Abstract
Single-stranded DNA sequences that are highly specific for a target ligand are called aptamers. While the incorporation of aptamer sequences into stem-loop molecular beacons has become an essential tool in optical biosensors, the design principles that determine the magnitude of binding affinity and its relationship to placement of the aptamer sequence in the stem-loop architecture are not well defined. By controlled placement of the aptamer along the loop region of the molecular beacon, it is observed that the binding affinity can be tuned over 4 orders of magnitude (1.3 nM – 203 μM) for the Huizenga and Szostak ATP DNA aptamer sequence. It is observed that the Kd is enhanced for the fully exposed sequence, with reduced binding affinity when the aptamer is part of the stem region of the beacon. Analysis of the ΔG values indicate a clear correlation between the aptamer hybridized length in the stem and its observed Kd. The use of a nanometal surface energy transfer probe method for monitoring ATP binding to the aptamer sequence allows the observation of negative cooperativity between the two ATP binding events. Maintenance of the high binding affinity of this ATP aptamer and the observation of two separate Kd’s for ATP binding indicate NSET as an effective, nonmanipulative, optical method for tracking biomolecular changes.
Introduction
Nucleic acid sequences can form noncanonical structures by folding in the presence of small molecule ligands or following the binding of a protein.1−3 The realization that single-stranded DNA forms unique, complex, nucleic acid structures in response to ligand binding has led to the development of synthetic, single-stranded DNA sequences tailored to fold in the presence of a specific target. These synthetic sequences, termed aptamers, can be highly selective for a target ligand.4−8 The rapid advancement of the field stems from the use of combinatorial library screening to select aptamer sequences based on binding affinity (Kd) response, resulting in new aptamers appearing in the literature for a wide range of small molecules including thrombin, ATP, cocaine, HIV-1 REV, Hg2+, and prostate-specific membrane antigen (PSMA).4−15 Since their discovery, the use of aptamers to sense the binding of target molecules in solution or on cell surfaces has evolved rapidly for bioanalytical applications. Recently, the incorporation of gold nanoparticles (AuNPs) onto aptamers has further pushed aptamer technology into applications such as fluorescence, electrochemical, and plasmonic sensors to report binding.16−21
It is well-known that the fluorescent quenching of a dye by a AuNP is well described within the nanometal surface energy transfer (NSET) model, and thus has attracted considerable attention as an optical ruler in biophysics.22 NSET with longer R0 values (R0 >100 Å) makes separation of the donor and acceptor more attainable than FRET studies, which can be advantageous for molecular beacon fluorescent sensors. In fact, earlier fluorescent biosensors utilizing pyrene-appended aptamers have shown perturbation of target binding due to interference of the binding site by dye-labeling in the binding pocket.23 NSET allows the positioning of the donor and acceptor labels away from the binding pocket.
While the AuNP based fluorescent assays are very sensitive, the difficulty in tuning the binding affinity to match the experimental conditions has limited their broader applicability. To increase the long-term applicability of nanoparticle based aptamer sensors, the sensor response has to be generalizable to the experimental conditions in biophysics. The ability to systematically tune a previously identified aptamer’s binding affinity to better match experimental conditions (concentration range) without redesigning the aptamer or relying on single point mutations is not well developed.24 When the aptamer is bound to a AuNP, the response of the aptamer can be influenced by the aptamer structure and any interaction with the AuNP surface, which can impact sensitivity as well as specificity of future sensors. The interaction of the AuNP with the aptamer can be minimized by surface passivation as previously demonstrated,2,25 but the effect of aptamer design on response is far less understood. It has been observed in molecular beacon stem-loop architectures that the aptamer Kd values are affected by stem-loop size, stem length, stem composition, and single point mutations in and away from the target’s binding site.24,26,27 Several studies have also indicated that the aptamer binding affinity in optical sensors, including nonhairpin optical probes, is strongly impacted by the degree of duplex formation over the aptamer sequence. The higher Kd values are attributed to the thermodynamics associated with overcoming the duplexed regions or target accessibility to the binding sites on the aptamer.17,27 Developing a deeper understanding of the underlying consequences of positioning the single-stranded aptamer sequence into a stem-loop molecular beacon framework could allow more judicious design of AuNP aptasensors.
