Abstract
Database searching has allowed the identification of a number of previously unreported single and multidomain isoform members of the Arabidopsis cyclophilin gene family. In addition to the cyclophilin-like peptidyl-prolyl cis-trans isomerase domain, the latter contain a variety of other domains with characterized functions. Transcriptional analysis showed they are expressed throughout the plant, and different isoforms are present in all parts of the cell including the cytosol, nucleus, mitochondria, secretory pathway, and chloroplast. The abundance and diversity of cyclophilin isoforms suggests that, like their animal counterparts, plant cyclophilins are likely to be important proteins involved in a wide variety of cellular processes. As well as fulfilling the basic role of protein folding, they may also play important roles in mRNA processing, protein degradation, and signal transduction and thus may be crucial during both development and stress responsiveness.
Cyclophilins are ubiquitous proteins (Galat, 1999) present in all subcellular compartments, which are involved in a wide variety of processes including protein trafficking and maturation (Shieh et al., 1989; Ferreira et al., 1996), receptor complex stabilization (Leverson and Ness, 1998), apoptosis (Lin and Lechleiter, 2002), receptor signaling (Brazin et al., 2002; Yurchenko et al., 2002), RNA processing (Krzywicka et al., 2001), and spliceosome assembly (Bourquin et al., 1997; Mortillaro and Berezney, 1998; Horowitz et al., 2002). Cyclophilin A was first identified and purified while screening for the intracellular protein target of cyclosporin A (CsA), a fungal metabolite with potent immunosuppressive activity (Handschumacher et al., 1984). Cyclophilins belong to a family of immunosuppressant receptors called immunophilins, which also includes the FK506-binding proteins (FKBPs; Harding et al., 1989) and the parvulins (Dolinski and Heitman, 1997). Immunophilin-drug complexes block T-cell activation by inhibiting the phosphatase activity of calcineurin, resulting in inhibition of T-cell activation by blocking the expression of a number of immunoresponsive genes (Molkentin et al., 1998). Immunophilins possess peptidyl-prolyl cis-trans isomerase (PPiase) activity, catalyzing the rotation of X-Pro peptide bonds from a cis to trans conformation, a rate-limiting step in protein folding (Brandts et al., 1975) and of widespread importance since over 90% of proteins contain trans prolyl imide bonds (Stewart et al., 1990).
Plant cyclophilins were first identified in 1990 with the isolation of cyclophilin cDNA sequences from tomato (Lycopersicon esculentum), maize (Zea mays), and oilseed rape (Brassica napus; Gasser et al., 1990). To date, 14 cyclophilin-like proteins have been reported in Arabidopsis: 5 cytoplasmic cyclophilins (rotamase cyclophilin [ROC] 1-3, ROC5-6; Hayman and Miernyk, 1994; Chou and Gasser, 1997), 2 endoplasmic reticulum (ER) localized isoforms (ROC7 and AtCYP5; Jackson and Soll, 1999; Grebe et al., 2000), 6 chloroplast isoforms (Lippuner et al., 1994; Peltier et al., 2002; Schubert et al., 2002), and the Arabidopsis homolog of human Cyclophilin 40 (Cyp40; Berardini et al., 2001). ROC1-7 predicted proteins range in size from 18 to approximately 28 kD and show significant sequence identity, ranging from 60% to 90% between members. While only one chloroplast cyclophilin, ROC4, has been shown to be targeted to the stromal compartment (Lippuner et al., 1994; Schubert et al., 2002), the five remaining chloroplast isoforms have been isolated in independent proteomic analyses of the thylakoid lumen (Peltier et al., 2002; Schubert et al., 2002); among these is the Arabidopsis homolog of the previously characterized spinach (Spinacia oleracea) cyclophilin TLP40 (Fulgosi et al., 1998; Vener et al., 1999).
With the exception of ROC4, which is only expressed in photosynthetic tissue, a number of reported cyclophilins show ubiquitous spatial expression patterns (Lippuner et al., 1994; Chou and Gasser, 1997; Jackson and Soll, 1999; Berardini et al., 2001). Furthermore, cyclophilin expression has been shown to be induced by both biotic and abiotic stresses including HgCl2 (Marivet et al., 1992), viral infection, ethephon (an ethylene releaser), salicylic acid, salt stress, heat and cold shock (Marivet et al., 1994, 1995; Scholze et al., 1999; Godoy et al., 2000), light (Chou and Gasser, 1997; Luan et al., 1994), drought (Sharma and Singh, 2003), wounding, fungal infection, abscisic acid, and methyl jasmonate (Godoy et al., 2000; Kong et al., 2001).
Yeast 2-hybrid experiments have allowed the identification of specific interactions between plant cyclophilins and a number of proteins. ROC7 binds the regulatory subunit of the protein phosphatase 2A (PP2A) RCN1; however, unlike PP2B, PP2A activity is not affected by CsA (Jackson and Soll, 1999). ROC6 binds strongly and specifically to the Agrobacterium tumefaciens T-complex trafficking protein VirD2, suggesting a potential role of cyclophilins in T-DNA transfer (Deng et al., 1998). Furthermore, CsA inhibits A. tumefaciens-mediated transformation of both Arabidopsis and tobacco (Nicotiana tabacum; Deng et al., 1998). AtCYP5 interacts with GNOM, a GTP exchange factor that acts on ADP ribosylation factor-type G-proteins, an important component required for the coordination of cell polarity along the apical-basal embryo axis (Grebe et al., 2000). The chloroplast localized cyclophilin ROC4 binds thioredoxin m in vitro (Motohashi et al., 2001). Thus far however, the only in vivo evidence of cyclophilin function comes from analysis of a loss-of-function mutant of Arabidopsis Cyp40. The Cyp40 protein has been identified in yeast (Mayr et al., 2000), mammals, and plants (Owens-Grillo et al., 1996) and consists of a conserved N-terminal cyclophilin domain and a C-terminal tetratricopeptide (TPR) domain; the TPR domain has been shown to be required for association with the Hsp90 chaperone complex (Owens-Grillo et al., 1995, 1996; Duina et al., 1996). The Cyp40 mutant shows both a reduction in leaf number and an alteration in leaf morphology and is thus functionally implicated in the vegetative maturation of the shoot (Berardini et al., 2001).
By far the best-characterized plant cyclophilin is the atypical multidomain spinach thylakoid lumen cyclophilin TLP40. TLP40 is a 40 kD lumenal protein that contains a poorly conserved carboxy-terminal cyclophilin domain, a predicted amino-terminal Leu zipper and a phosphatase-binding domain. TLP40 plays a key role in turnover of the D1 photosystem II protein by regulating its dephosphorylation (Fulgosi et al., 1998; Vener et al., 1999).
Progress has also been made in the characterization of the FKBP-type plant PPiases; chloroplast localized FKBP13 may be involved in sequestering the Rieske subunit of the cytochrome b6f complex (Gupta et al., 2002), whereas FKBP42 (Kamphausen et al., 2002) and FKBP72 (Carol et al., 2001; Faure et al., 1998; Vittorioso et al., 1998) mutants show marked developmental abnormalities. It is clear therefore that PPiases play a number of important roles in plant biology, but we do not yet know the number of genes, their sequences, or their expression patterns. Analysis of the Arabidopsis genome sequence has revealed that the FKBP-type gene family is composed of 22 members, including both single and multidomain isoforms (Schubert et al., 2002). In the study presented here, database searches were carried out to determine the extent of the Arabidopsis cyclophilin gene family. This paper describes the identification, classification, and transcriptional analysis of novel genes encoding cyclophilin-like proteins which, combined with the previously identified isoforms, constitute the largest cyclophilin gene family so far described in any organism, consisting of 29 members.
RESULTS
The Arabidopsis Genome Contains a Large Cyclophilin Gene Family
A comprehensive database survey (see “Materials and Methods”) identified a further 15 genes encoding putative proteins containing cyclophilin-like domains in addition to the 14 Arabidopsis cyclophilin genes that have been previously reported. Cyclophilin isoforms previously identified by a proteomic approach (Peltier et al., 2002; Schubert et al., 2002) were also analyzed to verify their predicted coding regions. To conform to nomenclature adopted for this family with that used in other organisms, we propose an alteration in the gene name from ROC (Chou and Gasser, 1997; Jackson and Soll, 1999) to AtCYP, with specific designation provided by the molecular weight, and genes encoding isoforms with similar molecular weight numbered appropriately. In the case of proteins possessing cleavable signal peptides, the nomenclature represents the molecular weight of the mature protein predicted by TargetP. Following experimental elucidation of the mature molecular weight, nomenclature may be adjusted in the future.
Where possible, sequence analysis was carried out to confirm predicted cyclophilin gene cDNA sequences present on the Munich Information Center for Protein Sequences (MIPS) database. Expressed sequence tag (EST) cDNAs were obtained for 11 cyclophilins, 4 full-length cDNAs were generated by reverse transcription (RT)-PCR, and 5 sequences were deduced from predicted cDNA/mRNA sequences present within the MIPS database (see “Materials and Methods”). For the majority of intron-containing genes, the predicted splice sites were correct on the MIPS database, but a number of genes were erroneously annotated; in particular, AtCYP18-1, AtCYP21-2, AtCYP37, AtCYP57, AtCYP59, AtCYP71, and AtCYP95 annotations contained predicted splice site errors resulting in deletions and out of frame mutations (data not shown). Corrected sequences and novel gene nomenclatures were reported to the MIPS database.
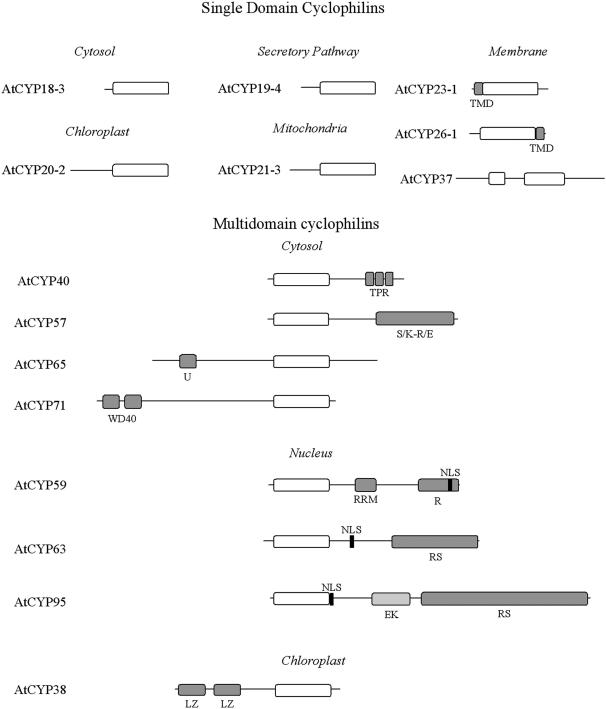
Table I shows the classification of the Arabidopsis cyclophilin gene family indicating, where applicable, previous gene designations. All gene sequences were identified from the Columbia genome sequence, the accession used by the Arabidopsis Genome Initiative. ATCYCP, a cyclophilin isolated and sequenced from a Landsberg erecta derived cDNA library (Bartling et al., 1992) whose authenticity has been disputed (Chou and Gasser, 1997), was not present within the MIPS database and is therefore not included within this classification. To rationalize the classification, all previously reported sequences (Saito et al., 1995; Chou and Gasser, 1997; Jackson and Soll, 1999; Berardini et al., 2001) have been appropriately renamed and have been further subdivided into single and multidomain isoforms. The single domain cyclophilins are characterized by a conserved cyclophilin domain, whereas the multidomain members contain additional conserved domains that have been characterized in other proteins (Fig. 1).
