Abstract
Our goal is to construct a robust physical map for maize (Zea mays) comprehensively integrated with the genetic map. We have used a two-dimensional 24 × 24 overgo pooling strategy to anchor maize expressed sequence tagged (EST) unigenes to 165,888 bacterial artificial chromosomes (BACs) on high-density filters. A set of 70,716 public maize ESTs seeded derivation of 10,723 EST unigene assemblies. From these assemblies, 10,642 overgo sequences of 40 bp were applied as hybridization probes. BAC addresses were obtained for 9,371 overgo probes, representing an 88% success rate. More than 96% of the successful overgo probes identified two or more BACs, while 5% identified more than 50 BACs. The majority of BACs identified (79%) were hybridized with one or two overgos. A small number of BACs hybridized with eight or more overgos, suggesting that these BACs must be gene rich. Approximately 5,670 overgos identified BACs assembled within one contig, indicating that these probes are highly locus specific. A total of 1,795 megabases (Mb; 87%) of the total 2,050 Mb in BAC contigs were associated with one or more overgos, which are serving as sequence-tagged sites for single nucleotide polymorphism development. Overgo density ranged from less than one overgo per megabase to greater than 20 overgos per megabase. The majority of contigs (52%) hit by overgos contained three to nine overgos per megabase. Analysis of approximately 1,022 Mb of genetically anchored BAC contigs indicates that 9,003 of the total 13,900 overgo-contig sites are genetically anchored. Our results indicate overgos are a powerful approach for generating gene-specific hybridization probes that are facilitating the assembly of an integrated genetic and physical map for maize.
In the absence of a complete annotated genome sequence, an integrated genetic and physical map is the most comprehensive resource for understanding genome structure and function. An integrated map allows a starting point for elucidating genome organization and comparative mapping studies. Positional cloning, an approach once regarded as impractical in a complex plant genome, becomes feasible with an integrated genetic and physical map. Perhaps most importantly, a genetically anchored physical map allows a targeted approach to whole genome sequencing that capitalizes on previously existing genetic mapping information.
A high-resolution genetic map is essential for an integrated map that allows localization of genetic markers to a small interval on the physical map. The determinants of resolution on a genetic map are 2-fold: the availability of genetic markers in sufficient numbers to allow high density along a chromosome and the mapping of a population with adequate numbers of recombinational breakpoints to allow resolution of very tightly linked genetic markers. The availability of molecular markers and mapping populations in recent years has allowed polymorphic, high-resolution genetic maps to be constructed for a large number of plants and animals (O'Brien, 1993).
Construction of whole-genome physical maps, particularly for large genomes, has until recently been problematic for a variety of reasons. An absolute requirement for the development of a physical map is a DNA cloning system that can faithfully accommodate DNA fragments of sufficient size that will yield a detailed molecular fingerprint. A low-cost, high-throughput DNA fingerprinting methodology is also needed for identifying overlapping DNA fragments that can be assembled into contigs. Yeast artificial chromosome (YAC) vectors were the first cloning vectors that allowed very large (300- to 1,000-kb) DNA fragments to be cloned (Burke et al., 1987). Unfortunately, YACs proved to be subject to rearrangements and interstitial deletions, and these characteristics compromised their usefulness (Green et al., 1991; Selleri et al., 1992). Despite these obstacles, YAC libraries have been constructed for rice (Oryza sativa; Saji et al., 2001), mouse (Mus musculus; Burke et al., 1991; Haldi et al., 1996), human (Homo sapiens; Chumakov et al., 1995), and maize (Edwards et al., 1992). The development of the bacterial artificial chromosome (BAC) cloning system (Shizuya et al., 1992) allowed for stable cloning of large (100- to 200-kb) DNA fragments, with the added advantage of being generally more amenable to high-throughput applications, including DNA sequencing. For these reasons, BACs have largely replaced YACs as the cloning system of choice for physical map construction. A wide variety of BAC libraries have been prepared from plant and animal species (http://www.chori.org; http://www.genome.clemson.edu). Physical maps based on BAC fingerprinting have been developed for rice (Chen et al., 2002), sorghum (Sorghum bicolor; Klein et al., 2000), Arabidopsis (Mozo et al., 1999), and humans (International Human Genome Mapping Consortium, 2001).
Correlation of genetic and physical maps to derive an integrated map relies extensively on the use of molecular markers since they can be placed with precision on both types of maps. In the absence of a comprehensive BAC end sequence database to generate sequenced-tagged connectors to genetically mapped molecular markers, map integration methodologies generally rely upon DNA hybridization or PCR approaches. Efficient pooling strategies coupled with PCR have been effective in identifying BACs or YACs of interest from large libraries (Green and Olson, 1990; Bruno et al., 1995; Klein et al., 2000). These approaches rely on condensing the library into pools representing overlapping groups of clones and usually require a large number of PCRs to address a single target sequence. Pooled BACs are very effective when small numbers of target sequences need to be addressed to BACs but become cumbersome when trying to determine BAC addresses for large numbers of target sequences. Hybridization-based strategies take a different approach to the problem of trying to address large numbers of probes to the physical map. Hybridization probes can be pooled and hybridized in groups of intersecting rows and columns to high-density BAC filters, followed by a deconvolution process that establishes BAC-probe addresses (Evans and Lewis, 1989; Asakawa et al., 1997). PCR amplicons or cDNA inserts are individually radiolabeled, then pooled and hybridized as a group to high-density filters. A serious drawback to this approach is both uniform radiolabeling of all the probes in the pool and the presence of repeat elements or conserved motifs in the labeled probes that often confound hybridization results by hybridization to sequences that are scattered throughout the genome. It is not uncommon for cDNA or genomic probes that appear to be single copy on Southern hybridizations to identify BACs on high-density filters that correspond to multiple genomic loci.
