Abstract
Cohesion between sister chromatids in mitotic and meiotic cells is promoted by a ring-shaped protein structure, the cohesin complex. The cohesin core complex is composed of four subunits, including two structural maintenance of chromosome (SMC) proteins, one α-kleisin protein, and one SA protein. Meiotic cells express both mitotic and meiosis-specific cohesin core subunits, generating cohesin complexes with different subunit composition and possibly separate meiotic functions. Here, we have analyzed the in vivo function of STAG3, a vertebrate meiosis-specific SA protein. Mice with a hypomorphic allele of Stag3, which display a severely reduced level of STAG3, are viable but infertile. We show that meiocytes in homozygous mutant Stag3 mice display chromosome axis compaction, aberrant synapsis, impaired recombination and developmental arrest. We find that the three different α-kleisins present in meiotic cells show different dosage-dependent requirements for STAG3 and that STAG3-REC8 cohesin complexes have a critical role in supporting meiotic chromosome structure and functions.
Keywords: chromosome, cohesin, meiosis, recombination, synaptonemal complex
See also: T Winters et al (June 2014)
Introduction
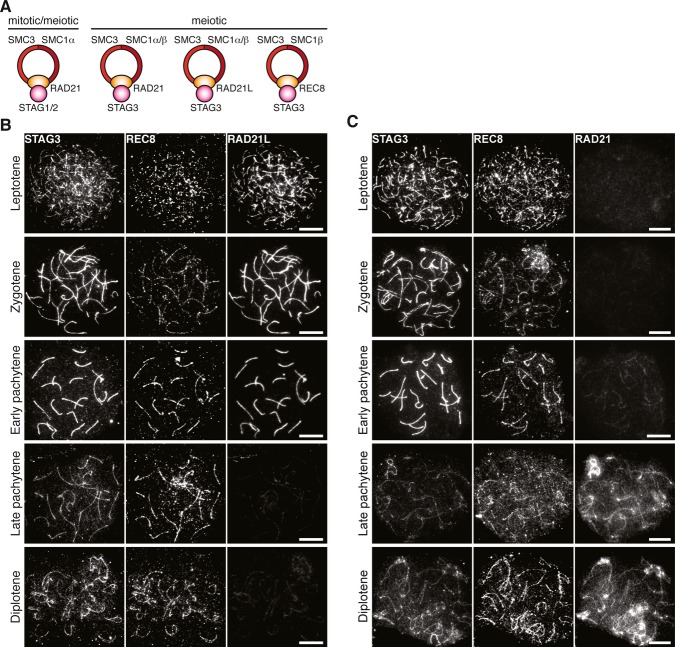
Cohesion between sister chromatids is critical for the accuracy of the chromosome segregation process during cell division (Peters et al, 2008; Nasmyth & Haering, 2009). Cohesion is established during replication of chromosomes in mitotic and meiotic cells through chromosomal loading of a conserved ring-shaped protein structure, the cohesin complex. The cohesin core complex in mitotic cells is composed of four subunits, two SMC proteins (structural maintenance of chromosomes), Smc1 and Smc3, one α-kleisin protein (Rad21, also called Scc1 or Mcd1), and one SA protein (stromal antigen, also called Scc3). Smc1 and Smc3 together generate a ring-like structure that is sealed through the association of Rad21 to the globular terminal domains of the SMC proteins. Destruction of Rad21, through proteolytic cleavage at the metaphase-to-anaphase stage of mitosis, removes cohesion between sister chromatids, allowing their individual segregation into separate daughter cells. Two different SA proteins have been characterized in mitotic cells of vertebrates, STAG1 and STAG2 (also called SA1 and SA2). The binding of SA proteins to Rad21 generates two separate cohesin complexes, contributing to centromere cohesion (STAG2), as well as telomere cohesion (STAG1) (Canudas & Smith, 2009). Inactivation of Stag1 in mice results in embryonic lethality, whereas heterozygous Stag1 mutant mice display a short lifespan and an enhanced level of tumorigenesis (Remeseiro et al, 2012). Four meiosis-specific cohesin core components have been identified in vertebrates, SMC1β, RAD21L, REC8, and STAG3 (also called SA3) (Pezzi et al, 2000; Prieto et al, 2001, 2002; Revenkova et al, 2001; Eijpe et al, 2003; Lee et al, 2003; Gutierrez-Caballero et al, 2011; Ishiguro et al, 2011; Lee & Hirano, 2011). Expression studies of the meiosis-specific and the mitotic cohesin core proteins during meiosis in mice have revealed both constitutive, as well as transient, expression patterns. In agreement with these findings, biochemical experiments support the presence of at least six different cohesin complexes in meiotic cells (Fig 1A) (Ishiguro et al, 2011; Jessberger, 2011; Lee & Hirano, 2011).
Figure 1. Chromosomal localization of meiotic cohesin components.
A Composition of different cohesin complexes during mitosis and meiosis.
B, C Chromosomal localization of STAG3 during prophase I. Nuclear spreads of wild-type spermatocytes were immunostained for STAG3, REC8, and RAD21L (B) or for STAG3, REC8, and RAD21 (C). Note that the late pachytene spermatocyte in (C) is at a later stage than the one in (B). Bars, 10 μm.
The functions of the different cohesin complexes during meiosis are yet poorly understood. It is known that cohesin core proteins contribute to the assembly of a chromosome axis in meiotic cells (a cohesin axis) that holds sister chromatids together and from which chromatin loops emanate (Klein et al, 1999; Pelttari et al, 2001). The cohesin axis is necessary for the assembly of the synaptonemal complex (SC), a meiosis-specific protein structure (Klein et al, 1999; Llano et al, 2012). The SC is essential for synapsis of homologous chromosomes, and for the formation of crossovers in mammalian cells, recombination events that ensure that the homologous chromosomes are faithfully segregated at the first meiotic division (de Vries et al, 2005; Kouznetsova et al, 2009, 2011). The SC is composed of three distinct protein substructures, axial elements (AEs), transverse filaments, and a central element (Zickler & Kleckner, 1999; Page & Hawley, 2004). The AE of the SC assembles on the sister chromatid axis at the leptotene stage of the first meiosis (meiosis I), overlapping with the cytological localization of the cohesin axis. Binding of transverse filament proteins to the AEs at the zygotene stage of meiosis I regulates assembly of a central element in between the aligned homologous chromosomes (Costa et al, 2005; Hamer et al, 2006; Schramm et al, 2011). Longitudinal polymerization of the transverse filaments and the central element of the SC along the aligned homologous chromosomes then results in synapsis, that is, close pairing of the homologous chromosomes at the pachytene stage of meiosis I. The SC is dissolved at the diplotene stage of meiosis I, after which the homologous chromosomes remain attached to each other through chiasmata, physical linkages resulting from the crossover recombination process (Petronczki et al, 2003). Cohesin complexes are removed from the sister chromatids in two steps, at the metaphase–anaphase transition of meiosis I, when arm cohesion is lost, and at the metaphase–anaphase transition at meiosis II, when centromere cohesion is lost (Petronczki et al, 2003).
