Abstract
Regulatory T cells (Tregs) control autoreactive T cells by inhibiting activation-induced proliferation and cytokine expression. The molecular mechanisms responsible for the inactivation of effector T cells by Tregs remain yet to be fully characterized. We report that T-helper cells stimulated in the presence of Tregs quickly activate NFAT1 and have increased NFAT1-dependent expression of the transcription repressor Ikaros. NFAT1 deficiency or dominant-negative Ikaros compromises Treg-mediated inhibition of T-helper cells in vitro and in vivo. Thus, our results place NFAT-dependent mechanisms as general regulators of T-cell tolerance and show that Treg-mediated suppression of T-helper cells results from the activation of NFAT-regulated gene expression.
Subject Categories: Immunology; Signal Transduction
Keywords: Ikaros, NFAT, regulatory T cell
Introduction
A major mechanism of peripheral tolerance is suppression by regulatory T cells (Tregs) 1. Several subtypes of Tregs have been defined based on the specific developmental processes involved in their generation. tTregs develop in the thymus and constitute approximately 5–10% of the total peripheral CD4+ T-cell population 1. In contrast to tTregs, peripheral pTregs develop in peripheral lymphoid tissues from naïve CD4+ progenitors 2. Several models have been proposed to explain how Tregs mediate suppression of T-cell activation 3, including secretion of immunomodulatory cytokines 4,5 and cell-to-cell contacts mediated by proteins such as CTLA-4 6 or LAG-3 7. Regardless of which mechanisms are in play in any given context, the general outcome is an inhibition of proliferation and cytokine secretion. The molecular events that occur within the suppressed T cell that are ultimately responsible for inhibiting T-cell activation remain yet to be fully characterized.
The family of NFAT transcription factors is composed of four calcium-/calcineurin-regulated members, NFAT1-4, and a more distantly related NFAT5 8. In T cells, engagement of the TCR causes an elevation of intracellular calcium and the activation of calcineurin, which in turn dephosphorylates NFAT proteins leading to their nuclear translocation. NFAT proteins play major roles not only during T-cell activation but also in the induction of T-cell tolerance 9,10.
Here, we show that NFAT1 is required for effective Treg-mediated suppression of T-helper (Th) cell activation. When T cells are activated in the presence of Tregs, they upregulate the NFAT1-dependent expression of Ikaros, which is necessary to achieve efficient suppression. Our results support a central role for NFAT1 in the regulation of Treg-mediated suppression of Th cells.
Results and Discussion
NFAT1-deficient T-helper cells show resistance to Treg-mediated suppression
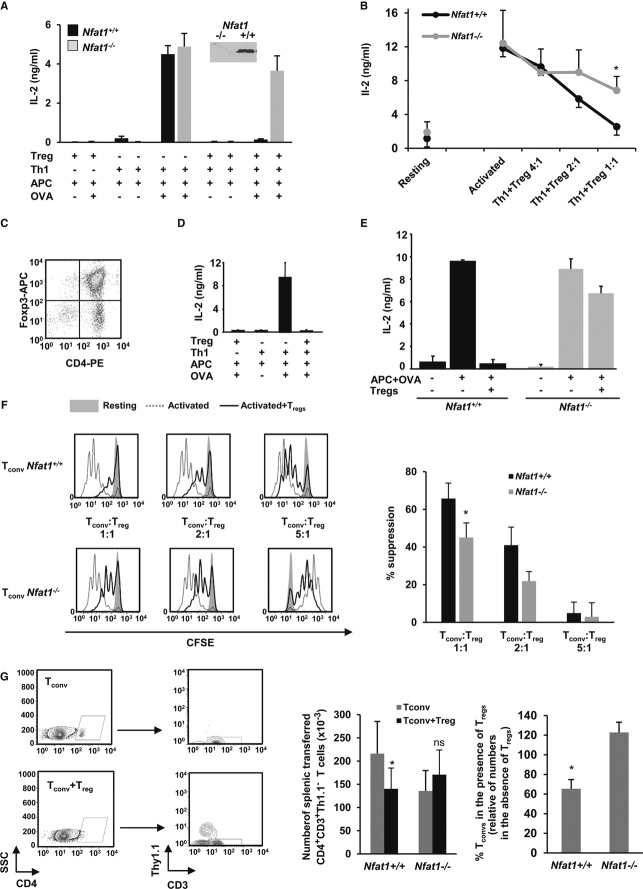
Previous studies have shown that NFAT1 directs the expression of a specific set of genes responsible for the inhibition of Th1 cell effector functions in response to tolerizing stimuli 9–11. To determine whether NFAT1 could regulate Treg-mediated suppression of effector Th-cell activation, we assessed the ability of Tregs to suppress NFAT1-deficient Th1 cells. In vitro differentiated Th1 cells from wild-type (WT) or Nfat1−/− DO11.10 mice were stimulated for 48 h with ovalbumin323–339 peptide-loaded antigen-presenting cells (APC-OVA) in the presence or absence of WT CD4+CD25+Foxp3+ Tregs isolated from BALB/c mice. Whereas WT T cells were efficiently suppressed by Tregs, T cells from Nfat1−/− mice were resistant to Treg-mediated suppression (Fig 1A). Similar experiments using OT-II mice and Tregs from B6 Foxp3-RFP mice confirmed a significant decrease (~ 3-fold) in the susceptibility to Treg-mediated suppression in Th1 cells lacking NFAT1, ensuring that this was not a strain-specific effect (Fig 1B).
