Abstract
UBL5 is an atypical ubiquitin-like protein, whose function in metazoans remains largely unexplored. We show that UBL5 is required for sister chromatid cohesion maintenance in human cells. UBL5 primarily associates with spliceosomal proteins, and UBL5 depletion decreases pre-mRNA splicing efficiency, leading to globally enhanced intron retention. Defective sister chromatid cohesion is a general consequence of dysfunctional pre-mRNA splicing, resulting from the selective downregulation of the cohesion protection factor Sororin. As the UBL5 yeast orthologue, Hub1, also promotes spliceosome functions, our results show that UBL5 plays an evolutionary conserved role in pre-mRNA splicing, the integrity of which is essential for the fidelity of chromosome segregation.
Keywords: pre-mRNA splicing, sister chromatid cohesion, sororin, ubiquitin-like protein, UBL5
Introduction
Protein modification by ubiquitin and ubiquitin-like modifiers (UBLs) plays key regulatory roles in numerous aspects of cell biology 1,2. Eukaryotic cells express more than a dozen UBLs, which share similar three-dimensional structures with ubiquitin 1. While the functions of several UBLs are well understood, others remain poorly characterized. Among these are UBL5 (known as Hub1 in yeast), which is unique among UBLs in that it lacks the C-terminal glycine used for covalent conjugation to target proteins 3,4. However, UBL5 displays strong sequence conservation across eukaryotes, suggesting that it has a fundamentally important cellular function. Studies of Hub1 in the yeasts revealed that it is required for pre-mRNA splicing. S. pombe Hub1 is an essential gene, and loss of Hub1 protein results in pre-mRNA splicing defects, likely reflecting its interaction with the spliceosomal protein Snu66 (known as SART1 in mammalian cells) and perhaps other splicing factors 5,6. In S. cerevisiae, Hub1 binds directly to Snu66 via a specific sequence motif in Snu66 termed HIND (Hub1-interaction domain) 7. While ablation of Hub1 is not lethal and does not significantly affect general pre-mRNA splicing in S. cerevisiae, it selectively impairs alternative splicing of SRC1, the only known alternatively spliced gene in this species 7. The function of UBL5 in higher eukaryotes remains largely unexplored.
Following DNA replication, sister chromatids are kept tightly connected until anaphase by cohesin, a multisubunit complex composed of SMC1, SMC3, RAD21, and SA1/2, which has a pivotal role in ensuring faithful chromosome segregation 8,9. In mammalian cells, loading of cohesin onto chromatin is initiated in telophase. During DNA replication, establishment of ‘cohesive’ cohesion is dependent on the ESCO1/2 acetyltransferases, which acetylate SMC3 to promote recruitment of Sororin, a key positive regulator of sister chromatid cohesion 9–11. Sororin protects cohesion during the S and G2 phases by antagonizing WAPL, a cohesion resolution factor 12,13. In early mitosis, polo-like kinase 1 phosphorylates SA2 to dissociate the bulk of cohesin from chromosome arms, while centromeric cohesion is protected by Shugoshin (SGO1) in conjunction with the phosphatase PP2A through Sororin until anaphase onset 9,14,15.
Here, we discovered that human UBL5 is essential for chromosome cohesion maintenance. UBL5 associates with the pre-mRNA splicing machinery and supports its functional integrity, which is in turn required for cells to prevent premature sister chromatid separation by ensuring proper splicing and expression of the cohesion factor Sororin. Our findings establish for the first time the cellular function of UBL5 in higher eukaryotes and reveal a strong requirement of pre-mRNA splicing integrity for sister chromatid cohesion in human cells.