The recently developed overgo probe (Ross et al., 1999) represents a significant improvement in the pooled hybridization approach. This is due to its small 40-bp size, which can be radiolabeled to high specific activity and thus likely hybridize in a locus-specific fashion. Overgo probes are designed to 40-bp regions of cDNA or genomic clones that have been prescreened to mask out all known repeat elements. This assumes the availability of a comprehensive repeat element database that allows repeat masking of the target sequence. Two 24-mer oligos are designed that, when annealed, form an 8-bp double-stranded segment with two 16-base 3′overhangs suitable for filling in with radiolabeled nucleotides using the Klenow fragment of DNA polymerase I. Key to the success of this approach is double labeling of both DNA strands with both [α−32P]dCTP and [α−32P]dATP to create a probe with very high specific activity. Overgo probes have been used to construct physical maps in mouse (Cai et al., 1998, 2001), human (Han et al., 2000), and rice (Chen et al., 2002).
The Maize Mapping Project (MMP; http://www.maizemap.org) is focused on developing an integrated genetic and physical map for the genome of maize (Coe et al., 2002). We have developed a high-resolution genetic map by placing more than 1,800 molecular markers (Sharopova et al., 2002) on an intermated B73 × Mo17 mapping population specifically developed to have an enhanced number of recombination breakpoints per individual (Lee et al., 2002). Concurrently, 465,000 BACs (approximately 25×) from HindIII, EcoRI, and MboI libraries have been HindIII fingerprinted and assembled into contigs (http://www.genome.arizona.edu/fpc/maize/). As part of an ongoing effort to integrate the genetic and physical map, a partnership was formed with the MMP, Incyte Genomics, and the DuPont Agricultural Biotechnology group to anchor a common set of 10,648 maize unigenes to both the MMP (B73) and DuPont (Mo17) physical maps. In this article, we present the anchoring of 9,371 overgo probes derived from a joined Public/DuPont cornsensus expressed sequence tagged (EST) unigene set (www.agron.missouri.edu/files_dl/MMP/Cornsensus/) to 165,888 EcoRI and HindIII BACs (approximately 10×) that are included in the MMP physical map. Our results indicate that the overgo approach is an efficient strategy for unambiguously anchoring cDNAs to the physical map. Currently, these physically anchored cDNAs are serving as the foundation for single nucleotide polymorphism (SNP) discovery and genetic mapping, which is allowing targeted anchoring of BAC contigs to the high-resolution genetic map.
RESULTS
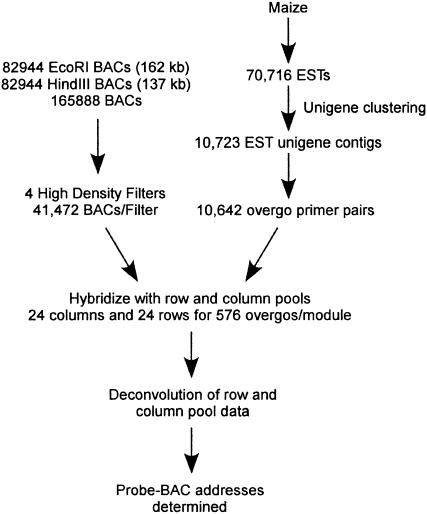
A total of 10,642 40-bp overgo probes designed to maize cDNA unigenes were used in hybridizations to 165,888 EcoRI and HindIII BACs that had been gridded onto high-density filters. A 6 × 6 array pattern was used that could accommodate 165,888 BACs in a four-filter set. Each BAC was double spotted to give a total of 82,944 BACs per filter (Fig. 1). A multiplex pooling strategy (Evans and Lewis, 1989) using row and column pools of 24 overgos was used. In theory, this approach should allow BAC addresses for 576 (24 × 24) overgo probes to be assigned in 48 hybridizations (24 row pools + 24 column pools), with each hybridization containing a pool of 24 radiolabeled overgo probes.
Figure 1.
Flowchart for determination of overgo-BAC addresses. Approximately 70,716 maize ESTs were clustered to derive 10,723 EST unigene contigs, which were masked for repeat sequences prior to the identification of 10,642 40-bp regions that were suitable for overgo development. A total of 165,888 BACs evenly divided between HindIII and EcoRI libraries were spotted in duplicate to four high-density nylon filters. A two-dimensional, 24 × 24 overgo pooling strategy was used to allow identification of 576 overgo-BAC addresses upon deconvolution of the pooled hybridization data. For further details, see “Materials and Methods.”
In practice, the success of an overgo pooling approach relies on being able to design overgo probes with uniform hybridization characteristics to 40-bp regions that are devoid of repeated sequences and are therefore unique sequence-tagged sites (STSs). To design 10,642 overgo sequences, 70,716 maize EST sequences were clustered into 10,723 EST unigene assemblies and designated as the Public/DuPont cornsensus EST unigene set (for details, see “Materials and Methods”). A total of 165,254 BACs from the EcoRI and HindIII libraries were identified for 9,371 overgo hybridization probes (Table I). This represents an overall success rate of 88% (9,371/10,643) for overgos in identifying at least one BAC. More than 96% of the successful overgos (Fig. 2) identified two or more BACs. Only 6% of the overgos identified more than 25 BACs in the two pooled libraries. All BACs reported as positive for a particular overgo probe were confirmed by two positive hybridizations in both a row pool and an intersecting column pool. On average, overgos hit 8.0 and 10.5 BACs in the HindIII (4.6×) and EcoRI (5.4×) libraries, respectively. These inflated average numbers of hits per BAC, relative to the estimated genome coverage, were due to the presence of a small number of overgo probes that hybridized to many genomic loci.
Table I.
Two-dimensional hybridization results
| Library | HindIII | EcoR1 | Total |
|---|---|---|---|
| Total Number of Overgos Hybridized | 10,643 | 10,643 | 10,643 |
| Overgos Identifying at Least One BACa (Overgo % Success) | 8,879 (83.4%) | 8,977 (84.3%) | 9,371 (88.0%) |
| Total Number of Overgo-BAC Hits | 70,695 | 94,559 | 165,254 |
| Library-Specific Overgo Hitsb | 408 | 492 | 900 |
| Average Number of BAC Hits/Overgo | 8.0 | 10.5 | 17.6 |
All overgo-BAC hits reported in this article are those that could be confirmed by duplicate hybridizations in both a row pool and column pool. Data in this table include all overgo-BAC hits.
A small number of overgos hit BACs in only one of the two libraries.