Figure 1. NFAT1-deficient T cells are more resistant to suppression by Tregs.
A, B DO11.10 (A) or OTII (B) WT or Nfat1−/− Th1 cells were activated with APC-OVA for 48 h with or without WT CD4+CD25+FoxP3+ Tregs (mean ± s.e.m. of 5 (A) or 3 (B) experiments, t-test, *P < 0.05). Inset: immunoblot for NFAT1.
C FACS of Tregs from BALB/c mice expanded in vitro for 1 week.
D, E IL-2 production in DO11.10 WT (D, E) or Nfat1−/− (E) Th1 cells stimulated with APC-OVA for 48 h with or without expanded Tregs (ratio 1:1) (mean + s.e.m. of 2 and 3 independent experiments performed in triplicate, respectively).
F C57BL6 WT or Nfat1−/− naïve CD4+ T cells (Tconv) labeled with CFSE were stimulated with 0.5 μg/ml anti-CD3 antibody and APCs for 72 h with or without Tregs at indicated ratios. Graph shows % of inhibition of CFSE dilution from 3 experiments (mean + s.e.m., t-test, *P < 0.05).
G Rag1−/− mice were adoptively transferred with 2.5 × 106 naïve WT or Nfat1−/− CD4+Thy1.2+ T cells (Tconv) with or without 5 × 105 Foxp3+RFP+Thy1.1+ Tregs. Seven days later, CD4+CD3+Thy1.1− Tconv in spleens was measured by FACS. Graphs show numbers of cells and relative percentage of suppression of Tconv expansion (mean + s.e.m. from 8 experiments, Wilcoxon, *P < 0.05).
Source data are available online for this figure.
Tregs activate and expand when encountering their cognate antigen in vivo, and CD28 signaling appears crucial to maintain efficient Treg-suppressive activity 12,13. CD4+CD25+ T cells expanded for 1 week (Fig 1C) remained suppressive (Fig 1D), but still failed to effectively inhibit Nfat1−/− T cells (Fig 1E). As we had seen in anergic cells 10,14, effector NFAT1-deficient Th1 cells were less susceptible to Treg-mediated suppression. Further analyses using naïve CD4+ T cells confirmed that the requirement for NFAT1 also extended to this T-cell population (Fig 1F). To confirm the NFAT1 requirement in vivo, we measured the ability of Tregs to suppress the homeostatic proliferation of CD4+ T cells transferred into lymphopenic hosts. One week after adoptively transferring CD4+CD25− T cells into Rag1−/− hosts, the number of donor T cells in the spleen was reduced by more than 40% when Tregs were co-transferred. However, Nfat1−/− T-cell expansion was not similarly affected (Fig 1G).
To confirm the role of NFAT1 in the regulation of the susceptibility of T cells to Tregs, Nfat1−/− DO11.10 Th1 cells were transduced with retrovirus expressing NFAT1. Positively infected T cells were re-stimulated with APC-OVA in the presence of Tregs. Nfat1−/− Th1 cells regained susceptibility to Tregs upon restoration of NFAT1 expression (Fig 2A). Similar results were obtained when cells were transiently transfected with a plasmid expressing NFAT1 (Fig 2B). Altogether, these results indicated that NFAT1 activity in Th cells is required to effectively respond to Tregs. It is possible, however, that other NFAT proteins might also cooperate with NFAT1 in this program. NFAT4 has been shown to exert a negative control on T-cell responses, and NFAT1/NFAT4-deficient mice present a more severe lymphoproliferative phenotype than NFAT1-deficient mice 15,16. Furthermore, cells lacking these two NFAT proteins have also been shown to be resistant to Treg-mediated suppression 17. Our data show, however, that T cells that lack just NFAT1 show no major alterations in their responses to TCR engagement, while rendering them less susceptible to Treg-mediated suppression.
