA pathway (illustrated in Fig. 2A), we took advantage of the observation that defects in piRNA biogenesis (e.g., loss of Armi) cause severe reductions in Piwi protein levels, presumably because unloaded Piwi is unstable (Fig. 2C; Olivieri et al. 2010; Saito et al. 2010). Interestingly, depletion of Mago, Tsu, RnpS1, or Acn, but not Btz, also causes a strong reduction of Piwi levels in soma and the germline (Fig. 2C,D [see also B for a summary of knockdown strategies]). The clonal knockdown approach in somatic cells allowed us to show that the other nuclear EJC core factor, eIF4AIII, is also required for Piwi accumulation (Fig. 2C).
Figure 2.
The EJC is required for piwi mRNA and protein expression. (A) Illustration of Piwi-mediated transposon silencing. piRNA precursor transcripts are expressed from heterochromatic piRNA clusters (e.g., flamenco) and exported to cytoplasmic piRNA biogenesis sites. Upon loading with a piRNA, Piwi enters the nucleus, where it guides transcriptional silencing of TEs. Defective piRNA biogenesis (e.g., caused by loss of Armi) prevents Piwi loading, which renders it unstable. (B) Illustration of applied RNAi knockdown strategies. Indicated factors were depleted in either clones of somatic cells (left) or all germline cells (right). Confocal images in C and D represent surface sections and cross-sections through stage 3–6 egg chambers, respectively. (C,D) Confocal sections of egg chambers with clones of somatic follicle cells (C) or germline cells (D) expressing shRNAs against EJC factors stained for Piwi. Bars, 10 μm. Piwi levels are greatly reduced in cells depleted for EJC factors except Btz. shRNAs against GFP and armi served as controls. (E) Confocal sections through stage 9–10 egg chambers expressing shRNAs against the indicated genes in the germline stained for piwi exon8 (green) and DNA (magenta). Bars, 50 μm. Egg chambers from piwi[06843] germline clones served as negative control. Cytoplasmic piwi staining is lost upon Acn or RnpS1 knockdown but not upon armi knockdown. Nuclear staining (individual nurse cell nuclei enlarged) of piwi reflects nascent transcripts and thus active transcription.
Because the EJC is known to control RNA fate, we hypothesized that loss of the EJC impairs expression or specification of piRNA precursor transcripts, which are transcribed from discrete, often heterochromatic loci termed piRNA clusters (Senti and Brennecke 2010). However, depletion of the EJC in OSCs results in only moderate and insignificant changes in steady-state RNA levels of flamenco, the major somatic piRNA cluster (Brennecke et al. 2007). Also, integrity of the Yb body, the cytoplasmic piRNA processing center in somatic ovarian cells (Szakmary et al. 2009; Olivieri et al. 2010; Saito et al. 2010), does not depend on the EJC (Supplemental Fig. S2).
We therefore analyzed piwi expression itself using fluorescent in situ hybridization (FISH). In wild-type egg chambers, an exon8 antisense probe detected abundant piwi transcript in the cytoplasm of nurse cells (Fig. 2E). In addition, signal within nurse cell nuclei was observed in strong foci that presumably reflect nascent transcripts at the sites of transcription. Both signals are absent in piwi[06843] mutant egg chambers (germline clones), where a P element inserted into the first intron perturbs piwi transcription. Consistent with the loss of Armi resulting in post-translational destabilization of Piwi protein (Olivieri et al. 2010; Saito et al. 2010), piwi transcript patterns in the cytoplasm and nucleus are normal in Armi-depleted egg chambers (Fig. 2E). In contrast, depletion of Acn or RnpS1 results in the severe reduction of cytoplasmic piwi transcript levels, while nuclear foci are unaffected, indicating that primary piwi transcription is normal (cf. the signals in nurse cell nuclei in Fig. 2E). Thus, depleting EJC activity impairs the processing, export, or stability of piwi transcripts.
EJC factors are required for piwi intron4 splicing
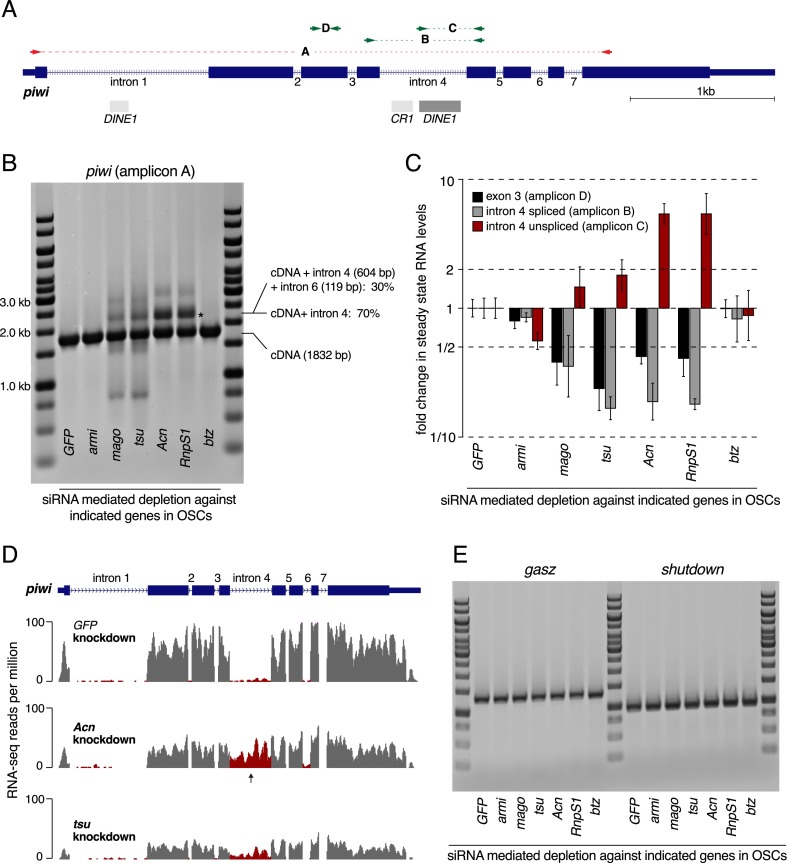
Because the nuclear EJC and RnpS1 are required for faithful splicing of numerous mRNAs (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010), we analyzed piwi splicing patterns by RT–PCR using total RNA from OSCs depleted for individual EJC factors by RNAi. A primer pair mapping to the first and the last piwi exon exclusively amplifies the expected spliced wild-type piwi cDNA from control and Armi-depleted cells (Fig. 3A,B, amplicon A).
Figure 3.