Table I.
Nomenclature, accessions, and available resources of the Arabidopsis cyclophilin family
| Namea | AGI IDb | kD /aa | ESTsc | BACd | Subcellular Location/ Target Pe | Alignedf | RFWH/ PPiaseg | % Identity/ Similarityh |
|---|---|---|---|---|---|---|---|---|
| Single Domain Cyclophilins | ||||||||
| AtCYP18-1 | At1g01940 | 17.5/160 | 2 | AC020622 | Cytosol | Full | RFWY/− | 53/67 |
| AtCYP18-2 | At2g36130 | 18.2/164 | 3 | AC007135 | Cytosol | Full | RFSH/− | 53/65 |
| AtCYP18-3 (ROC1)1,2,3 | At4g38740 | 18.4/172 | 42 | AL035656 | Cytosol | Full | RFWH/− | 67/79 |
| AtCYP18-4 (ROC5,AtCYP1)1,4 | At4g34870 | 18.4/172 | 47 | AL079347 | Cytosol | Full | RFWH/− | 69/80 |
| AtCYP19-1 (ROC3)1 | At2g16600 | 18.5/173 | 18 | AC005825 | Cytosol | Full | RFWH/− | 68/79 |
| AtCYP19-2 (ROC6)1,3 | At2g21130 | 18.5/174 | 2 | AL138642 | Cytosol | Full | RFWH/− | 67/80 |
| AtCYP19-3 (ROC2)1 | At3g56070 | 18.9/176 | 2 | AL391711 | Cytosol | Full | RFWH/− | 70/76 |
| AtCYP19-4 (Cyp5)5,6 | At2g29960 | 21.5/201 | 4 | AL161586 | SP (0.98) | Full | RFWH/− | 62/72 |
| AtCYP20-1 (ROC7)7 | At5g58710 | 21.9/204 | 7 | AB020755 | SP (0.95) | Full | RFWH/− | 62/71 |
| AtCYP20-28 | At5g13120 | 28.3/259 | 10 | AL391711 | TL (0.96) | Full | RFWH/+ | 65/76 |
| AtCYP20-3 (ROC4)1,9 | At3g62030 | 28.2/260 | 21 | AL138642 | Stroma (0.93) | Full | RFWH/+ | 67/73 |
| AtCYP21-1 | At4g34960 | 24.6/224 | 3 | AL022023 | SP (0.97) | Full | RFWH/− | 52/67 |
| AtCYP21-2 | At3g55920 | 24.5/228 | 0 | AL163832 | SP (0.98) | Full | RFWH/− | 59/70 |
| AtCYP21-3 | At2g47320 | 26/230 | 9 | AC002337 | Mitochondria (0.71) | Full | RKDL/− | 27/48 |
| AtCYP21-4 | At3g66654 | 26.4/236 | 8 | AC036106 | Mitochondria (0.75) | Full | RKDL/− | 24/43 |
| AtCYP22-1 | At2g38730 | 21.5/199 | 2 | AC005499 | Cytosol | Full | RFWH/− | 55/68 |
| AtCYP23-1 | At1g26940 | 25.5/226 | 2 | AC005508 | SP (0.97) | 1-203 | RRHY/− | 28/40 |
| AtCYP26-1 | At3g22920 | 26/232 | 0 | AP001300 | Cytosol | 1-175 | HLQH/− | 46/59 |
| AtCYP26-2 | At1g74070 | 34.4/317 | 1 | AC016662 | TL (0.82) | 1-297 | KVEV/− | 24/38 |
| AtCYP288 | At5g35100 | 31.4/290 | 6 | AF058825 | TL (0.59) | 1-254 | KLQN/− | 29/43 |
| AtCYP378 | At3g15520 | 45.1/461 | 5 | AC024081 | TL (0.82) | 1-308 | LSLL/− | 7/8 |
| Multidomain Cyclophilins | ||||||||
| AtCYP388 | At3g01480 | 48/437 | 10 | AC009325 | TL (0.97) | 243-437 | REIY/+ | 8/10 |
| AtCYP40 (CYP40)10 | At2g15790 | 40.5/361 | 0 | AC018722 | Cytosol | 1-181 | RNHH/− | 56/69 |
| AtCYP57 | At4g33060 | 57.1/504 | 3 | AL031804 | Cytosol | 1-179 | RFWH/− | 48/64 |
| AtCYP59 | At1g53720 | 58.8/506 | 1 | AC024260 | Nucleus | 1-170 | TFYH/− | 40/53 |
| AtCYP63 | At3g63400 | 63.5/566 | 5 | AL163818 | Nucleus | 1-182 | RFHH/− | 60/71 |
| AtCYP65 | At5g67530 | 65/595 | 6 | AB013390 | Cytosol | 335-500 | RNHH/− | 48/62 |
| AtCYP71 | At3g44600 | 70.7/631 | 3 | AL353818 | Cytosol | 466-631 | RNWH/− | 51/68 |
| AtCYP95 | At4g32420 | 94.6/837 | 9 | AL034567 | Nucleus | 1-171 | RSQN/− | 46/60 |
Original nomenclature shown in parentheses. References: 1, Chou and Gasser (1997); 2, Lippuner et al. (1994); 3, Deng et al. (1998); 4, Hayman and Miernyk (1994); 5, Saito et al. (1995); 6, Grebe et al. (2000); 7, Jackson and Soll (1999); 8, Schubert et al. (2002); 9, Motohashi et al. (2001); 10, Berardini et al. (2001).
Systematic nomenclature provided by MIPS (http://mips.gsf.de/proj/thal/) from the AGI sequencing project.
Number of ESTs in the MIPS database is indicated.
Bacterial artificial chromosome (BAC) numbers are for genomic DNA deposited in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/).
Subcellular location and prediction score obtained using TargetP (http://www.cbs.dtu.dk/services/TargetP/; SP, secretory pathway; TL, thylakoid lumen), and PredictNLS (http://cubic.bioc.columbia.edu/cgi/var/nair/resonline.pl).
Refers to Figure 2.
Amino acid present at positions required for PPiase activity and CsA binding (Arg-225, Phe-230, Trp-332, His-337; see Fig. 2) experimentally determined PPiase activity.
Values are compared to human cyclophilin A.
Figure 1.
Primary domain structure of representative isoforms of the SD and MD Arabidopsis cyclophilin proteins. TMD, transmembrane domain; TPR, tetratricopeptide repeat; S/K-R/E, Ser/Lys-Arg/Glu-rich region; U, U-box domain; WD40, WD40 repeat; RRM, RNA recognition motif; R, Arg- rich region; NLS, nuclear localization signal; EK, Glu-Lys-rich region; RS, Arg-Ser rich region; LZ, Leu zipper.
Single Domain Cyclophilins
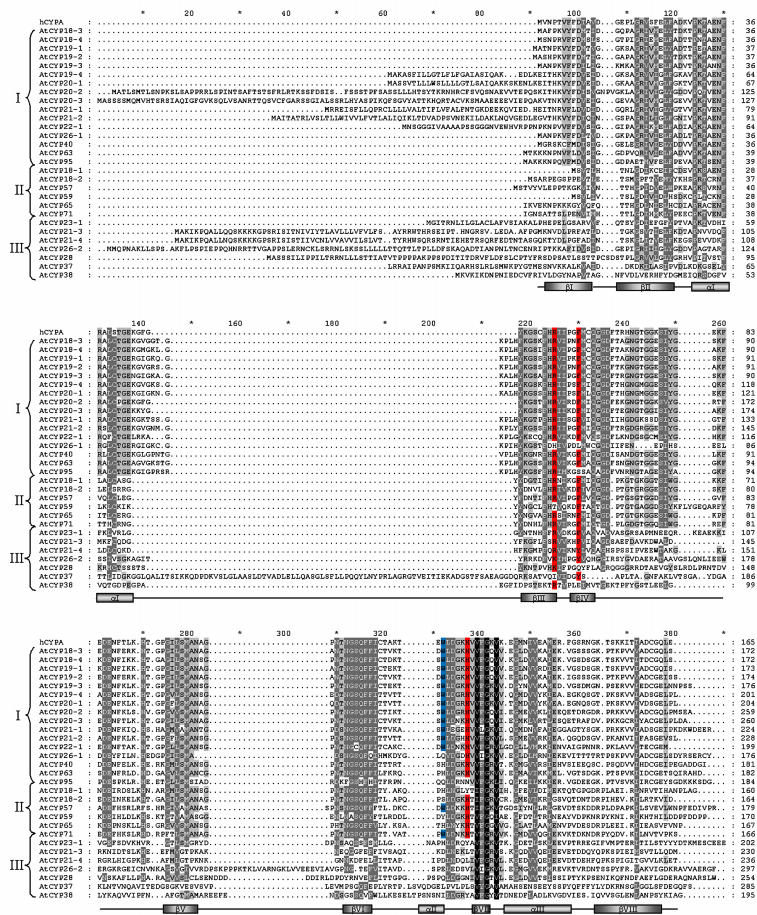
Figure 2 shows a multiple sequence alignment of the cyclophilin domains of both single domain (SD) and multidomain (MD) members of the cyclophilin gene family; truncated versions of a number of proteins were used to improve the alignment, the exact region of aligned sequence being indicated in Table I. Sequence and secondary structural details of the extensively characterized human cyclophilin A isoform (hCYPA) are shown as an external reference for comparison to the Arabidopsis cyclophilin sequences.
Figure 2.
Multiple alignment of the deduced protein sequences of the Arabidopsis cyclophilin family. Multidomain isoform sequences have been truncated to improve the alignment (see Table I for details of sequence used). Backgrounds indicate percentage of amino acid similarity: black, 100%; dark gray, 80%; light gray, 60%. Amino acids in red represent those required for PPiase catalysis, those in blue for CsA binding (Zydowsky et al., 1992). Secondary structure features are those of human Cyclophilin A (Kallen et al., 1991). Roman numerals on the left-hand side correspond to the cladistic subdivision used in Figure 3.