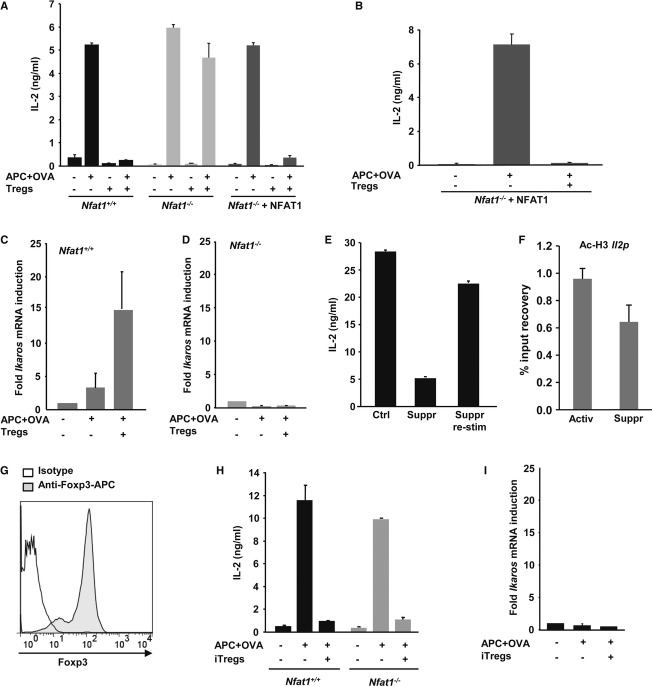
Figure 2. Suppressed CD4+ T cells upregulate the NFAT1-dependent expression of Ikaros.
A Th1 cells from WT or Nfat1−/− DO11.10 mice transfected with a control (WT and Nfat1−/− cells) or NFAT1-expressing (Nfat1−/− cells) retrovirus were stimulated with APC-OVA for 48 h with or without WT Tregs (1:1). IL-2 production determined by ELISA (mean + s.e.m. of three experiments).
B Nfat1−/− DO11.10 Th1 cells transfected with a plasmid expressing NFAT1 were activated as described in (A) (mean + s.e.m. of three experiments).
C, D WT (C) and Nfat1−/− (D) DO11.10 Th1 cells were stimulated with APC-OVA with or without Tregs (1:1) for 12 h. Th1 cells were reisolated and RNA obtained. Results (mean + s.e.m. of three experiments) show Ikaros mRNA fold induction relative to resting cells.
E IL-2 expression in DO11.10 Th1 cells activated as described in (A). Th1 cells were reisolated, rested for 4 h, and restimulated (mean + s.e.m. of three experiments).
F WT and Nfat1−/− Th1 cells were stimulated with APC-OVA in the presence or absence of Tregs (1:1) for 24 h. Th1 cells were reisolated and Histone H3 acetylation at the Il2 promoter measured by ChIP (mean + s.e.m. of three experiments).
G FACS analysis of Foxp3 expression in iTregs.
H WT and Nfat1−/− DO11.10 Th1 cells were stimulated with APC-OVA in the presence or absence of iTregs (1:1) for 48 h. IL-2 values are mean + s.e.m. from three experiments.
I Th1 cells were activated for 12 h as in (H). Th1 cells were reisolated and RNA obtained. Results (mean + s.e.m. of three experiments) show Ikaros mRNA fold induction relative to resting cells.
Source data are available online for this figure.
Suppressed CD4+ T cells upregulate the expression of Ikaros
We and others have previously identified genes that inhibit T-cell activation that are also expressed in an NFAT-dependent manner in tolerized T cells 18. We showed that the expression of one such gene, Ikaros, was also upregulated (~ 15-fold) in suppressed Th1 cells (Fig 2C). The NFAT1 dependence of Ikaros was further confirmed by the inability of Nfat1−/− T cells to upregulate its expression when activated in the presence of Tregs (Fig. 2D). Expression of genes that are known to maintain T cells in an unresponsive state suggested the possibility that Tregs might stably suppress effector T cells. In fact, it has been previously reported that Tregs can anergize naïve CD4+ T cells 19 and Tregs have been shown to induce anergy in arthritogenic self-reactive T cells in vivo 20. To test this hypothesis, DO11.10 Th1 cells were stimulated with APC-OVA in the presence of Tregs for 24 h. Th1 cells were then purified and rested for 4 h before being re-stimulated. Previously suppressed T cells regained responsiveness when re-stimulated in the absence of Tregs (Fig 2E), indicating that while Treg-induced NFAT-dependent expression of specific genes inhibited T-cell activation, it was not sufficient to induce a long-lasting unresponsive state.
Ikaros mediates silencing of Il2 expression in anergic T cells by inducing epigenetic modifications that maintain the Il2 promoter locus in a closed state 21–24. We assessed whether those modifications were also induced by Tregs. Our data revealed that the presence of Tregs resulted in a slight deacetylation of the Il2 locus, a much smaller effect than reported in anergic cells 23 (Fig 2F). It is likely, thus, that additional signals to those transduced by the presence of Tregs might be required to induce a long-lasting, stable state of unresponsiveness in responder T cells. Accordingly, we could not detect upregulation of several other anergy-associated genes (data not shown), suggesting that specific differences in the programs of gene expression activated in anergic and suppressed T cells might determine divergences between these two forms, passive versus dominant, of T-cell tolerance 25.