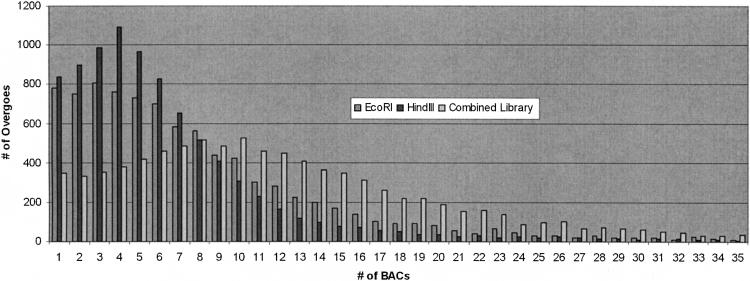
Figure 2.
Distribution of BAC hits per overgo. Both the EcoRI (162 kb; 5.4×) and HindIII (137 kb; 4.6×) libraries are shown to illustrate the distribution of BAC hits per overgo for 9,371 overgo hybridizations across the two libraries. Pooled EcoRI and HindIII BAC libraries (10×) are also shown and reveal 345 overgos that identify only one BAC in the combined libraries. Overall, repeat masking of EST assemblies was effective, as only approximately 5% of the overgo probes hit more than 25 BACs in either the EcoRI or HindIII libraries. In the pooled libraries, only 13% and 4% of the overgos hit more than 25 or 50 BACs, respectively.
In the initial undertaking of this project, we thought it important to utilize BAC libraries prepared using different restriction enzymes (EcoRI and HindIII) to ensure good genome coverage. Examination of each of the two BAC libraries separately reveals that 492 overgos hit only EcoRI BACs, and 408 overgos hit only HindIII BACs. A total of 1,272 overgos had no BAC hits in either the EcoRI or HindIII libraries (Table I). A small number of these failures are attributable to poor overgo design, failed radiolabeling reactions, and overgos designed to genomic regions that contained intron/exon junctions. It is also possible that there are segments of the maize genome that are not represented in the subset of BACs used in this study. The majority of overgos that failed to identify a single BAC address were due to a repeat element contained within a single overgo probe in a pool of overgos. This resulted in filter images that contained too many BAC hits to be reliably scored. A single repeat-containing overgo will result in the loss of BAC addresses for 47 overgo probes since it is present in both a row pool and column pool. The identity of the repeat-containing overgo can be inferred from the intersection of the row and column pools, but the BAC address information cannot be recovered for the other 46 overgo probes in the row and column pools. This represents one of the only, but potentially serious, drawbacks to the 24 × 24 multiplexing approach used in this study.
We found it informative to view the overgo-BAC hits as a distribution of the number of BAC hits per overgo (Fig. 2). In the initial stages of this project, this allowed us to monitor the effectiveness of overgo selection from EST regions that had been masked for repeats. If repeat masking is effective, the majority of overgos should hybridize to a small number of BACs reflecting one or two genomic loci. Some hybridization to two or more genomic loci was expected given the highly duplicated nature of the maize genome. Repeat masking was clearly effective, as 86% and 92% of the overgos hit 15 or fewer BACs in the EcoRI or HindIII libraries, respectively. Pooling the EcoRI and HindIII libraries to become an approximately 10× resource, 80% of the overgos identified less than 20 BACs. Despite repeat masking, a small number of overgos (404; approximately 5%) identified a disproportionate number of BACs (50 or more) in the combined BAC libraries. These 404 overgos accounted for 51,648 of the 164,254 (31%) total overgo-BAC hits. This suggests that these overgos contained repeats that facilitated their hybridization to multiple regions in the genome.
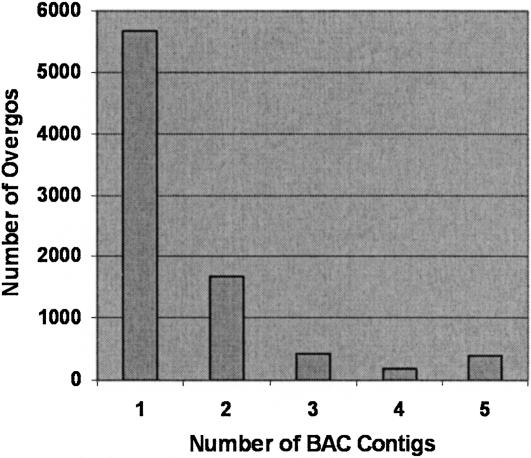
The locus-specific overgos used in this study are contributing to both contig assembly and placement on the high-resolution genetic map (Fig. 3). A high-confidence and robust data set for overgo-contig addresses was generated by requiring that all overgo-contig relationships have at least two BAC hits in the contig. This removed any spurious overgo-contig or low confidence associations and established 13,900 overgo-contig associations. Nearly 68% of the overgos hybridized to BACs contained within a single contig, and 20% hybridized to BACs contained within two contigs. Only 7% of the overgos hit BACs that were distributed over four or more contigs. Those overgos hitting multiple contigs likely represent a mix of unmerged contigs, duplicated genomic segments, and overgos designed to multicopy regions.
Figure 3.
Distribution of overgo-BAC hits across multiple contigs. A total of 292,039 HindIII BAC fingerprints (approximately 15×) and overgo markers identifying 25 or fewer BACs were assembled into 4,518 contigs using the automated assembly feature of FPC. Cutoff values used were 10−12, 10−11, 10−10, and 10−9 for zero, one, two, and three shared overgo markers, respectively. Contigs with greater than 5 Questionable clones were tested at higher stringencies of 10−13 and 10−14. Manual editing of the 4,518 contigs using the FPC contig end joining tool in conjunction with shared markers and selectively reduced cutoffs down to 10−8 reduced the contigs number to 3,488. To establish a robust data set, overgos were required to hit two or more BACs in a given contig, thereby removing single BAC hits in contigs. Using these filtering criteria, 13,900 overgo-contig associations were derived for 8,353 overgos. Approximately 88% of the overgo markers hybridized to BACs contained within one or two contigs, while only 7% hybridized to four or more contigs.