The functions of the vertebrate meiosis-specific cohesin core components SMC1β, REC8, and RAD21L in cohesion, synapsis, and recombination have been studied in mice (Bannister et al, 2004; Revenkova et al, 2004; Xu et al, 2005; Herran et al, 2011; Llano et al, 2012; Ishiguro et al, 2014). Rec8-null mice were found to display shortened chromosome axes, inter-sister SC assembly and impaired DNA double-strand break (DSB) repair in meiotic cells (Bannister et al, 2004; Xu et al, 2005), whereas Rad21L-null mice displayed fragmented chromosome axes, non-homologous synapsis and impaired DSB repair (Herran et al, 2011). In contrast, Rec8 and Rad21L double-null mice failed to form meiotic chromosome axes and did not assemble AEs or SCs (Llano et al, 2012; Ishiguro et al, 2014). Finally, inactivation of Smc1β has been shown to result in shortened chromosome axes, extensions of chromatin loops, and incomplete synapsis, and to affect sister chromatid cohesion and recombination, as well as telomere integrity (Revenkova et al, 2004; Novak et al, 2008; Adelfalk et al, 2009). We have now investigated the in vivo function of the only known meiosis-specific SA component, STAG3. Phenotypic analysis of mice with a hypomorphic allele of Stag3 reveals that STAG3 is of critical importance for stabilization of REC8 cohesin complexes and their association with the meiotic chromosome axes. Loss of REC8 in homozygous Stag3 mutant mice from the meiotic chromosome axes, but not RAD21L or RAD21, results in chromosome axis compaction and synapsis failure. Thus, α-kleisins show a different dosage-dependent requirement for STAG3, contributing to a functional diversification among the different cohesin complexes present in meiotic cells.
Results
The localization pattern for STAG3 on the chromosome axes mimics the distribution of three different α-kleisins
STAG3 has been shown to interact with the α-kleisin subunit of the cohesin core complex in meiotic cells (Fig 1A) (Ishiguro et al, 2011; Lee & Hirano, 2011). To assess the expression pattern and localization of STAG3 relative to the three different α-kleisin subunits of the cohesin core complex during the prophase stage of meiosis I (prophase I), nuclear spreads of mouse spermatocytes were immunostained for STAG3 and REC8, as well as either RAD21L or RAD21. STAG3, REC8, and RAD21L, but not RAD21, were found to be localized at the axes of meiotic chromosomes at the leptotene stage of meiosis I (Fig 1B and C). STAG3 and RAD21L accumulated in a uniform manner along the axes of chromosomes at the zygotene and early pachytene stages of meiosis I. The intensity of the immunofluorescence signals for STAG3 was drastically reduced at the late pachytene stage of meiosis I, temporally coinciding with a loss of RAD21L from the axes of chromosomes (Fig 1B). Instead, a non-continuous STAG3 pattern emerged, closely resembling the axial patterns observed collectively for REC8 and RAD21 at the late pachytene and diplotene stages of meiosis I (Fig 1B and C). In summary, our results suggest that STAG3 is part of several different cohesin complexes distinguished by the presence of different α-kleisin core subunits.
Homozygous Stag3 mutant mice that express a severely reduced level of STAG3 are viable but infertile
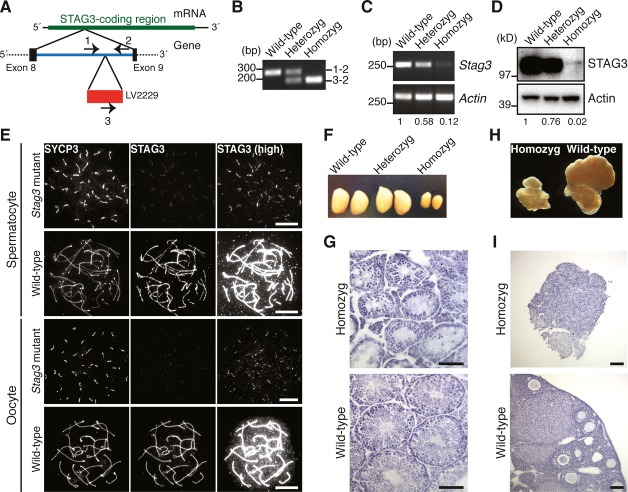
To assess the function of STAG3 in meiotic cells, we have used a transgenic mouse line generated by a transgene-based random mutagenesis protocol. In the mouse line, a transgene was inserted in between exons 8 and 9 within the Stag3 gene locus (Fig 2A and B). We have characterized this mutant mouse strain and examined mRNA and protein expression levels in the testis of Stag3 mutant animals by RT-PCR and immunoblotting experiments. Mice homozygous for the mutation showed an approximately 10-fold lower level of Stag3 mRNA in testicular cells (Fig 2C). In agreement with this, the STAG3 protein levels were drastically reduced (˜20–50-fold) in adult and juvenile testes (Fig 2D, Supplementary Fig S1A and B) and in embryonic ovaries (embryonic day 16.5, E16.5) (Supplementary Fig S1C). A severely reduced level of STAG3 was also found to be associated with the axis of meiotic chromosomes in homozygous Stag3 mutant mice (Fig 2E).
Figure 2. STAG3 is required for gametogenesis.
- Structure of the Stag3 mutant allele. The transgene (LV2229) is inserted within the intron between exon 8 and exon 9 of the Stag3 gene. The positions of PCR primers for genotyping are indicated.
- PCR analysis for genotyping. Genomic DNA from wild-type, heterozygous mutant, and homozygous mutant mice were analyzed using the primer pairs indicated in (A).
- RT-PCR analysis of RNA extracted from testes of wild-type, heterozygous mutant, and homozygous mutant mice. The signals were quantified, and relative values for Stag3 mRNA abundance normalized to Actin are presented below the panels.
- Immunoblotting analysis of testicular protein extracts from wild-type, heterozygous mutant, and homozygous mutant mice. The signals were quantified, and relative values for STAG3 normalized to actin are presented below the panels.
- Immunostaining of wild-type and homozygous Stag3 mutant spermatocytes and oocytes. Nuclear spreads were stained for SYCP3 (an AE protein labeling the meiotic chromosome axis) and STAG3. High-exposure images of STAG3 staining (high) are shown to highlight residual STAG3 in the mutant. Bars, 10 μm.
- Testes from 8-week-old wild-type, heterozygous Stag3 mutant, and homozygous Stag3 mutant mice.
- Testicular histology of 8-week-old wild-type and homozygous Stag3 mutant mice. Testis sections were stained with hematoxylin and eosin. Bars, 100 μm.
- Ovaries and oviducts from 8-week-old wild-type and homozygous Stag3 mutant mice.
- Ovarian histology of 8-week-old wild-type and homozygous Stag3 mutant mice. Ovary sections were stained with hematoxylin and eosin. Bars, 100 μm.
Source data are available online for this figure.
The homozygous Stag3 mutant mice were found to develop normally; however, both males and females were infertile. Consistent with this, the sizes of the testes and the ovaries of homozygous Stag3 mutant mice were smaller than observed for wild-type mice (Fig 2F and H). Histological analysis revealed that spermatids and ovarian follicles were absent in the homozygous Stag3 mutant testes and ovaries, respectively (Fig 2G and I). Our results therefore show that the integration of a transgene into the Stag3 gene locus has generated a hypomorphic allele of Stag3, severely disrupting expression of STAG3 in homozygous Stag3 mutant mice, resulting in arrested gametogenesis and male and female infertility.
