Abstract
Autophagosome formation is promoted by the PI3 kinase complex and negatively regulated by myotubularin phosphatases, indicating that regulation of local phosphatidylinositol 3-phosphate (PtdIns3P) levels is important for this early phase of autophagy. Here, we show that the Caenorhabditis elegans myotubularin phosphatase MTM-3 catalyzes PtdIns3P turnover late in autophagy. MTM-3 acts downstream of the ATG-2/EPG-6 complex and upstream of EPG-5 to promote autophagosome maturation into autolysosomes. MTM-3 is recruited to autophagosomes by PtdIns3P, and loss of MTM-3 causes increased autophagic association of ATG-18 in a PtdIns3P-dependent manner. Our data reveal critical roles of PtdIns3P turnover in autophagosome maturation and/or autolysosome formation.
Subject Categories: Autophagy & Cell Death; Membrane & Intracellular Transport
Keywords: autolysosome, autophagy, Caenorhabditis elegans, MTM-3, PtdIns3P
Introduction
In the process of macroautophagy (hereafter called autophagy), cytosolic materials are sequestered within double-membrane autophagosomes, which mature to fuse with lysosomes where cargoes are degraded 1. Autophagosome biogenesis is controlled by four protein complexes. The Atg1/Atg13 complex and the class III PI3 kinase Vps34 complex are required for induction and nucleation of the isolation membrane or phagophore, while two ubiquitin-like conjugation systems (Atg5-Atg12, Atg16, and Atg8-PE) act sequentially to elongate the isolation membrane, leading to autophagosome formation 2. The Atg18/Atg2 and Atg1 complexes regulate retrieval of Atg9, which supplies membrane for autophagosome formation 2. Except for Atg8/LC3-II, Atg proteins dissociate from sealed autophagosomes, which then fuse directly with vacuoles in yeast, but interact with endocytic compartments in higher eukaryotes before fusing with lysosomes 3. Consistent with this, essential endocytic regulators such as Rab GTPases and ESCRT proteins play important roles in autophagosome maturation, while Beclin1-binding proteins UVRAG and Rubicon regulate both endocytic transport and autophagy 4.
Phosphatidylinositol 3-phosphate (PtdIns3P) regulates various cellular processes by recruiting specific protein effectors to target membranes. Upon autophagy induction, PtdIns3P is produced at the PAS in yeast or a subdomain of the ER or ER–mitochondria contact sites in higher eukaryotes and promotes autophagosome biogenesis by recruiting PtdIns3P-binding effectors such as Atg18/WIPI and DFCP1 2,5–7. In addition to the PI3 kinase Vps34, which generates PtdIns3P, recent studies indicate that myotubularin phosphatases, which convert PtdIns3P to PI, also play a role in autophagy. There is evidence that both Jumpy/MTMR14 and MTMR3 negatively regulate early events in the mammalian autophagy pathway 8,9. These studies suggest that at the initiation step, the local PtdIns3P level is tightly controlled by both PI3 kinase and phosphatase to ensure that autophagy occurs at an appropriate level. PtdIn3P regulation in autophagosome initiation has been extensively studied, but little is known about how it is controlled in autophagosome maturation into autolysosomes. Ymr1, the only myotubularin phosphatase in yeast, promotes dissociation of Atg proteins from autophagosomes and thus positively regulates autophagy progression 10. It is unclear whether the distinct effect of human and yeast myotubularin phosphatases on autophagy is due to intrinsic species-specific differences in autophagy, or because PtdIns3P turnover has opposing effects if it occurs at different stages of autophagy.
Caenorhabditis elegans contains three active myotubularin phosphatases (MTM-1, 3, and 6) and two inactive members (5 and 9) 11. MTM-1 is important for apoptotic cell clearance, while MTM-6 and 9 regulate endocytic transport 12–15. The cellular functions of MTM-3 and 5 are unclear. Here, we identified C. elegans MTM-3 as a positive regulator of autophagy. Our data indicate that PtdIns3P turnover catalyzed by myotubularin phosphatase plays important roles in autophagosome maturation and/or autolysosome formation.