In this manuscript, we systematically study the influence of the location of a 25-mer ATP single-stranded DNA aptamer on the binding affinity for ATP when incorporated into the loop region of a 57-mer stem-loop molecular beacon. The influence of target site exposure in the loop is investigated by sliding the aptamer sequence along the loop circumference, resulting in a variable degree of single-stranded exposure for the ATP aptamer. The design allows the influence of ligand accessibility and duplex formation to be investigated. The chosen ATP aptamer is based upon the Huizenga and Szostak 25-mer ATP DNA aptamer sequence whose structure and ATP binding properties were previously analyzed by NMR.10,28 The aptamer binds two ATP molecules per sequence and induces a folded structure. For the analysis of the influence of aptamer position within the loop, the aptamer beacon incorporates a 3 nm AuNP at the 3′ end as an energy acceptor and a fluorescein dye at the 5′ end as an excited state donor. The use of the AuNP-FAM energy transfer pair allows the binding affinity to be measured and the change in distance following ATP binding to be monitored using nanometal surface energy transfer techniques.29 The change in the photoluminescent (PL) intensity data with increasing ATP concentration allows the change in Kd and the change in aptamer sequence location in the loop to be correlated. The results clearly demonstrate that the binding affinity of ATP to the aptamer is directly dependent on degree of sequence exposure.
The highest binding affinities are observed with full exposure of the aptamer sequence in the loop, while duplex formation reduces binding affinity most likely due to the thermodynamics of DNA base pairing. Importantly, the positioning of the aptamer allows several orders of magnitude variability in the Kd values, indicating that for a given aptamer, the Kd can be systematically tuned by placement in the stem-loop architecture. No differences in binding affinity were observed for partial blocking of the 5′ vs 3′ aptamer sequence ends in this study. Hill plot analysis of the aptamer concentration dependence on the PL intensity reveals negative cooperativity between sequential ATP binding events. In addition, the specificity of the aptamer for ATP over UTP and GTP is maintained, as well as similar responsiveness to AMP vs ATP. Inspection of the thermodynamic stabilities of the loop and stem region indicates that a nonlinear correlation exists between the measured binding affinity and length of aptamer duplexed region in the stem-loop architecture. The results of the study establish a guideline for systematically manipulating Kd values in a stem-loop molecular beacon designed around the thermodynamics of the aptamer construct.
Results and Discussion
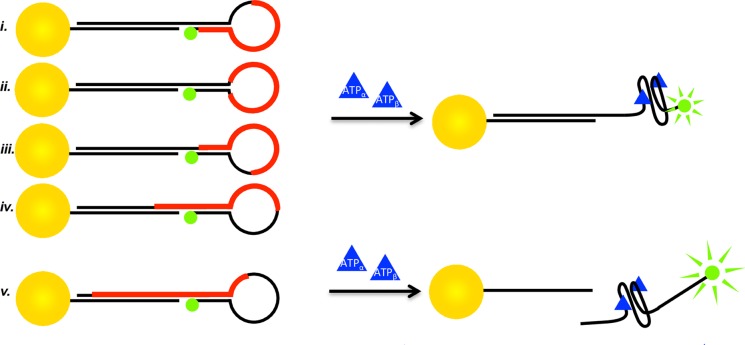
The stem-loop construct contains a 25-mer Huizenga-Szostak ATP aptamer, wherein the aptamer sequence is moved along the stem-loop region from the 5′ toward the 3′ end, resulting in partial blocking of the ATP target site due to base pairing when the loop is closed. The five individual 57-mer stem-loop constructs utilized in the study are shown in Figure 1 and Table 1. In Table 1, the 25-mer Huizenga-Szostak aptamer sequence in each of the five constructs is red and italicized for clarity. To distinguish between the loop and stem region, the loop is highlighted by a dashed underline, while the stem is highlighted by a solid underline. As a control, the stem-loop studies were compared to a single-stranded sequence containing the aptamer (sequence vi) that cannot form a stem-loop structure (Supporting Information Figure SF2).