RAD21L, RAD21, and AE proteins, but not REC8, localize to axial structures formed in meiotic cells in homozygous Stag3 mutant animals
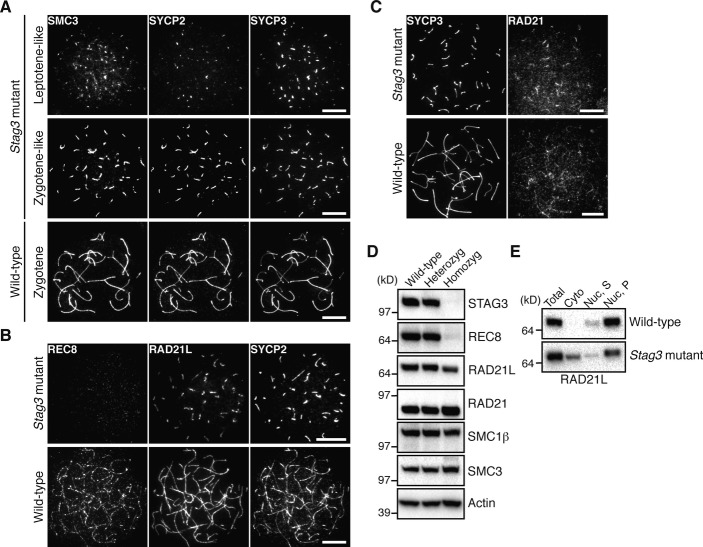
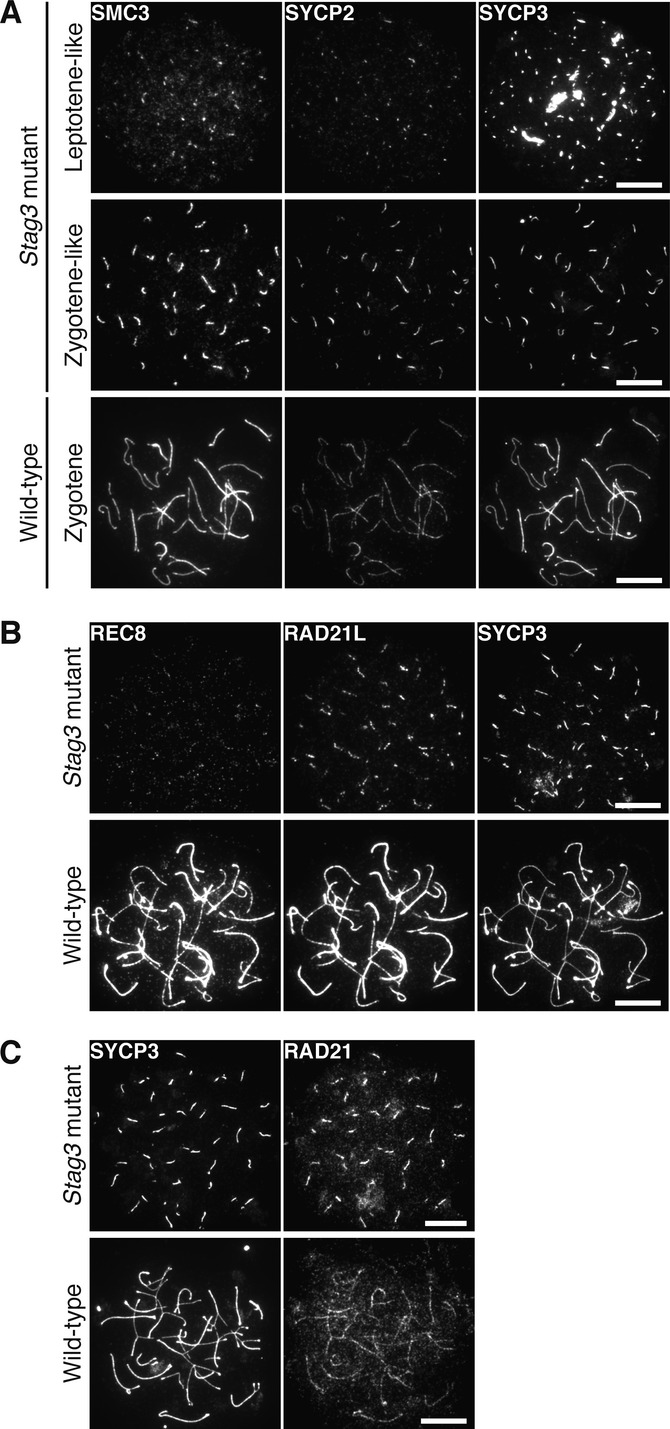
Cohesin complexes and AE proteins contribute to the formation of a chromosome axis during prophase I. To find out the role for STAG3 in axis organization, nuclear spreads of homozygous Stag3 mutant spermatocytes and embryonic oocytes (E16.5) were immunolabeled with antibodies against SMC3 and the AE proteins SYCP2 and SYCP3. SMC3 was found to co-localize with SYCP2 and SYCP3 along the axial structures formed in homozygous Stag3 mutant spermatocytes and oocytes (Fig 3A and Fig 4A) having either short (leptotene-like) or more extended axial structures (zygotene-like). Immunolabeling of homozygous mutant spermatocytes and oocytes with antibodies against the three different α-kleisin proteins expressed in meiotic cells, REC8, RAD21L, and RAD21, revealed that RAD21L and RAD21 co-localized with the axial structures labeled by SYCP2 (or SYCP3), while little or no REC8 signal was observed on the axial structures in homozygous mutant meiocytes (Fig 3B and C, Fig 4B and C). Consistent with this, immunoblotting analysis showed that the level of REC8, but not of RAD21, SMC1β or SMC3, was severely reduced in homozygous mutant testicular cells (Fig 3D). The level of RAD21L was slightly reduced in homozygous Stag3 mutant testes, albeit to a much lesser extent than REC8 (Fig 3D). Fractionation of testis extracts and subsequent immunoblotting analysis showed that RAD21L accumulated in the cytoplasmic fraction of homozygous mutant testes, a situation not seen in wild-type testes (Fig 3E). We conclude that STAG3 is essential for the stability of REC8 and the accumulation of this protein on the meiotic chromosome axes.
Figure 3. STAG3 is required for REC8 stabilization and accumulation onto the meiotic chromosome axes in spermatocytes.
- Immunostaining of wild-type and homozygous Stag3 mutant spermatocytes for SMC3, SYCP2, and SYCP3. Bars, 10 μm.
- Immunostaining of wild-type and homozygous Stag3 mutant spermatocytes for REC8, RAD21L, and SYCP2. Bars, 10 μm.
- Immunostaining of wild-type and homozygous Stag3 mutant spermatocytes for SYCP3 and RAD21. Bars, 10 μm.
- Immunoblotting analysis of testicular protein extracts from wild-type, heterozygous Stag3 mutant, and homozygous Stag3 mutant mice.
- Testicular protein extracts were separated into cytoplasmic (Cyto) and nuclear fractions. The nuclear fraction was further fractionated into soluble (Nuc, S) and insoluble (Nuc, P) fractions.
Source data are available online for this figure.
Figure 4. STAG3 is required for REC8 accumulation onto the meiotic chromosome axes in oocytes.
- Immunostaining of wild-type and homozygous Stag3 mutant E16.5 oocytes for SMC3, SYCP2, and SYCP3. Bars, 10 μm.
- Immunostaining of wild-type and homozygous Stag3 mutant E16.5 oocytes for REC8, RAD21L and SYCP3. Bars, 10 μm.
- Immunostaining of wild-type and homozygous Stag3 mutant E16.5 oocytes for SYCP3 and RAD21. Bars, 10 μm.