The SD cyclophilins contain a single conserved cyclophilin domain and are further distinguished by the absence (AtCYP18-1, AtCYP18-2, AtCYP18-3, AtCYP18-4, AtCYP19-1, AtCYP19-2, and AtCYP19-3) or presence (AtCYP19-4, AtCYP20-1, AtCYP20-2, AtCYP20-3, AtCYP21-1, AtCYP21-2, AtCYP21-3, AtCYP21-4, AtCYP23-1, AtCYP28, AtCYP26-2, and AtCYP37) of an N-terminal targeting sequence. AtCYP23-1 and AtCYP26-1 represent unique isoforms within the SD cyclophilins, as they possess predicted transmembrane domains located at the N- and C-termini, respectively (Fig. 1).
Considerable sequence identity exists between AtCYP18-3 and AtCYP19-2 and between AtCYP18-4 and AtCYP19-1 at both the DNA and protein level. As previously suggested (Chou and Gasser, 1997), these pairs are likely to have resulted from a gene duplication event. The majority of the SD cyclophilins contain an 8 to 11 amino acid insertion in the surface exposed extended loop region (Fig. 3A) located between α-helix-I and β-sheet-III (hereafter referred to as the α-I/β-III junction); however, AtCYP18-1, AtCYP18-2, AtCYP21-3, and AtCYP21-4 lack this insertion. Within the α-I/β-III junction, the SD chloroplast isoforms AtCYP20-2, AtCYP20-3, AtCYP26-2, and AtCYP28 possess short insertions of only 3 to 4 residues. AtCYP26-2 and AtCYP28 constitute the most significantly diverged SD cyclophilin isoforms, sharing only 24% to 29% sequence identity with hCYPA; this is largely due to the amino acid insertions 251 to 258 and the residues located between β-sheets V and VI. AtCYP37 is the largest and least conserved SD isoform showing poor sequence conservation throughout the cyclophilin domain (7% identity to hCYPA) as well as a unique 75 amino acid insertion located within the α-I/β-III junction.
Figure 3.
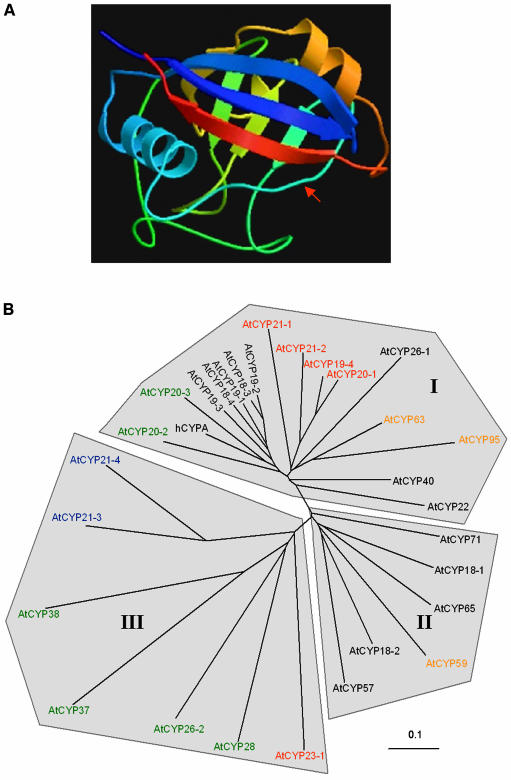
A, Structure of human cyclophilin A as elucidated by x-ray diffraction analysis. The variable insertion region corresponding to the α-I/β-III junction is indicated by the red arrow (Kallen et al., 1991; Ke, 1992; image obtained from the Swiss-3D image database at the ExPASY website [http://ca.expasy.org]). B, Unrooted cladogram of the Arabidopsis cyclophilin protein family and human cyclophilin A obtained using the sequence alignment of Figure 2. The branch lengths are proportional to divergence, with the scale of 0.1 representing 10% change. Colors represent predicted cellular location (black, cytosol; green, chloroplast; blue, mitochondria; red, secretory pathway; orange, nucleus).
The neural-network-based targeting prediction program TargetP is a powerful tool that allows the discrimination of subcellular destination based on conserved sequence features present within the N terminus of a given protein. Thirteen out of twenty-one SD cyclophilin isoforms are predicted to be targeted to distinct cellular subcompartments; their predicted location and TargetP scores are shown in Table I. The N-terminal sequences of AtCYP20-2 and AtCYP20-3 have properties of known chloroplast transit peptides, being rich in both Ser and Ala residues and deficient in acidic residues such as Asp and Glu (von Heijne et al., 1989). Both isoforms score highly for targeting to the chloroplast, theoretical data having been experimentally corroborated for both proteins (Lippuner et al., 1994; Schubert et al., 2002). AtCYP26-2 and AtCYP28 both possess N-terminal twin Arg motifs followed by a number of basic residues, which are transit peptide sequence characteristics of proteins targeted to the lumenal compartment of the thylakoid membrane via the TAT pathway (Schnell, 1998). While AtCYP26-2 shows a high localization prediction score for targeting to the thylakoid lumen, only AtCYP28 and AtCYP37 have been isolated from this compartment (Schubert et al., 2002). TargetP predicts high scores for ER localization (Emanuelsson et al., 2000) of the signal peptide containing isoforms AtCYP19-4, AtCYP20-1, AtCYP21-1, AtCYP21-2, and AtCYP23-1 (Table I) while AtCYP21-3 and AtCYP21-4 are predicted to be mitochondrial proteins. The high degree of sequence identity shared by AtCYP21-3 and AtCYP21-4 within the N-terminal presequence suggests that one may have occurred following gene duplication of the other. Although AtCYP22 also contains an N-terminal extension, it did not score highly for targeting to any subcellular compartment. A potentially significant feature of the N-terminal signal peptides of the predicted secretory pathways (SP) cyclophilins is the presence of a number of Leu residues; within the first 35 residues, AtCYP20-1, AtCYP21-1, and AtCYP21-2 contain between 8 and 11 such amino acids. Human CypB also contains a Leu-rich N terminus and has been shown to localize to both the SP and nucleus (Price et al., 1994).
Multidomain Cyclophilins
Nine Arabidopsis genes encode putative MD cyclophilins. AtCYP40 is the only Arabidopsis cyclophilin whose function has been demonstrated in vivo using a reverse genetics approach; its primary structure is characterized by three C-terminal tetratricopeptide repeats and a conserved cyclophilin domain (Berardini et al., 2001; Fig. 2). AtCYP38 is the Arabidopsis homolog of the extensively characterized spinach TLP40 (Fulgosi et al., 1998).
In addition to AtCYP38 and AtCYP40, six MD cyclophilins with complex deduced primary amino acid sequences were identified. Four of these (AtCYP57, AtCYP59, AtCYP63, and AtCYP95) contain sequence motifs characteristic of RNA-interacting proteins, such as the RNA recognition motif (RRM), the Glu-Lys (EK) domain, the Arg/Ser (RS)-rich domain, and the S/K-R/E-rich region (see Fig. 1; Birney et al., 1993; Fu, 1995; Weighardt et al., 1999), suggesting that cyclophilins may be involved in the regulation of RNA processing.
The smallest, AtCYP57, possesses an N-terminal cyclophilin domain, a charged central domain rich in Asp and Glu residues, and a C-terminal S/K-R/E-rich region (Weighardt et al., 1999). The deduced AtCYP59 protein belongs to the previously described family of cyclophilin-RNA interacting proteins or CRIPS that are conserved in fission yeast, plants, and metazoans (Krzywicka et al., 2001). In addition to the N-terminal cyclophilin domain, AtCYP59 possesses a central RRM domain and a charged C-terminal region rich in Arg residues. The presence of a nuclear localization signal (Mattaj and Englmeier, 1998) suggests AtCYP59 is a nuclear protein. AtCYP63 and AtCYP95 show considerable similarities within their domain organization; a central Ser-rich region separates the N-terminal cyclophilin domain and the C-terminal RS region. Both these isoforms are also characterized by the presence of a nuclear localization signal adjacent to the cyclophilin domain. Detailed analysis of AtCYP95 reveals the presence of up to 15 conserved S[R/K]SPXRX repeats (not shown) separated by Arg and Ser residues within the RS domain, as well as a central EK-domain rich in Glu and Lys residues.
In addition to the cyclophilin domain, AtCYP65 contains a 60 amino acid C-terminal region possessing significant homology to the U-box domain, a highly conserved modified RING-finger domain lacking the full complement of zinc binding residues, and present in the C-terminus of the yeast E4 ubiquitination factor UFD2 (Johnson et al., 1995). Analysis of the AtCYP71 predicted protein sequence revealed the presence of two N-terminal WD40 repeats similar to those found in the photomorphogenesis related protein COP1 (Schwechheimer and Deng, 2001).
Phylogenetic Analysis of the Arabidopsis Cyclophilins
The structure of human cyclophilin A (Fig. 3A) consists of an eight-antiparallel stranded β-barrel capped by two well-defined α-helices (Kallen et al., 1991). Mutagenesis studies have identified Trp-121 (Trp-332; Fig. 2) as a residue required for CsA binding but not involved in PPiase enzymatic activity (Liu et al., 1991; Zydowsky et al., 1992). Moreover, mutations in Arg-55 (Arg-225; Fig. 2), Phe-60 (Phe-230; Fig. 2), and His-126 (His-337; Fig. 2) result in less than 1% of the isomerase activity of the wild-type enzyme (Zydowsky et al., 1992). Exploring the phylogenetic relationships between cyclophilin proteins allows prediction of how sequence divergence may have contributed to functional specification. In particular, existing data obtained by mutagenesis allows the prediction of those isoforms that are likely to have retained PPiase activity and CsA-binding capability.
While Arabidopsis cyclophilins show significant variation in primary structure (7%–70% identical to hCYPA), the hydrophobic region located in and around β-sheet VII constitutes a universally conserved region of the catalytic domain. Compared to cyclophilin sequences analyzed from a wide variety of other organisms (Galat, 1999), unique features of plant cyclophilins include insertions within β-I/β-II, β-IV/βV (Fig. 2, residues 251–258), and βVI/α-II junctions. Figure 3B shows an unrooted cladogram generated from the sequence alignment of Figure 2. The phylogenetic tree can be primarily subdivided into three clades, distinguished by the sequence variability within a number of key motifs located within the cyclophilin domain.