Figure 1.
Scheme of the NSET stem-loop beacons and their response to ATP. The red bars in the stem-loop structures are indicative of the aptamer sequence location in the stem-loop architectures. Sequences i–iv remain a single construct upon ATP binding whereas sequence v contains enough aptamer sequence overlap in the stem to induce displacement of the flare strand upon ATP binding.
Table 1. NSET Stem-Loop Beacon Sequences Used in This Studya.

Red, italicized text is indicative of the conserved 25-base-pair ATP aptamer sequence. A dotted underline represents the loop portion in the stem-loop structure while a bold underline represents the 6-base-pair stem. Amine-terminated 15-base-pair linkers are listed below the sequence, aligned at the appropriate complementary positions.
The stem-loop construct is designed (Figure 1) to detect the conformational changes in the ATP-induced aptamer binding by monitoring the change in the fluorescent intensity of the appended fluorescein when quenched in the vicinity of the AuNP. A 3 nm AuNP with low loading (1:1 AuNP:DNA) was chosen to promote high efficiency of the aptamer binding to ATP, as observed in previous studies where hybridization of DNA on AuNP surfaces was most efficient at low primer loading levels and on surfaces of high curvature.31,32 The AuNP acts as a dark acceptor of the fluorescein excited state via a long-range NSET mechanism. The NSET method is similar to FRET techniques, wherein the separation distance between the fluorescein and AuNP are reported by the change in emission intensity of the fluorescein. The quenching can be described within an NSET model allowing the ensemble-averaged distances to be calculated by comparison to theoretical values. Unique to NSET, the R0 (critical distance for 50% energy transfer) is a longer value of 110.6 Å for a 3 ± 0.5 nm AuNP-FAM acceptor donor pair, allowing the fluorescein (FAM) and AuNP to be bound distally from the aptamer site to minimize impact on the observed aptamer folding behavior.29 The impact of small molecule dyes, quantum dots, and EPR tags on the folding of biopolymers is an area of open discussion in the biophysics community. Studies on AuNPs below 4 nm approach the size of the footprint of DNA. Earlier studies have shown that no impact on folding is observed for substrate induced folding of RNA or protein induced folding in DNA when bound to a small AuNP surface, presumably due to the increased distance of separation from the binding site allowable by NSET probes.1,2 It is also important to note that nonspecific DNA interactions with the AuNP surface are not expected because previous groups report little to no DNA adsorption onto nanoparticle surfaces with a ligand-passivated, neutral surface.25,33 Typical DNA adsorption studies that report high levels of single-stranded adsorption utilize citrate-capped AuNPs where the citrate molecule is displaced to accommodate DNA laying down on the surface.34 The AuNPs used in this study are DHLA-capped, thus the surface is thiol-coated, and it is assumed that the small particle size and the stable ligand coating make these AuNPs resistant to nonspecific adsorption.
The PL NSET efficiencies and calculated distances for the aptamer sequences at I0 (no ATP) and Imax (high ATP concentration) are listed in Supporting Information (SF Table 1.) In the absence of ATP, the aptamer forms a closed stem-loop hairpin conformation positioning the AuNP and the FAM in close proximity, resulting in partial quenching of the FAM in the AuNP near field. The intensity of the fluorescein emission in the absence of ATP for the closed stem-loop yields a separation distance of AuNP to FAM of 77 Å (E = 19%) confirming loop formation in the absence of ATP. Exposure of the aptamer to ATP induces opening of the loop and formation of a folded DNA conformer that repositions the FAM and AuNP to cause the change in fluorophore emission, tracking the binding of ATP. The fluorescein intensity at high concentrations of ATP yields a separation distance consistent with the NMR predicted folded structure, wherein the AuNP to FAM separation distance is 96–109 Å (∼44% E), confirming formation of the ATP induced folded aptamer in the presence of ATP.28 A distal range is reported here because TEM analysis indicates a ±0.5 nm Gaussian distribution in AuNP sizes, which results in an NSET R0 that is better described as a weighted probability to account for the nanoparticle size (Supporting Information). By incorporating the measured population distribution for particle size coupled to the change in extinction coefficient which scales as r3, the R0<avg> value for the prepared AuNPs in the study is 110.6 Å. Although it should be noted that the correction to R0 for the distribution results in only a minimal perturbation to the R0<calc,3 nm> for the 3 nm average size (R0<calc, 3 nm> = 110.5 Å). Moreover, while the ATP-bound NMR structure is available, its orientation to the surface could vary. Photoluminescent saturation is therefore used to establish the ATP saturation limit for ATP binding.