Results and Discussion
Loss of MTM-3 causes defects in autophagy-related processes
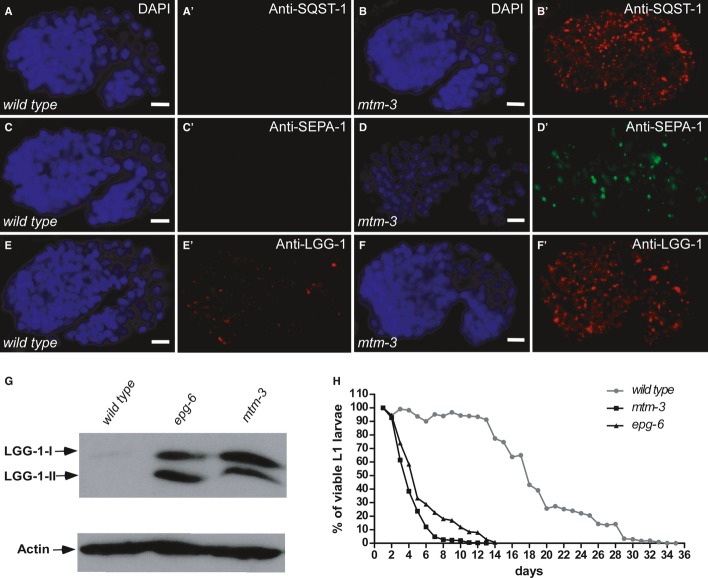
We examined autophagy-related processes in mutants that are defective in mtm-1, 3, 5, 6, and 9 (Supplementary Fig S1A). The C. elegans p62 homolog, SQST-1 (SeQueSTosome-related protein), associates with various protein aggregates and is removed by autophagy during embryogenesis 16. SQST-1 is weakly expressed and diffusely localized in the cytoplasm of wild-type embryos, but forms numerous aggregates in autophagy mutants 16. We found that SQST-1 aggregates were not detected in wild type but accumulated significantly in tm4475, a deletion mutant of mtm-3 that removes all but the first 66 amino acids of the MTM-3 protein (Fig 1A-B′; Supplementary Figs S1A and S2A and B′; Supplementary Methods). Loss of mtm-1, 5, 6, or 9 did not cause accumulation of SQST-1 (Supplementary Fig S1A–F; Supplementary Methods). Similarly, aggregates of SEPA-1, an autophagy substrate that mediates aggregation and autophagic degradation of PGL granules (somatic PGL-1- and PGL-3-positive granules) in C. elegans embryos, were not observed in late embryonic stages in wild-type or mtm-1, 5, 6, and 9 mutants, but persisted in mtm-3(tm4475) embryos (Fig 1C and D′; Supplementary Figs S1G–K and S2C and D′) 17. Other autophagy substrates, including C17E4.2 and C33D9.6, also accumulated ectopically in mtm-3 embryos (Supplementary Fig S2E-H′) 18. Thus, loss of mtm-3, but not other myotubularin phosphatases, affects degradation of various autophagic substrates. The C. elegans Atg8/LC3 homolog, LGG-1, which is essential for autophagosome biogenesis, associates with autophagosome membranes and their precursors. In wild type, LGG-1 puncta are mostly seen around the 100–200-cell stage and decrease as embryos develop 16 (Supplementary Fig S1E and E′). In mtm-3 mutants, however, we observed significantly increased LGG-1 puncta in late embryonic stages and accumulation of both LGG-1-I and LGG-1-II (lipid-conjugated form of LGG-1), suggesting a defect in autophagy (Fig 1F and G). LGG-1 puncta were normal in mtm-1, 5, 6, or 9 mutant embryos (Supplementary Fig S1L–P). In addition to degrading protein aggregates, autophagy activity is required for the survival of newly hatched L1 larvae in the absence of food 19. Like other autophagy-defective mutants, loss of mtm-3 severely affected the survival of starved L1 larvae, reducing the mean life span to 3.5 days from 18.0 days in wild type (Fig 1H). These data further suggest that loss of mtm-3 function impairs autophagy. Recombinant MTM-3 displayed significant phosphatase activity toward PtdIns3P and PtdIns(3,5)P2 in vitro (Fig 2J; Supplementary Methods). Expressing wild-type but not catalytically inactive MTM-3 completely rescued the autophagy phenotypes of mtm-3(lf), indicating that MTM-3 acts as a lipid phosphatase to promote autophagy (Fig 2J; Supplementary Fig S3A–C and H). Expression of human MTMR3 driven by the mtm-3 promoter efficiently rescued the autophagy defects in mtm-3 mutants (Supplementary Fig S3D and H), suggesting that human MTMR3 can substitute for MTM-3 in C. elegans autophagy. Overexpression of MTM-3 did not cause LGG-1 accumulation, consistent with a positive role of it in autophagy (Supplementary Fig S3E–I).
Figure 1. Loss of mtm-3 impairs autophagy.
A-F′ Confocal fluorescent images of wild-type (A, C, E) and mtm-3(tm4475) (B, D, F) embryos at the 1.5-fold stage stained by DAPI (A–F) and anti-SQST-1 (A′, B′), anti-SEPA-1 (C′, D′) or anti-LGG-1 (E′, F′) antibodies. Scale bars: 5 μm.
G Western blot analysis of LGG-1-I and LGG-1–II (lipidated) in wild type, epg-6, and mtm-3.
H mtm-3 and epg-6 mutant L1 larvae have shortened mean and maximum life span in the absence of food. At least 300 animals were scored each day.
Source data are available online for this figure.
Figure 2. GFP::MTM-3(C459S) associates with autophagic structures.
