Abstract
Shigella flexneri, the etiological agent of bacillary dysentery, invades the human colonic epithelium and causes its massive inflammatory destruction. Little is known about the post-translational modifications implicated in regulating the host defense pathway against Shigella. Here, we show that SUMO-2 impairs Shigella invasion of epithelial cells in vitro. Using mice haploinsufficient for the SUMO E2 enzyme, we found that sumoylation regulates intestinal permeability and is required to restrict epithelial invasion and control mucosal inflammation. Quantitative proteomics reveals that Shigella infection alters the sumoylation status of a restricted set of transcriptional regulators involved in intestinal functions and inflammation. Consistent with this, sumoylation restricts the pro-inflammatory transcriptional response of Shigella-infected guts. Altogether, our results show that the SUMO pathway is an essential component of host innate protection, as it reduces the efficiency of two key steps of shigellosis: invasion and inflammatory destruction of the intestinal epithelium.
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Post-translational Modifications, Proteolysis & Proteomics
Keywords: inflammation, proteomics, Shigella flexneri, SUMO, transcription, Ubc9
Introduction
Since its discovery, the post-translational modification by SUMO (Small Ubiquitin-like MOdifier) has been shown to regulate a wide range of cellular processes 1,2. SUMO modification is mechanistically similar to ubiquitin but is carried out by a distinct set of enzymes. Unlike ubiquitin, the SUMO modification system has only a single SUMO E1-activating enzyme (SAE1/SAE2) and a SUMO E2-conjugating enzyme (UBC9). Among the three SUMO paralogs, SUMO-2 and SUMO-3 can form polymeric chains 3, with one potential outcome being poly-SUMO-dependent polyubiquitylation, resulting in proteasomal degradation of the modified substrate. At the organism level, SUMO modification is essential, as mouse embryos deficient for the Ubc9 die early in development 4. However, only one expressed allele of Ubc9 is sufficient to maintain basic functions, as Ubc9+/− mice are viable with no observable phenotype 4. Similar to other post-translational modifications, sumoylation plays a role in pathogenic infections 5. Although the interplay between sumoylation and viral infection has been well established 6,7, the link between SUMO and bacteria is less well understood. To date, it has been shown that Listeria monocytogenes infection leads to a global decrease in the amount of SUMO conjugates, triggered by the pore-forming toxin LLO which induces the degradation of Ubc9 8.
We decided to study the interplay between sumoylation and the Gram-negative enterobacterium Shigella flexneri. This bacterium is the etiological agent of bacillary dysentery, an acute colitis responsible for heavy mortality of children in developing countries 9. The pathogenesis of Shigella involves the rupture, invasion and inflammatory destruction of the colonic epithelial lining that results in the destruction of the mucosa 10,11. The interplay between epithelial cell invasion and inflammation is central in the development of shigellosis 12,13.
Here, we demonstrate that SUMO modification impairs the invasive capacity of Shigella in vitro and is required to protect the host against the pathogen in vivo. Mice harboring loss of one Ubc9 allele is more susceptible to Shigella and exhibits a phenotype marked by increased epithelial cell permeability, massive mucosal invasion and a dramatic inflammatory response. Using quantitative proteomics, we show that infection with an invasive versus a non-invasive strain of Shigella alters the SUMO modification status of host cell proteins, especially affecting key transcriptional regulators involved in gut physiology and inflammatory response. Thus, SUMO-dependent regulation of a subset of transcriptional regulators may participate to the host–pathogen interaction upon Shigella infection. Altogether, these data show the essential role of sumoylation in the host capacity to control Shigella invasion.