As a control and in an effort to ensure that free, unlabeled AuNPs in solution do not impact the conformation of the folded aptamer, analysis of the fluorescein intensity following the addition of free AuNP (up to 2 nM excess AuNP) in solution is provided (Supporting Information Figure SF6). No change in PL saturation was observed. Binding assays for UTP and GTP on selected sequences are available in Supporting Information (Figure SF7) and demonstrate that specificity for ATP over other triphosphate nucleosides is not impacted by the binding of the AuNP or the position of the aptamer in the loop. Control studies on AMP binding relative to ATP are also shown in the Supporting Information, wherein similar binding affinities are observed to be consistent with literature results.23,35,36
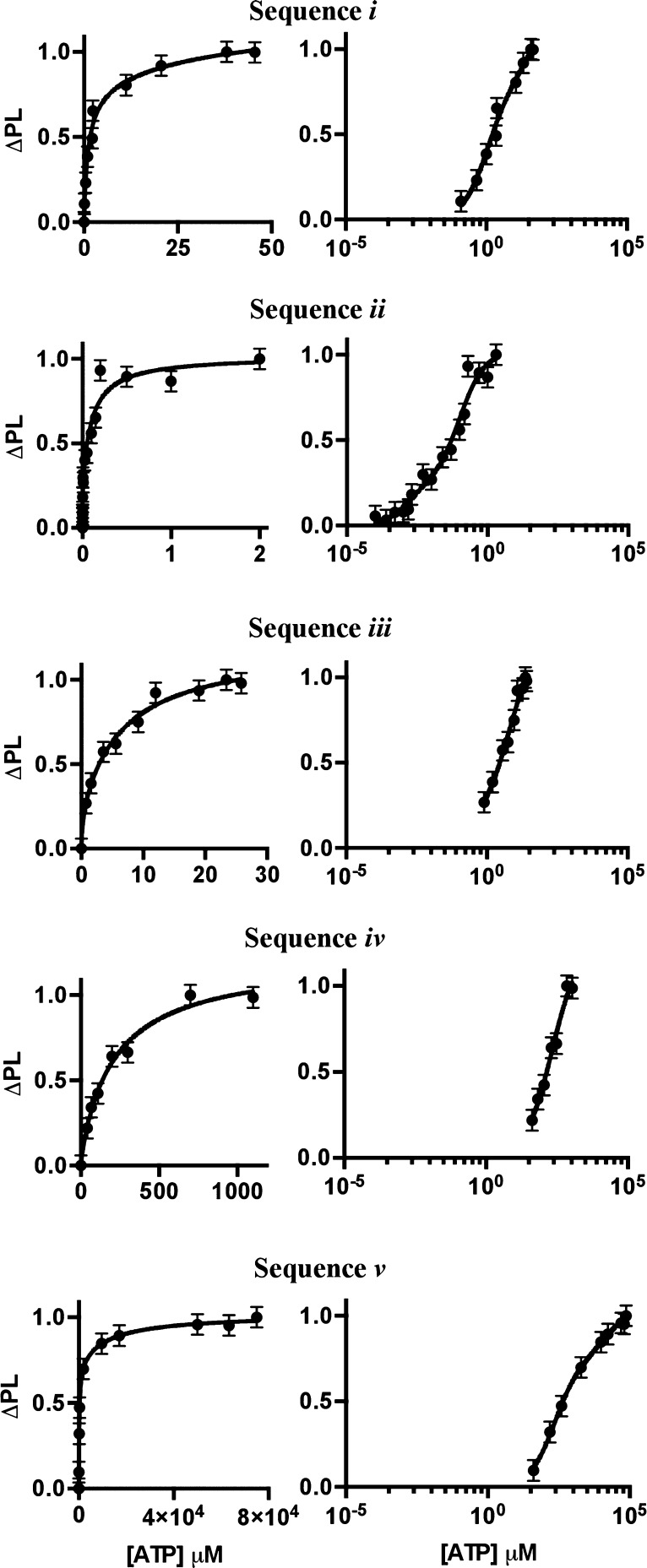
The binding affinities for the five stem-loop constructs were measured by analyzing the ATP concentration-dependent change in fluorescein photoluminescence (ΔPL) at 1 nM aptamer (Figure 2). The data is plotted as a saturation curve and as a log plot of ΔPL vs [ATP] in Figure 2, and as the raw PL data in Supporting Information Figure SF3. Consistent with the proposed structural folding of the aptamer following opening of the stem-loop upon ATP binding, the observed PL intensity increases asymptotically with increasing ATP concentration. The maximum intensity change (ΔPL) reflects the final conformation of the aptamer after ATP-induced folding. In analogy to the NMR studies, the binding affinity for the ATP is extracted by fitting the ΔPL data to a two-step ATP binding model
| 1 |