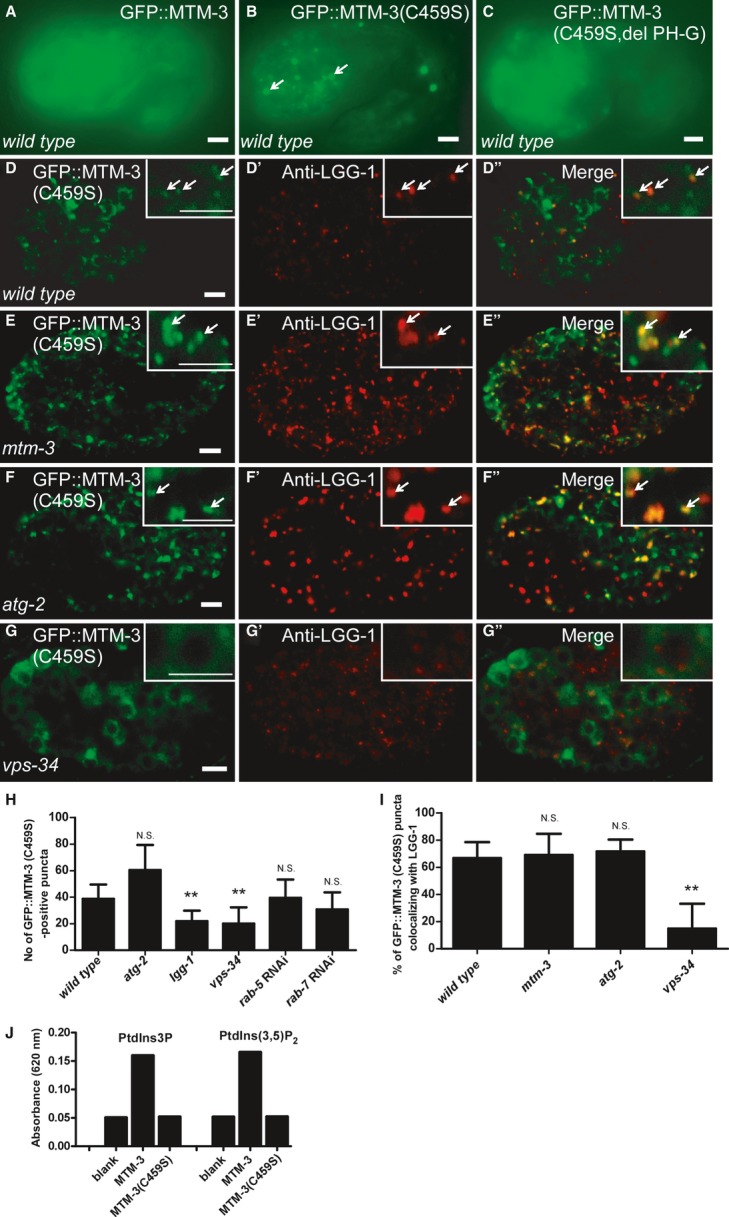
A–C Fluorescent images of wild-type embryos (1.5-fold stage) expressing GFP::MTM-3 (A), GFP::MTM-3(C459S) (B), or GFP::MTM-3(C459S, del PH-G) (C).
D-G″ Confocal fluorescent images of wild-type (D-D″), mtm-3 (E-E″), atg-2 (F-F″), and vps-34 (G-G″) embryos at the 200-cell stage expressing GFP::MTM-3(C459S) and stained by anti-LGG-1 antibody. Insets show magnified views. Arrows indicate overlapping GFP and LGG-1 puncta. Scale bars: 5 μm.
H, I Number of GFP::MTM-3(C459S)-positive puncta and co-localization of GFP::MTM-3(C459S)- and LGG-1-positive puncta in various strains [mtm-3(tm4475), atg-2(bp576), lgg-1(bp500), vps-34(h797)]. At least 10 (I) and 15 (H) embryos were scored per strain. Data are shown as mean ± SD. Data from different mutant backgrounds were compared with wild type. **P < 0.0001; N.S.: not statistically different.
J MTM-3 but not MTM-3(C459S) exhibits phosphatase activity toward PtdIns3P and PtdIns(3,5)P2 in vitro.
Source data are available online for this figure.
mtm-6 and 9 are required for endocytosis by the C. elegans macrophage-like coelomocytes 12, but in mtm-3(tm4475) mutants, GFP secreted from body wall muscle cells was efficiently endocytosed and transported by coelomocytes, which contained normal endosomes and lysosomes as in wild type (Supplementary Fig S4A and B′). Uptake, trafficking and degradation of the C. elegans yolk protein VIT-2 were also normal in mtm-3(tm4475) worms (Supplementary Fig S4C–H′). Thus, endocytosis is not obviously affected in mtm-3 mutants.