Results and Discussion
SUMO-2 and SAE2 impair Shigella flexneri infectivity in vitro
To investigate the impact of sumoylation on the pathogenicity of Shigella flexneri, we first evaluated the capacity of bacteria to enter host cells overexpressing affinity-tagged forms of SUMO in HeLa cells. Western blot analysis with antibodies specific for the protein A component of the TAP tag revealed that the levels of TAP-SUMO-1 and TAP-SUMO-2 were comparable (see Fig 1A, left panel). Immunoblot experiments confirmed the overexpression of the different SUMO-1 and SUMO-2 moieties (Fig 1A, middle and right panels). Using these stable cell lines, we performed gentamicin assays to measure the pool of intracellular bacteria after one hour of infection. As shown in Fig 1B, SUMO-2 overexpression resulted in a sixfold decrease in Shigella entry into cells. Taking an alternative approach, a similar assay was performed in cells transfected with a siRNA directed against the SUMO E1 activating enzyme SAE2. SAE2 depletion (Fig 1C) resulted in decreased sumoylation (SUMO-1 and SUMO-2) and increased the expression of free forms of the two SUMO paralogs (Supplementary Fig S1A and B). As a control, we verified that SAE2 depletion had no impact on protein ubiquitylation (Supplementary Fig S1C). Results presented in Fig 1D clearly show that the decrease in SUMO conjugation favored Shigella entry into host cells (3.4-fold increase). To further characterize the role of sumoylation in the bacterial invasive capacity and spreading, we performed plaque assays on the TAP-SUMO cells (Fig 1E and F). The number of lysis plaques following M90T infection appeared dramatically reduced in TAP-SUMO-2 cells compared to TAP and TAP-SUMO-1 cells (Fig 1E and F). As expected, no lysis plaque was observed when the cells were infected with the non-invasive mutant mxiD, which is mutated in the type III secretion system (Fig 1E). In agreement with Fig 1B, and contrary to what was reported in the case of Listeria monocytogenes 8, SUMO-1 overexpression did not affect the invasive capacity of Shigella. Since the molecular entry mechanisms of these two pathogens are fundamentally different (i.e. trigger versus zipper mechanisms) 14, it is not surprising that such discrepancy is observed.
Figure 1. SUMO regulates Shigella flexneri infectivity in vitro.
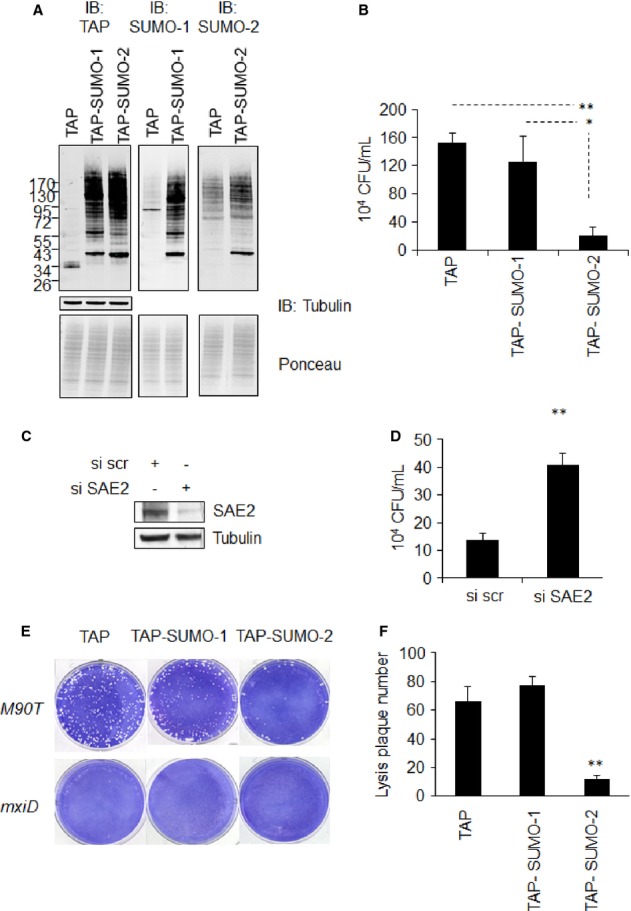
A Immunoblot analysis of sumoylation in HeLa cells expressing TAP, TAP-SUMO-1 or TAP-SUMO-2, n = 3.
B Gentamycin assay showing Shigella flexneri entry properties. Values of intracellular bacteria (M90T) are expressed as colony-forming unit and are relative to cells infected with mxiD non-invasive Shigella (n = 3 in three technical replicates). Error bars indicate standard deviation. *P < 0.05; **P < 0.01.
C Immunoblot analysis of HeLa cell lysates using siRNA targeting SAE2.
D Gentamycin assay as in (B) in Hela cells transfected with siRNA against SAE2 and scrambled siRNA as a control (n = 3). Error bars indicate standard deviation. **P < 0.01.