The presence of a meiotic chromosome axis in homozygous Stag3 mutant meiocytes suggests that the axis forms independently of STAG3, or alternatively that the residual level of STAG3 present in homozygous meiocytes contributes to axis formation. We asked whether the other SA proteins, STAG1 and STAG2, contribute to axis formation in homozygous Stag3 mutant meiocytes. Nuclear spreads of spermatocytes and oocytes were immunostained for STAG1 or STAG2; however, similar to the situation in wild-type cells (Prieto et al, 2002), neither STAG1 nor STAG2 accumulated on the chromosome axes in homozygous mutant spermatocytes (Fig 5A and B) or oocytes (Supplementary Fig S2A and B). Furthermore, immunoprecipitation of meiotic cohesin complexes in wild-type and homozygous Stag3 mutant spermatocytes using antibodies against RAD21L or SMC1β failed to detect STAG1 or STAG2 in meiotic cohesin complexes (Fig 5C and D). We conclude that STAG1 or STAG2 does not contribute to meiotic chromosome axis formation in homozygous Stag3 mutant meiocytes. Instead, it is very likely that the residual level of STAG3 present in homozygous Stag3 mutant meiocytes contributes to meiotic chromosome axis formation in these cells (see Discussion).
Figure 5. STAG1 and STAG2 do not accumulate at the meiotic chromosome axes in homozygous Stag3 mutant spermatocytes.
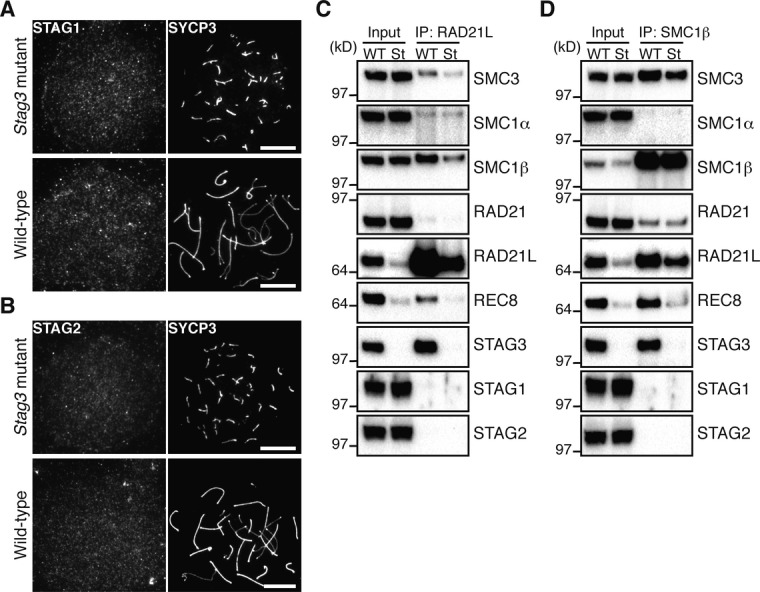
A Immunostaining of wild-type and homozygous Stag3 mutant spermatocytes for STAG1 and SYCP3. Bars, 10 μm.
B Immunostaining of wild-type and homozygous Stag3 mutant spermatocytes for STAG2 and SYCP3. Bars, 10 μm.
C, D Immunoprecipitation using nuclear extracts from 15-day-old wild-type (WT) and homozygous Stag3 mutant (St) testes. The extracts were immunoprecipitated with the antibody against RAD21L (C) or SMC1β (D). 4% of input samples (input) and the immunoprecipitates (IP) were subjected to immunoblotting using antibodies against cohesin subunits.
Source data are available online for this figure.
Synapsis is impaired in homozygous Stag3 mutant meiocytes
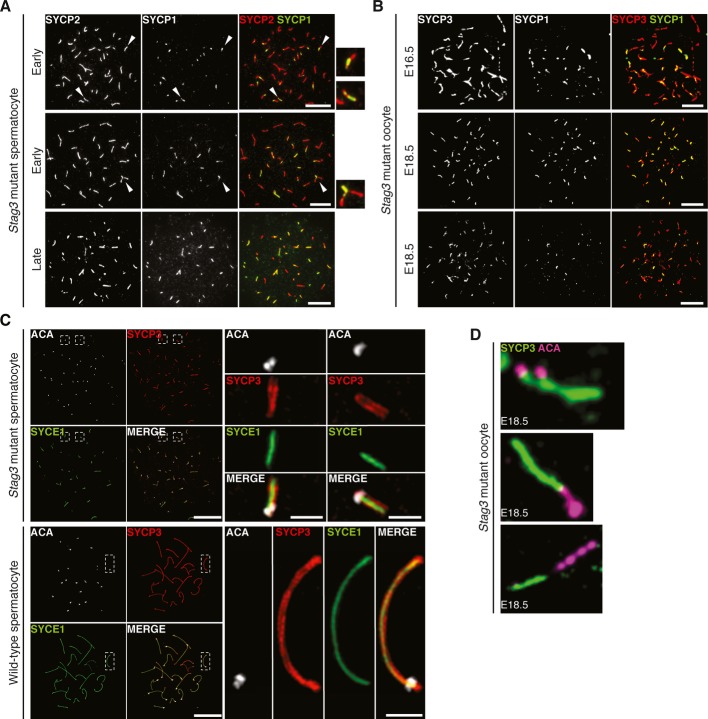
In order to further study the meiotic process in homozygous Stag3 mutant spermatocytes and oocytes (E16.5 and E18.5), mutant meiocytes were co-stained with antibodies against the AE protein SYCP2 (or SYCP3) and the transverse filament protein SYCP1, the latter a marker for synapsis (Fig 6A and B). We found little evidence for synapsis in the homozygous mutant meiocytes. In rare cases, closely aligned axial structures labeled by SYCP1, possibly representing homologous or non-homologous synapsis events, were observed (Fig 6A, indicated by an arrowhead in the middle panel), suggesting that the temporally most advanced homozygous Stag3 mutant spermatocytes could reach an early zygotene-like stage of meiosis I. However, this SYCP1 pattern was identified in only 37% (n = 76) of the homozygous mutant spermatocytes, and in most of these cases, only one such event was detected per cell (82%, n = 28). Analysis of the distribution of SYCP1 in homozygous Stag3 mutant E18.5 oocytes (representing a stage of meiosis at which most wild-type oocytes have reached pachytene) revealed no evidence of further progression of homologous alignment or synapsis (Figure6B).
Figure 6. STAG3 is essential for chromosome synapsis.
- Immunostaining of homozygous Stag3 mutant spermatocytes for SYCP2 and SYCP1. Enlarged views of the axes (arrowheads) are shown to the right. Bars, 10 μm.
- Immunostaining of homozygous Stag3 mutant E16.5 and E18.5 oocytes for SYCP3 and SYCP1. Bars, 10 μm.
- SIM analysis of wild-type and homozygous Stag3 mutant spermatocytes immunostained for SYCP3, SYCE1, and ACA. Enlarged views of axial structures from mutant spermatocytes and homologous chromosomes from wild-type spermatocytes are shown to the right. Scale bars are 10 μm and 1 μm for whole and close-up images, respectively.
- Immunostaining of homozygous Stag3 mutant E18.5 oocytes for SYCP3 and ACA displaying different types of aberrant chromosome structures.
In support of the asynapsis phenotype, FISH (fluorescence in situ hybridization) showed a separation of homologous chromosomes into univalents, resulting in two chromosome 17 signals in homozygous Stag3 mutant meiocytes and two X-chromosome signals in homozygous Stag3 mutant oocytes (Supplementary Fig S3A and B). Furthermore, labeling of homozygous Stag3 mutant and wild-type spermatocytes using ACA, a centromere marker, identified on average 38 ACA foci (n = 10) in homozygous Stag3 mutant spermatocytes, whereas 21 ACA foci were seen in wild-type pachytene spermatocytes (Fig 6C). Evidence of centromere separation (2.0 ± 1.2 events per oocytes) was seen in homozygous mutant E18.5 oocytes and spermatocytes (Fig 6C and D), together with stretched or deformed centromeres (5.2 ± 3.3 events per oocyte) (Fig 6D). We conclude that whereas synapsis is impaired in STAG3 homozygous Stag3 mutant meiocytes, cohesion between sister chromatids is to a large extent retained, even though centromere cohesion appears to be weakened in the mutant.