Naïve T cells can also be differentiated into Tregs in the periphery when activated in the presence of specific signals and cytokine environments 26. To assess whether iTregs also rely on NFAT1-mediated mechanisms, WT and Nfat1−/− Th1 cells were co-cultured with in vitro generated iTregs (Fig 2G) and stimulated with APC-OVA. Contrary to what we had seen with tTregs, iTregs efficiently suppressed both WT and Nfat1−/− T cells (Fig 2H). Interestingly, iTregs could not induce upregulation of Ikaros (Fig 2I). The mechanisms involved in iTreg- and tTreg-mediated suppression appear, thus, to be different, and the requirement for NFAT1 is likely specific for tTregs.
Early nuclear NFAT1 translocation occurs in T cells stimulated in the presence of Tregs
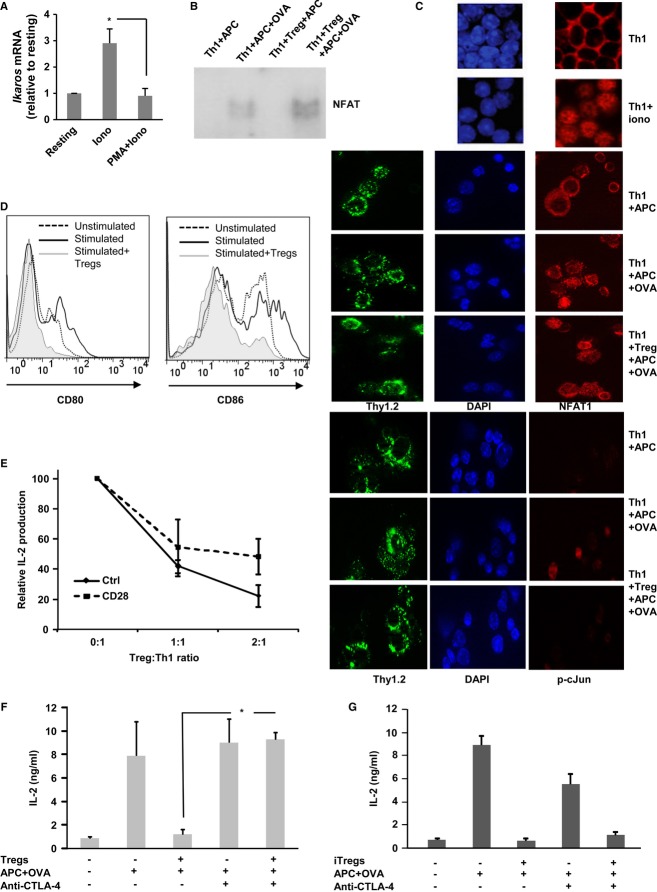
Expression of anergy-inducing genes occurs in response to the activation of NFAT1 in the absence of its major transcriptional partner in activated T cells, AP-1 10. Indeed, specific activation of NFAT using the calcium ionophore, ionomycin, induced a marked upregulation of Ikaros expression, which was prevented when costimulatory pathways were also activated with PMA (Fig 3A). We, thus, determined whether NFAT1 would be activated in T cells stimulated in the presence of Tregs. Nuclear extracts prepared from DO11.10 Th1 cells activated for 4 h with APC-OVA in the presence of Tregs were analyzed by EMSA, which showed that nuclear NFAT was present in T cells during Treg-mediated suppression (Fig 3B). To confirm these results in B6 mice, OTII-Thy1.2+ Th1 cells were stimulated in the presence or absence of Thy1.1+ Tregs. After 4 h, cells were analyzed by immunofluorescence. NFAT1 remained cytosolic in resting cells but translocated into the nucleus in Th1 cells that had been activated either in the presence or absence of Tregs (Fig 3C). As mentioned above, it is NFAT1 in the absence of AP-1 activation that induces the expression of tolerance-associated genes, and immunofluorescence analysis revealed that Tregs also prevented c-Jun phosphorylation in T cells (Fig 3C). Although Tregs have been shown to inhibit signaling events downstream of the TCR, including the calcium/NFAT pathway 27, our data show that early after stimulation in the presence of Tregs, T cells can still activate NFAT but show reduced cJun activation. The presence of an active NFAT protein without concomitant activation of AP-1 should allow suppressed T cells to upregulate the NFAT-dependent AP-1-independent expression of Ikaros 10.
Figure 3. Early activation of NFAT1 in suppressed T cells.