Results and Discussion
UBL5 is required for cell proliferation and sister chromatid cohesion
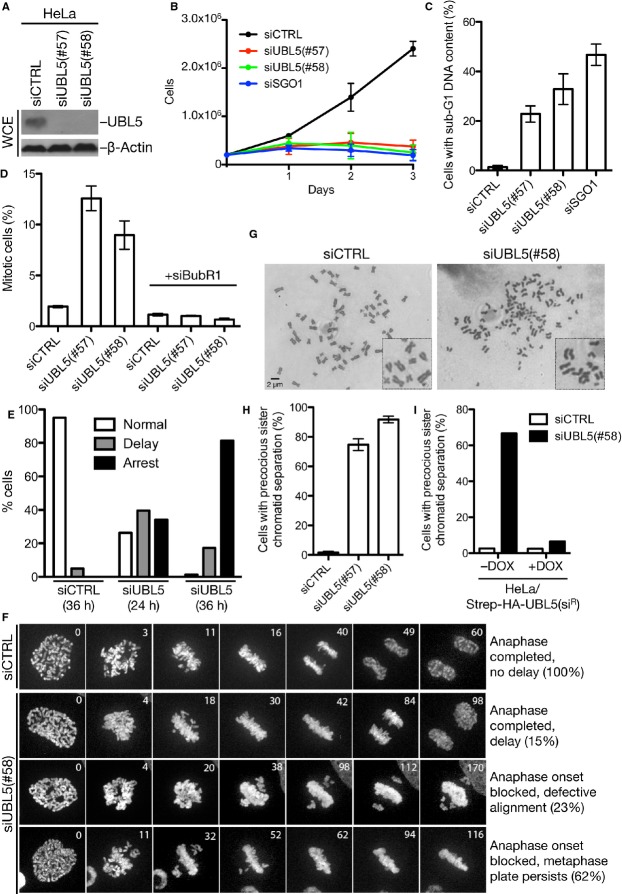
To investigate the function of human UBL5, we knocked down its expression using independent siRNAs (Fig 1A). Loss of UBL5 had a profound impact on cellular fitness and survival, causing a strong block to cell proliferation and enhanced cell death, as evidenced by accumulation of cells with sub-G1 DNA content (Fig 1B and C, and Supplementary Fig S1A). Cell cycle profiles of UBL5-depleted cells also revealed a marked increase in the proportion of mitotic cells, which could be efficiently reversed by the ablation of spindle checkpoint components such as BubR1 16 (Fig 1D, and Supplementary Fig S1B). This suggested that UBL5 loss impairs proper chromosome alignment in mitosis. Indeed, time-lapse microscopy showed a prominent delay or block to anaphase onset in UBL5-depleted cells, in some cases resulting from an inability of chromosomes to align properly at the metaphase plate while in the majority of cells chromosome congression appeared overtly normal (Fig 1E and F, and Supplementary Movie S1). To better understand this defect, we prepared metaphase chromosome spreads from these cells. Strikingly, UBL5 knockdown led to premature loss of sister chromatid cohesion in a large fraction of mitotic cells (Fig 1G and H, and Supplementary Fig S1C). This defect could be seen with several independent siRNAs and was fully reversed by ectopic expression of siRNA-resistant UBL5 (Fig 1H and I), demonstrating that it was a specific consequence of UBL5 loss. Depletion of factors required for protecting centromeric cohesion in mitosis, such as SGO1 17, impaired proliferation and stimulated cell death to an extent comparable with UBL5 depletion (Fig 1B and C). We conclude that UBL5 plays a crucial role in sister chromatid cohesion and cell proliferation in human cells.
Figure 1. Human UBL5 is required for cell proliferation and sister chromatid cohesion maintenance.
A Whole-cell extracts (WCE) of HeLa cells transfected with non-targeting (CTRL) or UBL5 siRNAs for 48 h were immunoblotted with anti-UBL5 antibody.
B HeLa cells were counted at indicated times after transfection with control, UBL5, or SGO1 siRNAs [mean ± SD (error bars); N = 3].
C Sub-G1 DNA content of HeLa cells transfected with indicated siRNAs for 72 h was measured by flow cytometry analysis of propidium iodide-labeled cells [mean ± SD (error bars); N = 3].
D Mitotic indices of HeLa cells transfected with indicated siRNAs for 48 h were quantified by flow cytometry analysis of the proportion of phospho-H3(Ser10)-positive cells [mean ± SD (error bars); N = 3].
E Mitotic duration from nuclear envelope breakdown to chromatin decondensation in single live HeLa/H2B-mCherry cells transfected with control (n = 20) or UBL5 siRNA (n = 80) was monitored by time-lapse microscopy and classified as normal (45–60 min), delayed (longer than 60 min), or arrested (no decondensation).
F Time-lapse analysis of mitotic chromosome dynamics in live HeLa/H2B-mCherry cells transfected with control or UBL5 siRNA. Selected time frames of representative mitotic progression patterns and their frequencies are shown.
G Representative images of metaphase spreads from cells transfected with non-targeting (CTRL) or UBL5 siRNA. See also Supplementary Fig S1.
H Quantification of precocious sister chromatid separation in HeLa cells after knockdown of UBL5 [mean ± SD (error bars); N = 3]. At least 100 metaphase spreads were counted.