E Analysis of Shigella invasive properties using lysis plaque formation assay in SUMO-expressing HeLa cells. Cells were infected with Shigella flexneri M90T and non-invasive mxiD mutant as a control (n = 4).
F Graphical representation of lysis plaque number. Error bars indicate standard deviation. **P < 0.01.
Source data are available online for this figure.
Sumoylation regulates Shigella pathogenesis in vivo
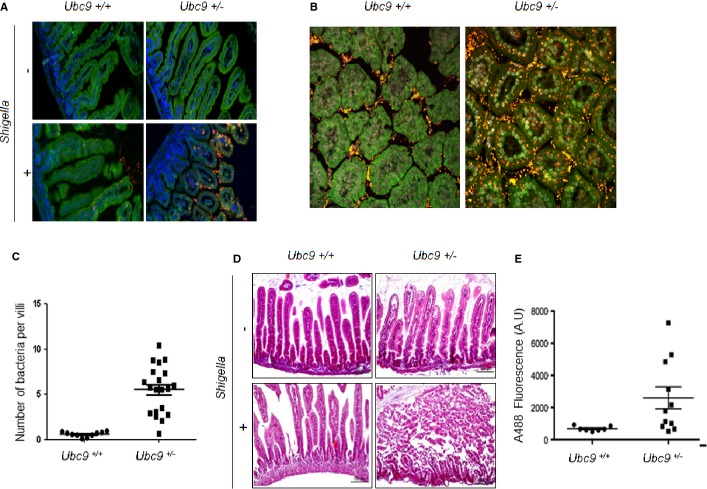
While the data presented here indicate that SUMO modification restricts the invasion and the growth of Shigella in cultured human cells, it was important to establish that this was also the case in vivo. As adult mice are refractory to Shigella infection, we used the previously reported newborn mouse model of intragastric (i.g.) infection 15. The impact of sumoylation on infection was tested using Ubc9+/− mice compared to Ubc9+/+ mice. We inoculated i.g. 4-day-old newborn mice for 3 h with the invasive isolate M90T or used physiological water as a control. By performing immunofluorescence on tissue sections using an anti-LPS antibody, we observed that intestinal mucosae of Ubc9+/− mice contain significantly more bacterial foci than that of Ubc9+/+ mice (Fig 2A and B). Indeed, there was a fivefold difference in the mean number of bacteria per villus, indicating a massive epithelial invasion in intestines with reduced amount of Ubc9 (Fig 2C). We next analyzed mucosal lesions on tissue sections stained with hematoxylin–eosin (Fig 2D). In agreement to what has been previously published, infection by the Shigella strain M90T triggers a shortening of the intestinal villi and a thickening of the lamina propria in wild-type animals (see Fig 2D, left panel). This mild phenotype contrasts with the one observed in Ubc9+/− mice which show a massive destruction of the villi characteristic of a strong inflammatory phenotype (Fig 2D, right panel). These results show that Ubc9, and therefore sumoylation, play a critical role in Shigella infectivity in vivo. This phenotype seems to be specific to infection as no difference was observed in the two genotypes treated with physiological water (Fig 2D, upper panel).
Figure 2. Ubc9+/− mice display hypersensitivity to Shigella flexneri and increased intestinal permeability.
A, B Epifluorescence (A) and confocal microscopy analysis (B) of staining of the intestinal epithelium on paraffin sections after Shigella infection. Physiological water was used as a control. Bacteria are stained using an anti-LPS antibody (in red), auto-fluorescence of the intestinal tissue is shown (in green), and nuclei are counterstained with DAPI (in blue).
C, D Intestinal invasiveness (C) and hematoxylin–eosin staining (D) of the intestinal epithelium on paraffin sections, original magnifications (40× ), (n = 3 per group).
E Intestinal permeability measured by serum containing dextran-FITC of 4 days newborn Ubc9+/+ versus Ubc9+/− mice. FITC fluorescence is quantified by measuring absorbance at 488 nm and graphically represented in arbitrary unit (AU), n = 6 for each experimental condition.