The nature of the SYCP1 staining pattern of the axial structures in homozygous Stag3 mutant meiocytes is enigmatic, considering that synapsis is abrogated in the mutant cells. The axial structures found in homozygous Stag3 mutant meiocytes were labeled at one of the two ends by ACA (Fig 6C) and at both end by TRF1 (a telomere protein) (Supplementary Fig S4A), strongly arguing that the axial structures observed in mutant meiocytes represent compacted univalent chromosomes. Furthermore, the central element proteins SYCE1, SYCE2, and TEX12 accumulated in a pattern similar to SYCP1 along the axial structures found in homozygous Stag3 mutants (Supplementary Fig S4B-D), suggesting that an SC-like structure assembles on the univalent in mutant meiocytes. To further describe the organization of the SC-like structures in mutant spermatocytes, the localization of SYCE1 in wild-type and homozygous Stag3 mutant spermatocytes was analyzed using super-resolution structured illumination microscopy (SIM), a method with a lateral resolution (XY) of 100–130 nm. We found as expected SYCE1 to be localized in between the AEs (labeled by SYCP3) of the SC in wild-type cells (Fig 6C, Supplementary Fig S5B). Importantly, a tripartite structure with SYCE1 surrounded by AEs was also observed in homozygous Stag3 mutant spermatocytes (41% of the analyzed axial structures, n = 376) (Fig 6C, Supplementary Fig S5A). These results show that SC-like structures assemble on univalents in homozygous Stag3 mutant spermatocytes, similar to what has been observed in Rec8-null spermatocytes (Xu et al, 2005). In summary, our results show that STAG3 supports meiotic chromosome axis organization and integrity, promotes initiation and progression of synapsis, and prohibits SC along the axis of asynapsed chromosomes. The reduced expression of STAG3 in homozygous Stag3 mutant meiocytes, however, does not abolish cohesion between sister chromatids.
Meiotic recombination is initiated but not completed in homozygous Stag3 mutant meiocytes
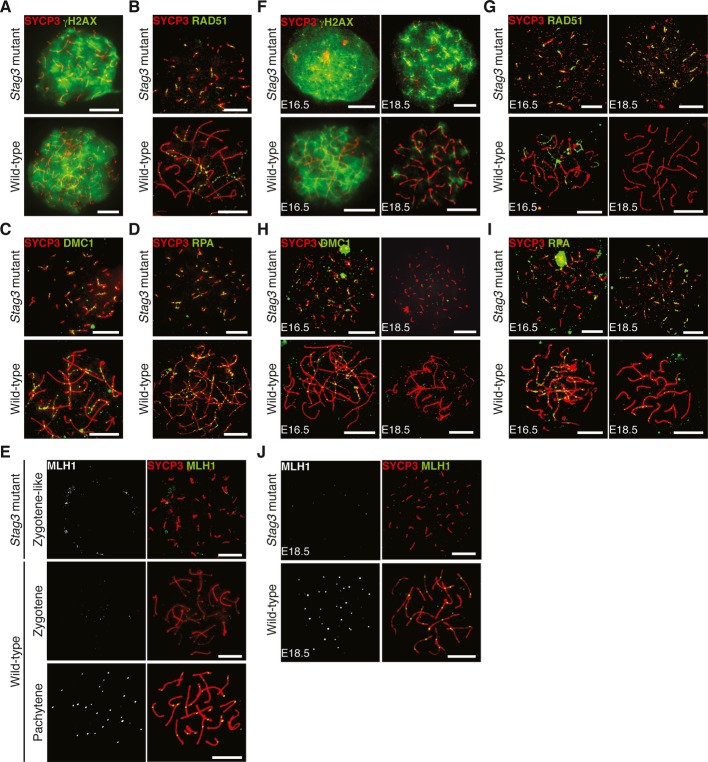
Meiotic recombination is initiated by programmed DSBs. The DSBs are repaired by homologous recombination using the homologous chromosome as a template, a process that gives rise to crossovers and chiasma formation. We have examined how impaired STAG3 expression affects the meiotic recombination process. First, DSB formation was monitored by the presence of γH2AX, a phosphorylated histone variant that is a marker of DSBs. Wild-type leptotene and homozygous Stag3 mutant leptotene-like spermatocytes and oocytes (E16.5) displayed similar γH2AX patterns, strongly suggesting that the DSB formation process is initiated in the homozygous mutant meiocytes (Fig 7A and F). We next examined the expression of RAD51 and DMC1, two repair proteins that mediate strand invasion following DSB formation. Similar to wild-type cells, RAD51 (Fig 7B and G) and DMC1 (Fig 7C and H) were detected on the meiotic chromosome axes labeled by the AE protein SYCP3 in homozygous Stag3 mutant spermatocytes and oocytes (E16.5). Also RPA, a repair protein that is loaded at recombination sites subsequently to RAD51 and DMC1, localized to the meiotic chromosome axes in homozygous mutant spermatocytes and oocytes (Fig 7D and I). We analyzed the expression of γH2AX, RAD51, DMC1, and RPA also in E18.5 homozygous mutant oocytes and found that γH2AX, RAD51, and RPA continued to accumulate on the axial structures present in homozygous mutant cells (Fig 7F–I). This was in contrast to the situation in wild-type E18.5 oocytes, where γH2AX staining and the number of RAD51 and RPA foci were reduced relative to the situation in E16.5 oocytes. A deviation to this pattern was seen for DMC1, for which foci no longer were detectable in homozygous mutant E18.5 oocytes (Fig 7H). Finally, we analyzed the expression of MLH1, a marker for late recombination events representing crossover sites. One or two MLH1 foci were found to localize to the axis of synapsed meiotic chromosomes at the pachytene stage of meiosis I in wild-type spermatocytes and oocytes (E18.5), but not at the zygotene stage of meiosis I (Fig 7E). In contrast, no MLH1 foci were detected at the axial structures found in homozygous Stag3 mutant spermatocytes (Fig 7E) or in homozygous mutant E18.5 oocytes (Fig 7J). We conclude that STAG3 is required for progression, but not for the initiation of the meiotic recombination process.
Figure 7. Meiotic recombination is initiated but not completed in homozygous Stag3 mutant spermatocytes and oocytes.
A–E Immunostaining of wild-type and homozygous Stag3 mutant spermatocytes for γH2AX (A), RAD51 (B), DMC1 (C), RPA (D), and MLH1 (E). Bars, 10 μm.
F–J Immunostaining of wild-type and homozygous Stag3 mutant oocytes for γH2AX (F), RAD51 (G), DMC1 (H), RPA (I), and MLH1 (J). The chromosome axes were labeled by SYCP3. Bars, 10 μm.
Discussion
In mammals, there are four known meiosis-specific cohesin core subunits, SMC1β, RAD21L, REC8, and STAG3. Mouse strains deficient for SMC1β, RAD21L, and REC8 have previously been generated, and their functions analyzed (Bannister et al, 2004; Revenkova et al, 2004; Xu et al, 2005; Herran et al, 2011; Llano et al, 2012). We have here studied the in vivo function of STAG3 in homozygous mutant mice carrying a hypomorphic allele of Stag3, resulting in a severely reduced level of STAG3 expression in meiocytes. We find that homozygous Stag3 mutant mice fail to complete meiosis and that both males and females are infertile. We have used these homozygous mutant mice to investigate the role of STAG3 in chromosome axis organization, synapsis, and recombination during mammalian meiosis.