I HeLa cells inducibly expressing siRNA-resistant (siR) Strep-HA-UBL5 were transfected with control or UBL5 siRNA in the absence or presence of doxycycline (DOX). Two days later, cells were treated with nocodazole for 3 h and harvested for chromosome analysis.
UBL5 associates with components of the pre-mRNA spliceosome
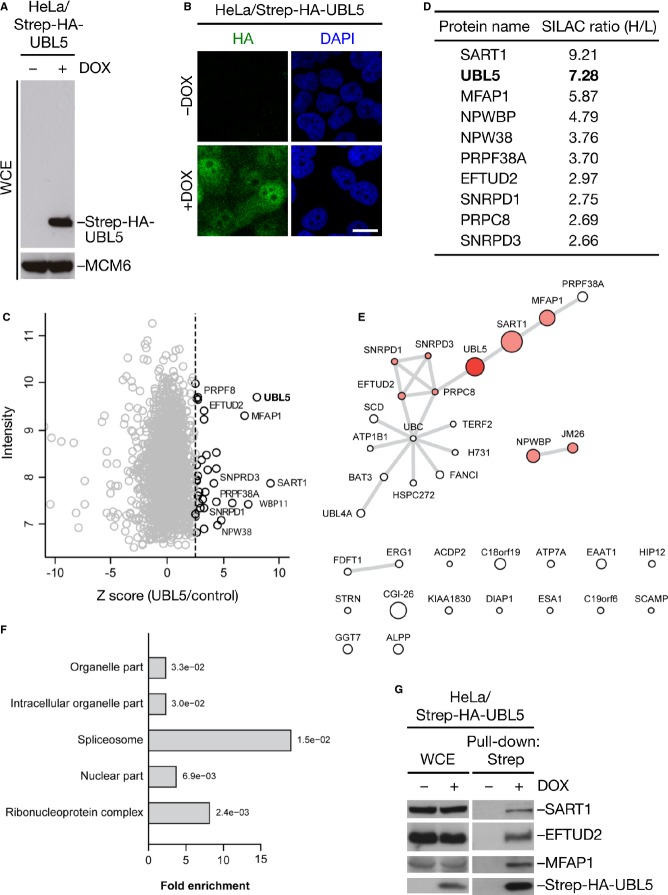
To understand how UBL5 promotes sister chromatid cohesion, we mapped UBL5-interacting proteins using quantitative mass spectrometry. Ectopic UBL5 localized mainly to the nucleus, and no formation of high-molecular weight, covalently conjugated UBL5 species was evident (Fig 2A and B). Using SILAC labeling 18, we identified a range of potential UBL5 interactors (Fig 2C and D, and Supplementary Table S1). While we did not find any known cohesion factors, many of the highly enriched proteins co-purifying with ectopic UBL5 were components of the spliceosome, including SART1, PRPC8, and EFTUD2 (Fig 2C–E). Indeed, gene ontology (GO) term analysis revealed strong and selective enrichment of spliceosome and ribonucleoprotein complex factors among putative UBL5-interacting factors (Fig 2F). Robust interactions between UBL5 and several splicing factors including SART1 and EFTUD2 were further validated biochemically (Fig 2G). These findings suggest that an involvement in pre-mRNA splicing is a major function of human UBL5 that is thus conserved from yeast. However, the mechanistic basis of Hub1/UBL5 interactions with splicing factors may differ to some extent between organisms. To this end, we found that a D22A mutation in UBL5, the corresponding mutation in Hub1 of which abrogates binding to Snu66 7, did not impair UBL5 binding to SART1, and the UBL5 D22A mutant rescued the cohesion defect resulting from loss of endogenous UBL5 as efficiently as wild-type UBL5 (Fig 1I, and Supplementary Fig S2A–C).
Figure 2. UBL5 preferentially associates with spliceosomal proteins in human cells.
A Whole-cell extracts (WCE) of stable HeLa/Strep-HA-UBL5 cells induced or not with doxycycline (DOX) for 48 h were analyzed by immunoblotting with antibodies to HA and MCM6 (loading control).
B HeLa/Strep-HA-UBL5 cells treated as in (A) were fixed and immunostained with HA antibody. Scale bar, 10 μm.