MTM-3 is widely distributed and can be recruited to autophagic structures
We generated a GFP::MTM-3 reporter driven by the mtm-3 promoter (Pmtm-3GFP::MTM-3), which efficiently rescued the autophagy phenotype in mtm-3 mutants (Supplementary Fig S3B and H). GFP::MTM-3 was diffuse in the cytoplasm of almost all cells during embryogenesis (Fig 2A; Supplementary Fig S3K–L′). In larvae and adults, MTM-3 was found in various cell types including pharyngeal cells, vulva muscle cells, intestine and cells in the tail region (Supplementary Fig S3M–P). GFP::MTM-3 expression was unaltered in autophagy mutants, suggesting that MTM-3 itself is not removed by autophagy (Supplementary Fig S3Q–T). To examine whether MTM-3 is recruited to autophagosomes or their precursors, we constructed a GFP reporter of MTM-3(C459S), a catalytically inactive substrate-trapping form of MTM-3 (Fig 2J). GFP::MTM-3(C459S) was expressed at a higher level than the wild-type protein and formed punctate structures that overlapped well with endogenous LGG-1 puncta (Fig 2B, D-D′′ and I; Supplementary Fig S3J). Loss of lgg-1, which disrupts autophagosome biogenesis, but not rab-5 or rab-7, which affect endosome formation, significantly reduced the number of GFP::MTM-3(C459S) puncta, suggesting that MTM-3(C459S) localizes to autophagic structures (Fig 2H). Loss of VPS-34, the PI3 kinase that produces PtdIns3P on autophagic structures, dramatically reduced GFP::MTM-3(C459S) puncta and their co-localization with LGG-1 (Fig 2G–I). Moreover, GFP::MTM-3(C459S, del PH-G), which lacks the phosphoinositide-binding PH-GRAM domain of MTM-3, failed to form puncta (Fig 2C) 20. These data suggest that MTM-3 is recruited to autophagosomes by PtdIns3P. Mutation of atg-2, which blocks completion of autophagosomes, did not affect MTM-3(C459S) puncta or their co-localization with LGG-1 (Fig 2F–F′′, H and I).
MTM-3 acts downstream of ATG-9, ATG-18, and ATG-2 in the aggrephagy pathway
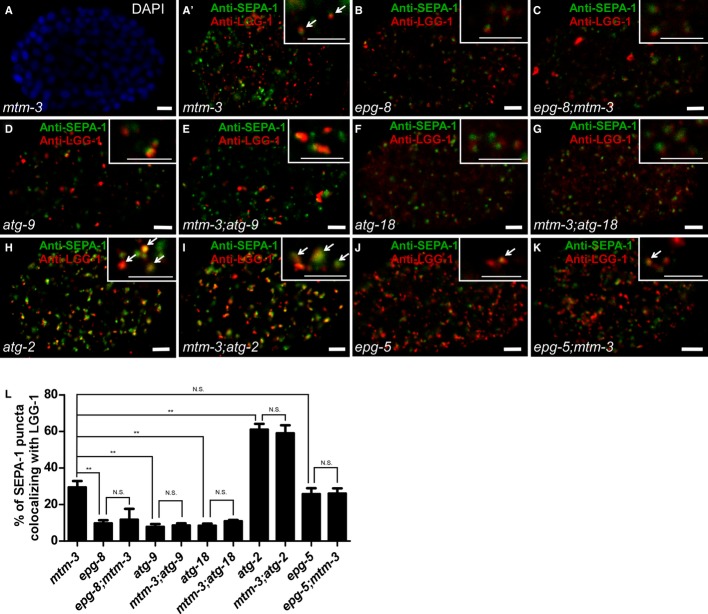
Autophagy proteins act in a stepwise pathway to remove various protein aggregates (aggrephagy) during C. elegans embryogenesis. The C. elegans Atg1 kinase complex UNC-51-EPG-1-EPG-9 acts first, followed by the PI3 kinase complex EPG-8-VPS-34-BEC-1. ATG-18/WIPI1/2 acts earlier than the ATG-2-EPG-6 complex, which regulates progression from isolation membrane to autophagosome. EPG-5/mEPG-5 functions downstream of ATG-2-EPG-6 to regulate autolysosome formation, while CUP-5/TRPML1 is required for degradation of autophagic cargo in autolysosomes 21. To determine where MTM-3 acts in this pathway, we examined the morphology and distribution of PGL granules (detected by anti-SEPA-1 antibodies) and LGG-1 puncta in mtm-3, atg, or epg single mutants and atg/epg;mtm-3 double mutants. In mtm-3(tm4475) embryos, PGL granules and LGG-1 puncta are spherical and dispersed in the cytoplasm, with 31.6% of PGL granules overlapping with LGG-1 (Fig 3A, A′ and L; Supplementary Fig S5A–A′′′). epg-8 encodes a highly divergent functional homolog of Atg14, a component of the Vps34 PI3K complex 21. epg-8 mutants contained spherical PGL granules and LGG-1 puncta which were mostly separated (Fig 3B and L; Supplementary Fig S5B–B′′′) 22. Similarly, in atg-9 mutants, the fewer and bigger LGG-1 puncta were mostly separable from PGL granules (Fig 3D and L; Supplementary Fig S5D–D′′′) 23. The PtdIns3P-binding protein ATG-18/WIPI1/2 acts early in autophagosome formation and loss of its function caused accumulation of PGL granules and LGG-1 puncta, which were close but did not overlap (Fig 3F and L; Supplementary Fig S5F–F′′′) 23. PGL granules and LGG-1 puncta in epg-8;mtm-3, mtm-3;atg-9, or mtm-3;atg-18 double mutants resembled those in epg-8, atg-9, or atg-18 single mutants, suggesting that epg-8, atg-9, and atg-18 are epistatic to mtm-3 in the aggrephagy pathway (Fig 3C, E, G and L; Supplementary Fig S5C–C′′′, E–E′′′ and G–G′′′). The ATG-2 protein forms a complex with EPG-6/WIPI3/4 to regulate formation of autophagosomes from omegasomes. epg-8, atg-9, and atg-18 are