Clade I
Clade I comprises the 17 cyclophilin isoforms distributed within the upper half of the phylogenetic tree. From Figure 2, we can see that these isoforms share a number of residues constituting the conserved sequence region which forms a surface exposed extended loop corresponding to the α-I/β-III junction (Kallen et al., 1991; Ke, 1992; Fig. 3A). With the exception of AtCYP20-2 and AtCYP20-3, the closest homologs of hCYPA, all clade I isoforms possess 9 to 11 amino acids within this region, specifically, between Glu-137 and Phe/Tyr-218. The significant degree of sequence conservation found within this region suggests that it may have been present in early eukaryotes, with subsequent deletions accounting for the observed sequence variations (Chou and Gasser, 1997). Despite a few exceptions, residues important in CsA-binding and PPiase activity are largely conserved in clade I, suggesting that they are catalytically active isoforms. Additional sequence motifs unique to this clade are the highly conserved residues Arg-131, the dipeptide Val-Phe/Tyr-98-99 located within β-sheet I, and the tripeptide Thr-Gly-Glu-135-137, positioned immediately prior to the α-I/β-III junction.
Within clade I, the SD cytoplasmic isoforms AtCYP18-3, AtCYP18-4, AtCYP19-1, AtCYP19-2, and AtCYP19-3 are most closely related to each other and are also closely related to hCYPA (67%–70%/76%–80% identity/similarity); primary sequence features characteristic of these isoforms include residues Thr-123, Cys-232, and Cys-319, the latter also shared by AtCYP20-2 and AtCYP20-3. Two unique amino acid substitutions in AtCYP19-3 (G108K, E128N) account for its divergence. AtCYP20-2 and AtCYP28 share a three residue insertion (105–107), which forms a partially extended loop between β-sheets I and II. Although AtCYP20-2 and AtCYP20-3 share significant sequence homology with the cytoplasmic SD cyclophilins, they also share two conserved amino acids (Glu-88, Thr-94) and the N-terminal signal peptide with the predicted SP isoforms AtCYP19-4, AtCYP20-1, AtCYP21-1, and AtCYP21-2, which form a separate branch within clade I. AtCYP21-1 shows the most divergence within the branch containing the SP cyclophilins due to substitutions in conserved residues, namely G244S, G245S, D334E, and F340L. However, with substitutions in seven highly conserved residues (L133F, G220E, S221C, G234S, G245C, S314C, and G335N), AtCYP22-1 is most phylogenetically diverged within clade I.
Despite considerable sequence conservation within the N terminus, AtCYP26-1 shows significant phylogenetic diversion due to two unique deletions (residues 242–245 and 281–283) and sequence divergence in conserved residues in the region corresponding to β-sheets III and IV, in particular, R225H and F230L substitutions of the amino acids implicated in reaction catalysis. Trp-332 is also substituted for a Gln residue, whereas the third residue implicated in PPiase activity, His-337, is conserved.
Having retained the insertion in the α-I/β-III junction, the three MD isoforms AtCYP40, AtCYP63, and AtCYP95 also group within clade I, but are more diverged, with identity to hCYPA ranging from 46% to 60%. AtCYP40 divergence is due to the substitutions in conserved residues such as V97C, S221N, and W332H, although the three residues required for PPiase activity are conserved. Of the putative nuclear isoforms, AtCYP63 is the most conserved, possessing substitutions W332H and G112Q, whereas AtCYP95 contains extensive conversions throughout conserved regions of the cyclophilin domain (G112E, F230S, M231S, G234A, G250A, A280S, N281I, G283D, T311K, N312F, Q315H, F317H, G335R, R/K336N, H337N, and L/M348K).
Clades II and III
While clade I isoforms form a distinct subdivision within this large gene family, clades II and III may be differentiated by more subtle sequence characteristics. Clade II isoforms show hCYPA identity values ranging from 40% to 53% and comprise the two smallest SD cyclophilin proteins AtCYP18-1 and AtCYP18-2, as well as four MD isoforms (AtCYP57, AtCYP59, AtCYP65, and AtCYP71) that lack the conserved Val-97 and Phe-98 residues characteristic of members of clade I. A number of conserved residues suggest that these isoforms may have evolved relatively recently, in particular Tyr-217, Pro-238, and Thr-338 are present in all members, as well as a distinctive deletion at position 237. With the exception of AtCYP18-1 and AtCYP59, which possess R225T and H337Y substitutions respectively, residues required for cis-trans catalysis have been retained; however, the CsA-binding residue Trp-332 is conserved in only two (AtCYP57 and AtCYP71) of the six isoforms. AtCYP59 is the most divergent member of this clade due to a seven amino acid insertion between positions 250 and 257.
Branch lengths of clade III clearly indicate that it contains the least conserved cyclophilin isoforms, with hCYPA amino acid identity values ranging between 7% and 29%, a result of numerous substitutions of conserved residues throughout the cyclophilin domain. Isoforms forming this group are poorly interrelated and display original sequence features such as the 50 residue insertion within the α-I/β-III junction of AtCYP37, deletions (AtCYP21-3), and insertions in the β-IV/β-V (AtCYP23-1), β-V/β-VI (AtCYP26-2, AtCYP28, AtCYP37, AtCYP38), and β-VI/α-II (AtCYP37 and AtCYP38) junctions. All clade III isoforms except AtCYP37 retain Arg-225; Phe-230 is present in AtCYP21-3 and AtCYP23-1 and substituted by a related tyrosine in AtCYP21-4 and AtCYP26-2. Both the CsA-binding residue Trp-332 and the catalytic residue His-337 are absent in all clade III isoforms. As exemplified by the AtCYP38 spinach homolog TLP40 (Fulgosi et al., 1998), these highly diverged isoforms may have evolved to carry out more specific functions within the cell.
Expression Analysis of the Arabidopsis Cyclophilins
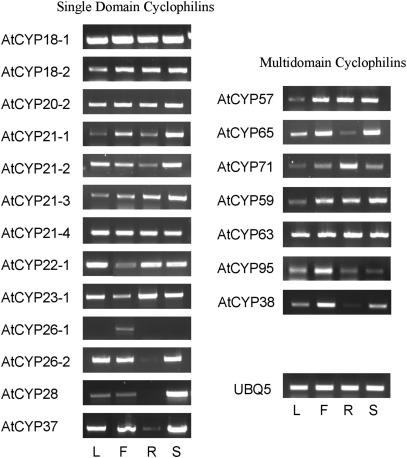
Transcriptional analysis carried out for previously reported Arabidopsis cyclophilin genes shows that they are expressed throughout the plant (Lippuner et al., 1994; Chou and Gasser, 1997; Jackson and Soll, 1999; Berardini et al., 2001). To determine the expression patterns of the newly identified cyclophilin genes, qualitative RT-PCR analysis was carried out on RNA extracted from leaf, flower, root, and stem tissue (Fig. 4). All PCR products were purified and sequenced to verify the specificity of each reaction. The primers and annealing conditions used for each gene are presented in Table II. While AtCYP20-2 is expressed in all tissues, the other chloroplast cyclophilins show considerably reduced expression levels in the root, suggesting that their expression is mainly restricted to photosynthetic tissue. With the exception of AtCYP26-1, which is only expressed in floral tissue, transcripts of the remaining isoforms were identified in all tissues tested.
Figure 4.
RT-PCR analysis of the expression patterns of the newly identified cyclophilin genes in tissues of Arabidopsis (Columbia). Figure 4 shows ethidium bromide-stained gels of PCR amplification using cDNA produced from leaf (L), flower buds (F), roots (R), and stems (S). The UBQ5 gene was used as a control for amplification.
Table II.
Primer pairs and conditions used for RT-PCR analysis of cyclophilin expression
| Gene Name | Primer Sequences (F,R[5′-3′]) | AT |
|---|---|---|
| °C | ||
| AtCYP18-1 | CTTTGCACACGAACCTGGGTGA, CAGCAAGTGGATTGGCGTGGA | 60 |
| AtCYP18-2 | ATGTCGGCAAGACCTGAAGGAA, CTGTCGGTGTTATCGGTTTGGA | 60 |
| AtCYP19-4 | GCCATAGCTTCAATTCAGGCAA, GGAAGTTCTCCACTGTCTGCAA | 60 |
| AtCYP20-2 | CCTTCACACGTCTTATACCA, CCTTCTATTACTTGCCCGAA | 60 |
| AtCYP21-1 | CTCTTCGTCACACTTGCTCTCA, AACATTAGCTTTGGGCACTCCA | 60 |
| AtCYP21-2 | CCGTCTCGGATAAGCATAACCA, TGGCGACTTTGGTTGATACTGA | 60 |
| AtCYP21-3 | AACCTATCGCCATTGGTCCCA, CCTATTGGCGATTTCGGTTGG | 60 |
| AtCYP21-4 | AAGCAAGAAGAAGAAGGGTCCA, TCATCCGTATCCACTTCTTCGA | 60 |
| AtCYP22-1 | CTTCTTCTCGTTGCCCTAACGA, AGGGATCTCTCCAGAATCAGCA | 60 |
| AtCYP23-1 | CCATTGCTAAAGCTCTGCCTCA, TCTCTGTCTTTCGACCTCCACA | 60 |
| AtCYP26-1 | AAACCCGCTGGTCGGATCGTGA, CCACCGCAGTCCGCAATCACCA | 63 |
| AtCYP26-2 | GGTTATGTCCAACACGGCGGAA, CACCAAACCACCATGGCTGCAA | 60 |
| AtCYP28 | GGTCAGTATTTTCTCGCCGGAA, TTTAGCAACCACCAGAGTAGGA | 58 |
| AtCYP37 | GAGATTTCCGCATGCGTCTGCA, GGCGTTCTGTAGAGAGGTTCGA | 60 |
| AtCYP38 | GGCAAGTAGAACATTGCAGCAA, TGTCCCAAAAGCGTTGAAAGGA | 60 |
| AtCYP57 | ATCAAGTGCTCAGCAGAAGGAAGGA, GGCAAGAGATTTTCCAGAC | 55 |
| AtCYP59 | AAGAACTTGGTGCTCAGGAACTGGA, GGACTTCTCCTCTGAATCTTCTCA | 60 |
| AtCYP63 | GTGATGGGAAGCATAGGAAGAGGAA, AGAAGGAGTATGGTACTTCCGAGCA | 60 |
| AtCYP65 | ACAGAATGGGGAGGCGCTAAATCCA, TTCCACCTCCACCATGTAAAGCAA | 60 |
| AtCYP71 | TGCCACCTGTAGTAGAAGAAGAGGA, AATGGGTTCGTTGGAACCAGAACGA | 60 |
| AtCYP95 | AAGTCCTAGATGGAATGGGGGCAGA, CAATCGAATAGGACTCCTAGATCACA | 55 |
F, forward primer; R, reverse primer; AT, annealing temperature.