C Mass spectrometry (MS)-based analysis of UBL5-interacting proteins. HeLa cells expressing Strep-HA-UBL5 or not were cultured in heavy (H) or light (L) SILAC medium, respectively. Strep-HA-UBL5 and associated proteins enriched on Strep-Tactin Sepharose were analyzed by MS. Plot shows Z-scores (from SILAC H/L ratios) and total intensity of identified proteins. Selected prospective UBL5 interactors associated with the spliceosome are highlighted. See Supplementary Table S1 for full results.
D Selected proteins with high SILAC (H/L) ratios identified in the experiment in (C).
E Functional interactions of all proteins with a Z-score above 2.5 in the UBL5 pull-down experiment were obtained from the STRING database and visualized as a network. Node size corresponds to the Z-score. Known spliceosome components are highlighted in red.
F Gene ontology (GO) enrichment analysis of putative UBL5-interacting proteins. Significance of the enrichment is indicated for each term.
G Extracts of stable HeLa/Strep-HA-UBL5 cells induced or not with doxycycline (DOX) for 48 h were subjected to Strep-Tactin pull-down followed by immunoblotting with indicated antibodies.
Loss of UBL5 causes a global decrease in pre-mRNA splicing efficiency
We next analyzed the potential role of UBL5 in pre-mRNA splicing in human cells. To do this, we performed RNA-Seq analysis to map global transcriptome changes resulting from loss of UBL5 function, using three independent UBL5 siRNAs that all gave rise to a strong cohesion defect or SART1 siRNA. We observed excellent correlation between two replicates of each knockdown condition, as well as between different UBL5 siRNAs (Supplementary Fig S3A and B, and Supplementary Table S2). Examination of these RNA-Seq data revealed a striking increase in the number of reads mapping to introns and exon–intron junctions when UBL5 or SART1 were knocked down, as illustrated by the FASN gene (Fig 3A and B). Prompted by this, we performed a more elaborate genome-wide analysis of alternative splicing 19, showing that a significantly elevated proportion of alternative splicing events could be classified as intron retention (IR) in UBL5-depleted cells (Fig 3C). This could reflect compromised splicing efficiency, which should manifest as an increase in the fraction of overall gene expression, calculated as Isoform Fraction (IF) values, originating from transcripts with IR. To test this, we analyzed the distribution of IF values originating from the subset of transcripts containing IR. Indeed, depletion of UBL5 or SART1 causes a marked increase in this distribution (Fig 3D). Overall, the median IF value of transcripts with IR increased approximately 2.5-fold when UBL5 or SART1 was knocked down (Supplementary Fig S3C), further supporting the notion that splicing efficiency is strongly compromised in cells lacking UBL5 or SART1.
Figure 3. UBL5 depletion impairs pre-mRNA splicing by increasing intron retention.
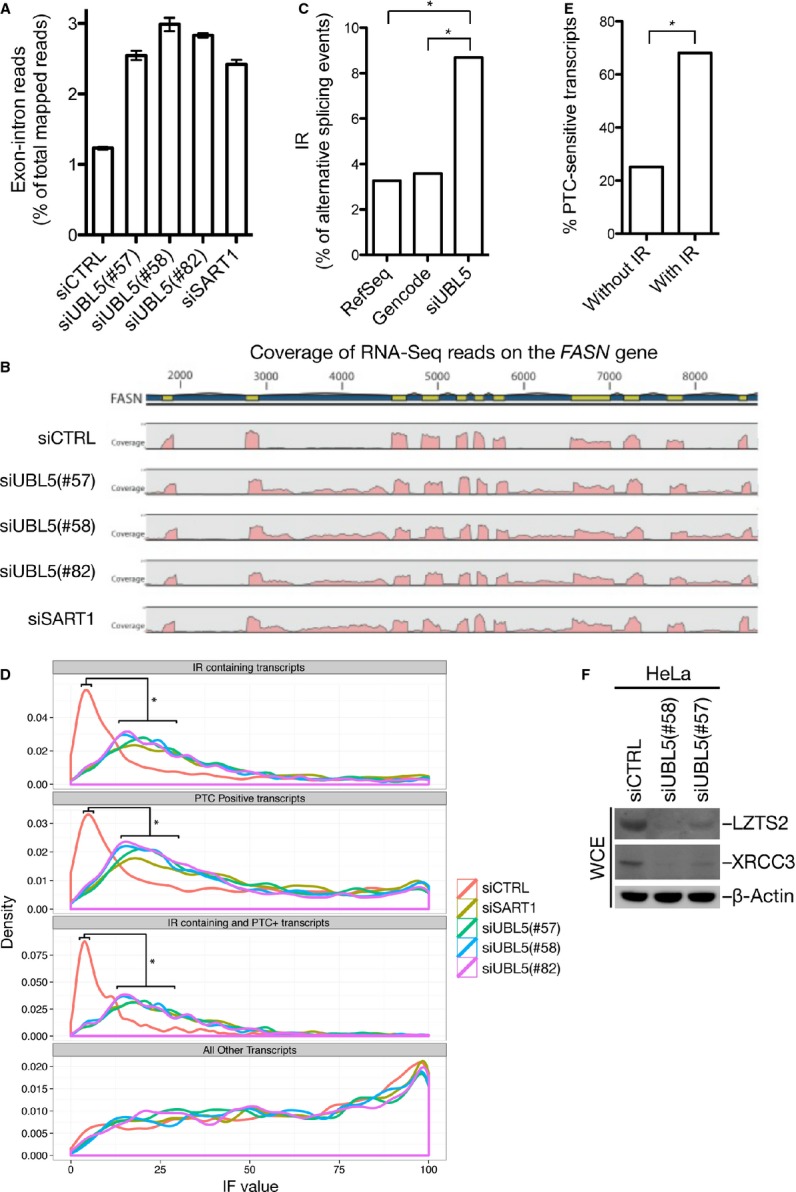