epistatic to atg-2 and epg-6 in the aggrephagy pathway 23. atg-2 mutants contained numerous PGL granules and LGG-1 puncta which were enlarged and irregular in shape; over 60% of the PGL granules co-localized with LGG-1 puncta (Fig 3H and L; Supplementary Fig S6A–A′′′) 23. The morphology and distribution of PGL granules and LGG-1 puncta in mtm-3;atg-2 double mutants resembled that in atg-2 single mutants (Fig 3A′, H, I and L; Supplementary Fig S6B–B′′′), suggesting that atg-2 is epistatic to mtm-3 in the aggrephagy pathway. epg-5 encodes a metazoan-specific autophagy gene, which acts downstream of the ATG-2-EPG-6 complex 23. We observed very similar patterns of PGL granules and LGG-1 puncta in mtm-3, epg-5, and epg-5;mtm-3 embryos (Fig 3A′, J and K; Supplementary Fig S6C–D′′′). The co-localization of PGL granules with LGG-1 puncta was also indistinguishable in mtm-3 (31.6%), epg-5 (25.7%) or epg-5;mtm-3 double mutants (26.0%) (Fig 3L). Together, these data indicate that MTM-3 acts at a similar step to EPG-5 in the aggrephagy pathway, and this step is downstream of EPG-8, ATG-9, ATG-18, and ATG-2.
Figure 3. MTM-3 acts downstream of EPG-8, ATG-9, ATG-18, and ATG-2 in the aggrephagy pathway.
A–K Confocal fluorescent images of 200-cell-stage embryos from the indicated strains stained by both anti-SEPA-1 and anti-LGG-1 antibodies. Insets show magnified views. Arrows indicate overlapping SEPA-1 and LGG-1 puncta. The wild type contains no visible SEPA-1 aggregates at this stage and is therefore not shown. Scale bars: 5 μm.
L The percentage of SEPA-1 puncta co-localizing with LGG-1 in each strain. At least 10 embryos were scored per strain. Data are shown as mean ± SD. **P < 0.0001; N.S.: not statistically different.
Source data are available online for this figure.
Autophagosomes in mtm-3 mutants are not fused with lysosomes
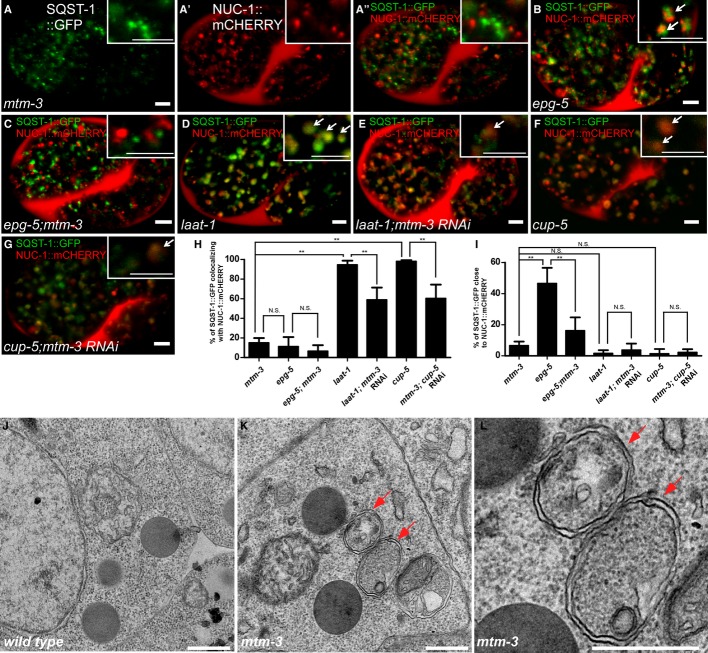
EPG-5 is suggested to be involved in the formation of functional autolysosomes 21,23. We examined autolysosome formation in mtm-3, epg-5, and epg-5;mtm-3 worms. In mtm-3(tm4475) embryos, 78.6% of SQST-1::GFP aggregates did not co-localize with the lysosomal reporter NUC-1::mCHERRY 24, suggesting that autophagic substrates are not within autolysosomes (Fig 4A–A′′ and H). In epg-5 mutants, 42.5% of SQST-1::GFP puncta were clearly separated from NUC-1::mCHERRY, while 46.4% stayed very close to but did not overlap with lysosomes (Fig 4B, H and I). The close association of SQST-1::GFP with lysosomes in epg-5 mutants was mostly suppressed in epg-5;mtm-3 double mutants, with 77.7% of SQST-1::GFP being separated from lysosomes, a phenotype resembling that in mtm-3 single mutants (Fig 4C, H and I). These data suggest that MTM-3 acts earlier than EPG-5 in autolysosome formation. Loss of function of laat-1, which encodes a lysosomal lysine/arginine transporter, severely affects lysosome function, causing accumulation of autophagic cargo in autolysosomes 25. CUP-5, the C. elegans functional ortholog of the mammalian lysosomal channel protein MLN1/TRPML1, regulates lysosome biogenesis and loss of cup-5 impairs degradation of autolysosomal contents 26–28. We found that 94% and 98% of SQST-1::GFP puncta co-localized with lysosomes in laat-1 and cup-5 mutant embryos, respectively, indicating accumulation of autophagy substrates in autolysosomes (Fig 4D, F and H). Lysosomal accumulation of SQST-1::GFP in laat-1 and cup-5 mutants was significantly reduced when mtm-3 was inactivated by RNAi (Fig 4E, G and H), suggesting that loss of mtm-3 impairs autolysosome formation. Similar phenotypes were observed when autolysosome formation was examined using GFP::LGG-1 and NUC-1::mCHERRY (Supplementary Fig S7). To corroborate this, we examined autophagosomes by transmission electron microscopy (TEM). In mtm-3(tm4475) mutant embryos, double-membrane autophagosomes, but not autolysosomes, were readily observed, whereas none of these autophagic structures were easily seen in wild type (Fig 4J–L). These data indicate that loss of mtm-3 function impairs autolysosome formation.