During the prophase I stage of meiosis in eukaryotes, cohesin complexes define a chromosome axis that acts as a structural scaffold for AE assembly (Klein et al, 1999; Pelttari et al, 2001; Molnar et al, 2003; Severson et al, 2009; Llano et al, 2012; Ishiguro et al, 2014). We show here that homozygous Stag3 mutant spermatocytes and oocytes exhibit shortened chromosome axes. This is similar to what has been observed in Smc1β and Rec8 knockout mice (Bannister et al, 2004; Revenkova et al, 2004; Xu et al, 2005; Novak et al, 2008), revealing an important structural role for cohesin complexes composed of the core subunits SMC1β, REC8, and STAG3 in chromosome axis organization. Cohesin complex proteins contribute to both DSB formation and to DSB repair by homologous recombination (Klein et al, 1999; Kugou et al, 2009; Kim et al, 2010). We find that the early recombination events, including DSB formation and loading of recombination proteins at DSBs, take place in homozygous Stag3 mutant meiocytes. The accumulation and persistence of RAD51 and RPA foci in homozygous mutant spermatocytes and oocytes, together with the absence of MLH1 in these cells, show that progression of the recombination process is disrupted in homozygous Stag3 mutant meiocytes. The disappearance of DMC1 foci and the persistence of RAD51 foci in homozygous mutant E18.5 oocytes mimic the situation in mouse SC mutants deficient in SC assembly and synapsis (Kouznetsova et al, 2011). Thus, this suggests that the continued presence of DMC1 on the chromosome axes during prophase I of mammalian meiosis is dependent on synapsis initiation, as shown for DMC1 in yeast meiotic cells (Schwacha & Kleckner, 1997).
Synapsis defects have been reported in mutants for different meiosis-specific cohesin core subunits. Rad21L-null oocytes show fully synapsed bivalents, whereas Rad21L-null spermatocytes display non-homologous chromosome synapsis (Herran et al, 2011). In Smc1β-null meiocytes, chromosome synapsis proceeds completely or partially between homologous chromosomes, and only occasionally asynapsed chromosomes (univalents) are observed (Revenkova et al, 2004). In contrast, chromosomes in Rec8-null meiocytes fail to undergo synapsis (Bannister et al, 2004; Xu et al, 2005). The synapsis phenotype observed in homozygous Stag3 mutant meiocytes is most similar to the one described for Rec8-null meiocytes, including a loss of homologous chromosome alignment and synapsis. It has been shown in Rec8-null spermatocytes that the central element of the SC assembles between axial cores of sister chromatids instead of between the homologous chromosomes (Xu et al, 2005). In agreement with this, we show here that SYCE1, a central element protein, assembles in between AEs composed of the sister chromatid cores of univalents in homozygous Stag3 mutant spermatocytes. Our results therefore suggest that STAG3-REC8 cohesin complexes promote synapsis between homologous chromosomes, while the same complexes also inhibit SC assembly to take place between sister chromatids.
Rec8-null, Rad21L-null, and Smc1β-null spermatocytes are eliminated at the zygotene to early pachytene stages of meiosis I (Bannister et al, 2004; Revenkova et al, 2004; Xu et al, 2005; Herran et al, 2011), similar to what we observe for homozygous Stag3 mutant spermatocytes. The situation is more complex when the corresponding mutations in cohesin core subunits are analyzed in oocytes. We find that homozygous Stag3 mutant females are infertile, similar to Rec8-null females and Smc1β-null females, whereas Rad21L-null females are fertile (Bannister et al, 2004; Revenkova et al, 2004; Xu et al, 2005; Herran et al, 2011). Furthermore, homozygous Stag3 mutant oocytes fail to form MLH1 foci, whereas MLH1 foci are formed in Smc1β-null oocytes (Revenkova et al, 2004). The severity of the phenotypes for the different cohesin mutants thus correlates with the extent to which synapsis is being impaired in the mutant oocytes, suggesting that the quality of the synapsis process is carefully monitored during meiosis.
The changes in the temporal and spatial distribution of STAG3, which occur along the meiotic chromosome axis during the prophase I stage (Fig 1), strongly support the notion that STAG3 interacts with the three different α-kleisin subunits present in mammalian meiotic cells (Ishiguro et al, 2011; Lee & Hirano, 2011). Remarkably, while a reduction in STAG3 levels in homozygous mutant spermatocytes has a dramatic effect on the protein levels of REC8, the same is not true for RAD21L and RAD21. Our data suggest that STAG3 in a dosage-dependent manner contributes to the stabilization of REC8 cohesin complexes associated with prophase I chromosomes, a stabilization process that could be regulated by phosphorylation of STAG3 (Prieto et al, 2001; Lee & Hirano, 2011; Fukuda et al, 2012), possibly analogous to the role of STAG2 in regulating cohesin complex dissociation from chromosome arms during mitosis (Hauf et al, 2005; McGuinness et al, 2005). It is not clear how a reduced level of STAG3 selectively affects the stability of REC8, but not the other α-kleisin subunits. However, one possibility is that STAG3 has a lower binding affinity for REC8 rendering this protein unstable at reduced levels of STAG3. The drastic reduction in REC8 protein levels in homozygous Stag3 mutant meiocytes most likely explains the similarity in phenotypes observed between Rec8-null and homozygous Stag3 mutant mice, including female infertility, shortened chromosome axes, impaired synapsis, and accumulation of the SC onto the univalents (Bannister et al, 2004; Xu et al, 2005), phenotypes not observed in meiocytes in Rad21L knockout mice (Herran et al, 2011).
Mouse mutant meiocytes that do not express either RAD21L or REC8 still form axial meiotic chromosome structures, whereas a simultaneous loss of both proteins in meiocytes (that still express RAD21) obliterates meiotic chromosome axis formation (Bannister et al, 2004; Xu et al, 2005; Herran et al, 2011; Llano et al, 2012; Ishiguro et al, 2014). Our results strongly suggest that the presence of a residual level of STAG3 in homozygous Stag3 mutant meiocytes contributes to the formation of STAG3-RAD21L cohesin complexes. The STAG3-RAD21L complexes then support the formation of short cohesin-based meiotic chromosome axes in homozygous mutant meiocytes, axial structures that then in turn promote AE assembly. Our results do not support a compensatory role for STAG1 or STAG2 in the formation of the meiotic chromosome axis in homozygous Stag3 mutant meiocytes. During the revision process of this paper, two publications emerged analyzing the same homozygous Stag3 mutant mouse strain as analyzed by us (Caburet et al, 2014; Llano et al, 2014). It was shown in these papers that homozygous Stag3 mutant female and male mice are infertile and that residual axial structures labeled by cohesin complex proteins and SC proteins remain in mutant meiocytes. The authors of the two studies claim that the homozygous Stag3 mutant mouse model used in their studies represents a complete loss-of-function (null) model, in which meiocytes are deficient for STAG3 expression. This statement is based on Northern blotting analysis of mRNA expression and immunofluorescence staining of meiocytes, neither method sufficiently sensitive to exclude a low level of expression of STAG3. In contrast, we show here using RT-PCR and Western blotting analysis that a low level of STAG3 expression remains in homozygous Stag3 mutant meiocytes. The residual presence of STAG3 in the homozygous Stag3 mutant mice shown in our study is in agreement with the formation of axial structure labeled by cohesin complex proteins and SC proteins in mutant meiocytes. Furthermore, as shown by Winters et al using a Stag3-deficient mouse model (Winters et al, 2014), complete loss of STAG3 expression eliminates formation of the meiotic chromosome axes and the AEs, verifying that STAG3 (but not STAG1 or STAG2) is essential for association of α-kleisins with the meiotic chromosome axes. In summary, our results show that STAG3-REC8 and STAG3-RAD21L cohesin complexes jointly contribute to the formation of the meiotic chromosome axis, that REC8 and RAD21L cohesin complexes display a different dosage-dependent requirement for STAG3, and that STAG3-REC8 cohesin complexes contribute to chromosome axes organization, synapsis, and progression of meiotic recombination.