A Proportion of reads spanning exon–intron junctions determined by RNA-Seq analysis of HeLa cells treated with control (CTRL), UBL5, or SART1 siRNAs for 48 h [mean ± SEM (error bars); N = 2]. Supplementary Table S2 shows full RNA-Seq results.
B Density of RNA-Seq reads (pink) mapping to exons (yellow) and introns (blue) in the FASN gene, representative of genes displaying intron retention.
C Frequency of intron retention (IR) among all classified alternative splicing events, obtained via spliceR from RefSeq, Gencode, and UBL5 transcriptomes. Asterisk (*) denotes statistically significant changes (P < 2.2 × 10−16, Fisher’s exact test).
D Significantly differentially expressed transcripts were extracted and divided into the four indicated, overlapping subsets. The distribution of IF values from each condition was plotted. Asterisk (*) denotes changes that are statistically significant between control and all other samples (P < 1 × 10−4, pairwise Mann–Whitney U-test).
E Significantly differentially expressed transcripts were extracted, and the frequency of NMD sensitivity was compared between transcripts with and without IR. Asterisk (*) denotes statistically significant changes (P < 2.2 × 10−16, Fisher’s exact test).
F Whole-cell extracts (WCE) of HeLa cells transfected with non-targeting (CTRL) or UBL5 siRNAs for 48 h were immunoblotted with indicated antibodies.
IR can lead to introduction of premature stop codons (PTCs), sensitizing transcripts to nonsense-mediated mRNA decay (NMD) 20. We found that knockdown of UBL5 or SART1 led to an increase in the subset of transcripts containing PTCs (Fig 3D). This significant shift toward higher IF values was even more pronounced in the subset of transcripts that contain both IR and PTC (Fig 3D), where an approximately threefold increase in the median IF value was evident (Supplementary Fig S3C). This may be explained by the high probability of a PTC being introduced by intron retention, as previously suggested 21. Indeed, we noted a higher fraction of NMD-sensitive isoforms among transcripts containing IR (Fig 3E). To confirm that these RNA-Seq data reliably predict proteins whose expression is deregulated in UBL5-depleted cells, we monitored the protein levels of XRCC3, whose mRNA level is significantly decreased, as well as LZTS2, which is significantly upregulated on the overall transcript level but displays marked isoform switching from a PTC-negative to a PTC-positive transcript (Supplementary Table S3). We found that the expression of both proteins was indeed downregulated in cells lacking UBL5, as expected (Fig 3F).
Together, these data demonstrate that UBL5 has a crucial role in supporting pre-mRNA splicing integrity in human cells and that functional ablation of UBL5 deregulates this process by increasing IR, affecting a large number of transcripts. The precise mechanistic basis of this remains to be established, however, as we have not observed any unique features of retained introns in UBL5-depleted cells.