DISCUSSION
We present results to show that the Arabidopsis genome contains 29 cyclophilin genes, the largest cyclophilin family identified in any organism to date. The encoded cyclophilins have been classified according to whether they have the single cyclophilin domain or whether they possess additional functional domains. Within these two broad classes, cyclophilins have been grouped either according to the possession of conserved sequences that provide clues as to their function, or in terms of their location in the cell. Analysis of sequences of the cyclophilin genes in comparison to related proteins and cyclophilin counterparts in other organisms allows suggestions to be made of their biological function.
Multidomain Cyclophilins Containing RNA-Interaction Domains
In addition to the cyclophilin domain, AtCYP57, AtCYP59, AtCYP63, and AtCYP95 possess domains characteristic of proteins involved in RNA processing. Although they all possess RNA-interacting domains, it is likely that they participate at different stages within the mRNA processing pathway; the NLS signal prediction program shows that only AtCYP59, AtCYP63, and AtCYP95 contain predicted nuclear localization motifs in the form of poly-Arg/Lys extensions. The absence of any NLS-like motifs in AtCYP57 does not discount the possibility that it may be a nuclear protein. The S/K-R/E-rich region found in AtCYP57 has also been found in the hnRNP A1 associated protein (HAP), where it has been shown to play a crucial role in mediating its interaction with the human heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1; Weighardt et al., 1999). Although hnRNP A1 function is mainly associated with the nucleus, it does not possess a classic NLS signal and is shuttled into the cytoplasm following inhibition of transcription (Krecic and Swanson, 1999). Functions related to RNA processing have been previously documented for proteins containing PPiase domains; the human parvulin Par14/EPVH associates with the preribosomal ribonucleoprotein complex (Fujiyama et al., 2002) and the Escherichia coli trigger factor is a 50S ribosome-associated FKBP that catalyzes peptidyl prolyl cis-trans isomerization of nascent polypeptides (Stoller et al., 1995; Scholz et al., 1997). One possibility is that the hnRNP-binding domain present in AtCYP57 may mediate ribosomal association while the cyclophilin domain folds newly translated proteins.
The RRM found in AtCYP59 is composed of 80 to 90 amino acids and exists as one or more copies; it is found in proteins involved in posttranscriptional processes where it mediates binding to various RNAs (pre-mRNA, mRNA, pre-rRNA, cpRNAs, and small nuclear RNAs) to execute both housekeeping functions and regulatory mechanisms (Albà and Pagès, 1998). RRM containing proteins constitute a large gene family in Arabidopsis, being composed of approximately 200 members (Lorkovic and Barta, 2002), but are also found in animals, fungi, and cyanobacteria (Burd and Dreyfuss, 1994). KIN241, an 83-kD Paramecium CRIP, shows significant sequence identity to AtCYP59, is localized in the nucleus, and is involved in cell morphogenesis (Krzywicka et al., 2001).
The RS-rich domains found in AtCYP63 and AtCYP95 are characteristic of SR-proteins (splicing factors containing Ser-Arg repeats; Fig. 1), which are involved in both constitutive and alternative splicing; during the early stages of spliceosome assembly they promote interactions across intron-exon boundaries for selection of appropriate splice sites, as well as mediating the function of spliceosomal enhancers (Fu, 1995; Graveley, 2000). SR-proteins containing cyclophilin domains have been identified in a number of eukaryotes. A conserved mammalian SR-cyclophilin interacts with a CDC28/cdc2-like kinase (Nestel et al., 1996) as well as the phosphorylated C-terminal domain (CTD) of RNA polymerase II (Bourquin et al., 1997); it is present within splicing factor-rich nuclear speckles, components of the nuclear matrix, where it may aid in the formation of macromolecular complexes that associate with elongating RNA polymerase II and assist in the processing of nascent transcripts (Mortillaro and Berezney, 1998). Interestingly, AtCYP95 also possesses an EK-rich region that has been shown to inhibit splicing at early stages of spliceosome assembly (Li et al., 2002), suggesting that AtCYP95 function comprises a complex regulatory mechanism.
The identification of four MD cyclophilins containing motifs related to RNA processing is significant in view of evidence that the Arabidopsis genome encodes over 400 proteins containing Pro-Pro-Arg repeats (Arabidopsis Genome Initiative, 2000), tripeptide sequences characteristic of proteins involved in RNA stabilization and processing; the cyclophilin domain may play an important role in stabilizing these Pro-containing domains. In contrast, yeast, fruitfly (Drosophila melanogaster), and Caenorhabditis elegans have a combined total of 10 proteins containing this signature.
Other MD Cyclophilins
AtCYP65 is unique among the MD cyclophilins in that it contains a predicted C-terminal U-box, suggesting an involvement in the ubiquitination-dependent protein degradation pathway. The U-box is a modified form of the RING-finger motif that lacks metal chelating residues (Freemont, 1993; Saurin et al., 1996) and that mediates the interaction of E4s with E2 ubiquitin conjugated enzymes. Thirty-seven other U-box proteins have been identified in the Arabidopsis genome (Azevedo et al., 2001). The Brassica gene ARC1, encoding the only functionally characterized U-box protein, has been shown to be required for self-incompatibility (Stone et al., 1999). However, a homolog of AtCYP65 has been identified in the nematode Brugia malayi where it is expressed in body wall striated cells in early stages of larval development (Page and Winter, 1998). The human homolog of AtCYP65, designated Cyp60, was identified by its interaction with the C-terminus of the proteinase inhibitor eglin-c and has been shown to be nuclear-localized (Wang et al., 1996), although nothing is known about its precise function.
As well as a cyclophilin domain, AtCYP71 contains two regions within its N terminus, showing significant homology with WD40 repeat consensus sequences. Although many proteins containing WD40 repeats exist in Arabidopsis, very few of them have been characterized. The photomorphogenesis-related protein COP1 contains seven WD40 repeats that mediate interaction with the basic region leucine/zipper (bZIP) transcription factor HY5 to target it for degradation by the 26S proteasome (Schwechheimer and Deng, 2001). The TRANSPARENT TESTA GLABRA 1 protein also contains several WD40 repeats and is required for trichome initiation (Szymanski et al., 2000). Database searches reveal the presence of human and yeast homologs of AtCYP71, although their function is yet to be elucidated.
SD Cytoplasmic Cyclophilins
The abundance of cytoplasmic isoforms is indicative of potential specialization within the SD cyclophilins. The high degree of sequence identity found within the SD cytoplasmic cyclophilins makes prediction of their putative function a complex task. Transcriptional data shows that their expression occurs in all tissues within the mature plant (Chou and Gasser, 1997; Jackson and Soll, 1999), suggesting that all isoforms may be required within the cell at any one time. If correlated with transcript abundance, the number of reported ESTs allows indirect determination of the quantitative requirements of each isoform; AtCYP18-3 and AtCYP18-4 show a 2- to 20-fold higher EST number compared to the other SD cytoplasmic isoforms (data from MIPS database, 08/05/2003). This is corroborated by expression data showing that AtCYP18-4 transcript levels are significantly higher than AtCYP19-3 (Chou and Gasser, 1997). The comparatively high expression levels of AtCYP18-3 and AtCYP18-4 suggest that they may act as the principal housekeeping cyclophilin isoforms that are either broad-range protein foldases or PPiases specifically required for the assembly of an abundant protein. Although closely related, AtCYP19-1, AtCYP19-2, and AtCYP19-3 may carry out more specific roles, hence their apparently reduced expression level. No evidence for AtCYP19-1 and AtCYP19-3 function exists, but a yeast 2-hybrid screen identified a specific interaction between AtCYP19-2 and the T-DNA 5′ endonuclease VirD protein (Deng et al., 1998). Furthermore, CsA has been shown to inhibit Agrobacterium-mediated transformation in both tobacco and Arabidopsis. The significance of this relationship in vivo is unknown, although AtCYP19-2 may be necessary for the initial stages of T-DNA integration into the genome (Deng et al., 1998). Compared to the SD isoforms described above, significant sequence divergence was noted in the remaining SD cytoplasmic cyclophilins AtCYP18-1 and AtCYP18-2, most notably the deletion in the surface exposed α-I/β-III junction, causing them to group with the MD cyclophilins in clade II. No evidence exists for the function of AtCYP18-1 and AtCYP18-2; however, the deletion constitutes a structural alteration that may confer functional specificity to these two isoforms, possibly through their interaction with a distinct subset of proteins.
Chloroplast Cyclophilins
Two independent proteomics analyses of the Arabidopsis thylakoid lumen have shown that, being comprised of 15 isoforms, immunophilins make up one of the largest families of proteins within this subcompartment (Peltier et al., 2002; Schubert et al., 2002). Seven of the nine FKBPs have been experimentally assigned to the thylakoid lumen whereas two remain as yet predicted to be present in this compartment. In addition to one isoform (AtCYP20-3) that is targeted to the stroma (Lippuner et al., 1994), the remaining five chloroplast cyclophilins are targeted to the thylakoid lumen. Although AtCYP20-3 has been clearly identified as a stromal protein (Lippuner et al., 1994; Schubert et al., 2002), the inability of CsA to block import of Rubisco and the 33-kD subunit of the oxygen evolving complex in an in vitro import assay suggests that PPiase activity may not be involved in this process (although CsA access into the chloroplast in these experiments was not assessed). The SD isoform AtCYP20-2 and its spinach homolog TLP20 have been shown to possess CsA-sensitive PPiase activity (Edvardsson et al., 2003; P.G.N. Romano, unpublished data), and have been isolated from both lumenal and thylakoid membrane fractions (Peltier et al., 2002; Schubert et al., 2002; Edvardsson et al., 2003), suggesting that this protein may be recruited to the membrane as part of its function.
AtCYP26-2 is the only chloroplast cyclophilin whose location is based solely upon prediction; however, judging by the high score obtained, it is very likely to be a lumenal protein, but it may be synthesized at low levels or following specific environmental or developmental cues. Its putative location is in part corroborated by its expression pattern that, as expected for most chloroplast localized proteins, suggests significantly reduced expression in nonphotosynthetic tissue (see Fig. 4). Both AtCYP37 and AtCYP38 have been identified as lumenal proteins by proteomic analysis (Schubert et al., 2002; Peltier et al., 2002). In vitro studies have shown that the AtCYP38 homolog TLP40 associates with and regulates the activity of a D1 phosphatase within the thylakoid membrane, an essential process in the coordination of proteolysis and integration of newly synthesized D1 subunits during PSII protein turnover (Fulgosi et al., 1998; Vener et al., 1999). Although, as expected, the W332I substitution present in both TLP40 and AtCYP38 results in a CsA-insensitive protein, it is interesting to note that PPiase activity is retained despite substitutions in two key amino acids (F230E and H337Y).