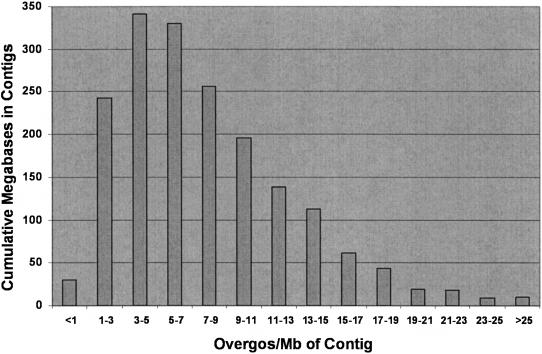
The distribution of overgo hits across 165,888 BACs from the two libraries suggests genomic domains of both high and low gene density for the 10,642 overgos used in this study (Table II). Approximately 54% (89,888/165,888) of the BACs used in this study hybridized to one or more overgos, while 46% of the BACs had no overgo hits. In our current Finger Print Contig (FPC) assembly, 1,483 of the 3,488 contigs (258 Mb of the total 2,050 Mb in BAC contigs) were not hit by overgos (data not shown). These 1,483 contigs may be regions of the maize genome that are gene poor, at least for this large but incomplete data set. The majority of the BACs (79%, 70,649/89,888) identified by overgo probes hybridized to one or two overgos. There were, however, 1,436 BACs (approximately 1%) that were hit by six or more overgo probes, which suggests that these 100- to 200-kb genomic regions must be gene dense. To gain greater insight on the density of 13,900 overgo-contig sites across 2,005 contigs containing 1,792 Mb, each contig was evaluated for overgo density, which was defined as the number of overgos per megabase of contig (Fig. 4). The value obtained for each of the 2,005 contigs was placed in an overgo density category, and the total number of megabases in each category was summed to generate a total for each of the 14 categories. Overgo density varied greatly across contigs ranging from less than one overgo per megabase for 19 large contigs containing 30.6 Mb to more than 20 overgos per megabase for 34 contigs containing 36.9 Mb. Of the 1,792 Mb accessed with the 10,642 overgos used in this study, 926 Mb contained an overgo density in the range of three to nine overgos per megabase.
Table II.
Distribution of multiple overgo hits on single BACs
| Overgo Hits/BAC | HindIII | EcoR1 | Total |
|---|---|---|---|
| 0 | 41,308 | 34,692 | 76,000 |
| 1 | 23,846 | 23,153 | 46,999 |
| 2 | 10,352 | 13,298 | 23,650 |
| 3 | 4,434 | 6,424 | 10,858 |
| 4 | 1,832 | 3,015 | 4,847 |
| 5 | 737 | 1,361 | 2,698 |
| 6 | 286 | 588 | 874 |
| 7 | 100 | 257 | 357 |
| 8 | 33 | 88 | 121 |
| 9 | 12 | 49 | 61 |
| 10 | 2 | 11 | 13 |
| 11–15 | 2 | 8 | 10 |
| Total BACs Hit | 41,636 | 48,252 | 89,888 |
The distribution of overgo-BAC hits or overgo-contig hits for 9,300 overgos was evaluated to identify potential regions of high or low gene density. There were 89,888 BACs that were hit by one or more overgos. Approximately 78% of the BACs hit (70,649) by overgos were hit by one or two overgos. A small number of BACs (205/165,888) were hit by eight or more overgos, indicating gene-dense 100- to 200-kb regions in the maize genome.
Figure 4.
Distribution of overgo density in BAC contigs The 2,005 contigs containing one or more overgos were evaluated for overgo density, placed in density categories (x axis), and cumulative sums calculated (y axis) to generate a distribution of overgo density in BAC contigs. Overgo density is defined as the number of overgos per megabase of contig. Overgo density varied up to 10-fold across contigs, with some contigs containing less than three overgos per megabase, while others contained 30 to 40 overgos per megabase. A small group of 16 contigs totaling approximately 9 Mb (data not shown) was very overgo dense with more than 40 overgos per megabase.
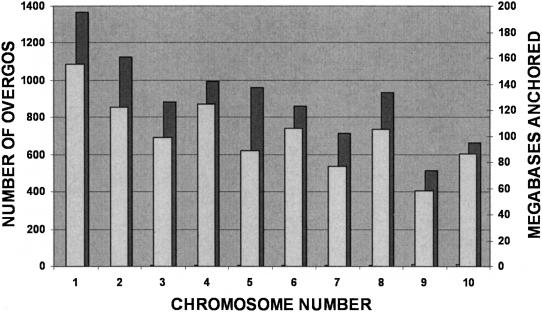
Current efforts to anchor BAC contigs to our high-resolution map using a variety of anchoring approaches have resulted in 1,022 Mb of genetically anchored contigs spread across the 10 chromosomes of maize (Fig. 5). A total of 9,003 overgo-contig sites (65%) are anchored to the high-resolution genetic map. While the total number of megabases anchored to each of the 10 chromosomes varied, the total number of genetically anchored overgos was roughly proportional to the total number of megabases anchored for each chromosome. The average genome-wide overgo density in anchored contigs was 8.8 overgos per megabase, with the individual chromosome values ranging from 7.7 to 10.7 overgos per anchored megabase. About 4,900 overgo-contig sites remain associated with 770 Mb of unanchored contigs and will allow a targeted approach to anchoring these contigs to the genetic map.
Figure 5.
Overgo distribution across maize chromosomes. A total of 13,900 overgo locations are distributed across 2,005 contigs encompassing 1,792 Mb. All overgo locations are confirmed by at least two BAC hits in a contig. There are 9,003 overgo locations (65%) distributed across 735 genetically anchored BAC contigs encompassing 1,053 Mb. Genetically anchored overgo locations are distributed across all 10 chromosomes. There are 4,897 of the 13,900 overgo locations on BAC contigs that remain unanchored to a chromosome. These overgo locations represent sites for targeted genetic marker development that will allow a rational approach toward genetic anchoring of unanchored BAC contigs.
Six overgos, which were associated with finished genomic sequence, were queried to determine the fidelity of overgo hybridization. Two overgos were queried against two BACs that had been placed on the physical map by direct HindIII fingerprinting. Four overgos were queried against YAC 334B07 containing the adh1 region (SanMiguel et al., 1996) that had been completely sequenced and placed on the physical map using fragments generated from a simulated HindIII digest. Homology of the 40-base overgo with the queried genomic sequence varied from the expected complete match (40 bases over 40 bases) to a match of 28 bases over a 32-base region. Equally important was whether homology to the cornsensus unigene from which an overgo was derived could be detected in the queried sequence. This would indicate that overgo hybridization could be relied upon to reflect the unigene from which it was derived. In all cases examined (data not shown), extensive homology was found in the expected linear order and DNA strand orientation.