A Th1 cells were activated in with 1 μM ionomycin or 20 nM PMA + 500 nM ionomycin for 6 h. Expression of Ikaros was assessed by qPCR. Values (fold induction relative to resting cells) are mean + s.e.m. from three experiments (t-test. *P < 0.05).
B Nuclear extracts from DO11.10 Th1 cells stimulated with APC-OVA in the presence or absence of Tregs (1:1). DNA–protein complexes containing NFAT proteins were analyzed by EMSA.
C NFAT1 subcellular localization and c-Jun phosphorylation were determined by immunofluorescence in resting OT-II Th1 (Thy1.2)+ cells or cells treated with ionomycin or stimulated with APC-OVA in the presence or absence of Thy1.1+ CD4+CD25+ Tregs for 4–6 h. DAPI was used to stain nuclei.
D Splenic DCs were loaded with OVA and cocultured for 24 h with OT-II Th1 cells in the presence or absence of Foxp3-RFP+ Tregs. Expression of CD80 and CD86 was determined in CD11c+CD4− DC populations.
E Th1 cells were activated with plate-bound anti-CD3 with or without soluble anti-CD28 in the presence of increasing numbers of Tregs. Results (mean ± s.e.m. from three experiments) show relative expression of IL-2 compared with the levels detected in the absence of Tregs.
F, G IL-2 production in DO11.10 Th1 cells stimulated with APC-OVA in the presence or absence of Tregs (E) or iTregs (F) (1:1) for 48 h. Where indicated, blocking antibodies for CTLA-4 were also added. Graph shows mean + s.e.m. of three experiments (one-way ANOVA, *P < 0.05).
Source data are available online for this figure.
It has been shown that Tregs can prevent the expression of B7 proteins on dendritic cells (DCs) to generate costimulation-deficient cells through mechanisms that involve CTLA-4 6,28. To determine whether this mechanism could explain our results, we analyzed the expression of CD80 and CD86 in DCs in suppression reactions and saw that Tregs induced a clear downregulation of CD80 and CD86 (Fig 3D). Providing direct costimulation using anti-CD28 antibodies resulted in reduced efficacy of Treg-mediated suppression (Fig 3E). Furthermore, as previously reported 29,30, suppression assays in the presence of a blocking anti-CTLA-4 antibody confirmed that blockade of CTLA-4 prevented Treg-mediated suppression (Fig 3F); the blocking antibodies had however no effect on the activity of iTregs (Fig 3G). Our data do not exclude that signals directly transmitted by Tregs to effector T cells may also mediate the activation of regulatory gene expression. Supporting this possibility, upregulation of Grail was described when naïve T cells were activated just with anti-CD3 antibodies in the presence of Tregs 19. Also, direct engagement of B7 proteins on effector T cells by CTLA-4 expressed on Tregs has been proposed as a mechanism of suppression 31.
Ikaros activity is required for optimal Treg-mediated suppression
Ikaros has been shown to inhibit IL-2 expression in CD4+ T cells 21,22. Thus, we assessed Ikaros’ role in the Treg-mediated suppression of IL-2 expression. Our results showed that Th1 cells transfected with a plasmid expressing a dominant-negative form of Ikaros (IK-DN) mimicked NFAT1-deficient T cells and became more refractory to Tregs (Fig 4A). Similar results were obtained using CD4+CD25− T cells isolated from transgenic mice expressing IK-DN (Fig 4B). Furthermore, WT Tregs could not suppress wasting induced by the transfer of naïve IK-DN T cells into Rag1−/− hosts as efficiently as the one caused by transfer of WT T cells (Fig 4C). These results confirmed that the activity of Ikaros in CD4+ T cells is required for efficient Treg-mediated suppression. While necessary, this, however, may not be the only mechanism that represses Il2 transcription in suppressed T cells. Treg-mediated activation of ICER/CREM complexes in effector T cells has also been shown to participate in the inhibition of IL-2 production 32. The possibility remains that these complexes may also regulate Treg-mediated suppression by directly modulating NFAT activity in suppressed T cells 33.
Figure 4. Treg-mediated suppression requires Ikaros activity.