The potential importance of peptidyl-prolyl cis-trans isomerization in chloroplast function cannot be overstated given the abundance of these enzymes within the thylakoid lumen. Most recently, the precursor of the lumenal AtFKBP13 has been shown to interact with the Rieske Fe-S protein, a component of the cytochrome b6f complex, suggesting association occurs along the import pathway. Antisense AtFKBP13 lines have increased Rieske levels suggesting it may act as a suppressor of Rieske accumulation (Gupta et al., 2002). In view of this evidence, it is likely that (1) each isoform may exert a certain degree of substrate specificity, (2) that interaction may not necessarily be exclusive to the thylakoid lumen but may occur throughout the import pathway, and (3) that the PPiase reaction may not always be required for interaction. Cyclophilin interactions with both thioredoxin m (Motohashi et al., 2001) and peroxiredoxins (Lee et al., 2001) in vitro, important chloroplast signal transduction and antioxidative components respectively, are consistent with the view that they may also play a regulatory role. AtCYP20-3 has recently been shown to contain multiple internal disulphide bonds that are reduced by dithiothreitol in the presence of thioredoxin m (Motohashi et al., 2001). As suggested elsewhere, cyclophilins may play a crucial role in sequestering and protecting unassembled lumenal proteins which are prone to degradation, such as those comprising the oxygen evolving complex (Schubert et al., 2002).
Where PPiase activity is a crucial component of the association, certain isoforms may fold newly imported soluble or membrane associated lumenal proteins; they may act individually, or as part of a multisubunit chaperone complex including additional proteins such as heat-shock proteins, as seen in mitochondria (see below). For example, the rapid and large-scale changes in the composition of thylakoid components during acclimation of plants to environmental change (Anderson, 1986) may be in part mediated by cyclophilins; PPiase activity may be required for the rapid and efficient integration of newly synthesized intrinsic membrane proteins. Pro residues are characteristic components of interhelical loop domains of chlorophyll-binding LHCII proteins, protruding on both the membrane and lumenal side. Mutations in these Pro residues are known to cause significant reduction in the integration efficiency of this chlorophyll binding protein (Heinemann and Paulsen, 1999).
Mitochondrial Cyclophilins
In fungi, mitochondrial cyclophilin accelerates folding of newly imported proteins within the matrix as part of a complex that includes the chaperones Hsp60 and Hsp70 (Matouschek et al., 1995; Rassow et al., 1995). Mammalian cyclophilin D has been shown to reversibly associate with the permeability transition pore, which is located at the contact site between inner and outer membranes, where it regulates pore opening and closure (Nicolli et al., 1996; Scorrano et al., 1997; Crompton, 2000). Pore opening forms the basis of the mitochondrial permeability transition (MPT), a complex and tightly regulated process resulting in mitochondrial swelling and apoptosis (Zoratti and Szabo, 1995). Early biochemical evidence for the presence of a plant mitochondrial cyclophilin comes from work showing the presence of CsA-sensitive PPiase activity in purified pea (Pisum sativum) mitochondria (Breiman et al., 1992). More recently, experiments in isolated potato (Solanum tuberosum) mitochondria suggest that regulation of plant MPT is similar to its animal counterpart and that a cyclophilin is involved; calcium induced mitochondrial swelling is inhibited by CsA (Arpagaus et al., 2002). Arabidopsis contains two genes encoding mitochondrial cyclophilins. The two isoforms could be heterogeneously distributed between the mitochondrial membrane and matrix and carry out distinct functions within these two compartments.
Secretory Pathway Cyclophilins
In addition to the previously identified isoforms AtCYP19-4 and AtCYP20-1, the Arabidopsis genome was found to encode three further cyclophilin proteins with predicted targeting to the SP. AtCYP19-4 contains a putative ER-targeting signal peptide at its N terminus, but no typical ER-retention signal in its C-terminus (Saito et al., 1995). Fluorescence from an AtCYP19-4-GFP fusion has been detected mainly around the nucleus, indicating translocation into the ER (Saito et al., 1995). Furthermore, AtCYP19-4 has been shown to interact with the N terminus of a guanine nucleotide exchange factor that acts on an ADP ribosylation factor-type G-protein called GNOM, which is involved in coordination of cell polarity along the apical-basal embryo axis. AtCYP19-4 is expressed in all tissues and partitions between cytosolic and membrane fractions, most probably, as shown earlier, to ER subcompartments, where it may modulate GNOM activity (Grebe et al., 2000).
AtCYP20-1 has been identified in a yeast 2-hybrid screen in which the regulatory subunit RCN1 of a Ser/Thr-specific protein phosphatase was used as bait. Interaction occurs via a DENFKL domain, which is conserved within certain members of the cyclophilin family. However, AtCYP20-1 binding does not alter phosphatase activity. Interestingly, plants expressing an antisense AtCYP20-1 gene show an elongated root phenotype, suggesting that the AtCYP20-1-RCN1 complex may be required for control of root cell proliferation (Jackson and Soll, 1999).
Although numerous cyclophilins containing ER signal peptides have been identified in a wide variety of organisms, little is known about their function either in vivo or in vitro. With the exception of AtCYP23-1, the Arabidopsis SP cyclophilins show 100% conservation in the four residues implicated in substrate binding and catalysis, suggesting that PPiase activity is an active component of their function. AtCYP23-1 shows only 28% sequence identity compared to hCYPA and is the only SP isoform containing a predicted transmembrane domain; of the four residues mentioned above, only Arg-225 is conserved, suggesting PPiase activity may be compromised in this isoform. Given their ubiquitous expression patterns, it is likely that all SP isoforms are present simultaneously, and thus, as previously shown (Jackson and Soll, 1999; Grebe et al., 2000), they may associate with specific substrates in vivo. Such specificity is exemplified by work carried out with the inhibitor CsA, which suggests a role for these proteins in mediating calcium-derived signals (Smaili et al., 2001). Recent studies suggest that cyclophilins are able to mediate transcriptional regulation by way of their enzymatic activity; human CypB is localized in the ER and extracellular space and has been shown to be present in the nucleus as part of the prolactin signaling complex, where it acts as a transcriptional activator, catalyzing the amino-terminal isomerization and subsequent inactivation of the repressor Stat5 (Rycyzyn and Clevenger, 2002). Signal peptide sequence homology observed between CypB and the putative SP cyclophilins described here suggests that they may be able to translocate to the nucleus and fulfill similar regulatory roles.
CONCLUSIONS
We have carried out a comprehensive sequence analysis of every Arabidopsis cyclophilin gene and provided basic spatial expression data and resource information. In addition to providing accurate sequence data, the analysis highlights the caution with which predicted mRNA sequence information must be approached, with approximately one-quarter of genes being erroneously annotated on the MIPS database. The presence of previously reported functional motifs means that functional predictions are possible for the MD isoforms; in a similar way, existing evidence for chloroplast and mitochondrial cyclophilin function gives us an insight into their putative function. With regards to cytoplasmic isoforms, we may assume that subtle functional differentiation may have been brought about by unique amino acid substitutions, deletions, and insertions leading to micro- and macromodifications within the three-dimensional structure, thus conferring substrate specificity. With particular reference to the MD cyclophilins, the sequence analysis predicts regulatory roles in the development of plants and their dynamic response to the environment. The scope of the latter in plants may explain why the gene family is so much larger in the Arabidopsis genome than that found to date in any other sequenced genome. The analysis presented here provides the foundation for the identification of the physiological roles and the molecular mechanisms of this complex and important family of proteins.
MATERIALS AND METHODS
Plant Growth and Maintenance
Arabidopsis (Col-0) was grown under short days (8 h light, 20°C; 16 h dark, 15°C) at a photon flux density of 100 μmol m−2 s−1 (color 84 fluorescent tubes, Philips TLD 35 W, or Philips PL-L 55 W heat-filtered through 2 mm polyacetate; Philips, Eindhoven, The Netherlands). For RNA extraction, leaves and roots were harvested from 5-week-old plants.
DNA Sequencing
Using an archetypal Arabidopsis cyclophilin domain (AtCYP18-3), The Arabidopsis Information Resource (TAIR) and National Center for Biotechnology Information (NCBI) databases were searched using the BLAST algorithm for any sequence containing cyclophilin-like domains. Following the identification of novel genes in the Munich Information Centre for Protein Sequence (MIPS) database, cDNAs were obtained from ABRC (AtCYP20-2 [F14217], AtCYP22-1 [190K19], AtCYP21-4 [32E3], AtCYP71 [G10H7], AtCYP95 [124N3]) and Riken BRC (AtCYP18-2 [pda02396], AtCYP21-3 [pda03027], AtCYP38 [pda02423], AtCYP63 [pda02459], AtCYP65 [pda04293]). The AtCYP59 RIKEN BRC clone (pda02588) contained a coding region lacking a stop codon or 3′ extension, thus the predicted C-terminal sequence was obtained by RT-PCR amplification using additional forward (5′-GACGGATCATATTGCCAA-3′) and reverse (5′-TCATCTATCCCTTCTCTCATG-3′) primers. Due to the unavailability of ESTs, the putative coding region of the following genes was isolated by RT-PCR using the predicted cDNA sequences from the MIPS entries and the following primers: AtCYP18-1 (5′-ACAATGTCGGTAACTTTGC-3′, 5′-CAGCAAGTGGATTGGCGTGGA-3′), AtCYP21-1 (5′-ATGCGTAGAGAGATCTCG-3′, 5′-TCATCTCTCTTCATCCCA-3′) AtCYP21-2 (5′-ATGGCAATTACGGCGACT-3′, 5′-TTACAAAGAAACTTCTCCACTAGCA-3′), AtCYP57 (5′-ATGTCGACGGTGTACGTG-3′, 5′-GGCAAGAGATTTTCCAGAC-3′). We were unable to clone full-length sequences for AtCYP26-1 and therefore its predicted sequence is based on RT-PCR products and sequenced cDNA data available within the MIPS database. Sequences for AtCYP23-1, AtCYP26-2, and AtCYP28 were derived from Riken RAFL19-56-O08, EMBL-AY062660.1, and GenBank P82869, respectively. Sequencing was obtained with a 373A DNA sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA) and Taq DyeDeoxy terminator cycle sequencing (ABI, Sunnyvale, CA). Sequencing data was collected by the Krebs Biomolecular Synthesis Laboratory, Department of Molecular Biology and Biotechnology, University of Sheffield, United Kingdom.
Phylogenetic Analysis
Edited sequences were aligned using ClustalW (Thompson et al., 1994) and displayed with Genedoc v.2.6.02 (Nicholas et al., 1997; Nicholas and Nicholas, 1997). To construct the phylogenetic tree, the .dnd file generated in ClustalW was displayed in Treeview (Page, 1996). Domains were identified using the Conserved Domain Architectural Retrieval Tool at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi).