where B(1)max represents the maximum ΔPL for the first binding event, B(2)max represents the maximum ΔPL for the second binding event, and Kd(1) and Kd(2) are the binding affinities for the individual events. The Kd values are listed in Table 2 and the fits are shown in Figure 2. The Kd values obtained from the two-state binding model were statistically analyzed using an F-test and Akaike’s test of the DOF/sum of squares. Plots of the two- vs one-state model for each sequence are shown in the Supporting Information (Figure SF4). For the stem-loop conformers, analyzing the sum of the squares relative to the degrees of freedom yields F-test values of >1 with the Akaike’s test values <0 (Table 2). The F-test values of >1 coupled to negative AIC values confirms the validity of the more complex two-state model for the studied sequences. Details of the statistical equations and the values for the single-stranded control are available in the Supporting Information (SF Table 2). The control sequence, which cannot form a stem-loop structure, exhibits an F-test value of zero and a positive AIC value, and therefore is fit to the single-binding model.
Figure 2.
ATP saturation curves of the five NSET stem-loop beacons fit to eq 1. The left-hand column displays the relationship between the ΔPL intensity and micromolar ATP concentration. The right-hand column utilizes the same fit in eq 1 but graphs the ATP concentration as a log function to demonstrate the detectable concentration range of each NSET stem-loop beacon.
Table 2. Table of the Kd Values with the Reported 95% Confidence Limits (2σ) Extracted from eq 1, the Hill Coefficients (h) Extracted from eq 2, and the F-test and Akaike’s Values Extracted from Statistical Analyses.