Materials and Methods
Mice
C56BL/6J was used for the analysis of chromosomal localization of STAG3. The Stag3 mutant mouse line, Stag3TgTn(sb-cHS4, Tyr)2312COve, was obtained from the Mutant Mouse Regional Resource Center (MMRRC; Stock number, 036275-JAX). The line was generated by mutagenesis on an FVB/N background (Paul A Overbeek, Baylor College of Medicine) (Caburet et al, 2014). All mice were used in accordance with regulations provided by the animal ethics committee.
Antibodies
The following antibodies were used: guinea pig anti-SYCP2, anti-SMC1β, anti-STAG3, anti-REC8, and anti-SYCP1 antibodies (Kouznetsova et al, 2005); guinea pig anti-SYCE1, anti-SYCE2, and anti-TEC12 antibodies (Hamer et al, 2006); mouse and rabbit anti-RAD21L antibodies and mouse and rabbit anti-REC8 antibodies (Ishiguro et al, 2011); rabbit anti-SYCP3 antibody (Liu et al, 1996); mouse and rabbit anti-γH2AX antibodies (#05-636 and #07-164) from Millipore; mouse anti-SYCP3 (sc-74569) and rabbit anti-RAD51 (sc-8349) antibodies and goat anti-DMC1 (sc-8973) and anti-TRF1 (sc-5475) antibodies from Santa Cruz Biotechnology; goat anti-STAG1 (ab4457) and anti-STAG2 (ab4463) antibodies and rabbit anti-SMC3 (ab9263), anti-SYCP1 (ab15090), anti-RAD21 (ab154769) and anti-SMC1α (ab21583) antibodies from Abcam; mouse anti-actin (clone SC-40, A4700) antibody from Sigma; rabbit anti-RPA antibody from P. Moens; mouse anti-MLH1 (clone G168-15, 551092) antibody from BD Biosciences; human anit-centromere antiserum (ACA, 15-234-0001) from Antibodies Inc.
RT-PCR
Total RNA was isolated from testes using Trizol reagent (Invitrogen). Equal amounts of the RNA were reverse transcribed into cDNA by oligo-dT using Superscript II reverse transcriptase (Invitrogen). PCR was performed using the primer pairs, Stag3 (5′-GGA CCA TGT CTT TCT CCA GCC-3′ and 5′-TAG AGC TGC TTT AGG CTC AGG-3′) and β-actin as described previously (Fukuda et al, 2010).
Immunoblotting and immunoprecipitation
Testicular and ovarian extracts were prepared as follows. Testes or ovaries were homogenized in a buffer containing 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride (PMSF) and the complete protease inhibitors (Roche). Fractionation of testis extracts and immunoprecipitation were performed as described previously (Fukuda et al, 2012). Proteins were separated on 4–12% NuPAGE Bis-Tris gels (Invitrogen) in MOPS running buffer (Invitrogen) or 4–15% Mini-PROTEAN TGX gels (Bio-Rad) in Tris–glycine running buffer and were subsequently transferred onto PVDF membranes (Millipore or Bio-Rad). Signals were detected with horseradish peroxidase-conjugated secondary antibodies and visualized by ECL Prime (GE Healthcare). The quantitative evaluation of the bands was carried out with Image Lab software (Bio-Rad).
Immunofluorescence staining
For preparation of nuclear spreads, a drying-down technique was used (Peters et al, 1997). For immunohistochemistry, testis and ovary sections were prepared as described previously (Fukuda et al, 2010). Slides were viewed at room temperature using Leica DMRA2 and DMRXA microscopes. Images were captured with a Hamamatsu digital charge-coupled device camera C4742-95 and processed with Openlab 3.1.4 software (Improvision) and Adobe Photoshop. Super-resolution imaging was performed on a Zeiss 780LSM Elyra PS.1 system. Images were captured with an Andor iXon DU 897 camera and a 100x (1.46 NA, oil) objective. Laser wavelengths used were 488, 561 and 642 nm, an EMCCD-gain of 20 and 5 rotations. Images were processed with an integrated Zeiss ELYRA S system software (Zen 2011 SP2 Black). FISH was performed as described previously (Wang & Hoog, 2006). Meiocytes were detected by morphology and SYCP3 staining. We utilized a chromosome painting probe recognizing chromosome 17 (Chrombios) and a point-probe hybridizing with DXMit190 loci located on chromosome X (ID Labs Inc.) according to the manufacturer's instructions.
Acknowledgments
We thank K. Ishiguro and Y. Watanabe for antibodies. We are grateful to S. Valentiniene and J.G. Liu for technical support. We acknowledge support with super-resolution 3DSIM imaging by Hans Blom and Hjalmar Brismar from the National Advanced Light Microscopy facility at Science for Life laboratory. This work was supported by grants from the Swedish Cancer Society, the Swedish Research Council, Torsten och Ragnar Söderbergs Stiftelser, Karolinska Institutet, and the Japan Society for the Promotion of Science (25891015).