Finally, we looked for a functional feature of the Ubc9+/− mice which could explain the increased bacterial uptake. We searched for possible dysregulation of the intestinal permeability in Ubc9+/− newborn mice. Briefly, a solution of dextran-FITC was administrated to newborn mice that were shortly after sacrificed. The level of dextran-FITC, which can potentially cross the intestinal barrier and be released from the intestine, is measured in the serum 16. As shown in Fig 2E, Ubc9+/− mice display significantly higher levels of dextran-FITC in the serum when compared to control animals. Therefore, Ubc9+/− mice display an increased intestinal permeability that could account for the higher Shigella uptake. This role of SUMO in the maintenance of intestinal structure and function is in agreement with our previous findings that conditional knock-out of Ubc9 leads to a rapid death of animals due to intestinal failure with profound alteration of the polarized organization and mechanical stability of the enterocytes 17. The leaky epithelial barrier of Ubc9+/− mice, despite sufficient to maintain tissue homeostasis in steady-state conditions, is likely to facilitate the invasive process upon bacterial infection. Altogether, these results show that sumoylation plays a protective role in Shigella infection in vivo. Moreover, this indicates that Ubc9+/− newborn mice are a new model of hypersensitivity to this pathogen.
Identification of targets of SUMO-2 by SILAC-based quantitative proteomics
To gain mechanistic insight into SUMO-dependent host–pathogen interactions, we next considered the possibility that Shigella infection may alter the SUMO modification status of host proteins. To investigate this, we used a quantitative proteomics approach to study changes to the host SUMO-2 conjugate proteome during Shigella infection. We used the above-described HeLa cells that overexpress TAP-SUMO-2 in conjunction with SILAC (Stable Isotope Labeling of Amino acids in Culture) 18. A triple label approach was taken as described in Fig 3A. A crude extract was also analyzed to control for total protein changes among conditions. By this design, comparisons between TAP only and TAP-SUMO-2 (M/L and H/L ratios) gives an indication of the TAP-SUMO-2 conjugate sub-proteome, and comparisons between M90T and mxiD infections (M/H) can reveal changes specific to invasive infection. Mass spectrometry coupled to quantitative data analysis quantified 2,222 proteins from crude cell lysates and 852 proteins from the TAP-purified samples. The mass spectrometry data are available via the PRIDE repository (dataset identifier PXD001100) 19. 567 proteins could be identified in both crude and purified samples (Supplementary Fig S2). As shown on the scatter plot in Fig 3B in which each dot represents a single protein, the distribution of log2 ratios is broader in the TAP-purified samples than in the crude lysates, with the 448 putative TAP-SUMO-2 conjugates apparent as a high ratio cluster distinct from the contaminants. This group compares favorably (77%) with two previously published TAP-SUMO-2 proteomes 18,20 (Supplementary Fig S2C), leaving 102 proteins being identified in this study as putative novel TAP-SUMO-2 substrates.
Figure 3. A SILAC-based quantitative proteomics experiment to identify putative SUMO-2 substrates and their regulation upon infection by Shigella flexneri.
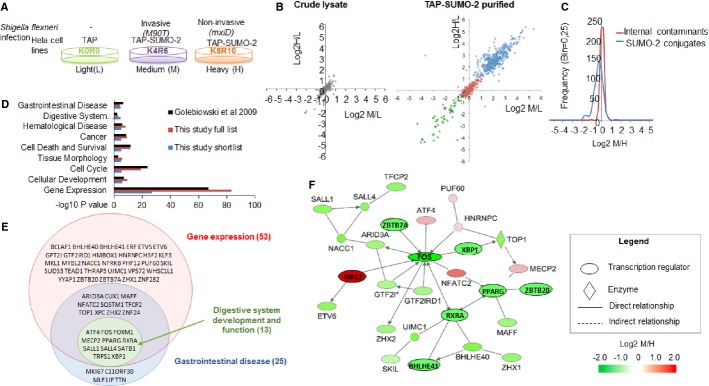
A Overview of the quantitative experiment comparing TAP-expressing with TAP-SUMO-2-expressing HeLa cells that were infected with invasive strain of Shigella (M90T) or infected with non-invasive Shigella mutant (mxiD) for 1 h (n = 1).
B Scatter plots comparing M/L and H/L ratios proteins identified in crude cell lysates (left) and from the TAP-SUMO-2 purified samples (right). In the TAP-SUMO-2 plot, each identified proteins is shown as either external contaminant (in green), internal contaminant (in red) or TAP-SUMO-2 conjugate (in blue).