Figure 4. Loss of mtm-3 impairs autolysosome formation.
A–G Confocal fluorescent images of 1.5-fold-stage embryos from the indicated strains expressing both SQST-1::GFP and NUC-1::mCHERRY. Insets show magnified views. Arrows indicate overlapping or closely associated GFP and mCHERRY. Scale bars: 5 μm. The wild type contains no visible SQST-1::GFP puncta at this stage and is therefore not shown.
H, I The percentage of SQST-1::GFP-positive puncta that overlap with (H) or are close to (I) NUC-1::mCHERRY-positive structures in each strain. At least 10 embryos were scored per strain. Data are shown as mean ± SD. **P < 0.0001; N.S.: not statistically different.
J–L TEM analysis of embryos in wild type (I) and mtm-3(tm4475) (J, K). Double-membrane autophagosomes (arrows) were observed in mtm-3 but not wild-type embryos. Scale bars: 500 nm.
Source data are available online for this figure.
Loss of MTM-3 increases the association of ATG-18 with autophagic structures
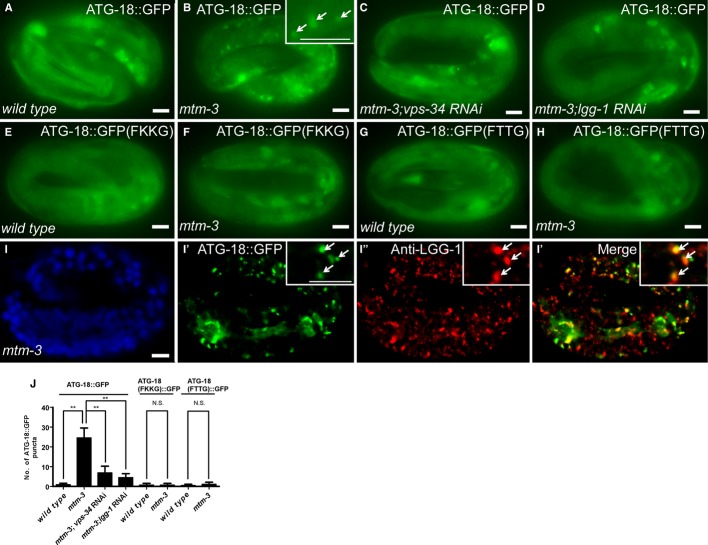
As a PtdIns3P phosphatase, MTM-3 may modulate the PtdIns3P level on autophagic structures. We examined the localization of ATG-18::GFP, the PtdIns3P-binding effector, in wild-type and mtm-3 mutants 23. ATG-18::GFP was diffuse in the cytoplasm of wild type but formed puncta in mtm-3 mutants (Fig 5A, B and J; Supplementary Fig S8E). ATG-18 remained diffuse in other autophagy mutants like atg-2 or epg-5, indicating that the ATG-18::GFP puncta seen in mtm-3(lf) are not simply protein aggregates formed in autophagy-defective mutants (Supplementary Fig S8A–D′). The ATG-18::GFP-positive structures in mtm-3 mutants co-localized well with puncta labeled by endogenous LGG-1 and were significantly reduced in number in mtm-3; lgg-1 RNAi worms (Fig 5D, I and J), indicating that they associated with autophagic structures. Loss of VPS-34 greatly reduced the number of ATG-18::GFP puncta, and two mutations in ATG-18, FKKG and FTTG, which abolish its binding to PtdIns(3)P, disrupted ATG-18 puncta formation in mtm-3 embryos (Fig 5C, E–H and J) 23. These data suggest that loss of mtm-3 elevates the PtdIns3P level on autophagic structures, leading to increased autophagic association of the PtdIns3P-binding effector ATG-18.
Figure 5. Loss of mtm-3 causes increased autophagic association of ATG-18.