| Seq | Kd1 (2σ) | Kd2 (2σ) | Hill (h) | F-test, AIC |
|---|---|---|---|---|
| i | 1.1 (1.2) μM | 35.3 (30.2) μM | 0.77 | 1.615, −0.307 |
| ii | 1.3 (2.4) nM | 102.3 (93.4) nM | 0.55 | 8.754, −10.502 |
| iii | 0.3 (0.6) μM | 6.9 (12.0) μM | 0.70 | 2.368, −1.818 |
| iv | 33.6 (5.4) μM | 232.8 (333.4) μM | 0.49 | 1.498, −0.473 |
| v | 203.0 (129.8) μM | 5486 (8248) μM | 0.73 | 7.940, −9.123 |
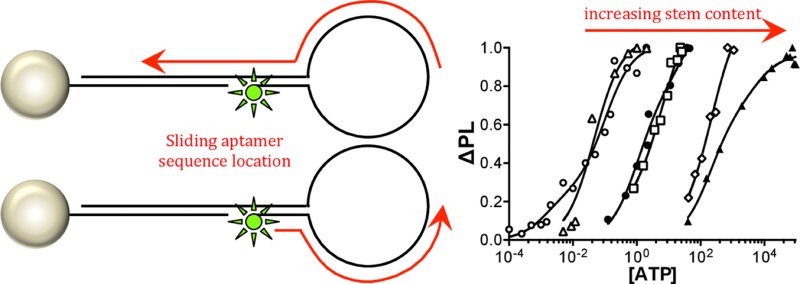
Comparison of the Kd values for the sequences reveals that increasing single-stranded exposure in the loop decreases the Kd value. A decreasing Kd value indicates stronger binding affinity. No significant preference in Kd value is observed if the 5′ or 3′ end of the aptamer is partially blocked (sequence i vs sequence iii). The measured Kd value of the fully exposed loop sequence (sequence ii) is consistent with the measured value for the single-stranded molecular beacon analogue in the Supporting Information. Likewise the sequence incorporated into the stem-loop configuration with significant duplex formation in the stem exhibits Kd values similar to previous reports in an analogous nanoflare design, supporting the effect of aptamer location on the observed Kd values.17,35 The results of the saturation curves imply that ATP accessibility to the single-stranded region is critical to the experimentally measured binding affinity with an observed tunability over 4 orders of magnitude.
Interestingly, NSET observes two ATP binding events similar to the reported NMR studies, but earlier FRET studies have been unable to distinguish the separate events. Whether the FRET studies’ shortcomings are due to the close proximity of the dye molecule tagged to the binding site, or due to cooperativity effects, is not well understood. To analyze cooperativity in the binding of ATP to the stem-loop constructs, concentration dependent data for ATP and the stem-loop construct were analyzed by a Hill coefficient expression
| 2 |
where h is the Hill coefficient representative of positively cooperative (h > 1), negatively cooperative (h < 1), or noncooperative behavior (h = 1) between ATP binding sites. The Hill plots of the NSET stem-loop beacons are shown in the Supporting Information (Figure SF5). Fitting the ΔPL data to the Hill equation (eq 2) yields values for h between 0.4 and 0.8 for the stem-loop constructs. A Hill value of <1 indicates the presence of negatively cooperative binding of the ATP molecules for the sequences in Figure 1. A negative cooperativity can be understood in terms of the initial binding of one ATP obstructing the binding of the second ATP in the aptamer. A Hill coefficient value of 2.04 is measured for the control sequence, which is indicative of near-simultaneous binding of the two ATP moieties. It is worth noting that a value of ∼2.0 in the control sequence would explain the inability to observe two distinct binding events in support of the F-test and AIC statistical analyses for the control.
It is believed that the folded aptamer is energetically favorable in the presence of the ATP, but the ATP binding is hindered by base pairing in the duplex region. The observed differences between a fully exposed single-stranded aptamer and an aptamer embedded in a stem-loop conformation should reflect thermodynamic competition between a Watson–Crick base-pairing configuration, and the induced denaturing of the duplex by ATP binding and folding of the single-stranded aptamer. Inspection of the thermodynamic values of the stem-loop conformers (Table 3) and correlating the values with Kd is informative. The −ΔG value is calculated by assuming a fully complexed aptamer region in the stem is formed with no nick in the 5′ stem of the molecular beacon, as shown in Figure 3.
Table 3. % Double-Stranded Portion of the Aptamer Sequence, the Melting Temperature of the Hairpin, the Gibbs Free Energy of the Hairpin, and the Gibbs Free Energy of the Duplexed Aptamer Sequence in Each NSET Stem-Loop Beacona.
| % d.s. | Thairpin °C | ΔGhairpin kcal/mol | ΔGduplex kcal/mol | |
|---|---|---|---|---|
| Seq i | 20% | 47.1 | –3.31 | –9.69 |
| Seq ii | 0% | 36.5 | –1.79 | -------- |
| Seq iii | 20% | 40.5 | –2.29 | –8.19 |
| Seq iv | 44% | 50.2 | –4.18 | –24.25 |
| Seq v | 68% | 33.9 | –1.34 | –34.92 |
Melting temperatures and ΔG values were calculated with IDT’s oligo analyzer software.