C Frequency histogram comparing M/H ratios (invasive/non-invasive) between the TAP-SUMO-2 putative substrates and the internal contaminants.
D IPA comparative analysis for functions and diseases associated with the 87 significantly regulated in M/H ratio putative TAP-SUMO-2 conjugates (blue), all 438 TAP-SUMO-2 putative conjugates identified in this study (red) and the 753 putative TAP-SUMO-2 conjugates identified by Golebiowski et al 18 (black).
E Overlap between enriched functional groups identified by IPA for the shortlisted group of 87 TAP-SUMO conjugates. Proteins are shown by gene name.
F Twenty-eight of the 53 transcriptional regulators from the shortlist formed an IPA functional network, implying that the modification state of functionally related proteins involved in gene expression is significantly different between cells invaded by Shigella in comparison with those that are not.
Shigella infection modulates a specific set of sumoylated transcriptional regulators
By studying the distribution of normalized log2 ratios of purified samples from invasive (M)/non-invasive (H), we observed that proteins were generally less modified by SUMO-2 under invasive infection (Fig 3C). However, on this global scale, the difference between non-invasive and invasive Shigella is relatively modest (median log2M/H = −0.33), so we next sought to identify individual proteins that were modified to a different extent when comparing invasive and non-invasive infections. 87 proteins had a significantly affected M/H ratio (MaxQuant Significance B < 0.1), among which, 22 were more modified by SUMO-2 upon invasive infection, and 65 less sumoylated compared to the non-invasive infection (Supplementary Table S1, and Supplementary Dataset S1). Consistent with previous TAP-SUMO proteomic studies proteomes 18,20, ‘Gene expression’ was the most enriched function in both the full TAP-SUMO-2 list as well as the significant shortlist (Fig 3D). In total, 53 of the 87 shortlisted proteins were defined as transcriptional regulators. Two further functional categories with relevance to this study were significantly enriched in the shortlist of TAP-SUMO-2 substrates: ‘Gastrointestinal disease’ (25/87) and ‘Digestive system development and function’ (13/87) (Fig 3D). Importantly, these groups are more significantly enriched in this study than in the other TAP-SUMO-2 studies, suggesting this to be a specific feature of Shigella infection. The vast majority of these proteins are also transcriptional regulators (Fig 3E), and 28 of the 53 shortlisted transcriptional regulators were capable of forming a functional network in IPA (Fig 3F), indicating a close relationship among them. Interestingly, many of the proteins (16) from the shortlist have already been linked with inflammation, with 8 of these representing known SUMO substrates including c-FOS and PPARγ that have already been associated with Shigella infection (Supplementary Table S2 and Fig 3F, names inside ovals). Remarkably, Shigella-induced changes in sumoylation status of most of the SUMO substrates previously known to play a role in inflammation are predicted to favor the inflammatory response. Most significantly, c-FOS, PPARγ and its heterodimeric partner RXRα have been shown to play key roles in inflammation. Indeed, it is well accepted that sumoylation generally acts as a repressor for transcription factors and activates transcriptional repressors 21–23. Interestingly, sumoylation was shown to downregulate c-FOS and RXRα transcriptional activity 24,25, whereas sumoylation of PPARγ represses the transcriptional activation of inflammatory response genes 26. Thus, the relatively lower levels of SUMO-modified c-FOS and PPARγ/RXRα observed in response to invasive Shigella infection is expected to contribute to the associated inflammatory phenotype. We analyzed by immunoblotting the sumoylation status of two of the transcriptional regulators associated with digestive functions present in the short list, c-FOS and SATB1 (special AT-rich sequence-binding protein-1) (Fig 3E). We confirmed, for both ectopically expressed substrates, the decrease in SUMO-2-modified forms induced by Shigella infection (Supplementary Fig S3). Hence, we conclude that invasive infection by Shigella leads to specific changes in host SUMO modification and particularly affects SUMO-2-modified transcriptional regulators involved in intestinal functions and inflammation response.