DISCUSSION
A total of 9,371 overgos identified a total of 165,254 BACs, representing an average of 17.6 BACs identified per overgo for a 10× library resource. The inflated 17.6 BACs per overgo for a 10× resource can be attributed to the presence of a small percentage of overgos hitting multiple loci. Removing BAC-overgo addresses for overgos identifying more than 25 BACs (12%) reduced the average number of BACs hits to 10.7. The overall success of 88% of the overgos identifying one or more BACs compares favorably with that obtained for mouse (92%) and human (91%; Cai et al., 1998; Han et al., 2000). A total of 1,272 overgos failed to identify any BACs in either the EcoRI or HindIII BAC libraries in this study. A total of 1,035 of the 1,272 failed overgos also failed to identify BACs in the 10× DuPont library (data not shown). This indicates that these overgo failures were most likely not due to a lack of genome coverage since the DuPont and MMP libraries represent 20× coverage using multiple enzymes for library construction from two diverse genotypes with B73 and Mo17 representing the Dent and Lancaster heterotic groups, respectively. Many of these failures common to both libraries were most likely due to BAC-overgo addresses that could not be identified from row and column pools whose images could not be scored due to the presence of a repeat. Overgos that did not radiolabel well likely also contributed to these failures, although the extent of failure due to low radioisotope incorporation is difficult to determine since individual overgo labeling reactions were pooled prior to removal of unincorporated nucleotides. Given the small 40-bp size of overgo probes, good incorporation of both radiolabeled nucleotides is essential for detectable hybridization. There were 237 overgos that failed to identify any BACs in our libraries but did identify at least one BAC in the DuPont libraries. It is possible that the genomic regions corresponding to these overgos are absent in the subset of HindIII and EcoRI BACs used in this study. They may be present in the MboI BAC library (approximately 7×), which was built subsequently and was used for HindIII fingerprinting and contig assembly. It is also possible that some of the open reading frames corresponding to these failed overgos could be absent in B73 but present in Mo17 since the ESTs that were used to derive the unigene set were derived from a wide range of germplasm. A recent comparative sequencing study of the bz1 region of two North American maize lines has revealed differences in both the genic organization and composition (Fu and Dooner, 2002). Given this finding, it is plausible that a collection of 10,643 EST unigene assemblies might contain unigenes that are not present across all lines of maize.
At the initiation of this project, we expected that a certain fraction of overgo probes would identify BACs contained in two or more contigs that represent duplicate genetic loci. What was difficult to anticipate was the extent to which this would occur. From the standpoint of unambiguous anchoring of contigs to the genetic map, overgos identifying BACs contained within a single contig are preferred, but overgos identifying BACs in two contigs can also be used with judicious analysis. Currently, those overgos hitting only one contig are the focus of our SNP discovery and mapping project targeted at unambiguous anchoring of BAC contigs to the intermated B73 × Mo17 genetic map. Alternatively, overgos identifying two or more contigs provide insight into the duplicate nature of the maize genome. Our finding that 20% of the overgos identified two contigs is consistent with the findings of Helentjaris et al. (1988) that 29% of the RFLP probes developed from low copy regions (PstI and cDNA) hybridized to more than one fragment on Southern blots. Genetic mapping of these duplicated loci has revealed extensive genome duplication in the maize genome. Completion of the integrated genetic and physical map will provide a unique opportunity to view genome duplication in maize on a comprehensive scale.
The 9,300 overgos used in this study allow establishment of STSs for 1,792 Mb of the total 2,050 Mb in BAC contigs. SNP discovery is being focused on EST unigenes corresponding to overgos that hit multiple BACs contained in a single contig. Genetic mapping of SNPs is being prioritized toward large contigs with no anchoring information. Ideally each contig would contain two genetic anchors, each serving to cross-validate the genetic location of the other in addition to allowing orientation of the contig on the genetic map. Since SNP mapping is cost limiting, new contig anchoring must be balanced against multiple SNP mapping within a contig to orient the contig on the genetic map. One of the advantages of this approach that we have taken to anchoring EST unigenes to the BAC physical map is that limited mapping resources can be used very efficiently by selective mapping of contigs. This is particularly useful for the large maize genome. Despite the large set of overgos used in this study, there are 258 Mb in 1,483 small contigs that were not identified by any overgos. These contigs may contain highly repetitive or centromeric regions that are difficult to assemble in FPC using HindIII fingerprints and will be difficult to anchor genetically.
The overgo density (overgos per megabase) varied across contigs. About 5,400 of the total 13,900 overgo-contig relationships fall into contigs that have between three and nine overgos per megabase. Unanchored contigs in this density category containing overgos that hybridize to a single contig are attractive candidates for SNP anchoring in that they are often large (>1 Mb) and contain multiple STSs for SNP development. Contigs containing either a low or high overgo density are of particular interest when taking a selective approach toward genetic anchoring of contigs. Much of our previous anchoring efforts have been driven by a cost-efficient approach using previously developed simple sequence repeat (SSR) markers on BAC pools. Contigs containing large numbers of overgos are selectively favored for identification by SSRs since most of our SSRs have been developed from ESTs in the cornsensus unigene set. It is not uncommon for an overgo-dense contig to have multiple genetically mapped SSRs associated with it and as such can be ignored for further mapping efforts. BAC contigs containing a low density of overgos (three or less) are more challenging anchoring targets since they require both an overgo that uniquely associates with that contig and has a SNP for the high-resolution B73 × Mo17 mapping population.