EJC factors are required for piwi int4 splicing. (A) Cartoon of the genomic piwi locus showing exons as blue boxes, introns as lines, TE fragments as gray boxes, and the locations of RT–PCR primers used in B–D. (B) Agarose/EtBr gel showing piwi RT–PCR products (primers in exon1 and exon8; amplicon A in A; size markers at the borders) amplified from total RNA isolated from OSCs depleted for the indicated genes. piwi cDNAs containing int4 or both int4 and int6 (indicated by an asterisk) are amplified along with wild-type piwi cDNA in mago, tsu, Acn, or RnpS1 knockdowns. (C) Shown are the fold changes in steady-state RNA levels of all piwi transcripts (exon3; black bars) and those piwi transcripts with spliced (gray bars) or unspliced (red bars) int4 amplified from total RNA isolated from OSCs depleted for the indicated genes. Values are normalized to rp49 levels, and averages of three biological replicates relative to control knockdowns are shown, with error bars indicating standard deviation. The levels of piwi transcripts with spliced and unspliced int4 are decreased and increased by twofold to fourfold in mago, tsu, Acn, or RnpS1 knockdowns, respectively. (D) RNA-seq profiles at the piwi locus obtained from polyA-selected RNA from OSCs depleted for the indicated genes (exonic reads in gray; intronic reads in red; arrows mark int4), showing the retention of int4 in Acn and tsu knockdowns. (E) Agarose/EtBr gel showing gasz and shutdown RT–PCR products (primers in the respective first and last exons) amplified from total RNA isolated from OSCs depleted for the indicated genes. Unlike piwi, the splicing of these transcripts is not affected by the depletion of EJC factors.
Depletion of Mago, Tsu, RnpS1, or Acn, but not Btz, results in additional, longer products, suggestive of intron inclusion, and shorter products that are indicative of exon skipping (Mago and Tsu only) (Fig. 3B). We focused on the major longer fragment, considering that it is observed in all four knockdown samples. Sequencing showed that it comprises the expected piwi cDNA plus the entire intron4 (n = 10 of 10); three clones contained intron6 in addition.
Intron retention in cells with reduced EJC activity was unexpected because loss of the EJC has previously been shown to be associated with skipping of exons, in particular those adjacent to very long introns (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010). We therefore confirmed the retention of piwi intron4 (int4) by qPCR. Compared with control cells, we observed an approximately fourfold decrease of the spliced species (amplicon B) and an approximately fourfold increase of the unspliced species (amplicon C) in Acn- or RnpS1-depleted cells (Fig. 3C). The accumulation of the unspliced species is evident but less pronounced in Mago- or Tsu-depleted cells, perhaps because depletion of these core factors has a broader impact on piwi splicing patterns or causes a general reduction in cell viability. We observed a similar degree of int4 retention also in ovaries depleted for EJC factors specifically in germline cells (Supplemental Fig. S3).
A highly similar intron retention pattern was found by sequencing polyA-selected RNAs: int4 is substantially retained, and int6 is slightly overrepresented in Acn- or Tsu-depleted OSCs (Fig. 3D). These data also confirm a general reduction in piwi steady-state RNA levels, especially in Tsu-depleted cells, presumably a consequence of nonsense-mediated decay (NMD) because inclusion of int4 adds a premature stop codon.
Intron retention is not a general phenotype in EJC-depleted cells; splicing of several other transcripts (e.g., of the piRNA pathway factors gasz or shutdown) is normal in the various OSC knockdowns tested (Fig. 3E). Thus, the reduced Piwi activity is probably due to a selective failure of piwi RNA splicing.
piRNA pathway defects in EJC-depleted ovaries are largely due to retention of piwi intron4
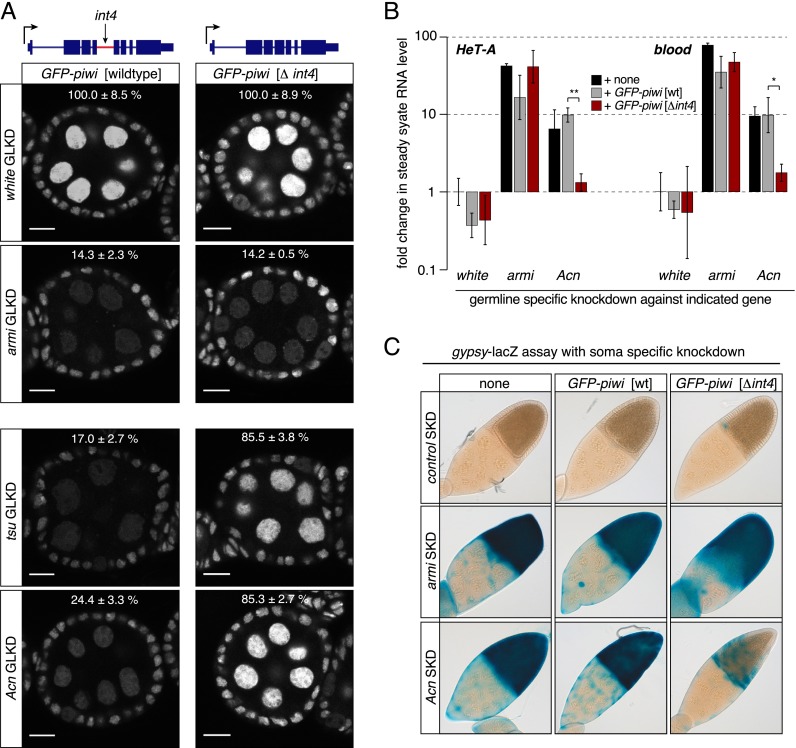
To check whether int4 retention is indeed the major block to Piwi expression in EJC-depleted cells, we tested whether removing the intron renders piwi EJC-independent. We generated transgenic flies that express an N-terminal GFP-Piwi fusion in the context of a fully functional genomic construct containing the entire piwi locus (GFP-piwi) (Sienski et al. 2012) and flies expressing an equivalent gene that lacks int4 (GFP-piwi[Δint4]).
As expected, GFP-Piwi levels from the former construct are sensitive to the depletion of EJC factors in ovarian somatic and germline cells (Fig. 4A; Supplemental Fig. S4A). In contrast, GFP-Piwi from GFP-piwi[Δint4] is insensitive to depletion of Tsu or Acn (Figs. 4A; Supplemental Fig. S4A) while remaining sensitive to reduced piRNA synthesis by Armi depletion. Retention of int4 is therefore causal to the reduced Piwi levels in EJC-depleted cells.
Figure 4.