Sumoylation regulates the pro-inflammatory transcriptional response to Shigella infection in vivo
We have observed that Ubc9 haploinsufficiency leads to a hyper invasive and inflammatory phenotype in vivo upon infection. At the molecular level, we demonstrated that invasive Shigella infection alters the SUMO modification status of a restricted set of transcription factors involved in gut functions and inflammation. To investigate how SUMO modification impacts on host transcription after Shigella infection, we studied gene expression in the infected intestine of Ubc9 animal models. To analyse the expression of relevant genes regulated by infection, we used a PCR array specific for anti-bacterial response containing probes for 84 relevant genes (Supplementary Table S3). Fig 4A and B shows volcano plots representing significantly dysregulated genes upon infection in three independent Ubc9+/− and Ubc9+/+ mouse intestines. Four genes (out of 84) were induced at least twofold (P-value < 0.05) upon infection in Ubc9+/+ animals (Fig 4A), whereas 17 genes were upregulated in Ubc9+/− animals (Fig 4B). When comparing the genes induced in Ubc9+/− versus Ubc9+/+ infected mucosae, 16 genes were found to be exclusively induced in Ubc9+/− intestines upon infection (Fig 4C), including a large number of inflammatory cytokines and chemokines. The level of sumoylation only regulates pro-inflammatory gene transcription upon infection as no statistical difference could be observed between Ubc9+/+and Ubc9+/− animals in non-infected conditions (Supplementary Fig S4). We confirmed these results by individual RT-qPCR on selected genes. Fig 4D shows that Ifn-γ, Cxcl-1 and Il-6 are significantly more induced in Ubc9+/− than in Ubc9+/+ infected mucosae. These results show that Ubc9 haploinsufficiency leads to enhanced transcriptional activation of inflammatory response genes upon Shigella infection. Upon infection, we identified a regulation of the sumoylation status of key transcriptional regulators likely to favor the induction of pro-inflammatory genes. Together, these data suggest that, in host cells, the concerted decrease in sumoylated forms of a functionally related set of substrates involved in intestinal homeostasis and inflammation contributes to proper anti-bacterial transcriptional response. In conclusion, we identified that SUMO is involved in the primary host response to invasive Shigella infection and is required for efficient protection against Shigella pathogenicity. The SUMO pathway appears to be involved in multiple steps of the host–pathogen interactions upon Shigella infection, and further studies will help to unravel the associated mechanisms of this critical interplay between SUMO and bacteria.
Figure 4. Defects in Ubc9+/− leads to aberrant induction of pro-inflammatory genes upon Shigella infection.
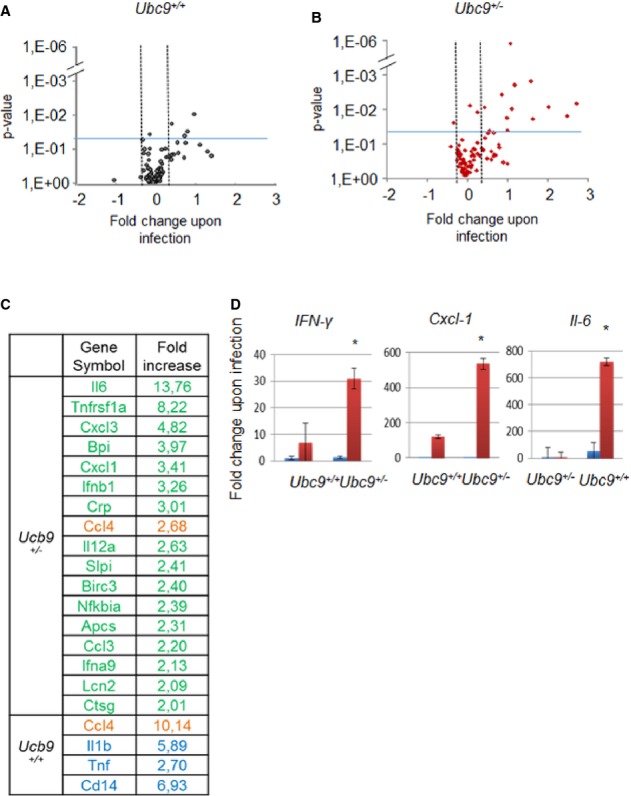
A–C Analysis of gene expression using ‘anti-bacterial response’ PCR arrays, n = 3. Volcano plots (A, B) showing statistically dysregulated gene upon Shigella infection. The graph represents fold change (> 2) ratio of genes in infected (M90T) compared to non-infected conditions (log2 scale). The vertical axis represents the statistically significant dysregulated genes upon infection (−log10 scale; P value < 0.05, see blue line on the graph). Data used for volcano plots originate from experiments performed in Ubc9+/+ (A) and Ubc9+/− (B) mice. Upregulated genes in Ubc9+/− (in green) and Ubc9+/+ (in blue) mice upon infection conditions are shown in (C). Common induced genes are indicated in orange.