Defective pre-mRNA splicing impairs sister chromatid cohesion through downregulation of Sororin
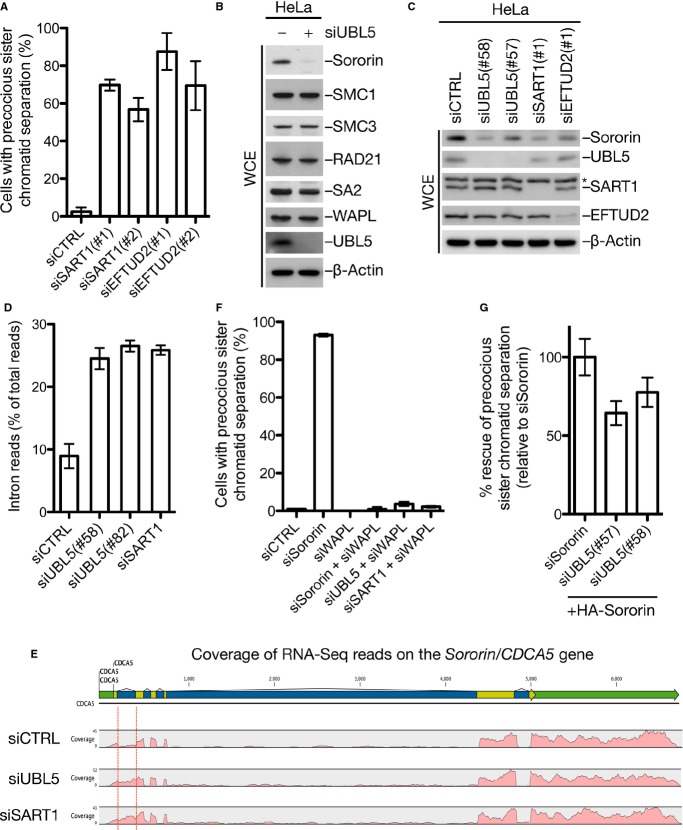
Based on the above findings, we surmised that UBL5 might exert its role in chromosome cohesion maintenance indirectly via its involvement in pre-mRNA splicing. In support of this idea, we found that knockdown of the UBL5-interacting splicing factors SART1 and EFTUD2 also strongly impaired sister chromatid cohesion in mitosis (Fig 4A, and Supplementary Fig S4A). Inspection of the MitoCheck database 22,23 of genes whose knockdown perturbs mitotic progression corroborated that depletion of UBL5 and other spliceosomal proteins gives rise to multiple mitotic defects, revealing a notable correlation between phenotypes resulting from knockdown of splicing factors and cohesin components (Supplementary Fig S4B). This suggests that the integrity of pre-mRNA splicing is essential for proper chromosome cohesion maintenance in human cells. To address the mechanistic basis of this requirement, we analyzed cohesion status in UBL5-depleted cells. The loading of cohesin onto chromatin upon exit from mitosis appeared overtly normal in these cells, and the localization of SGO1, which protects cohesion maintenance in mitosis, to kinetochores was not affected by UBL5 knockdown (Supplementary Fig S5A and B). Remarkably, however, we observed marked loss of Sororin but not other known cohesion factors in UBL5-depleted cells, and less Sororin was loaded onto chromatin upon mitotic exit (Fig 4B, and Supplementary Fig S5A and C). Sororin expression was also reduced in cells depleted of SART1 or EFTUD2 (Fig 4C), indicating that it was a consequence of compromised pre-mRNA splicing. Consistently, RNA-Seq analysis showed that knockdown of UBL5 or SART1 markedly enhanced IR in Sororin transcripts, primarily affecting the first intron (Fig 4D and E, and Supplementary Fig S5D). Unlike Sororin protein expression, however, the overall level of Sororin mRNA was normal in UBL5-depleted cells (Supplementary Fig S5D); thus, it is possible that the IR-containing Sororin transcripts are translated inefficiently or give rise to aberrant or unstable protein products. In line with a causal role of Sororin loss for the cohesion defect in UBL5- or SART1-depleted cells, we found that co-depletion of WAPL fully reversed this phenotype (Fig 4F, and Supplementary Fig S5E). Finally, we found that expression of an intron-less, and thus splicing-insensitive, Sororin cDNA in cells lacking UBL5 restored proper sister chromatid cohesion almost as efficiently as it did in cells depleted of endogenous Sororin (Fig 4G, and Supplementary Fig S5F). Together, these findings strongly suggest that precocious sister chromatid separation arising from deregulation of the pre-mRNA splicing machinery primarily reflects downregulation of Sororin. It is possible, however, that missplicing of factors outside the core cohesion machinery may also indirectly contribute to this defect.
Figure 4. Spliceosome dysfunction compromises sister chromatid cohesion through deregulation of Sororin.