A piwi transgene lacking int4 rescues transposon derepression in EJC-depleted cells. (A) Expression of wild-type GFP-piwi, but not of GFP-piwi Δint4, is dependent on EJC factors. Shown are confocal sections through egg chambers expressing GFP-piwi or GFP-piwi Δint4 depleted for the indicated genes in germline cells. Bars, 10 μm. Relative GFP levels (±standard deviation) from the indicated GFP-piwi transgenes in comparison with the white control knockdown are indicated (GFP fluorescence levels in nurse cell nuclei normalized to those in follicle cell nuclei from three egg chambers). (B) Displayed are fold changes in steady-state RNA levels of HeT-A and blood in ovaries depleted for the indicated genes in the germline and expressing the indicated GFP-piwi transgenes. RNA levels are normalized to rp49 levels, and averages of three biological replicates relative to control knockdowns are shown. Error bars indicate standard deviation. (*) P < 0.01; (**) P < 0.001. TE derepression caused by Acn knockdown is rescued by GFP-piwi Δint4 but not by wild-type GFP-piwi. (C) Shown are the X-gal stainings of egg chambers depleted for the indicated genes in somatic follicle cells and expressing the gypsy-lacZ reporter and the indicated GFP-piwi transgenes. GFP-piwi lacking intron4, but not wild-type GFP-piwi, greatly reduces the gypsy-lacZ expression induced by the depletion of Acn.
We further tested whether the EJC-independent GFP-piwi[Δint4] transgene rescues the TE derepression phenotype in EJC-depleted flies. We depleted Acn specifically in ovarian germline or somatic cells of flies carrying either the wild-type GFP-piwi or the GFP-piwi[Δint4] construct. TE derepression in the germline caused by the depletion of Acn, but not of Armi, is greatly rescued by the GFP-piwi[Δint4] transgene and not by the wild-type GFP-piwi transgene (Fig. 4B). The transgene also rescues TE derepression in the soma: In Acn-depleted somatic cells, expression of the gypsy-lacZ reporter is largely repressed in the presence of GFP-piwi[Δint4] (Fig. 4C).
The GFP-piwi[Δint4] construct fails to rescue germline TE derepression due to Tsu depletion (data not shown), perhaps because these flies also show reduced germline expression of AGO3 (Supplemental Fig. S4B), a second central Argonaute protein involved in the piRNA pathway. AGO3 pre-mRNA has very large introns and thus resembles transcripts known to require the EJC for efficient splicing. Indeed, AGO3 mRNA level is reduced upon Tsu but not Acn depletion. We did not detect misspliced AGO3 mRNA species, possibly due to efficient NMD (Supplemental Fig. S4C,D). We note that the GFP-piwi[Δint4] construct does reduce desilencing of mdg1 in Tsu-depleted follicle cells in which AGO3 is not expressed (Supplemental Fig. S4E). Taken together, our results indicate that missplicing of piwi int4 is the major cause of TE derepression in ovaries depleted for EJC components.
Splicing of piwi intron4 requires deposition of the EJC at a nearby splice junction
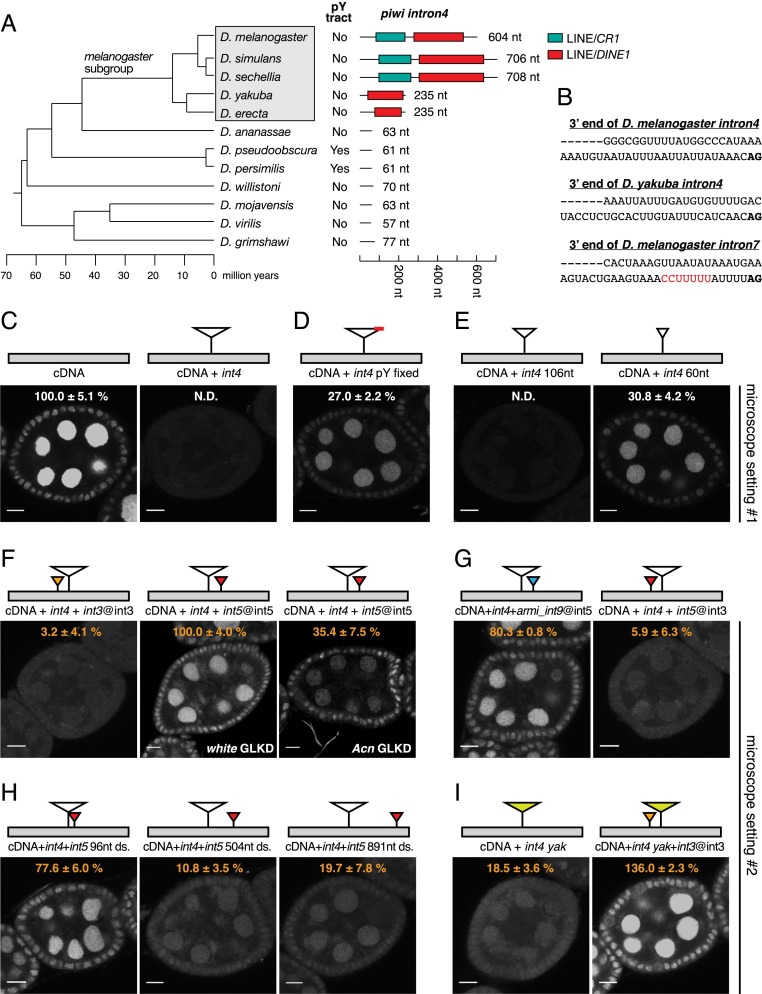
To understand why splicing of piwi int4 is sensitive to EJC levels, we compared the int4 sequence in Drosophila melanogaster with that in other Drosophilids (Fig. 5A). In the melanogaster subgroup, int4 harbors one or two TE remnants that extend the intron to ∼200–700 nt, whereas the intron is devoid of TE fragments and very short (∼60 nt) in the more distantly related species. Also, the 3′ end of piwi int4 lacks a robust polypyrimidine tract (pY; defined as at least seven consecutive pyrimidines) in nearly all of these species. Assembly of spliceosome components at 3′ splice junctions involves recognition of the pY tract by the U2 snRNP factor U2AF65 (Zamore et al. 1992). Introns with a poor pY tract can be spliced in vitro, but only if they are <90 nt (Guo et al. 1993).
Figure 5.