D Individual RT-qPCR on selected genes. Expression data upon infection are shown in red, while control conditions are in blue. For each gene, results are expressed as fold regulation relative to gene expression levels in non-infected Ubc9+/+ mice, (n = 3 per group and each n in three technical replicates). Error bars indicate standard deviation. **P < 0.01.
Materials and Methods
Bacterial strains and culture conditions
Shigella flexneri serotype 5a strains were isolated on congo red agar plates. The invasive wild-type strain M90T and its isogenic non-invasive derivative mxiD (impaired for the type-three secretion system) were used. For infection experiments, strains were cultured in BTCS medium (Difco) overnight at 37°C with agitation. Subcultures were performed for 3 h to reach the exponential phase and resuspended in DMEM medium (Invitrogen). For proteomic studies, M90T and corresponding mxiD-Afae strains were used.
Infectivity assays in vitro
Gentamycin assay was performed to quantify intracellular bacteria. Bacteria were added to HeLa cells at a multiplicity of infection of 50. After 45 min of infection, cells were washed and incubated with 10 μg/ml gentamicin for 1 h to kill extracellular bacteria. Infected cells were lysed in PBS/Triton X-100 0.2%. Quantification of intracellular bacteria was done by plating the lysate in triplicates. Cell to cell spreading of bacteria was measured by lysis plaque assay performed as previously described 27 with minor modifications. For enhanced visualization of the plaques, an overlay consisting of DMEM, 0.05% crystal violet (SIGMA) and 0.05% agarose was added on the day 3. Pictures were acquired on day 5, and the number of lysis plaques was measured using ImageJ software. Infectivity assays were done in three technical and biological replicates.
Newborn mice infection
3-day-old newborn mice Ubc9+/+ and Ubc9+/inv (indicated as +/− in this manuscript) were used 4. Mice were separated from their mothers 60 min before the experiment for starvation and then inoculated i.g. with 30 μl of the bacterial suspension (1 × 109 bacteria per μl) or sterile physiological water as a control. For i.g. inoculation of mice, an overnight culture at 37°C on GTCS plates was diluted in physiological water to obtain the bacterial suspension. Newborn mice were maintained at 30°C and monitored. Mice were sacrificed after 3 h and the intestine of newborn mice was removed. Samples of the proximal jejunum were collected for morphological analysis. The rest of the proximal jejunum was used for RNA extraction. Genotypes of the mice were determined post-mortem. Animal experiments were performed accordingly to the guidelines of the Institut Pasteur’s ethical committee for animal use in research (CETEA number 2013-0028).
Bioinformatic and Statistical analysis
Proteins were analyzed for functional enrichment using Ingenuity Pathway Analysis software (IPA). Means ± standard deviations are shown in the figures (1, 2 and 4), and P-values were calculated using a two-tailed two samples equal variance Student’s t-test, (*P < 0.05; **P < 0.01).
Acknowledgments
We acknowledge Ivan Matic and Michael Tatham for help with proteomics. We thank Jacob Seeler and Thierry Pedron for helpful discussions. This work was done as part of the RUBICON-FP6 program (grant to AD and RH). This work was supported by grants from LNCC (Equipe labellisée), Odyssey-RE, INCa, ANR and ERC. S.F. was supported by RUBICON and Sidaction.