There are three factors that contributed substantially toward the overall success of this approach to anchoring 9,371 EST unigenes to the physical map. First, the use of a 6 × 6 gridding pattern on the high-density filters allowed 41,472 BACs (each double spotted) to be placed on a single filter. This represents a 2.2-fold increase over the standard 4 × 4 pattern (18,432 BACs) that is more commonly used. While this did not reduce the time needed for scoring the filters, which is in itself a lengthy process, it did allow increased efficiencies in the use of laboratory materials and wet bench labor. Secondly, the use of a two-dimensional 24 × 24 pooling strategy that in theory allowed 576 overgo probes to be addressed in 48 filter hybridizations significantly improved efficiency. This approach allows a theoretical efficiency of 12 overgo probes to be addressed per hybridization (576/48). This is similar to the approach used for mouse chromosome 11, which used a 23 × 18 pooling strategy to address 412 MIT simple sequence length polymorphism markers in 41 hybridizations to give an efficiency of 10 overgo probes per hybridization (Cai et al., 1998). A variant of the two-dimensional approach was used for human chromosome 16q by using top pools of up to 236 overgos to identify the BACs of interest from a 12× library. Positive BACs were rearrayed and screened using an 8 × 24 two-dimensional pooling approach (Han et al., 2000). In our 24 × 24 pooling approach, the likelihood of any two of the 576 overgos in a module residing on the same BAC was very small because probes were not selected on the basis of chromosome location, as was done for the previous mouse and human studies. This simplified the deconvolution of the row and column hybridization data by greatly reducing the number of ambiguous positives due to two overgos in a module hitting the same BAC. Lastly, and perhaps most importantly, the use of overgos, which were designed to have uniform hybridization kinetics from regions devoid of repeated sequences, allowed the construction of locus-specific hybridization probes. Maize, like many crop plants, is thought to be an ancient allotetraploid (Anderson, 1945; Helentjaris, 1993). It is a general expectation that cDNA probes will hybridize to multiple loci, rendering them ineffective for unambiguously anchoring contigs to the genetic map. Complicating matters further is the presence of miniature inverted repeat elements in the 3′ untranslated regions of many maize genes (Casa et al., 2000). In general, repeat masking was successful, with only one or two repeat-containing overgos found per module. One module did, however, contain multiple repeats to the extent that only 59% of the overgos could be confirmed as identifying one or more BACs. This module and failed row and column pools from other modules were repeated to minimize losses due to repeat-containing overgos. In subsequent modules, the stringency of repeat masking was increased, and the percentage of overgo probes identifying at least one BAC address was restored to previous levels. Nonspecific overgo probes can limit multiplexing schemes to such an extent that even nonspecific probes occurring at 1% present a problem. One group has addressed this issue by developing a computer program (Primergo II) that uses characteristics of overgos found to contain repeats to reliably predict nonspecific overgo sequences prior to their use as overgo hybridization probes (Cai et al., 2001). This effectively eliminated loss of hybridization address information from overgo probes whose address could not have been deconvoluted due to the presence of a single repeat.
CONCLUSION
The use of overgo probes to create gene-specific hybridization probes in combination with efficient pooling strategies has proven to be a powerful approach in anchoring large numbers of cDNA unigenes to the maize BAC contig map. Overgo probes have allowed the identification of gene-rich regions and suggested that there may be large regions of the maize genome that are gene barren, at least for the 9,371 overgo probes analyzed. Currently, the overgo probes used in this study are providing STSs for a rational and highly efficient approach to anchoring BAC contigs to the genetic map via SNP discovery and mapping.
MATERIALS AND METHODS
Unigene Assembly, Repeat Masking, and Overgo Selection
As shown in the flowchart (Fig. 1), 70,716 public ESTs, all GenBank entries, were clustered (90% identical over 100 bp using a 70-bp sliding window) using the DoubleTwist CAT Assembler (DoubleTwist, Seattle). The vast majority of the EST sequences used in this study were from the Stanford Maize EST Project (http://www.maizegdb.org), which sampled maize ESTs from a wide variety of tissues (Fernandes et al., 2002). Approximately 1,500 proprietary full-length cDNA sequences provided by DuPont (Wilmington, DE) were included to monitor overclustering or underclustering by the CAT assembler. Clustering using these criteria resulted in 19,468 unigenes representing 12,131 contigs and 7,337 singletons. Using the criteria of 95% homology over 100 bp, the 19,468 Public unigenes were clustered with the DuPont/Pioneer unicorn set to derive 10,723 Public/DuPont EST clusters that contained at least one Public EST sequence. Each Public/DuPont cluster was queried to find the longest EST contig sequence within the cluster that could be used for repeat masking and overgo design. This identified 6,405 and 4,318 EST contigs that originated from the DuPont and Public unigene sets, respectively.
Repetitive and low complexity sequences were identified and masked prior to overgo selection using RepeatMasker (http://repeatmasker.genome.washington.edu), a software program developed at the University of Washington that identifies repeat elements in the query sequence using Cross_Match with a maize repetitive element database. Overgo Maker 40 used the masked EST contig sequences to design two 24-base oligos that were self-complementary over 8 bp so that when annealed, they could form a 40-bp overgo with two 16-base overhangs that could be filled in with Klenow and deoxynucleotide triphosphates. G/C content varied from 40% to 60% with an optimum of 50%. All 40-bp overgos derived from masked sequences were rechecked against the repeat database to identify any overgos with significant homology to repeats. Overgos were then loaded into 24 × 24 spreadsheets for oligo synthesis, which allowed the 24 × 24 module of 576 overgos to be delivered in six 96-well plates.
Filter Production and Preparation
Filters were prepared using 432 384-well plates evenly distributed between the EcoRI and HindIII BAC libraries constructed for the MMP. The first 216 plates were used from each library. A 6 × 6 gridding pattern that allowed 108 plates with 384 wells to be spotted onto a single Millipore Imobilon N+ nylon membrane (Bedford, MA) was used. The four corners were removed from 12 of the plates and refilled with an Escherichia coli culture containing the pBR325 plasmid that served as hybridization control, which allowed filter images to be accurately aligned in the imaging software. The pBR325-specific overgos were GTTGCCTTACTGGTTAGCAGAATG and CGCGTATCGGTGATTCATTCTGC. Each of the four bar-coded filters had a slightly different constellation of 48 hybridization control points that allowed them to be distinguished from one another. After gridding, membranes were carefully placed in Luria-Bertani broth agar plates (14 μg/mL chloramphenicol) with the bacteria side up. Agar-nylon plates were covered, inverted, and grown at 42° for 10 to 12 h. Nylon filters were removed and denatured (1.6 m NaCl and 0.5 m NaOH) two times for 4 min each, followed by neutralization (1.6 m NaCl, 1 m Tris, and 50 mm HCl) for 4 min. Filters were dried and treated with Proteinase K (100 mils at 1 mg/mL; Sigma, St. Louis) for 50 min at 42°C. Following hybridization, filters were stripped in 100 mils of 0.1× SSC and 0.1% SDS at 90°C for 10 min and stored at −20°C. Filters were used five times.