The EJC facilitates piwi int4 splicing from a nearby splice junction. (A) Architecture of piwi int4 from 12 Drosophila species (phylogenetic tree is at the left). Indicated are TE fragments (colored), intron length, and the presence of a pY tract. piwi int4 is enlarged in the melanogaster subgroup (boxed) due to insertions of TE fragments. (B) Depicted are the last 50 nt of the indicated introns, with pY tract in red. piwi int4 from D. melanogaster and D. yakuba have suboptimal pY tracts. (C–I) piwi int4 is not efficiently spliced when it is placed in isolation. The features of int4 that contribute to the inefficient splicing and the effects of additional flanking introns are examined. Shown are confocal sections through egg chambers expressing the indicated GFP-piwi transgenes (depicted as cartoons above each image). Bars, 10 μm. In F, shRNAs against white or Acn are expressed in germline cells (GLKD). The transgene cartoons represent the piwi cDNA (gray), melanogaster int4 (white large triangles), yakuba int4 (green large triangles), and additional indicated introns (small colored triangles). The indicated GFP levels (±standard deviation) were calculated by measuring GFP fluorescence in nurse cell nuclei of three egg chambers. Microscope settings were identical in C–E and F–I, respectively. GFP intensity was set to 100% for the left panel in C and the middle panel in F. For a global comparison, the intensity of GFP-piwi [cDNA + int4 + int5@int5] is 13.0 ± 2.0% of the intensity of GFP-piwi [cDNA].
This analysis raised the possibility that int4 is a suboptimal intron in the melanogaster subgroup. To test this idea, we generated transgenic flies harboring the aforementioned genomic GFP-piwi construct but lacking either all introns or all except int4. The intron-less construct drives readily detectable GFP-Piwi expression in ovarian somatic and germline cells (Fig. 5C). However, addition of int4 abolishes expression of GFP-Piwi almost completely (Fig. 5C), indicating that even wild-type cells cannot splice piwi int4 if it is present in isolation.
Replacing the 3′-most 50 nt of int4 with the equivalent part of piwi int7 that contains a clear pY tract (the pY-fixed construct) (Fig. 5B) largely rescues int4 splicing, as does shortening of int4 to 60 nt (Fig. 5D,E). A length reduction to 106 nt does not restore splicing, consistent with the aforementioned length cutoff of ∼90 nt for introns with poor pY tracts (Guo et al. 1993). These results suggest that piwi int4 is a very poorly defined intron due to the lack of an optimal pY tract coupled with its large size.
Because piwi int4 is efficiently spliced in the context of the wild-type locus but poorly spliced when isolated, perhaps prior splicing of flanking introns is required for int4 definition. We tested this idea directly by adding either int3 or int5 at their natural positions to the transgene that contains only int4. Addition of int3 has no measurable impact, but the presence of int5 results in significant GFP-Piwi expression to about half the level observed in the pY-fixed construct (Fig. 5F). Most importantly and as shown for the wild-type construct, GFP-Piwi expression from the int4/5 construct is EJC-dependent, being approximately threefold reduced in germline cells upon germline-specific depletion of Acn (Fig. 5F).
These results strongly favor a model in which splicing of int5 with subsequent EJC deposition facilitates int4 splicing. To understand why addition of int5 but not of int3 rescues int4 splicing, we analyzed the impact of intron identity, absolute intron distance to int4, and relative intron positioning (upstream versus downstream) on GFP-Piwi expression. Intron identity plays no role because an intron derived from the armi locus rescues expression as efficiently as piwi int5, while piwi int5 placed at the int3 position lacks rescuing activity (Fig. 5G). Increasing the distance between int4 and int5 from 200 nt (wild type) to 500 or 900 nt impairs rescue, while shortening the distance has no impact (Fig. 5H), suggesting that proximity to the splice junction is important. To test whether the upstream introns fail to rescue because they are too distant from the int4 3′end, we examined whether an upstream intron can rescue the splicing of a shorter int4 from Drosophila yakuba (235 nt), which, when isolated, is also splice-defective in wild-type cells (Fig. 5I). This shorter int4 is indeed rescued by the upstream presence of the melanogaster int3 (Fig. 5, cf. I and F), indicating that both upstream and downstream introns can rescue int4 splicing if placed nearby, presumably due to prior deposition of the EJC to a flanking exon–exon junction.
Splicing of first introns tends to depend on the EJC if followed by a large second intron
To investigate whether piwi int4 is indicative of a more general role of the EJC in efficient intron removal, we sequenced polyA-selected RNAs from OSCs depleted for Tsu, Acn, or GFP (Materials and Methods). We considered ∼1500 introns for which a significant quantitative analysis was possible (Materials and Methods).
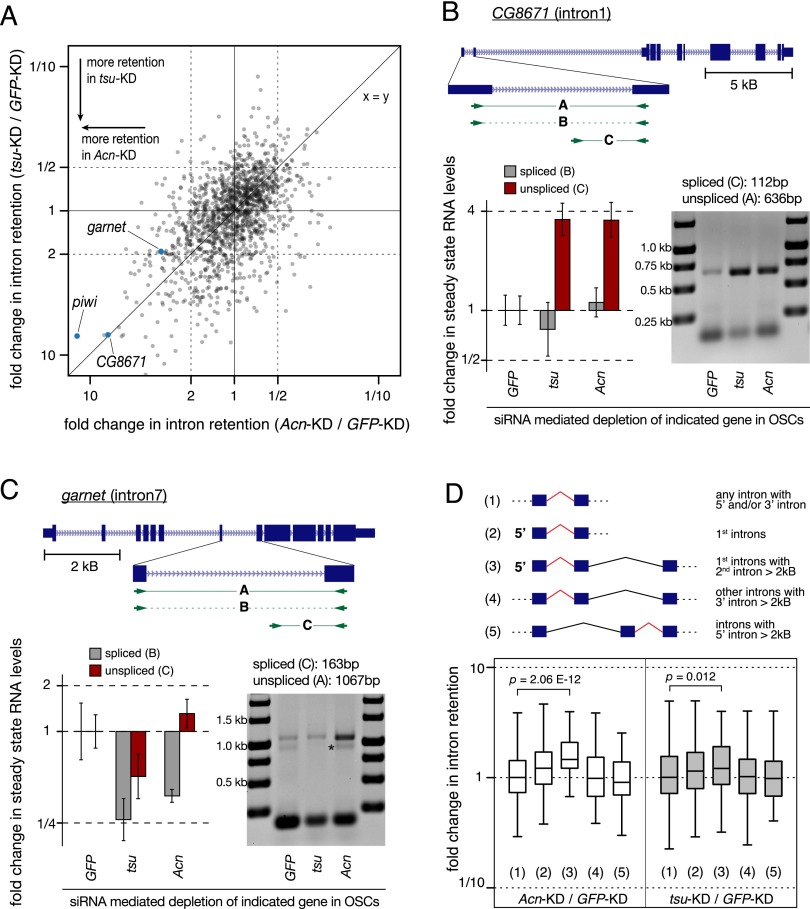
An analysis of intronic RNA sequencing (RNA-seq) reads indicates that splicing of most introns is EJC-independent (median changes in intron retention index upon tsu and Acn knockdown are both 1.00). Nevertheless, 286 introns exhibit pronounced retention characteristics (twofold to 10-fold higher than in control cells) in cells depleted for either Tsu or Acn (Fig. 6A). We verified the RNA-seq results for several EJC-dependent introns by performing RT experiments or by qPCR using amplicons that discriminate spliced and nonspliced transcripts (shown for CG8671 and garnet in Fig. 6B,C). Of note, piwi int4 is among the most affected intron transcriptome-wide.