Figure 3.
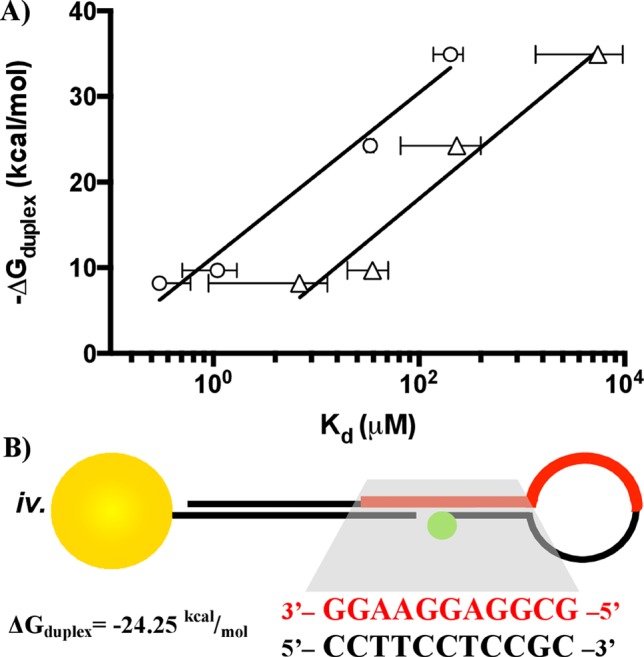
(A) Correlation between the aptamer Gibbs free energy of a pseudoduplex vs experimentally measured Kd1 (circles) and Kd2 (triangles) values of the aptamer. (B) The region of duplex sequence utilized for the −ΔGduplex calculation, depicting sequence iv as an example.
The ΔG for duplex formation indicates that with increasing ΔGduplex, the Kd value also increases, while ΔGhairpin does not appear to correlate. This implies that competitive binding of the ATP for the duplex region is the likely driving force for the observed increasing Kd values with increasing duplex content. To ensure that a change in aptamer specificity does not also contribute to the scaling of Kd with duplex content, AMP concentration studies were performed on sequences iii and iv. The NSET beacon binds AMP on sequences iii and iv nearly identically to ATP (Supporting Information Figure SF7). The results are consistent with experimental results on the Huizenga-Szostak apatmer which is observed to bind to ATP, AMP, and ADP with similar affinity.36,37 Coupled with the observation that 5′ and 3′ blockages of the aptamer sequence exhibit similar changes in binding affinity, this too supports the notion that only thermodynamic properties are responsible for the observed modulations in aptamer binding affinity.
The values of −ΔG and Kd should be logarithmically related based on the thermodynamic relationship between equilibrium constant and free energy change. A plot of −ΔGduplex vs Kd1 and Kd2 on a logarithmic scale is shown in Figure 3A. The graph demonstrates that a logarithmic correlation is observed consistent with thermodynamic expectations. Moreover, correlation to a logarithmic fit across all ATP binding events and for the limited AMP measurements implies that the kinetic expression of this system is not altered by duplex formation over the aptamer sequence, but rather the ratio of kon/koff changes from the thermodynamic hindrance of the duplex, shifting the equilibrium and the dissociation constant. Similar phenomena were observed previously in varying duplex lengths away from the aptamer sequence.37 The results of the thermodynamic plot supports the observation that the ATP aptamer site location in the stem-loop architecture provides a strategy to systematically vary Kd by intentionally including duplexed regions without affecting selectivity or cooperative binding behavior.