RT-PCR
Total RNA was prepared using an RNeasy RNA extraction kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. DNA was degraded and removed using DNA-free (Ambion Corp., Austin, TX). One μg of total RNA was used to generate the corresponding single-stranded cDNAs by using an oligo(dT)15 primer and AMV reverse transcriptase (Promega, Madison, WI) in the presence of an RNAse inhibitor (RNasin, Promega) in a 25 μl volume. The primers and annealing temperature used for PCR amplification of the cyclophilin genes are shown in Table II. PCR was carried out using standard protocols, using 1 to 5 μl of RT reaction and the following conditions: 94°C for 5 min (1 cycle); 1 min at 94°C, 1 min at 52°C to 65°C (see Table II), 1 min 15 s at 72°C (30–40 cycles); 7 min at 72°C (1 cycle). Control primers to amplify the ubiquitin gene (UBQ5, accession At3g62250) were 5′-GTGGTGCTAAGAAGAGGAAGA-3′ and 5′-TCAAGCTTCAACTCCTTCTTT-3′. All primers were designed to amplify products in the range 400 to 600 bp and, where appropriate, to span an exon-intron boundary. Expression analyses were carried out twice with independent RNA extracts, and the results of one of the experiments are shown.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AtCYP18-1 (AY568515), AtCYP18-2 (AY568516), AtCYP20-2 (AY568517), AtCYP21-1 (AY568518), AtCYP21-2 (AY568519), AtCYP21-3 (AY568520), AtCYP21-4 (AY568521), AtCYP22-1 (AY568522), AtCYP26-1 (AY568523), AtCYP38 (AY568524), AtCYP57 (AY568525), AtCYP59 (AY568526), AtCYP63 (AY568527), AtCYP65 (AY568528), and AtCYP95 (AY568529).
Acknowledgments
We thank the ABRC and Riken BRC for provision of cDNAs.
This work was supported by the Natural Environment Research Council and by the Biotechnology and Biological Sciences Research Council.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022160.
References
- Albà MM, Pagès M (1998) Plant proteins containing the RNA-recognition motif. Trends Plant Sci 3: 15–21 [Google Scholar]
- Anderson JM (1986) Photoregulation of the composition, function, and structure of thylakoid membranes. Annu Rev Plant Physiol Plant Mol Biol 37: 93–136 [Google Scholar]
- Arabidopsis Genome Initiative (2000) Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arpagaus S, Rawyler A, Braendle R (2002) Occurrence and characteristics of the mitochondrial permeability transition in plants. J Biol Chem 277: 1780–1787 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Santos-Rosa MJ, Shirasu K (2001) The U-box protein family in plants. Trends Plant Sci 6: 354–358 [DOI] [PubMed] [Google Scholar]
- Bartling D, Heese A, Weiler EW (1992) Nucleotide-sequence of a cDNA-encoding and Arabidopsis cyclophilin-like protein. Plant Mol Biol 19: 529–530 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Bollman K, Sun H, Poethig RS (2001) Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science 291: 2405–2407 [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res 21: 5803–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourquin JP, Stagljar I, Meier P, Moosman P, Silke J, Baechi T, Georgiev O, Schaffner W (1997) A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res 25: 2055–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandts JF, Halvorson HR, Brennan M (1975) Consideration of the possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry 14: 4953–4963 [DOI] [PubMed] [Google Scholar]
- Brazin KN, Mallis RJ, Fulton DB, Andreotti AH (2002) Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc Natl Acad Sci USA 99: 1899–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman A, Fawcett TW, Ghirardi ML, Mattoo AK (1992) Plant organelles contain distinct peptidyl-prolyl cis-trans isomerases. J Biol Chem 267: 21293–21296 [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621 [DOI] [PubMed] [Google Scholar]
- Carol RJ, Breiman A, Erel N, Vittorioso P, Bellini C (2001) PASTICCINO1 (AtFKBP70) is a nuclear-localised immunophilin required during Arabidopsis thaliana embryogenesis. Plant Sci 161: 527–535 [Google Scholar]
- Chou IT, Gasser CS (1997) Characterisation of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol Biol 35: 873–892 [DOI] [PubMed] [Google Scholar]
- Crompton M (2000) Mitochondrial intermembrane junctional complexes and their role in cell death. J Physiol (Lond) 529: 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WY, Chen LS, Wood DW, Metcalfe T, Liang XY, Gordon MP, Comai L, Nester EW (1998) Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc Natl Acad Sci USA 95: 7040–7045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski K, Heitman J (1997) Peptidyl-prolyl isomerases. In J Sambrook, S Tooze, eds, Guidebook to Molecular Chaperones and Protein Folding Catalysts. Oxford University Press, London, pp 359–369
- Duina AA, Chang HCJ, Marsh JA, Lindquist S, Gaber RF (1996) A cyclophilin function in Hsp90-dependent signal transduction. Science 274: 1713–1715 [DOI] [PubMed] [Google Scholar]
- Edvardsson A, Eshaghi S, Vener AV, Andersson B (2003) The major peptidyl-prolyl isomerase activity in thylakoid lumen of plant chloroplasts belongs to a novel cyclophilin TLP20. FEBS Lett 542: 137–141 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Faure JD, Gingerich D, Howell SH (1998) An Arabidopsis immunophilin, AtFKBP12, binds to AtFIP37 (FKBP interacting protein) in an interaction that is disrupted by FK506. Plant J 15: 783–789 [DOI] [PubMed] [Google Scholar]
- Ferreira PA, Nakayama TA, Pak WL, Travis GH (1996) Cyclophilin-related protein RanBP2 acts as chaperone for red/green opsin. Nature 383: 637–640 [DOI] [PubMed] [Google Scholar]
- Freemont PS (1993) The ring finger: a novel protein-sequence motif related to the zinc-finger. Ann N Y Acad Sci 684: 174–192 [DOI] [PubMed] [Google Scholar]
- Fu X-D (1995) The superfamily of arginine/serine-rich splicing factors. RNA 1: 663–680 [PMC free article] [PubMed] [Google Scholar]
- Fujiyama S, Yanagida M, Hayano T, Miura Y, Isobe T, Takahashi N (2002) Isolation and proteomic characterization of human parvulin-associating preribosomal ribonucleoprotein complexes. J Biol Chem 277: 23773–23780 [DOI] [PubMed] [Google Scholar]
- Fulgosi H, Vener AV, Altschmied L, Herrmann RG, Andersson B (1998) A novel multi-functional chloroplast protein: identification of a 40 kDa immunophilin-like protein located in the thylakoid lumen. EMBO J 17: 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galat A (1999) Variations of sequences and amino acid compositions of proteins that sustain their biological functions: an analysis of the cyclophilin family of proteins. Arch Biochem Biophys 371: 149–162 [DOI] [PubMed] [Google Scholar]
- Gasser CS, Gunning DA, Budelier KA, Brown SM (1990) Structure and expression of cytosolic cyclophilin peptidyl-prolyl cis-trans isomerase of higher-plants and production of active tomato cylophilin in Escherichia coli. Proc Natl Acad Sci USA 87: 9519–9523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy AV, Lazzaro AS, Casalongue CA, San Segundo B (2000) Expression of a Solanum tuberosum cyclophilin gene is regulated by fungal infection and abiotic stress conditions. Plant Sci 152: 123–134 [Google Scholar]
- Graveley BR (2000) Sorting out the complexity of SR protein functions. RNA 6: 1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld JU, Salchert K, Koncz C, Jurgens G (2000) A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12: 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Mould RM, He ZY, Luan S (2002) A chloroplast FKBP interacts with and affects the accumulation of Rieske subunit of cytochrome bf complex. Proc Natl Acad Sci USA 99: 15806–15811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschumacher RE, Harding MW, Drugge RJ (1984) Cyclophilin-a specific cytosolic binding protein for cyclosporin A. Science 226: 544–547 [DOI] [PubMed] [Google Scholar]
- Harding MW, Galat A, Uehling DE, Schreiber SL (1989) A receptor for the immunosuppressant FK-506 is a cis-trans peptidyl-prolyl isomerase. Nature 341: 758–760 [DOI] [PubMed] [Google Scholar]
- Hayman GT, Miernyk JA (1994) The nucleotide and deduced amino acid sequences of a peptidyl-prolyl cis-trans isomerase from Arabidopsis thaliana. BBA-Gene Struct Expr 219: 536–538 [DOI] [PubMed] [Google Scholar]
- Heinemann B, Paulsen H (1999) Random mutations directed to transmembrane and loop domains of the light-harvesting chlorophyll a/b protein: impact on pigment binding. Biochemistry-US 38: 14088–14093 [DOI] [PubMed] [Google Scholar]
- Horowitz DS, Lee EJ, Mabon SA, Misteli T (2002) A cyclophilin functions in pre-mRNA splicing. EMBO J 21: 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson K, Soll D (1999) Mutations in a new Arabidopsis cyclophilin disrupt its interaction with protein phosphatase 2A. Mol Gen Genet 262: 830–838 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PCM, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognises ubiquitin as a degradation signal. J Biol Chem 270: 17442–17456 [DOI] [PubMed] [Google Scholar]
- Kallen J, Spitzfaden C, Zurini MGM, Wider G, Widmer H, Wuthrich K, Walkinshaw MD (1991) Structure of human cyclophilin and its binding site for cyclosporine-A determined by X-ray crystallography and NMR-spectroscopy. Nature 353: 276–279 [DOI] [PubMed] [Google Scholar]
- Kamphausen T, Fanghanel R, Neumann D, Schulz B, Rahfeld JU (2002) Characterization of Arabidopsis thaliana AtFKBP42 that is membrane-bound and interacts with Hsp90. Plant J 32: 263–276 [DOI] [PubMed] [Google Scholar]
- Ke HM (1992) Similarities and differences between human cyclophilin-A and other beta-barrel structures-structural refinement at 1.63 angstrom resolution. J Mol Biol 228: 539–550 [DOI] [PubMed] [Google Scholar]
- Kong HY, Lee SC, Hwang BK (2001) Expression of pepper cyclophilin gene is differentially regulated during the pathogen infection and abiotic stress conditions. Physiol Mol Plant Pathol 59: 189–199 [Google Scholar]
- Krecic AM, Swanson MS (1999) hnRNP complexes: composition, structure and function. Curr Opin Cell Biol 11: 363–371 [DOI] [PubMed] [Google Scholar]
- Krzywicka A, Beisson J, Keller AM, Cohen J, Jerka-Dziadosz M, Klotz C (2001) KIN241: a gene involved in cell morphogenesis in Paramecium tetraurelia reveals a novel protein family of cyclophilin-RNA interacting proteins (CRIPs) conserved from fission yeast to man. Mol Microbiol 42: 257–267 [DOI] [PubMed] [Google Scholar]