Figure 6.
Transcriptome-wide analysis of EJC-dependent introns. (A) Scatter plot showing the fold changes in the retention of individual introns of OSC transcripts upon Acn or tsu knockdown relative to the control knockdown. Validated hits are indicated (blue dots). piwi int4 is among the most prominent introns that are retained both in Acn and tsu knockdowns. (B,C) Experimental validations of enhanced retention of CG8671 int1 and garnet int7 in tsu- or Acn-depleted OSCs using polyA-selected RNAs. Shown at the top is a cartoon of the respective locus (exons as boxes and introns as lines) with the location of the primers in RT–PCR. The bar charts indicate fold changes in steady-state levels of the indicated CG8671 (B) or garnet (C) transcripts. (Gray) Spliced; (red) unspliced. Amplicon identity is given above. The Agarose/EtBr gel images show RT–PCR products obtained with primers mapping to the flanking exons. The asterisk in C marks a nonspecific product. (D, top) The cartoon depicts the analyzed groups of introns, considering their location in the transcript and the identity of flanking introns. The box plot shows fold changes in intron retention for the groups defined above upon Acn or tsu depletion. First introns followed by a large second intron (group 3) are significantly more retained in EJC-depleted cells (P-values are calculated by the Wilcoxon rank-sum test).
As a poor pY tract is an apparent characteristic of piwi int4, we inspected whether EJC-dependent introns generally exhibit a weak pY tract. However, the group of introns harboring a poor pY tract (n = 272) (Materials and Methods) is not significantly more dependent on the EJC than the average intron (median changes in intron retention index upon tsu or Acn knockdown are 1.03 and 1.02, respectively; P-values > 0.5). Also, the suspicious TE insertions in piwi int4 do not seem to suggest a general pattern: TE remnants are only rarely found in tsu- or Acn-dependent introns (only four out of the 30 most affected introns harbor TE remnants).
Loss of the nuclear EJC has been shown to result in exon skipping for transcripts containing large introns (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010). This appears not to be the case for intron retention because the intron retention index is not significantly increased for either introns that are themselves >2 kb (n = 162; median changes in intron retention index upon tsu or Acn knockdown are 0.93 and 0.92, respectively) or introns within transcripts containing at least one intron >10 kb (n = 353; median changes in intron retention index upon tsu or Acn knockdown are 1.00 and 0.94, respectively).
Manual inspection of many gene loci containing an EJC-dependent intron indicated that affected introns are often first introns and that they are frequently followed by a large second intron (e.g., CG8671 in Fig. 6B; see also Supplemental Table S1). The population of first introns followed by a large second intron (>2 kb; n = 77) is significantly more likely to be inefficiently spliced in tsu-depleted (median = 1.21; P = 0.012) or Acn-depleted (median = 1.46; P = 2.06 × 10−12) cells compared with control cells (Fig. 6D). In comparison, the group of all first introns (n = 320) or internal introns immediately followed (n = 64) or preceded (n = 99) by a large intron shows no significantly different behavior in EJC-depleted cells compared with control cells (Fig. 6D). Together, our results demonstrate an unanticipated dependency of first introns that are followed by a large second intron on the EJC for their splicing.
Exon skipping and intron retention are genetically separable events
Both exon skipping and intron retention are defects in EJC-depleted cells, arguing that the two processes are mechanistically related. However, we noticed one clear difference between the two phenotypes: Loss of Acn strongly impacts intron retention yet was not previously reported to have an impact on exon skipping (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010).
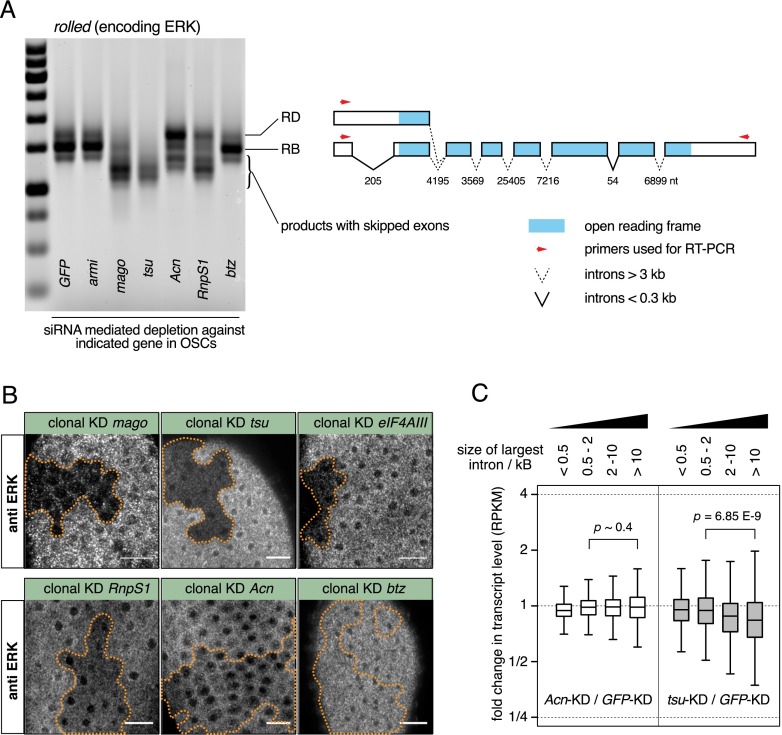
We therefore analyzed splicing patterns of the large intron-containing rolled and light transcripts in OSCs depleted for EJC core factors or RnpS1 or Acn. Consistent with previous reports, depletion of Mago or Tsu leads to severe exon skipping (Fig. 7A; Supplemental Fig. S5; Ashton-Beaucage et al. 2010; Roignant and Treisman 2010). The same is true, although to a lesser extent, for depletion of RnpS1 but not for Acn, whose depletion results in very mild exon skipping (Fig. 7A; Supplemental Fig. S5). Similarly, protein levels of the Rolled MAP kinase ERK are strongly reduced in vivo in cell clones depleted for EJC core factors (Mago, Tsu, and eIF4AIII), only mildly reduced upon RnpS1 depletion, and almost wild type in Acn-depleted cells (Fig. 7B). In contrast, loss of any involved EJC factor affects Piwi protein levels equally under comparable experimental conditions (Fig. 2C,D).
Figure 7.