A–H Fluorescent images of embryos at the 4-fold stage in the indicated strains expressing ATG-18::GFP (A–D), ATG-18(FKKG)::GFP (E, F), or ATG-18(FTTG)::GFP (G, H).
I–I″′ Confocal fluorescent images of 4-fold mtm-3 embryos expressing ATG-18::GFP (I′) and stained by DAPI (I) and anti-LGG-1 (I″) antibody. Insets show magnified views. Arrows indicate ATG-18 puncta labeled by anti-LGG-1 antibody. Scale bars: 5 μm.
J Number of wild-type or mutated ATG-18::GFP puncta in each strain. At least 15 embryos were scored per strain (mean ± SD). **P < 0.0001; N.S: not statistically different.
Source data is available online for this figure.
In summary, we identified MTM-3 as a positive regulator of autophagy in C. elegans. MTM-3 is recruited by and regulates PtdIns3P on autophagic structures to promote autophagosome maturation into autolysosomes (Supplementary Fig S8F). MTM-3 may hydrolyze PtdIns3P to trigger release of PtdIns3P-binding effectors from autophagosomes, thus allowing recruitment of proteins involved in next-step maturation or fusion with lysosomes. In yeast, myotubularin Ymr1 catalyzes PtdIns3P turnover to release Atg proteins from autophagosomes for fusion with vacuoles 10. Thus, PtdIns3P clearance from the autophagosomal surface is probably a prerequisite for autolysosome formation and myotubularin phosphatases play evolutionarily conserved roles in this process. It remains to be seen if this mechanism applies to mammalian autophagosomes.
Worm and yeast myotubularin phosphatases promote autolysosome formation, while human Jumpy and MTMR3 are suggested to negatively regulate autophagy at the initiation step (Supplementary Fig S8F). PtdIn3P turnover may therefore have distinct effects on autophagy at different stages (initiation versus maturation). It is important to determine how specific myotubularin phosphatases are temporally and spatially regulated to control PtdIns3P turnover at different stages and how this affects autophagy. Our observation that MTM-3 overexpression does not inhibit autophagy suggests that rate-limiting regulatory factors are involved in controlling recruitment, release or activity of MTM-3 on autophagic structures.
Materials and Methods
Quantification analysis
Fluorescent confocal images of embryos were collected to determine (1) the percentage of SQST-1::GFP or GFP::LGG-1 puncta that co-localize or are close to NUC-1::mCHERRY-positive structures and (2) the percentage of GFP::MTM-3(C459S)- or SEPA-1-puncta that co-localize with LGG-1 puncta. Puncta in whole embryos (2) or five different areas of the embryo (1) were quantified, and 10 embryos were scored in each strain. The number of GFP::MTM-3(C459S)- and ATG-18::GFP-positive structures in embryos was scored directly under the fluorescent microscope. At least 15 embryos were quantified in each strain.
Phosphatase assay
MTM-3 phosphatase activity was determined with a Malachite Green Assay kit (Echelon Biosciences Inc., USA) as instructed by the manufacturer. Briefly, 500 ng recombinant HIS-tagged MTM-3 or MTM-3(C459S) was incubated with 1 mM DiC8PtdIns3P or DiC8PtdIns(3,5)P2 in 25 μl reaction buffer (50 mM Tris-HCl, 100 mM NaCl, 2 mM CaCl2, 2 mM DTT) at 20°C overnight. One hundred microliters Malachite Green Solution was then added to each reaction and incubated for 20 min at room temperature. Absorbance was measured at 620 nm. Control reactions contained no MTM-3 protein (blank). Each reaction was performed in triplicate to get the mean absorbance. Two independent experiments were performed with similar results, and one of them is shown in Fig 2J.
Immunostaining
Mixed-stage embryos were fixed and incubated with anti-LGG-1 (1: 1,000) or anti-SEPA-1 (1:10,000) in blocking buffer at 4°C overnight. After washing with PBST (PBS + 0.2% Tween 20), samples were incubated with Texas red-conjugated and/or FITC-conjugated secondary antibodies (Jackson ImmunoResearch, 112-075-003 and 115-095-003) at a 1:200 dilution for 1 h at room temperature. The stained samples were washed and mounted in 15% VECTASHIELD mounting medium with DAPI (VECTOR) and visualized using a Zeiss LSM 510 Meta inverted confocal microscope (Carl Zeiss, Germany).
Statistical analysis
The standard deviation (SD) was used as the y error bar for bar charts plotted from the mean value of the data. Data derived from different genetic backgrounds were compared by Student’s two-way unpaired t-test. Data were considered statistically different at P < 0.05 (indicated in the figure legend).
See Supplementary Methods for a list of strains, imaging analysis, construction of reporters, RNAi and statistical analyses.
Acknowledgments
We thank Dr. G. Ou (Tsinghua University) for plasmids and Dr. Isabel Hanson for editing services. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40OD010440). This work was supported by the National Science Foundation of China (31325015), the National Basic Research Program of China (2013CB910100), and an International Early Career Scientist grant from the Howard Hughes Institute to XW.