Overgo Annealing and Labeling
Overgos were annealed by combining 4 pmol of each oligo to obtain a final volume of 8 μL that was heated in a thermocycler to 80°C for 5 min, 37°C for 10 min, and cooled to 4°C. Overgo labeling with [α−32P]dCTP and [α−32P]dATP was as described previously (Ross et al., 1999). Briefly, 7-μL overgo labeling master mix (2.8 μl oligo labeling buffer without dATP, dCTP, and random primers); 0.7 μL bovine serum albumin (2 mg/mL); 1.82 μL water; 0.28 μL Klenow (5 units/μL); 0.7 μL [α−32P]dCTP (3000 Ci/mmol); and 0.7 μL [α−32P]dATP (3,000 Ci/mmol) were added to 8 μL (4 pmol) of annealed overgo and incubated at 25°C for 2 h. Reactions were terminated with 5 μL of STE (0.1 m NaCl, 10 mm Tris, and 1 mm EDTA). Individual overgo labeling reactions were pooled in accordance to their row and column pools and passed over a Unifilter 800 (Whatman Polyfiltronics, Clifton, NJ) filled with Sephadex G-50 (Sigma) that had been equilibrated with TE (10 mm Tris and 1 mm EDTA). Percent incorporation was calculated for a small number of randomly chosen pools and was considered acceptable at 30% or greater.
Filter Hybridization and Washing
Each filter set was soaked in 2× SSC along with a piece of nylon mesh between each filter and rolled with the DNA side facing in and placed in a 38 mm × 300 mm glass hybridization tube containing 44 mL of Perfect Hyb solution (Sigma-Aldrich). Filters were prehybridized at 60°C with constant rotation for at least 2 h. Pooled overgo probes were denatured at 90°C for 10 min and added to 6 mL of Perfect Hyb solution, which was immediately added to the 44 mL of Perfect Hyb that had been used for prehybridization. Hybridization was for 12 to 16 h at 60°C. Filters were washed progressively for 1 h each at 60°C in 2× SSC and 0.1% SDS (wash 1), 1.5× SSC and 0.1% SDS (wash 2), and 0.5× SSC and 0.1% SDS (wash 3). Wash 1 was in the hybridization bottles, and washes 2 and 3 were done in trays. Filters were blotted dry and placed on phosphor imager plates for 3 h to overnight. All phosphorimaging was done on a Storm 820 PhosphorImager (Molecular Dynamics, Sunnyvale, CA), according to the manufacturer's instructions. All filter sets were stored at −20°C after scanning and stripping.
Image Analysis and Matrix Deconvolution
All image analysis was done using the screening feature of the Array Vision 4.0 software from Imaging Research (St. Catharines, Canada). A grid file was generated for each filter that reflected the 41,472 clone addresses and the unique spotting design of the 48 pBR325 positive controls, which are used to align the filter to the grid image. Array Vision automatically identifies the positive BAC clones and generates a text file. This information is frequently incorrect and is checked to ensure that the two spots identified are from the same 6 × 6 primary (18 unique BAC clones, each double spotted) and correspond to the same plate and well address. Spots that do not meet these criteria are deleted. To maintain high quality control standards, 25% of the scored filter images are selected at random and rescored by a second person. Data from the scored filters corresponding to a particular row or column pool are stored as a text file, with the 384 individual files making up a single module (48 sets × 8 filters/set; each filter has its own text file) of 576 probes being stored in a single Unix/Linux directory. The module of 576 overgo probes is then deconvoluted by running it within a predefined batch shell procedure, which is composed of six different Perl scripts that assign BAC addresses for each of the 576 overgo probes.
The cornsensus EST unigene contig assemblies have been submitted to GenBank under the accession numbers AY103536–AY112654. The complete list of unigenes used, overgo primer sequences, and singletons that have previously assigned nonsequential GenBank numbers are also available at www.agron.missouri.edu/files_dl/MMP/Cornsensus/ or http://www.maizemap.org/resources.htm. Individual BACs identified by overgos and/or complete BAC libraries are available from Clemson University Genomics Institute (http://www.genome.clemson.edu/) or Children's Hospital Oakland Research Institute (http://bacpac.chori.org).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY103536–AY112654.
Acknowledgments
We thank Susan Melia-Hancock and Loralynn Sullivan for excellent technical assistance. We also thank Mike McMullen and Perry Gustafson for helpful comments on the manuscript.
This work was supported by the National Science Foundation (grant no. 9872655).