Depletion of core EJC factors but not Acn results in exon skipping. (A) Agarose/EtBr gel image showing RT–PCR products of rolled transcripts amplified from total RNA from OSCs depleted for the indicated genes. Illustrated on the right is the rolled locus showing exons as boxes, with the ORF depicted in blue, introns as solid (<0.3 kb) and dashed (>3 kb) lines, and arrows as primers used in RT–PCR (modified from Ashton-Beaucage et al. 2010, © 2010, with permission from Elsevier). Each intron is labeled with their size (in nucleotides). rolled-RB is the major product in wild-type cells, and rolled-RD is an annotated rolled transcript with intron1 retained that still contains the wild-type ORF. Knockdown of mago, tsu, and, to a lesser degree, RnpS1 causes exon skipping of rolled, whereas the splicing is largely unaffected by the knockdown of Acn. (B) Confocal sections through the follicular epithelium of stage 3–6 egg chambers stained for ERK (encoded by rolled), showing that the depletion of mago, tsu, or eIF4AIII leads to a severe reduction in ERK protein level, which is moderately or only very mildly affected by the depletion of RnpS1 or Acn, respectively. Bars, 10 μm. Cells within the clone (boundaries marked by dashed line) express shRNAs against the indicated genes. (C) Box plot showing fold changes in the steady-state RNA level of transcript groups containing increasing intron length upon Acn (left) or tsu (right) depletion in OSCs. Grouping of transcripts was according to the largest intron (x in kilobases): from left, 0 < x = <0.5; 0.5 < x = 2; 2 < x = < 10; 10.0 < x. The level of transcripts with large introns is decreased upon depletion of tsu but not of Acn (P-values based on Wilcoxon rank-sum test).
As exon skipping results in reduced levels of affected transcripts (presumably due to NMD), depletion of the core EJC affects preferentially levels of transcripts harboring long introns (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010). We confirmed that transcript levels are increasingly more affected as a function of increasing intron length upon Tsu depletion in OSCs (Fig. 7C). Again, no such trend is detectable upon Acn depletion, confirming its limited impact on exon skipping.
Taken together, our observations indicate that exon skipping and intron retention are related yet genetically separable processes. While depletion of nuclear core EJC factors results in both phenotypes, depletion of Acn only has a clear effect on the intron retention phenotype.
Discussion
Genetic screens have uncovered a surprising link between the EJC and the piRNA pathway in Drosophila. We dissected the underlying molecular cause and uncovered several principles that connect the EJC to intron definition and splicing. The major outcome of our study is as follows:
(1) Loss of the nuclear EJC or the accessory factors RnpS1 or Acn impairs the D. melanogaster piRNA pathway due to impaired splicing of the piwi transcript.
(2) Deposition of the EJC to flanking introns, but not direct recruitment by the spliceosome, underlies the EJC’s impact on splicing.
(3) Many introns—preferentially first introns followed by large second introns—require the EJC for their splicing.
(4) Exon skipping and intron retention depend on related but distinct aspects of EJC function.
Roles of the EJC in the piRNA pathway
Defective splicing of piwi int4 is a major molecular defect underlying the involvement of the EJC in the D. melanogaster piRNA pathway (Figs. 3, 4). During evolution, one or two LINE fragments appear to have inserted into int4, which, together with its poor pY tract, renders it too long to be spliced efficiently. Based on these criteria, an involvement of the EJC in piwi expression appears restricted to the melanogaster subgroup (which branched from the ananassae subgroup ∼45 million years ago) (Fig. 5; Tamura et al. 2004). It is unclear whether a suboptimal int4 is simply tolerated given its rescue via the EJC or whether the dependence of int4 on the EJC provides a selective advantage.
In the case of Acn depletion, TE repression is nearly entirely restored by expression of an EJC-independent piwi transgene. Loss of Tsu or Mago, on the other hand, results in additional defects that impair TE silencing. It is currently unclear whether splicing of other piRNA pathway factor transcripts requires the core EJC. A strong candidate is AGO3, which is expressed as an ∼150-kb pre-mRNA with several large introns; indeed, AGO3 transcript levels are affected upon depletion of core EJC but not peripheral EJC factors (Supplemental Fig. S4).
EJC assembly to flanking splice junctions occurs prior to its contribution to splicing
Because deposition of the minimal EJC core (pre-EJC: eIF4AIII, Mago, and Tsu) at the upstream exon occurs shortly before or during exon ligation and therefore depends on a successful splicing event (Gehring et al. 2009), most studies on the EJC have focused on its role in controlling mRNA fate downstream from splicing. It came as a surprise that the EJC also impacts the process of splicing itself (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010). However, analyzing the molecular basis for this contribution to splicing has been difficult due to the complex architecture of previously described target transcription units (long genes with large introns harboring numerous TE insertions, often situated within the heterochromatin). In particular, it was unclear whether the role of the EJC in the splicing of transcripts containing large introns reflects a noncanonical role of EJC components in splicing or whether canonically deposited EJCs at flanking splice junctions aid the definition of flanking introns (discussed in Ashton-Beaucage and Therrien 2011).
The finding that the EJC is also required for splicing of int4 at the compact and euchromatic piwi locus provides a powerful system to address this important question. Our collective data strongly indicate that conventional assembly of the EJC at splice junctions can facilitate the subsequent splicing of neighboring introns that pose a challenge for the spliceosome, such as ones that are atypically large (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010) or lack strong splicing signals (this study). In the case of piwi int4, a neighboring EJC (upstream or downstream) can assist the splicing if it is bound within ∼250 nt of the 3′ splice site (Fig. 5F,H). Interestingly, this distance requirement resembles that of the minimum size of an intron in the intron definition model of splicing (Fox-Walsh et al. 2005) and indicates a direct role of the EJC in aiding splicing rather than a licensing model in which EJC deposition anywhere on the message promotes splicing of suboptimal introns. The EJC has been shown to interact with a number of SR proteins (Singh et al. 2012), suggesting that the EJC acts as an RNA-bound scaffold to recruit splicing machineries via SR proteins.
A temporal order in splicing instructed by the EJC?
Previous observations point to a preferential role of the EJC in the splicing of complex transcripts; namely those that contain unusually large introns, often harboring TE insertions. Why, however, would first introns followed by large second introns be disproportionally dependent on the EJC (Fig. 6D)? These first introns themselves are not unusually large and exceptional, yet their dependency on a flanking EJC suggests that they are intrinsically difficult to splice. An attractive, albeit speculative, model is that evolution has selected this dependency in order to splice first introns only after successful splicing of the large downstream intron. Large introns are likely to contain cryptic polyadenylation/cleavage sites, and their usage is suppressed by deposition of U1 onto the nascent transcript (Kaida et al. 2010). Delaying intron splicing to a time point after transcription and splicing of the large downstream intron would therefore increase gene expression fidelity. Interestingly, first introns have been reported to be less efficiently spliced compared with internal introns in Drosophila (Khodor et al. 2011).