A Precocious sister chromatid separation in HeLa cells transfected with indicated siRNAs [mean ± SD (error bars); N = 3]. At least 100 metaphase spreads were counted. Knockdown efficiency of siRNAs was analyzed by immunoblotting (Supplementary Fig S5A).
B Levels of cohesion factors in HeLa cells transfected with non-targeting (−) or UBL5 siRNAs were determined by immunoblotting.
C HeLa cells transfected with indicated siRNAs were processed for immunoblotting.
D Proportion of RNA-Seq reads from Sororin transcripts mapping to introns in cells transfected with indicated siRNAs [mean ± SEM (error bars); N = 2].
E Density of RNA-Seq reads (pink) mapping to exons (yellow) and introns (blue) in the Sororin (CDCA5) gene, showing retention of intron 1 (marked by red lines).
F Precocious sister chromatid separation in HeLa cells transfected with indicated siRNAs [mean ± SD (error bars); N = 3]. At least 100 metaphase spreads were counted. Knockdown efficiency of siRNAs was analyzed by immunoblotting (Supplementary Fig S5E).
G Sister chromatid cohesion status in metaphase spreads from cells transfected with indicated siRNAs and HA-Sororin cDNA [mean ± SD (error bars); N = 3]. Rescue efficiencies were normalized to the effect of expressing HA-Sororin in siSororin-treated cells. At least 100 metaphase spreads were counted. See also Supplementary Fig S5F.
Our results demonstrate that human UBL5 plays an evolutionarily conserved role in pre-mRNA splicing. Mechanistically how UBL5 underpins spliceosome integrity remains to be established, a challenging task given the exceedingly complex nature of this multimegadalton ribonucleoprotein complex. Because human UBL5 displays potential interactions with a range of proteins not known or predicted to function in a spliceosomal context (Supplementary Table S1), we consider it likely that it may also have functions outside pre-mRNA splicing.
Materials and Methods
Plasmids and siRNA
Full-length human UBL5 cDNA was inserted into pcDNA4/TO (Invitrogen) containing an N-terminal Strep-HA-tag, and the construct was verified by sequencing. Plasmid DNA and siRNA transfections were performed using GeneJuice (Novagen) and Lipofectamine RNAiMAX (Invitrogen), respectively, according to the manufacturer’s instructions.
Cell culture, flow cytometry, and chromosome spreads
Human U2OS and HeLa cells were cultured in DMEM containing 10% fetal bovine serum. HeLa/H2B-mCherry cells were a kind gift from Dr. Jakob Nilsson (NNF Center for Protein Research, University of Copenhagen, Denmark). Cell cycle profiles were determined by flow cytometry analysis using a FACSCalibur flow cytometer (BD Biosciences). To prepare chromosome spreads, cells treated with nocodazole for 3 h were collected and incubated in 0.075 M KCl solution for 20 min, fixed in methanol/acetic acid (3:1 ratio) and dropped onto glass slides, and stained with 5.8% Giemsa solution.
Immunochemical methods, immunofluorescence, and microscopy
Immunoblotting, Strep-Tactin pull-downs, chromatin enrichment, and immunofluorescence staining were done as described 24. To monitor mitotic progression in live cells, cells were grown in glass bottom culture dishes (Menzel Glaeser, Hounissen), and time-lapse 3D image stacks of single cells were acquired with a Zeiss Axio Observer confocal microscope using a 100× oil-immersion lens and a Yokogawa CSU-X1 spinning disk. Approximately 30 optical sections per single cell (delta z of 0.65 μm) were acquired at 1-min intervals for 60–180 min. For analysis of mitosis in larger cohorts of live cells, cells were cultured in CO2-independent F15 medium (Invitrogen) in ibiTreat microscopy chambers (ibidi, Germany). Time-lapse 3D image stacks of fields were acquired using a 40× oil-immersion lens and an Olympus IX71a epifluorescence microscope equipped with Delta Vision automatic acquisition software. Three focal planes (5 μm spacing) were acquired every 5 min for 24 h.