Author contributions
TF, NF, and CH conceived the study and wrote the paper. TF, NF, AA, AH, and AK performed the experiments.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Adelfalk C, Janschek J, Revenkova E, Blei C, Liebe B, Gob E, Alsheimer M, Benavente R, de Boer E, Novak I, Hoog C, Scherthan H, Jessberger R. Cohesin SMC1beta protects telomeres in meiocytes. J Cell Biol. 2009;187:185–199. doi: 10.1083/jcb.200808016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis. 2004;40:184–194. doi: 10.1002/gene.20085. [DOI] [PubMed] [Google Scholar]
- Caburet S, Arboleda VA, Llano E, Overbeek PA, Barbero JL, Oka K, Harrison W, Vaiman D, Ben-Neriah Z, Garcia-Tunon I, Fellous M, Pendas AM, Veitia RA, Vilain E. Mutant cohesin in premature ovarian failure. N Engl J Med. 2014;370:943–949. doi: 10.1056/NEJMoa1309635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Smith S. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol. 2009;187:165–173. doi: 10.1083/jcb.200903096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Speed R, Ollinger R, Alsheimer M, Semple CA, Gautier P, Maratou K, Novak I, Hoog C, Benavente R, Cooke HJ. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J Cell Sci. 2005;118:2755–2762. doi: 10.1242/jcs.02402. [DOI] [PubMed] [Google Scholar]
- Eijpe M, Offenberg H, Jessberger R, Revenkova E, Heyting C. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1beta and SMC3. J Cell Biol. 2003;160:657–670. [Google Scholar]
- Fukuda T, Daniel K, Wojtasz L, Toth A, Hoog C. A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Exp Cell Res. 2010;316:158–171. doi: 10.1016/j.yexcr.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Pratto F, Schimenti JC, Turner JM, Camerini-Otero RD, Hoog C. Phosphorylation of chromosome core components may serve as axis marks for the status of chromosomal events during mammalian meiosis. PLoS Genet. 2012;8:e1002485. doi: 10.1371/journal.pgen.1002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Caballero C, Herran Y, Sanchez-Martin M, Suja JA, Barbero JL, Llano E, Pendas AM. Identification and molecular characterization of the mammalian alpha-kleisin RAD21L. Cell Cycle. 2011;10:1477–1487. doi: 10.4161/cc.10.9.15515. [DOI] [PubMed] [Google Scholar]
- Hamer G, Gell K, Kouznetsova A, Novak I, Benavente R, Hoog C. Characterization of a novel meiosis-specific protein within the central element of the synaptonemal complex. J Cell Sci. 2006;119:4025–4032. doi: 10.1242/jcs.03182. [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herran Y, Gutierrez-Caballero C, Sanchez-Martin M, Hernandez T, Viera A, Barbero JL, de Alava E, de Rooij DG, Suja JA, Llano E, Pendas AM. The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J. 2011;30:3091–3105. doi: 10.1038/emboj.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro K, Kim J, Fujiyama-Nakamura S, Kato S, Watanabe Y. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep. 2011;12:267–275. doi: 10.1038/embor.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro K, Kim J, Shibuya H, Hernandez-Hernandez A, Suzuki A, Fukagawa T, Shioi G, Kiyonari H, Li XC, Schimenti J, Hoog C, Watanabe Y. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev. 2014;28:594–607. doi: 10.1101/gad.237313.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger R. Cohesin complexes get more complex: the novel kleisin RAD21L. Cell Cycle. 2011;10:2053–2054. doi: 10.4161/cc.10.13.15802. [DOI] [PubMed] [Google Scholar]
- Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Kouznetsova A, Novak I, Jessberger R, Hoog C. SYCP2 and SYCP3 are required for cohesin core integrity at diplotene but not for centromere cohesion at the first meiotic division. J Cell Sci. 2005;118:2271–2278. doi: 10.1242/jcs.02362. [DOI] [PubMed] [Google Scholar]
- Kouznetsova A, Wang H, Bellani M, Camerini-Otero RD, Jessberger R, Hoog C. BRCA1-mediated chromatin silencing is limited to oocytes with a small number of asynapsed chromosomes. J Cell Sci. 2009;122:2446–2452. doi: 10.1242/jcs.049353. [DOI] [PubMed] [Google Scholar]
- Kouznetsova A, Benavente R, Pastink A, Hoog C. Meiosis in mice without a synaptonemal complex. PLoS ONE. 2011;6:e28255. doi: 10.1371/journal.pone.0028255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugou K, Fukuda T, Yamada S, Ito M, Sasanuma H, Mori S, Katou Y, Itoh T, Matsumoto K, Shibata T, Shirahige K, Ohta K. Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol Biol Cell. 2009;20:3064–3076. doi: 10.1091/mbc.E08-12-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Iwai T, Yokota T, Yamashita M. Temporally and spatially selective loss of Rec8 protein from meiotic chromosomes during mammalian meiosis. J Cell Sci. 2003;116:2781–2790. doi: 10.1242/jcs.00495. [DOI] [PubMed] [Google Scholar]
- Lee J, Hirano T. RAD21L, a novel cohesin subunit implicated in linking homologous chromosomes in mammalian meiosis. J Cell Biol. 2011;192:263–276. doi: 10.1083/jcb.201008005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JG, Yuan L, Brundell E, Bjorkroth B, Daneholt B, Hoog C. Localization of the N-terminus of SCP1 to the central element of the synaptonemal complex and evidence for direct interactions between the N-termini of SCP1 molecules organized head-to-head. Exp Cell Res. 1996;226:11–19. doi: 10.1006/excr.1996.0197. [DOI] [PubMed] [Google Scholar]
- Llano E, Herran Y, Garcia-Tunon I, Gutierrez-Caballero C, de Alava E, Barbero JL, Schimenti J, de Rooij DG, Sanchez-Martin M, Pendas AM. Meiotic cohesin complexes are essential for the formation of the axial element in mice. J Cell Biol. 2012;197:877–885. doi: 10.1083/jcb.201201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano E, Gomez-H L, Garcia-Tunon I, Sanchez-Martin M, Caburet S, Barbero JL, Schimenti J, Veitia RA, Pendas AM. STAG3 is a strong candidate gene for male infertility. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu051. doi: 10.1093/hmg/ddu051. [DOI] [PubMed] [Google Scholar]
- McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Doll E, Yamamoto A, Hiraoka Y, Kohli J. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J Cell Sci. 2003;116:1719–1731. doi: 10.1242/jcs.00387. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, Hoog C. Cohesin Smc1beta determines meiotic chromatin axis loop organization. J Cell Biol. 2008;180:83–90. doi: 10.1083/jcb.200706136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Pelttari J, Hoja MR, Yuan L, Liu JG, Brundell E, Moens P, Santucci-Darmanin S, Jessberger R, Barbero JL, Heyting C, Hoog C. A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol Cell Biol. 2001;21:5667–5677. doi: 10.1128/MCB.21.16.5667-5677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Pezzi N, Prieto I, Kremer L, Perez Jurado LA, Valero C, Del Mazo J, Martinez AC, Barbero JL. STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking the Williams-Beuren syndrome deletion. FASEB J. 2000;14:581–592. doi: 10.1096/fasebj.14.3.581. [DOI] [PubMed] [Google Scholar]
- Prieto I, Suja JA, Pezzi N, Kremer L, Martinez AC, Rufas JS, Barbero JL. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat Cell Biol. 2001;3:761–766. doi: 10.1038/35087082. [DOI] [PubMed] [Google Scholar]
- Prieto I, Pezzi N, Buesa JM, Kremer L, Barthelemy I, Carreiro C, Roncal F, Martinez A, Gomez L, Fernandez R, Martinez AC, Barbero JL. STAG2 and Rad21 mammalian mitotic cohesins are implicated in meiosis. EMBO Rep. 2002;3:543–550. doi: 10.1093/embo-reports/kvf108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Carretero M, Martinez P, Drosopoulos WC, Canamero M, Schildkraut CL, Blasco MA, Losada A. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Eijpe M, Heyting C, Gross B, Jessberger R. Novel meiosis-specific isoform of mammalian SMC1. Mol Cell Biol. 2001;21:6984–6998. doi: 10.1128/MCB.21.20.6984-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- Schramm S, Fraune J, Naumann R, Hernandez-Hernandez A, Hoog C, Cooke HJ, Alsheimer M, Benavente R. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet. 2011;7:e1002088. doi: 10.1371/journal.pgen.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Severson AF, Ling L, van Zuylen V, Meyer BJ. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 2009;23:1763–1778. doi: 10.1101/gad.1808809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries FA, de Boer E, van den Bosch M, Baarends WM, Ooms M, Yuan L, Liu JG, van Zeeland AA, Heyting C, Pastink A. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005;19:1376–1389. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hoog C. Structural damage to meiotic chromosomes impairs DNA recombination and checkpoint control in mammalian oocytes. J Cell Biol. 2006;173:485–495. doi: 10.1083/jcb.200512077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters T, McNicoll F, Jessberger R. Meiotic cohesin STAG3 is required for chromosome axis formation and sister chromatid cohesion. EMBO J. 2014;33:1256–1270. doi: 10.1002/embj.201387330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.