A related hypothesis is that the EJC licenses splice events during alternative splicing. Alternatively spliced introns often have weak splice recognition sites (Clark and Thanaraj 2002; Garg and Green 2007), and it has been shown that splicing of neighboring introns influences the selection of alternative splice sites (Fededa et al. 2005; Glauser et al. 2011). It would be intriguing if the order of intron splicing impacts alternative splicing in an EJC-dependent manner, thereby increasing the transcript repertoire in higher organisms.
Exon skipping versus intron retention and the role of Acinus
Intron retention and exon skipping have two different molecular causes: defective intron and exon definition, respectively (De Conti et al. 2013). Nevertheless, depletion of core EJC factors (eIF4AIII, Mago, and Tsu) or of RnpS1 can give rise to both phenotypes, suggesting that they are linked (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010; this study). Indeed, we hypothesize that both phenotypes manifest after a first successful round of splicing. In the case of exon skipping, we propose that the reinforcement of the exon definition of the newly formed exon pair is compromised (see also Ashton-Beaucage and Therrien 2011). In the case of intron retention, we propose that EJC loss results in a failure to license a weak nearby splice site.
Despite these similarities, it appears that these two processes are at least to some extent distinct, as loss of Acn leads more or less exclusively to intron retention and only marginally to exon skipping (Fig. 7). Our genome-wide analysis shows a global correlation between intron retention levels in Acn- and Tsu-depleted cells (Fig. 6A). Also, Acn enhances the splicing of piwi int4 in an EJC-dependent manner (Fig. 5F). Assembly of the EJC core is therefore probably a prerequisite for Acn to help defining neighboring splice sites. Acn forms a stable complex with the SR protein RnpS1 to interact with the EJC (Tange et al. 2005) and has recently been implicated in the regulation of alternative splicing (Michelle et al. 2012). Further genetic and biochemical analyses on Acn (and RnpS1) should be the key to understanding their distinctive roles in intron definition and exon definition.
Materials and methods
Fly husbandry and strains
All flies were kept at 25°C. MTD-GAL4 and piwi[06843] strains were obtained from the Bloomington Drosophila Stock Center, tj-GAL4 strain was obtained from the Drosophila Genomics Resource Center. The lacZ sensor flies for Burdock and gypsy were described in Handler et al. (2013) and Sarot et al. (2004), respectively. shRNA lines were generated by cloning short hairpins (for sequences, see Supplemental Table S3) into the Valium-20 vector modified with a white selection marker (Ni et al. 2011) and inserting these into the attp2 landing site on chromosome 3 (Groth et al. 2004). GFP-Piwi flies were described in Sienski et al. (2012). Knockdown crosses with Vienna Drosophila Resource Center lines (Dietzl et al. 2007) were with a UAS-Dcr2; NGT Gal4; nosGal4 driver line (Handler et al. 2013). shRNA lines were used for the Act-Gal4 flip-out clones (Olivieri et al. 2010) except for fs(1)Yb, for which a Vienna Drosophila Resource Center line (GD25437) was used.
For all experiments, flies were aged for 4–7 d. Knockdown experiments were always accompanied by a negative control of the same genetic background (carrying an shRNA against either GFP or white). During the last 2 d, flies were kept on apple juice plates supplemented with yeast paste at medium density to promote egg laying and uniform ovary morphology.
Cell culture
OSCs were cultured as described (Niki et al. 2006; Saito et al. 2009). In each experiment, 3 × 106 to approximately 4 × 106 cells were transfected twice with 200 pmol of preannealed siRNAs using Cell Line Nucleofector kit V (Amaxa Biosystems, program T-029) and cultured for 4–5 d (2 d plus 2–3 d) before harvesting (see Supplemental Table S4 for siRNA sequences).
Pacman[GFP-piwi] constructs
The construction of GFP-piwi was described previously (Sienski et al. 2012). All other GFP-piwi Pacman plasmids were constructed via Gibson assembly (Gibson et al. 2009). D. yakuba genomic DNA was obtained from the Drosophila Species Stock Center. The 3′ end of D. melanogaster piwi intron4 was replaced with the sequence from piwi intron7 using gBlock DNA fragments (Integrated DNA Technologies). The resulting sequences of shortened intron4 are as follows: intron4_60nt, GTAAGACTTTAAACTATATTTAAAT-deletion-CCATAAAAAATGTAATATTTAATTATTATAAACAG; and intron4_106nt, GTAAGACTTTAAACTATATTTAAATTAACAAGCTCTTGTGTCGCAAAC-deletion-GCAGTTTAGGGCGGTTTTATGGCCCATAAAAAATGTAATATTTAATTATTATAAACAG.
Immunohistochemistry, RNA FISH, and image analysis
Rabbit α-Ago3 was described in Brennecke et al. (2007). Mouse monoclonal α-Piwi was raised against the first 150 N-terminal amino acids.
In situ hybridization was performed as described in Lecuyer et al. (2007). Digoxigenin (DIG)-labeled probes were in vitro transcribed from piwi exon8 cloned into pBluescriptII using the following primers: 5′-ACTGACaagcttTGCCTCAATGGATCTACAGCAAA-3′ and 5′- ACTGACggtaccCGAACTTGTTGCGAGACCAGATA-3′. In situ signal was developed using fluorescein tyramide conjugate (Perkin Elmer).
All fluorescent images were taken on an LSM 780 (Zeiss). GFP intensity was quantified by ImageJ version 1.48f (Figs. 4, 5; Supplemental Fig. S3).
RT–PCR analysis
Total RNA was isolated from OSCs or ovaries using Trizol (Ambion). cDNA was prepared via random priming of total RNA unless otherwise stated. qPCR was performed using standard techniques. All experiments were performed in biological triplicate with technical duplicates. Relative RNA levels were normalized to rp49 levels. Fold enrichments were calculated in comparison with respective RNA levels obtained from flies expressing a control hairpin or from control siRNA transfections into OSCs (Schmittgen and Livak 2008). Primer sequences for qRT–PCR and RT–PCR analyses are listed in Supplemental Tables S5 and S6.