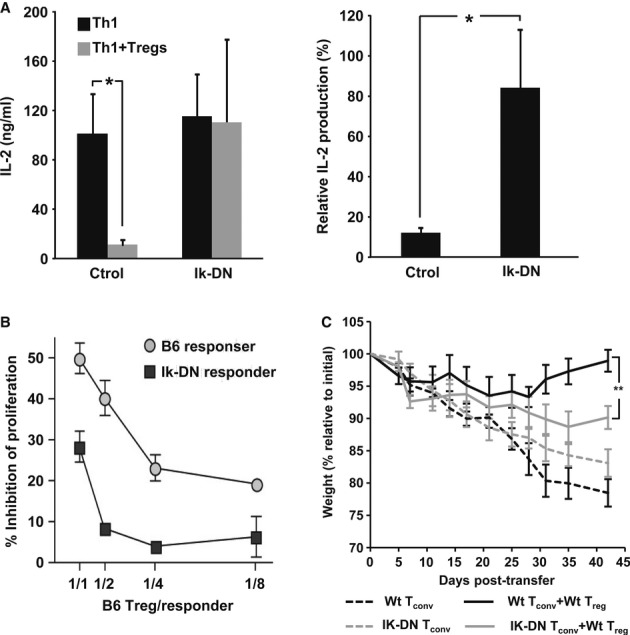
A Th1 cells were transfected with a plasmid expressing IK-DN or a control vector and activated in the presence of Tregs (1:1). Percent suppression of IL-2 expression was calculated (mean + s.e.m. from four experiments, t-test, *P < 0.05).
B B6 or B6-IkDN/+ CD4+CD25− T cells (Thy1.2+) were labeled with CFSE and activated with APC and soluble anti-CD3 in the presence of increasing amounts of CD4+CD25+ Treg from B6-Thy1.1+ mice. After 3 days, suppression of responder cell proliferation was determined by FACS, assessing the degree of inhibition of CFSE dilution (mean ± s.e.m.) from two experiments.
C CD25−CD4+ Tconv cells from WT or IK-DN B6 mice were transferred into B6-Rag1−/− mice. Twenty days later, mice received PBS or CD25+ CD4+ Treg purified from WT donors. Recipients were weighed (mean ± s.e.m.) and observed for symptoms of diarrhea approximately every 2 days (5 mice per group. one-way ANOVA **P < 0.01).
Source data are available online for this figure.
Overall, our results show that NFAT1-regulated gene expression is required for efficient Treg-mediated suppression of CD4+ T cells. These experiments duly correlate NFAT1 activation in suppressed T cells with the expression of T-cell inactivating genes and identify Ikaros as a regulator of suppression that is required for optimal inhibition of T-cell effector function, thereby providing a basis for how Tregs maintain Th cells in a suppressed state.
Materials and Methods
Mice
Strains description is available in Supplementary Methods.
T-cell isolation
CD4+ and CD4+CD25+ cells were isolated from the spleen and lymph nodes of mice using magnetic beads coupled to anti-CD4 or anti-CD25 antibodies (Life Technologies). Alternatively, Foxp3+ Tregs were isolated from Foxp3-RFP mice by sorting for CD4 and RFP expression.
In vitro Th1 differentiation
CD4+ T cells were differentiated in vitro as previously described 22. Details in Supplementary Methods.
Treg expansion
Tregs were stimulated with 0.25 μg/ml immobilized anti-CD3 and anti-CD28 for 24 h and cultured for 1 week in the presence of 200 U/ml murine IL-2 (Cell Signaling).
iTreg generation
Naive CD4+ T cells were stimulated with 0.5 μg/ml immobilized anti-CD3 and anti-CD28 in the presence of IL-2 (50 U/ml) and TGFβ-1 (3 ng/ml) (eBioscience) for 1 week.
In vitro suppression assays
DO11.10 or OTII effector T cells were stimulated for 24–72 h using T-cell-depleted splenocytes (APCs) loaded with 0.1–1 μM OVA in the presence of pre-activated Tregs isolated from BALB/c, C57BL6, or Foxp3-RFP mice (anti-CD3+ anti-CD28 for 24 h before the assay). Where indicated, 10 μg/ml anti-CTLA-4 antibody (UC10-4B9. Biolegend) was added. Alternately, CFSE-labeled CD4+CD25− T cells were activated with APCs and 0.5–4 μg/ml soluble anti-CD3 in the presence of different amounts of purified Tregs from C57BL/6 Thy1.1 or Foxp3-RFP mice, for 72 h. Responder cells were identified as CD4+Thy1.1− or CD4+RFP−, and suppression of proliferation was determined by assessing inhibition of CFSE dilution.
In vivo suppression assays
2.5 × 106 CD4+CD25− T cells from Thy1.2 WT or Nfat1−/− mice were adoptively transferred into Thy1.2 Rag1−/− B6 mice i.p. with or without 5 × 105 Thy1.1 Foxp3+RFP+ Tregs. Seven days post-transfer, splenic T cells were identified as CD3+CD4+Thy1.1−. Alternatively, to induce experimental colitis, 1 × 106 CD4+CD25− T cells from C57Bl/6 or B6-Ik-DN mice were adoptively transferred i.p. into Rag1−/− B6 recipients. Twenty days later, groups of six mice received PBS or 0.25 × 106, i.p. CD25+CD4+ WT Tregs. Recipients were weighed and observed for symptoms of diarrhea approximately every 2 days.