References
- Anderson E (1945) What is Zea mays? A report of progress. Chron Bot 9: 88–92 [Google Scholar]
- Asakawa S, Abe I, Kudoh Y, Kishi N, Wang Y, Kubota R, Kudoh J, Kawasaki K, Minoshima S, Shimizu N (1997) Human BAC library: construction and rapid screening. Gene 19: 69–79 [DOI] [PubMed] [Google Scholar]
- Bruno WJ, Knill E, Balding DJ, Bruce DC, Doggett NA, Sawhill WW, Stallings RL, Whittaker CC, Torney DC (1995) Efficient pooling designs for library screening. Genomics 26: 21–30 [DOI] [PubMed] [Google Scholar]
- Burke DT, Carle GF, Olson MV (1987) Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science 236: 806–812 [DOI] [PubMed] [Google Scholar]
- Burke DT, Rossi JM, Leung J, Koos DS, Tilghman SM (1991) A mouse genomic library of yeast artificial chromosome clones. Mamm Genome 1: 65. [DOI] [PubMed] [Google Scholar]
- Cai W, Chow C, Damani S, Gregory S, Marra M, Bradley A (2001) An SSLP marker-anchored BAC framework map of the mouse genome. Nat Genet 29: 133–144 [DOI] [PubMed] [Google Scholar]
- Cai W, Reneker J, Chow C, Vaishnav M, Bradley A (1998) An anchored framework BAC map of mouse chromosome 11 assembled using multiplex oligonucleotide hybridization. Genomics 54: 387–397 [DOI] [PubMed] [Google Scholar]
- Casa A, Brouwer C, Nagel A, Wang L, Zhang Q, Kresovich S, Wessler S (2000) The MITE family heartbreaker (Hbr): molecular markers in maize. Proc Natl Acad Sci USA 97: 10083–10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Presting G, Barbazuk WB, Goicoechea JL, Blackmon B, Fang G, Kim H, Frisch D, Yu Y, Sun S, Higgingbottom S, Phimphilai J, et al (2002) An integrated physical and genetic map of the rice genome. Plant Cell 14: 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumakov IM, Rigault P, Le Gall I, Bellanne-Chantelot C, Billault A, Guillou S, Soularue P, Guasconi G, Poullier E, Gros I (1995) A YAC contig map of the human genome. Nature Suppl 377: 175–297 [DOI] [PubMed] [Google Scholar]
- Coe EH Jr, Cone K, McMullen MD, Chen SS, Davis G, Gardiner J, Liscum E, Polacco M, Paterson A, Sanchez-Villeda H, Soderlund C, Wing RA (2002) Access to the maize genome: an integrated physical and genetic map. Plant Physiol 128: 9–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KJ, Thompson H, Edwards D, de Saizieu A, Sparks C, Thompson JA, Greenland AJ, Eyers M, Schuch W (1992) Construction and characterisation of a yeast artificial chromosome library containing three haploid maize genome equivalents. Plant Mol Biol 19: 299–308 [DOI] [PubMed] [Google Scholar]
- Evans GA, Lewis KA (1989) Physical mapping of complex genomes by cosmid multiplex analysis. Proc Natl Acad Sci USA 87: 5030–5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Brendel V, Gai X, Lal S, Chandler V, Elumalai R, Galbraith D, Pierson E, Walbot V (2002) Comparison of RNA expression profiles based on maize expressed sequence tag frequency analysis and micro-array hybridization. Plant Physiol 128: 896–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Dooner HK (2002) Intraspecific violation of genetic colinearity and its implications in maize. Proc Natl Acad Sci USA 99: 9573–9578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ED, Olson MV (1990) Systematic screening of yeast artificial-chromosome libraries by use of the polymerase chain reaction. Proc Natl Acad Sci USA 87: 1213–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ED, Riethman HC, Dutchik JE, Olson MV (1991) Detection and characterization of chimeric yeast artificial-chromosome clones. Genomics 11: 658–669 [DOI] [PubMed] [Google Scholar]
- Haldi ML, Strickland C, Lim P, VanBerkel V, Chen X, Noya D, Korenberg JR, Husain Z, Miller J, Lander ES (1996) A comprehensive large-insert yeast artificial chromosome library for physical mapping of the mouse genome. Mamm Genome 7: 767–769 [DOI] [PubMed] [Google Scholar]
- Han CS, Sutherland RD, Jewett PB, Campbell ML, Meincke LJ, Tesmer JG, Mundt MO, Fawcett JJ, Kim U, Deaven LL, Doggett NA (2000) Construction of a BAC contig map of chromosome 16q by two-dimensional overgo hybridization. Genome Res 10: 714–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helentjaris T (1993) Implications for conserved genomic structure among plant species. Proc Natl Acad Sci USA 90: 8308–8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helentjaris T, Weber D, Wright S (1988) Identification of the genomic location of duplicate nucleotide sequences in maize by the analysis of restriction fragment length polymorphisms. Genetics 118: 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Mapping Consortium (2001) A physical map of the human genome. Nature 409: 934–941 [DOI] [PubMed] [Google Scholar]
- Klein PE, Klein RR, Cartinhour SW, Ulanch PE, Dong J, Obert JA, Morishige DT, Schlueter SD, Childess KL, Ale M, Mullet JE (2000) A high throughput AFLP-based method for constructing integrated genetic and physical maps: progress toward a sorghum genome map. Genome Res 10: 789–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Sharopova N, Beavis WD, Grant D, Katt M, Blair D, Hallauer A (2002) Expanding the genetic map of maize with the intermated B73 x Mo17 (IBM) population. Plant Mol Biol 48: 453–461 [DOI] [PubMed] [Google Scholar]
- Mozo T, Dewar K, Dunn P, Ecker JR, Fischer S, Kloska S, Lehrach H, Marra M, Martienssen R, Meier-Ewert S, Altman T (1999) A complete BAC-based physical map of the Arabidopsis thaliana genome. Nat Genet 22: 271–275 [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, editor (1993) Genetic Maps: Locus Maps of Complex Genomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Ross MT, LaBrie S, McPherson J, Stanton VP Jr (1999) Screening large-insert libraries by hybridization. In A Boyl, ed, Current Protocols in Human Genetics. Wiley, New York, pp 5.6.1–5.6.52
- Saji S, Umehara Y, Antonio B, Yamane H, Tanoue H, Baba T, Aoki H, Ishige N, Wu JZ, Koike K, Matsumoto T, Sasaki T (2001) A physical map with yeast artificial chromosome (YAC) clones covering 63% of the 12 rice chromosomes. Genome 44: 32–37 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 737–738 [DOI] [PubMed] [Google Scholar]
- Selleri L, Eubanks JH, Giovannini M, Hermanson GG, Romo A, Djabali M, Maurer S, Mcelligott DL, Smith MW, Evans GA (1992) Detection and characterization of “chimeric” yeast artificial chromosome clones by fluorescent in situ suppression hybridization. Genomics 14: 536–541 [DOI] [PubMed] [Google Scholar]
- Sharopova N, McMullen MD, Schultz L, Schroeder S, Sanchez-Villeda H, Gardiner J, Bergstrom D, Houchins K, Melia-Hancock S, Musket T, Duru N, Polacco M, et al (2002) Development and mapping of SSR markers for maize. Plant Mol Biol 48: 463–481 [DOI] [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M (1992) Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA 89: 8794–8797 [DOI] [PMC free article] [PubMed] [Google Scholar]