RNA-seq
PolyA-plus or rRNA-depleted RNA from biological duplicates of OSCs treated with siRNA against GFP, tsu, or Acn was selected with Dynabeads Oligo(dT) (Invitrogen) from total RNA, fragmented, and reverse-transcribed with random hexamers. Strand-specific libraries were prepared using the UDG digestion-based strategy (Zhang et al. 2012) and cloned with NEBNext ChIP-seq library preparation reagent set for Illumina (New England Biolabs), and 50-bp paired-end reads were sequenced on a HiSeq2000 (Illumina) instrument. This yielded 10 million–20 million genome-mappable reads per sample. For the computational analyses, we first extracted high-quality bases from every read (6–50 nt) and mapped these to the Drosophila genome as well as to the transcript and intron annotations in FlyBase release 5.56, allowing up to three mismatches. Uniquely aligned reads were used for quantification of gene expression levels and intron levels. Transcripts that scored >10 RPKM (reads per kilobase per million) in GFP knockdown (n = 4203) and introns whose RPKM values were >10% of their transcript RPKM value in any of biological duplicates (n = 1478) were analyzed. The intron retention index was calculated as the intron RPKM level divided by the transcript RPKM level in the respective samples.
Statistical analysis
We used statistical packages implemented in R 2.15.3 for all calculations and plots. For data visualization in box plot format, we used standard features (horizontal bars represent median, the box depicts 25th and 75th percentile, and whiskers represent the 1.5 interquartile range). Statistical significances in Figures 6D and 7D were computed with Mann-Whitney U-test; other P-values were calculated using Student’s t-test.
Acknowledgments
We thank members of the Brennecke laboratory for support and comments on the manuscript; J. Cokcezade for fly injections; D. Jurczak for bioinformatics support; the Campus Science Support Facilities GmbH (CSF) Next-Generation Sequencing unit for Illumina sequencing; the Institute of Molecular Pathology (IMP)/Institute of Molecular Biotechnology (IMBA) Biooptics Unit for support; and the Vienna Drosophila Resource Center, the Drosophila Genomics Resource Center, and the Bloomington Stock Center for flies. We thank FlyBase for the continuous genome annotation efforts (version r5.56). All deep sequencing data sets are summarized in Supplemental Table S2. This work was supported by the Austrian Academy of Sciences, the European Community’s 7th Framework Programme (FP7/2007-2013; ERC grant no. 260711EU), and the Austrian Science Fund (FWF; Y 510-B12). R.H. is supported by an SFB grant from the FWF (no. F4303). D.I.H. is supported by the University College London and Cancer Research U.K.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.245738.114.
Freely available online through the Genes & Development Open Access option.
References
- Ashton-Beaucage D, Therrien M. 2011. The exon junction complex: a splicing factor for long intron containing transcripts? Fly (Austin) 5: 224–233 [DOI] [PubMed] [Google Scholar]
- Ashton-Beaucage D, Udell CM, Lavoie H, Baril C, Lefrançois M, Chagnon P, Gendron P, Caron-Lizotte O, Bonneil E, Thibault P, et al. . 2010. The exon junction complex controls the splicing of mapk and other long intron-containing transcripts in Drosophila. Cell 143: 251–262 [DOI] [PubMed] [Google Scholar]
- Bono F, Gehring NH. 2011. Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol 8: 24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Clark F, Thanaraj TA. 2002. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum Mol Genet 11: 451–464 [DOI] [PubMed] [Google Scholar]
- Czech B, Preall JB, McGinn J, Hannon GJ. 2013. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell 50: 749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Conti L, Baralle M, Buratti E. 2013. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA 4: 49–60 [DOI] [PubMed] [Google Scholar]
- Degot S, Le Hir H, Alpy F, Kedinger V, Stoll I, Wendling C, Seraphin B, Rio MC, Tomasetto C. 2004. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J Biol Chem 279: 33702–33715 [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. . 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Fededa JP, Petrillo E, Gelfand MS, Neverov AD, Kadener S, Nogues G, Pelisch F, Baralle FE, Muro AF, Kornblihtt AR. 2005. A polar mechanism coordinates different regions of alternative splicing within a single gene. Mol Cell 19: 393–404 [DOI] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ.. 2005. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci 102: 16176–16181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg K, Green P. 2007. Differing patterns of selection in alternative and constitutive splice sites. Genome Res 17: 1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. 2003. Y14 and hUpf3b form an NMD-activating complex. Mol Cell 11: 939–949 [DOI] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. 2009. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol 7: e1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Glauser DA, Johnson BE, Aldrich RW, Goodman MB. 2011. Intragenic alternative splicing coordination is essential for Caenorhabditis elegans slo-1 gene function. Proc Natl Acad Sci 108: 20790–20795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage ϕC31. Genetics 166: 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Lo PC, Mount SM. 1993. Species-specific signals for the splicing of a short Drosophila intron in vitro. Mol Cell Biol 13: 1104–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J. 2013. The genetic makeup of the Drosophila piRNA pathway. Mol Cell 50: 762–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell 6: 673–682 [DOI] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT 2nd, Rosbash M. 2011. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev 25: 2502–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. 2000a. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE. 2000b. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev 14: 1098–1108 [PMC free article] [PubMed] [Google Scholar]
- Li C, Lin RI, Lai MC, Ouyang P, Tarn WY. 2003. Nuclear Pnn/DRS protein binds to spliced mRNPs and participates in mRNA processing and export via interaction with RNPS1. Mol Cell Biol 23: 7363–7376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647 [DOI] [PubMed] [Google Scholar]
- Mayeda A, Badolato J, Kobayashi R, Zhang MQ, Gardiner EM, Krainer AR. 1999. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J 18: 4560–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelle L, Cloutier A, Toutant J, Shkreta L, Thibault P, Durand M, Garneau D, Gendron D, Lapointe E, Couture S, et al. . 2012. Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Mol Cell Biol 32: 954–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, Hannon GJ. 2013. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol Cell 50: 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. . 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8: 405–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki Y, Yamaguchi T, Mahowald AP. 2006. Establishment of stable cell lines of Drosophila germ-line stem cells. Proc Natl Acad Sci 103: 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. 2010. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell 143: 238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakashita E, Tatsumi S, Werner D, Endo H, Mayeda A. 2004. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol Cell Biol 24: 1174–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. 2004. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schwerk C, Prasad J, Degenhardt K, Erdjument-Bromage H, White E, Tempst P, Kidd VJ, Manley JL, Lahti JM, Reinberg D. 2003. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol Cell Biol 23: 2981–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Brennecke J. 2010. The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet 26: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T, Tange TO, Sonenberg N, Moore MJ. 2004. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol 11: 346–351 [DOI] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. 2012. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151: 750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Szakmary A, Reedy M, Qi H, Lin H. 2009. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J Cell Biol 185: 613–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol 21: 36–44 [DOI] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. 2004. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol 16: 279–284 [DOI] [PubMed] [Google Scholar]
- Tange TO, Shibuya T, Jurica MS, Moore MJ. 2005. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 11: 1869–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355: 609–614 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89: 357–364 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Theurkauf WE, Weng Z, Zamore PD. 2012. Strand-specific libraries for high throughput RNA sequencing (RNA-seq) prepared without poly(A) selection. Silence 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]