ELISA and immunofluorescence
See Supplementary Methods for detailed description.
Transfection and retroviral infection of T cells
T cells were transfected by electroporation using a nucleofector electroporator (Amaxa). Retrovirus expressing NFAT1 and conferring G418 resistance (from an IRES) were generated using the Phoenix ecotropic packaging cell line (A gift from Dr G. Nolan, Stanford University, CA). Viral supernatants were used to infect T cells by spin infection at a minimal ratio of five virions/cell. Cells were grown for 1 week in 500 μg/ml G418 to allow selection of positively infected T cells.
Semiquantitative real-time PCR (qPCR)
qPCRs were performed as previously described 22. Detailed description and primer sequences are provided in Supplementary Methods.
Electrophoretic mobility shift assays (EMSA)
Nuclear extracts were prepared from resting, stimulated, or suppressed cells. NFAT-specific oligonucleotides (5′-AGCTAGCTAGGAATATTCCTGGATGATC) were labeled with [32P]ATP and incubated with nuclear extracts (3–5 μg) and 100 ng/μl poly(dI-dC) for 20 min 14. DNA–protein complexes were resolved in polyacrylamide gel electrophoresis on a 4% non-denaturing gel.
Chromatin immunoprecipitation (ChIP) assays
ChIp assays suppressed and activated Th1 cells were performed as previously described 22. See Supplementary Methods for detailed description and primer sequences.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software). Differences between multiple groups were analyzed by one-way ANOVA with a Tukey’s post hoc test. Comparisons between data pairs were analyzed using a paired t-test (two-tail distribution) or a Mann–Whitney test as indicated in each figure.
Acknowledgments
This work was supported by National Institutes of Health grants AI059738, AI079363 (FM), and Training grants GM007288 (AJ and DS) and AI075625 (AJ).
Conflict of interest
The authors declare that they have no conflict of interest.
Author contributions
DSS, AJ, SB, RMT, SB, and EFZ performed the experiments. ADW and FM designed the studies. DSS and FM wrote the paper. All authors contributed to the analysis of the data and approved the manuscript.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Jin WW, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4(+)CD25(−) naive T cells to CD4(+)CD25(+) regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collison LW, Workman CJ, Kuo TT, Boyd K, Wang Y, Vignali KM, Cross R, Sehy D, Blumberg RS, Vignali DA. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:566–569. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR., Jr Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- Huang CT, Workman CJ, Flies D, Pan X, Marson AL, Zhou G, Hipkiss EL, Ravi S, Kowalski J, Levitsky HI. Role of LAG-3 in regulatory T cells. Immunity. 2004;21:503–513. doi: 10.1016/j.immuni.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- Heissmeyer V, Macian F, Im SH, Varma R, Feske S, Venuprasad K, Gu H, Liu YC, Dustin ML, Rao A. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol. 2004;5:255–265. doi: 10.1038/ni1047. [DOI] [PubMed] [Google Scholar]
- Macian F, Garcia-Cozar F, Im SH, Horton HF, Byrne MC, Rao A. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 2002;109:719–731. doi: 10.1016/s0092-8674(02)00767-5. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Soto-Nieves N, Macian F. Transcriptional regulation of T cell tolerance. Semin Immunol. 2007;19:180–187. doi: 10.1016/j.smim.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LS, Chodos A, Eggena M, Dooms H, Abbas AK. Antigen-dependent proliferation of CD4+ CD25+ regulatory T cells in vivo. J Exp Med. 2003;198:249–258. doi: 10.1084/jem.20030315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Huynh A, Whitcher G, Chang J, Maltzman JS, Turka LA. An obligate cell-intrinsic function for CD28 in Tregs. J Clin Invest. 2013;123:580–593. doi: 10.1172/JCI65013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Nieves N, Puga I, Abe BT, Bandyopadhyay S, Baine I, Rao A, Macian F. Transcriptional complexes formed by NFAT dimers regulate the induction of T cell tolerance. J Exp Med. 2009;206:867–876. doi: 10.1084/jem.20082731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Viola JP, Shaw KT, Luo C, Wallace JD, Bozza PT, Luk DC, Curran T, Rao A. An enhanced immune response in mice lacking the transcription factor NFAT1. Science. 1996;272:892–895. doi: 10.1126/science.272.5263.892. [DOI] [PubMed] [Google Scholar]