Author contribution
AD, PS and SF planned the project. AD, SF, PS and RH designed the experiments. SF, NL, JM, FG, AA and GJ performed the experiments. SF, NL and FG analyzed the data. SF, PS and AD wrote the paper and authors commented on it.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Hannoun Z, Greenhough S, Jaffray E, Hay RT, Hay DC. Post-translational modification by SUMO. Toxicology. 2010;278:288–293. doi: 10.1016/j.tox.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Békés M, Drag M. Trojan horse strategies used by pathogens to influence the small ubiquitin-like modifier (SUMO) system of host eukaryotic cells. J Innate Immun. 2012;4:159–167. doi: 10.1159/000335027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Boggio R, Passafaro A, Chiocca S. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J Biol Chem. 2007;282:15376–15382. doi: 10.1074/jbc.M700889200. [DOI] [PubMed] [Google Scholar]
- Ribet D, Hamon M, Gouin E, Nahori M-A, Impens F, Neyret-Kahn H, Gevaert K, Vandekerckhove J, Dejean A, Cossart P. Listeria monocytogenes impairs SUMOylation for efficient infection. Nature. 2010;464:1192–1195. doi: 10.1038/nature08963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77:651–666. [PMC free article] [PubMed] [Google Scholar]
- Sansonetti PJ. Rupture, invasion and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote-eukaryote cross-talks. FEMS Microbiol Rev. 2001;25:3–14. doi: 10.1111/j.1574-6976.2001.tb00569.x. [DOI] [PubMed] [Google Scholar]
- Sansonetti PJ, Tran Van Nhieu G, Egile C. Rupture of the intestinal epithelial barrier and mucosal invasion by Shigella flexneri. Clin Infect Dis. 1999;28:466–475. doi: 10.1086/515150. [DOI] [PubMed] [Google Scholar]
- Perdomo OJ, Cavaillon JM, Huerre M, Ohayon H, Gounon P, Sansonetti PJ. Acute inflammation causes epithelial invasion and mucosal destruction in experimental shigellosis. J Exp Med. 1994;180:1307–1319. doi: 10.1084/jem.180.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zychlinsky A, Perdomo JJ, Sansonetti PJ. Molecular and cellular mechanisms of tissue invasion by Shigella flexneri. Ann N Y Acad Sci. 1994;730:197–208. doi: 10.1111/j.1749-6632.1994.tb44249.x. [DOI] [PubMed] [Google Scholar]
- Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- Fernandez MI, Thuizat A, Pedron T, Neutra M, Phalipon A, Sansonetti PJ. A newborn mouse model for the study of intestinal pathogenesis of shigellosis. Cell Microbiol. 2003;5:481–491. doi: 10.1046/j.1462-5822.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- An G, Wei B, Xia B, McDaniel JM, Ju T, Cummings RD, Braun J, Xia L. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med. 2007;204:1417–1429. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarque MD, Nacerddine K, Neyret-Kahn H, Andrieux A, Danenberg E, Jouvion G, Bomme P, Hamard G, Romagnolo B, Terris B, et al. Sumoylation by Ubc9 regulates the stem cell compartment and structure and function of the intestinal epithelium in mice. Gastroenterology. 2011;140:286–296. doi: 10.1053/j.gastro.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Matic I, Mann M, Hay RT. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci Signal. 2011;4:rs4. doi: 10.1126/scisignal.2001484. [DOI] [PubMed] [Google Scholar]
- Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Girdwood DWH, Tatham MH, Hay RT. SUMO and transcriptional regulation. Semin Cell Dev Biol. 2004;15:201–210. doi: 10.1016/j.semcdb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Ouyang J, Gill G. SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics. 2009;4:440–444. doi: 10.4161/epi.4.7.9807. [DOI] [PubMed] [Google Scholar]
- Bossis G, Malnou CE, Farras R, Andermarcher E, Hipskind R, Rodriguez M, Schmidt D, Muller S, Jariel-Encontre I, Piechaczyk M. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol Cell Biol. 2005;25:6964–6979. doi: 10.1128/MCB.25.16.6964-6979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SJ, Chung SS, Rho EJ, Lee HW, Lee MH, Choi H-S, Seol JH, Baek SH, Bang OS, Chung CH. Negative modulation of RXRalpha transcriptional activity by small ubiquitin-related modifier (SUMO) modification and its reversal by SUMO-specific protease SUSP1. J Biol Chem. 2006;281:30669–30677. doi: 10.1074/jbc.M604033200. [DOI] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oaks EV, Wingfield ME, Formal SB. Plaque formation by virulent Shigella flexneri. Infect Immun. 1985;48:124–129. doi: 10.1128/iai.48.1.124-129.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.