Author contributions
YW and XW designed the experiments, analyzed the data, and wrote the manuscript. YW performed most of the experiments. SC and WZ performed some genetic experiments, and HZhao performed the EM analysis. SY, SM and HZhang provided reagents.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Noda T, Fujita N, Yoshimori T. The late stages of autophagy: how does the end begin? Cell Death Differ. 2009;16:984–990. doi: 10.1038/cdd.2009.54. [DOI] [PubMed] [Google Scholar]
- Mehrpour M, Esclatine A, Beau I, Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20:748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Dall’Armi C, Devereaux KA, Di Paolo G. The role of lipids in the control of autophagy. Curr Biol. 2013;23:R33–R45. doi: 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Matsunaga K, Taguchi-Atarashi N, Yoshimori T. Regulation of membrane biogenesis in autophagy via PI3P dynamics. Semin Cell Dev Biol. 2010;21:671–676. doi: 10.1016/j.semcdb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 2009;28:2244–2258. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 2010;11:468–478. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- Cebollero E, van der Vaart A, Zhao M, Rieter E, Klionsky DJ, Helms JB, Reggiori F. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol. 2012;22:1545–1553. doi: 10.1016/j.cub.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Fares H, Grant B, Li Z, Rose AM, Clark SG, Skolnik EY. Genetic analysis of the myotubularin family of phosphatases in Caenorhabditis elegans. J Biol Chem. 2003;278:34380–34386. doi: 10.1074/jbc.M303259200. [DOI] [PubMed] [Google Scholar]
- Dang H, Li Z, Skolnik EY, Fares H. Disease-related myotubularins function in endocytic traffic in Caenorhabditis elegans. Mol Biol Cell. 2004;15:189–196. doi: 10.1091/mbc.E03-08-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhankova M, Port F, Harterink M, Basler K, Korswagen HC. Wnt signalling requires MTM-6 and MTM-9 myotubularin lipid-phosphatase function in Wnt-producing cells. EMBO J. 2010;29:4094–4105. doi: 10.1038/emboj.2010.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Lu Q, Zhao D, Li W, Mapes J, Xie Y, Wang X. Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet. 2009;5:e1000679. doi: 10.1371/journal.pgen.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukomm LJ, Nicot AS, Kinchen JM, Almendinger J, Pinto SM, Zeng S, Doukoumetzidis K, Tronchère H, Payrastre B, Laporte JF, et al. The phosphoinositide phosphatase MTM-1 regulates apoptotic cell corpse clearance through CED-5-CED-12 in Caenorhabditis elegans. Development. 2011;138:2003–2014. doi: 10.1242/dev.060012. [DOI] [PubMed] [Google Scholar]
- Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, et al. Caenorhabditis elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in Caenorhabditis elegans. Cell. 2009;136:308–321. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Lin L, Yang P, Huang X, Zhang H, Lu Q. The scaffold protein EPG-7 links cargo-receptor complexes with the autophagic assembly machinery. J Cell Biol. 2013;201:113–129. doi: 10.1083/jcb.201209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs AL, Zhang H. Role of autophagy in Caenorhabditis elegans. FEBS Lett. 2010;584:1335–1341. doi: 10.1016/j.febslet.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Urbe S, Clague MJ. Analysis of phosphoinositide binding domain properties within the myotubularin-related protein MTMR3. J Cell Sci. 2005;118:2005–2012. doi: 10.1242/jcs.02325. [DOI] [PubMed] [Google Scholar]
- Lu Q, Wu F, Zhang H. Aggrephagy: lessons from Caenorhabditis elegans. Biochem J. 2013;452:381–390. doi: 10.1042/BJ20121721. [DOI] [PubMed] [Google Scholar]
- Yang P, Zhang H. The coiled-coil domain protein EPG-8 plays an essential role in the autophagy pathway in Caenorhabditis elegans. Autophagy. 2010;7:159–165. doi: 10.4161/auto.7.2.14223. [DOI] [PubMed] [Google Scholar]
- Lu Q, Yang P, Huang X, Hu W, Guo B, Wu F, Lin L, Kovács AL, Yu L, Zhang H, et al. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell. 2011;21:343–357. doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- Guo P, Hu T, Zhang J, Jiang S, Wang X. Sequential action of Caenorhabditis elegans Rab GTPases regulates phagolysosome formation during apoptotic cell degradation. Proc Natl Acad Sci USA. 2010;107:18016–18021. doi: 10.1073/pnas.1008946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Du H, Rutkowski R, Gartner A, Wang X. LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science. 2012;337:351–354. doi: 10.1126/science.1220281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Wang X, Lu Q, Ren H, Zhang H. CUP-5, the Caenorhabditis elegans ortholog of the mammalian lysosomal channel protein MLN1/TRPML1, is required for proteolytic degradation in autolysosomes. Autophagy. 2011;7:1308–1315. doi: 10.4161/auto.7.11.17759. [DOI] [PubMed] [Google Scholar]
- Fares H, Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat Genet. 2001;28:64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- Treusch S, Knuth S, Slaugenhaupt SA, Goldin E, Grant BD, Fares H. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc Natl Acad Sci USA. 2004;101:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.