- Lee SP, Hwang YS, Kim YJ, Kwon KS, Kim HJ, Kim K, Chae HZ (2001) Cyclophilin a binds to peroxiredoxins and activates its peroxidase activity. J Biol Chem 276: 29826–29832 [DOI] [PubMed] [Google Scholar]
- Leverson JD, Ness SA (1998) Point mutations in v-Myb disrupt a cyclophilin-catalysed negative regulatory mechanism Mol Cell 1: 203–211 [DOI] [PubMed] [Google Scholar]
- Li J, Barnard DC, Patton JG (2002) A unique glutamic acid-lysine (EK) domain acts as a splicing inhibitor. J Biol Chem 277: 39485–39492 [DOI] [PubMed] [Google Scholar]
- Lin DT, Lechleiter JD (2002) Mitochondrial targeted cyclophilin D protects cells from cell death by peptidyl prolyl isomerization. J Biol Chem 277: 31134–31141 [DOI] [PubMed] [Google Scholar]
- Lippuner V, Chou IT, Scott SV, Ettinger WF, Theg SM, Gasser CS (1994) Cloning and characterisation of chloroplast and cytosolic forms of cyclophilin from Arabidopsis thaliana. J Biol Chem 269: 7863–7868 [PubMed] [Google Scholar]
- Liu J, Chen CM, Walsh CT (1991) Human and Escherichia coli cyclophilins: Sensitivity to inhibition by the immunosuppressant cyclosporin A correlates with a specific tryptophan residue. Biochemistry 30: 2306–2310 [DOI] [PubMed] [Google Scholar]
- Lorkovic ZJ, Barta A (2002) Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res 30: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Albers MW, Schreiber SL (1994) Light-regulated, tissue-specific immunophilins in a higher-plant. Proc Natl Acad Sci USA 91: 984–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marivet J, Frendo P, Burkard G (1992) Effects of abiotic stresses on cyclophilin gene-expression in maize and bean and sequence analysis of cyclophilin cDNA. Plant Sci 84: 171–178 [Google Scholar]
- Marivet J, Margispinheiro M, Frendo P, Burkard G (1994) Bean cyclophilin gene-expression during plant development and stress conditions. Plant Mol Biol 26: 1181–1189 [DOI] [PubMed] [Google Scholar]
- Marivet J, Margispinheiro M, Frendo P, Burkard G (1995) DNA-sequence analysis of a cyclophilin gene from maize-developmental expression and regulation by salicylic acid. Mol Gen Genet 247: 222–228 [DOI] [PubMed] [Google Scholar]
- Matouschek A, Rospert S, Schmid K, Glick BS, Schatz G (1995) Cyclophilin catalyses protein folding in yeast mitochondria. Proc Natl Acad Sci USA 92: 6319–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L (1998) Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem 67: 265–306 [DOI] [PubMed] [Google Scholar]
- Mayr C, Richter K, Lilie H, Buchner J (2000) Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. J Biol Chem 275: 34140–34146 [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN (1998) A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93: 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortillaro MJ, Berezney R (1998) Matrin CYP, an SR-rich cyclophilin that associates with the nuclear matrix and splicing factors. J Biol Chem 273: 8183–8192 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci USA 98: 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestel FP, Colwill K, Harper S, Pawson T, Anderson SK (1996) RS cyclophilins: identification of an NK-TR(1)-related cyclophilin. Gene 180: 151–155 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr (1997) GeneDoc Analysis and visualization of genetic variation. http://www.cris.com/∼Ketchup/genedoc.shtml (January 2003)
- Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation. EMBNEW.NEWS 4: 14 [Google Scholar]
- Nicolli A, Basso E, Petronilli V, Wenger RM, Bernardi P (1996) Interactions of cyclophilin with the mitochondrial inner membrane and regulation of the permeability transition pore, a cyclosporin A-sensitive channel. J Biol Chem 271: 2185–2192 [DOI] [PubMed] [Google Scholar]
- Owens-Grillo JK, Hoffmann K, Hutchison KA, Yem AW, Deibel MR, Handschumacher RE, Pratt WB (1995) The cyclosporin A-binding immunophilin CyP-40 and the FK506-binding immunophilin Hsp56 bind to a common site on Hsp90 and exist in independent cytosolic heterocomplexes with the untransformed glucocorticoid receptor. J Biol Chem 270: 20479–20484 [DOI] [PubMed] [Google Scholar]
- Owens-Grillo JK, Stancato LF, Hoffmann K, Pratt WB, Krishna P (1996) Binding of immunophilins to the 90 kDa heat shock protein (hsp90) via a tetratricopeptide repeat domain is a conserved protein interaction in plants. Biochemistry 35: 15249–15255 [DOI] [PubMed] [Google Scholar]
- Page AP, Winter AD (1998) A divergent multi-domain cyclophilin is highly conserved between parasitic and free-living nematode species and is important in larval muscle development. Mol Biochem Parasit 95: 215–227 [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, von Heijne G, Van Wijk KJ (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ER, Jin MJ, Pati S, Walsh CT, McKeon FD (1994) Cyclophilin-B trafficking through the secretory pathway is altered by binding of Cyclosporin-A. Proc Natl Acad Sci USA 91: 3931–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J, Mohrs K, Koidl S, Barthelmess IB, Pfanner N, Tropschug M (1995) Cyclophilin-20 is involved in mitochondrial protein folding in cooperation with molecular chaperones Hsp70 and Hsp60. Mol Cell Biol 15: 2654–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycyzyn MA, Clevenger CV (2002) The intranuclear prolactin/cyclophilin B complex as a transcriptional inducer. Proc Natl Acad Sci USA 99: 6790–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Ishiguro S, Ashida H, Kawamukai M, Matsuda H, Ochihai H, Nakagawa T (1995) Cloning and sequence analysis of genes for cyclophilin from Arabidopsis thaliana. Plant Cell Physiol 36: 377–382 [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Borden KLB, Boddy MN, Freemont PS (1996) Does this have a familiar RING? Trends Biochem Sci 21: 208–214 [PubMed] [Google Scholar]
- Schnell DJ (1998) Protein Targeting to the Thylakoid Membrane. Annu Rev Plant Physiol Plant Mol Biol 49: 97–126 [DOI] [PubMed] [Google Scholar]
- Scholz C, Stoller G, Zarnt T, Fischer G, Schmid FX (1997) Cooperation of enzymatic and chaperone functions of trigger factor in the catalysis of protein folding. EMBO J 16: 54–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholze C, Peterson A, Diettrich B, Luckner M (1999) Cyclophilin isoforms from Digitalis lanata: sequences and expression during embryogenesis and stress. J Plant Physiol 155: 212–219 [Google Scholar]
- Schubert M, Petersson UA, Haas BJ, Funk C, Schröder WP, Kieselbach T (2002) Proteome map of the chloroplast lumen of Arabidopsis thaliana. J Biol Chem 277: 8354–8365 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Deng XW (2001) COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol 11: 420–426 [DOI] [PubMed] [Google Scholar]
- Scorrano L, Nicolli A, Basso E, Petronilli V, Bernardi P (1997) Two modes of inactivation of the permeability transition pore: the role of mitochondrial cyclophilin. Mol Cell Biochem 174: 181–184 [PubMed] [Google Scholar]
- Sharma AD, Singh P (2003) Effect of water stress on expression of a 20 kD cyclophilin-like protein in drought susceptible and tolerant cultivars of Sorghum. J Plant Biochem Biot 12: 77–80 [Google Scholar]
- Shieh BH, Stamnes MA, Seavello S, Harris GL, Zuker CS (1989) The NinaA gene required for visual transduction in Drosophila encodes a homolog of cyclosporin A-binding protein. Nature 338: 67–70 [DOI] [PubMed] [Google Scholar]
- Smaili SS, Stellato KA, Burnett P, Thomas AP, Gaspers LD (2001) Cyclosporin A inhibits inositol 1,4,5-trisphosphate-dependent Ca2+ signals by enhancing Ca2+ uptake into the endoplasmic reticulum and mitochondria. J Biol Chem 276: 23329–23340 [DOI] [PubMed] [Google Scholar]
- Stewart DE, Sarkar A, Wampler JE (1990) Occurrence and role of cis peptide bonds in protein structures. J Mol Biol 214: 253–260 [DOI] [PubMed] [Google Scholar]
- Stoller G, Rucknagel KP, Nierhaus KH, Schmid FX, Fischer G, Rahfeld JU (1995) A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J 14: 4939–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Arnoldo M, Goring DR (1999) A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286: 1729–1731 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Lloyd AM, Marks MD (2000) Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci 5: 214–219 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vener AV, Rokka A, Fulgosi H, Andersson B, Herrmann RG (1999) A cyclophilin-regulated PP2A-like protein phosphatase in thylakoid membranes of plant chloroplasts. Biochemistry 38: 14955–14965 [DOI] [PubMed] [Google Scholar]
- Vittorioso P, Cowling R, Faure JD, Caboche M, Bellini C (1998) Mutation in the Arabidopsis PASTICCINO1 gene, which encodes a new FK506-binding protein-like protein, has a dramatic effect on plant development. Mol Cell Biol 18: 3034–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG (1989) Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem 180: 535–545 [DOI] [PubMed] [Google Scholar]
- Wang BB, Hayenga KJ, Payan DG, Fisher JM (1996) Identification of a nuclear-specific cyclophilin which interacts with the proteinase inhibitor eglin c. Biochem J 314: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weighardt F, Cobianchi F, Cartegni L, Chiodi I, Villa A, Riva S, Biamonti G (1999) A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J Cell Sci 112: 1465–1476 [DOI] [PubMed] [Google Scholar]
- Yurchenko V, Zybarth G, O'Connor M, Dai WW, Franchin G, Hao T, Guo H, Hung HC, Toole B, Gallay P, Sherry B, Bukrinsky M (2002) Active site residues of cyclophilin A are crucial for its signaling activity via CD147. J Biol Chem 277: 22959–22965 [DOI] [PubMed] [Google Scholar]
- Zoratti M, Szabo I (1995) The mitochondrial permeability transition. BBA-Rev Biomembranes 1241: 139–176 [DOI] [PubMed] [Google Scholar]
- Zydowsky LD, Etzkorn FA, Chang HY, Ferguson SB, Stolz LA, Ho SI, Walsh CT (1992) Active site mutants of human cyclophilin-A separate peptidyl-prolyl isomerase activity from cyclosporin-A binding and calcineurin inhibition. Protein Sci 1: 1092–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]