- Ranger AM, Oukka M, Rengarajan J, Glimcher LH. Inhibitory function of two NFAT family members in lymphoid homeostasis and Th2 development. Immunity. 1998;9:627–635. doi: 10.1016/s1074-7613(00)80660-3. [DOI] [PubMed] [Google Scholar]
- Bopp T, Palmetshofer A, Serfling E, Heib V, Schmitt S, Richter C, Klein M, Schild H, Schmitt E, Stassen M. NFATc2 and NFATc3 transcription factors play a crucial role in suppression of CD4+ T lymphocytes by CD4+ CD25+ regulatory T cells. J Exp Med. 2005;201:181–187. doi: 10.1084/jem.20041538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baine I, Abe BT, Macian F. Regulation of T-cell tolerance by calcium/NFAT signaling. Immunol Rev. 2009;231:225–240. doi: 10.1111/j.1600-065X.2009.00817.x. [DOI] [PubMed] [Google Scholar]
- Ermann J, Szanya V, Ford GS, Paragas V, Fathman CG, Lejon K. CD4(+)CD25(+) T cells facilitate the induction of T cell anergy. J Immunol. 2001;167:4271–4275. doi: 10.4049/jimmunol.167.8.4271. [DOI] [PubMed] [Google Scholar]
- Martinez RJ, Zhang N, Thomas SR, Nandiwada SL, Jenkins MK, Binstadt BA, Mueller DL. Arthritogenic self-reactive CD4+ T cells acquire an FR4hiCD73hi anergic state in the presence of Foxp3+ regulatory T Cells. J Immunol. 2012;188:170–181. doi: 10.4049/jimmunol.1101311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Dure M, Paroder M, Soto-Nieves N, Puga I, Macian F. Interleukin 2 gene transcription is regulated by Ikaros-induced changes in histone acetylation in anergic T cells. Blood. 2007;109:2878–2886. doi: 10.1182/blood-2006-07-037754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RM, Chunder N, Chen CX, Umetsu SE, Winandy S, Wells AD. Ikaros enforces the costimulatory requirement for IL2 gene expression and is required for anergy induction in CD4(+) T lymphocytes. J Immunol. 2007;179:7305–7315. doi: 10.4049/jimmunol.179.11.7305. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Montagna C, Macian F. Silencing of the Il2 gene transcription is regulated by epigenetic changes in anergic T cells. Eur J Immunol. 2012;42:2471–2483. doi: 10.1002/eji.201142307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RM, Saouaf SJ, Wells AD. Superantigen-induced CD4+ T cell tolerance is associated with DNA methylation and histone hypo-acetylation at cytokine gene loci. Genes Immun. 2007;8:613–618. doi: 10.1038/sj.gene.6364415. [DOI] [PubMed] [Google Scholar]
- Sukiennicki TL, Fowell DJ. Distinct molecular program imposed on CD4+ T cell targets by CD4+ CD25+ regulatory T cells. J Immunol. 2006;177:6952–6961. doi: 10.4049/jimmunol.177.10.6952. [DOI] [PubMed] [Google Scholar]
- Shevach EM. From vanilla to 28 flavors: multiple varieties of T regulatory cells. Immunity. 2006;25:195–201. doi: 10.1016/j.immuni.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Oberle N, Weiss EM, Vobis D, Frischbutter S, Baumgrass R, Falk CS, Haag M, Brugger B, Lin H. Human regulatory T cells rapidly suppress T cell receptor-induced Ca(2+), NF-kappaB, and NFAT signaling in conventional T cells. Sci Signal. 2011;4:ra90. doi: 10.1126/scisignal.2002179. [DOI] [PubMed] [Google Scholar]
- Qureshi OS, Zheng Y, Nakamura K, Attridge K, Manzotti C, Schmitt EM, Baker J, Jeffrey LE, Kaur S, Briggs Z. Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4. Science. 2011;332:600–603. doi: 10.1126/science.1202947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read S, Malmstrom V, Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J Exp Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Tagami T, Yamazaki S, Uede T, Shimizu J, Sakaguchi N, Mak TW, Sakaguchi S. Immunologic self-tolerance maintained by CD25(+)CD4(+) regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J Exp Med. 2000;192:303–310. doi: 10.1084/jem.192.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paust S, Lu L, McCarty N, Cantor H. Engagement of B7 on effector T cells by regulatory T cells prevents autoimmune disease. Proc Natl Acad Sci U S A. 2004;101:10398–10403. doi: 10.1073/pnas.0403342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaeth M, Gogishvili T, Bopp T, Klein M, Berberich-Siebelt F, Gattenloehner S, Avots A, Sparwasser T, Grebe N, Schmitt E. Regulatory T cells facilitate the nuclear accumulation of inducible cAMP early repressor (ICER) and suppress nuclear factor of activated T cell c1 (NFATc1) Proc Natl Acad Sci U S A. 2011;108:2480–2485. doi: 10.1073/pnas.1009463108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor J, Habener JF. Role of transcriptional repressor ICER in cyclic AMP-mediated attenuation of cytokine gene expression in human thymocytes. J Biol Chem. 1998;273:9544–9551. doi: 10.1074/jbc.273.16.9544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.