Mass spectrometry and RNA-Seq analysis
Mass spectrometry-based identification of UBL5-interacting proteins was done as described 25. For RNA-Seq, total RNA was extracted using RNeasy Mini kit (Qiagen), and transcriptome libraries were generated using the TruSeq RNA Sample Prep kit v2 (Illumina) according to the manufacturer’s instructions. Second-generation sequencing of 100-nt single end reads was performed by the National High Throughput DNA Sequencing Centre (University of Copenhagen, Denmark) using an Illumina Hiseq2000. For initial read mapping assessment, all reads were aligned to the human genomic sequence and quantified using the CLC Genomic Workbench Software (CLC Bio). Reads were mapped with a minimum of 50 bases of consecutive matches, allowing for up to three mismatches. For all downstream bioinformatics analysis, reads were mapped to the human genome hg19 assembly, using TopHat v2.0.7 with default parameters. Full-length transcripts were obtained via Cufflinks v2.1.1 using UCSC hg19 annotations (provided by Illumina though the iGenomes package) as reference transcriptome, setting min-isoform-fraction parameter to 0.03. All full-length transcripts from RefSeq and Gencode were downloaded from the UCSC genome browser on August 8, 2013. Identification of alternative splicing was done with spliceR software based on full-length transcripts, as described 19. Raw RNA-Seq data can be downloaded from GEO (Gene Expression Omnibus; www.ncbi.nlm.nih.gov/geo/) under the accession number GSE59376.
Acknowledgments
We thank Drs. Hideki Yashiroda, Jan-Michael Peters, and Jakob Nilsson for providing reagents, and Kim Magnussen (University of Copenhagen) for help with sequencing of transcriptome libraries. This work was supported by grants from the Novo Nordisk Foundation, Danish Medical Research Council, and the Lundbeck Foundation.
Author contributions
YO and HV performed most experiments and analyzed data, supported by MM; KV-S, JW, AH and MR analyzed RNA-Seq data; PB and CC performed mass spectrometry experiments and data analysis; SB-J co-supervised the project and analyzed data; NM supervised the project, analyzed data, and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally T, Huang Q, Janis RS, Liu Z, Olejniczak ET, Reilly RM. Structural analysis of UBL5, a novel ubiquitin-like modifier. Protein Sci. 2003;12:1562–1566. doi: 10.1110/ps.0382803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelot TA, Cort JR, Yee AA, Semesi A, Edwards AM, Arrowsmith CH, Kennedy MA. Solution structure of the yeast ubiquitin-like modifier protein Hub1. J Struct Funct Genomics. 2003;4:25–30. doi: 10.1023/a:1024674220425. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Dittmar GA, Ohi MD, Uetz P, Jones N, Finley D. Ubiquitin-like protein Hub1 is required for pre-mRNA splicing and localization of an essential splicing factor in fission yeast. Curr Biol. 2004;14:2283–2288. doi: 10.1016/j.cub.2004.11.058. [DOI] [PubMed] [Google Scholar]
- Yashiroda H, Tanaka K. Hub1 is an essential ubiquitin-like protein without functioning as a typical modifier in fission yeast. Genes Cells. 2004;9:1189–1197. doi: 10.1111/j.1365-2443.2004.00807.x. [DOI] [PubMed] [Google Scholar]
- Mishra SK, Ammon T, Popowicz GM, Krajewski M, Nagel RJ, Ares M, Jr, Holak TA, Jentsch S. Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing. Nature. 2011;474:173–178. doi: 10.1038/nature10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Peters JM, Nishiyama T. Sister chromatid cohesion. Cold Spring Harb Perspect Biol. 2012;4:a011130. doi: 10.1101/cshperspect.a011130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell. 2005;18:185–200. doi: 10.1016/j.molcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Lafont AL, Song J, Rankin S. Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc Natl Acad Sci USA. 2010;107:20364–20369. doi: 10.1073/pnas.1011069107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010;143:737–749. doi: 10.1016/j.cell.2010.10.031. [DOI] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rankin S, Yu H. Phosphorylation-enabled binding of SGO1-PP2A to cohesin protects sororin and centromeric cohesion during mitosis. Nat Cell Biol. 2013;15:40–49. doi: 10.1038/ncb2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22:R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- Vitting-Seerup K, Porse BT, Sandelin A, Waage J. spliceR: an R package for classification of alternative splicing and prediction of coding potential from RNA-seq data. BMC Bioinformatics. 2014;15:81. doi: 10.1186/1471-2105-15-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Maquat LE. Organizing principles of Mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Hériché JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Hériché JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen M, Lukas C, Lukas J, Bekker-Jensen S, Mailand N. Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J Cell Biol. 2012;197:189–199. doi: 10.1083/jcb.201109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W, Wikström M, Choudhary C, Houtsmuller AB, Marteijn JA, et al. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol. 2013;201:797–807. doi: 10.1083/jcb.201212075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.