Conclusion
For the Huizenga-Szostak ATP aptamer in this study, the results clearly indicate that increasing duplex formation by positioning the sequence in the stem allows the binding affinities for ATP to be tuned over four orders of magnitude with a seven order range of detection, as shown by the saturation curves. The study shows a correlation between single-stranded exposure, lower Kd values, and a scaling with −ΔG values in a Huizenga-Szostak ATP aptamer. The advantage of using the stem-loop architecture and tuning the Kd values by positioning the aptamer target region in the loop is purely in the simplicity of the design parameters, not in adjusting the aptamer sequence itself. Earlier studies by Plaxco et al. show similar results for control of metal ion sensors using allosteric inhibition, where distal DNA duplex sequences predictably tune the binding affinities of Hg2+ ions two orders of magnitude and across detection ranges of three orders.38 This confirms an advantage in blocking portions of the aptamer sequence directly, as we see no change in ATP cooperative binding behavior while pushing the Kd’s and detection range to longer magnitudes. Another allosteric inhibition technique employed by Plaxco and colleagues tuned a cocaine aptamer with distal-site modifications where the Kd’s span three orders of magnitude and the detectable concentration range spans six.27 Changing the aptamer sequence itself can be risky, as single-site modifications in nucleic acid strands capable of forming secondary structures can unpredictably impede secondary structure formation.39,40
The observation of native biophysical constants for the NSET stem-loop designs indicates no perturbation of the ATP aptamer folding in the presence of the appended AuNP, which supports the utilization of NSET to monitor other complex biological events without manipulating native behavior. From a biophysical viewpoint the use of the NSET aptamer beacon allows multidimensional analysis of ATP aptamer binding. While previous optical studies have been unable to identify the cooperativity or distinguish individual ATP binding events, the NSET probe method clearly observed two distinct binding events with discrete structural motions.
The selection of aptamers against a target ligand leads to the aptamers with the lowest Kd values and, thus, aptamers are typically only useable in a narrow concentration range. The results of the study suggest that judiciously tuning the Kd value to expand the applicability of the aptamer can be achieved by site control of the aptamer in a stem-loop architecture without relying on mutation approaches or multiprobe platforms.26,27 The ability to tune the Kd value by changing the duplex content in a stem-loop architecture provides clear design criteria that can be more widely implemented, the results of which may have an advantage in sensor design, namely, the ability to rationally extend the detectable concentration range for a ligand in a previously isolated aptamer from library screening.
Materials
All reagents were purchased from Sigma-Aldrich and used as received. The aptamer and complementary DNA strands were purchased and purified by Midland Certified Reagent Company. Spherical, 3 nm dihydrolipoic acid (DHLA)-coated AuNPs were prepared by modification of literature protocol as described in Supporting Information.30
Aptamer Sequence
The 25-mer aptamer strand is incorporated into a 57-mer stem-loop construct with the aptamer sequence positioned sequentially around the structure. The sequences are listed in Table 1. The 57-mer DNA sequences contain a fluoroscein dye label coupled to a C6 phosphate spacer appended to the 5′ terminus of the single-stranded, aptamer-encoded DNA. The complementary 15-mer ssDNA sequence contains a 5′ C6-NH2 modification to allow coordination to the DHLA-AuNP through an EDC condensation reaction.
Construction of the NSET Aptamer Beacon
The aptamer hairpin is annealed prior to appendage to the AuNP by heating to 95 °C for 4 min then snap cooling to 4 °C to ensure hairpin formation. The annealing is carried out at pH 7.5 phosphate buffer (20 mM phosphate, 50 mM NaCl). Coordination of the aptamer hairpin to the AuNP is accomplished through condensation of the C6-NH2 to the free acid group on the DHLA-AuNP by reaction of DHLA-AuNPs (1 nmol) with 10 μL of a stock EDC solution (10 mg/mL), and 100 μL of stock NHSS solution (1 mg/mL) in 0.5 mL nanopure H2O at RT (3 h) to form the NHS intermediate. The NHS intermediate is coupled by addition of the annealed aptamer (3 nmol) and reacted for 3 h. Final concentrations obtained in the reaction are 1.3 μM and 3.8 μM for the AuNPs and aptamer, respectively. The AuNP–aptamer construct was purified via ethanol precipitation and analyzed by gel electrophoresis, UV–vis, and TEM (Supporting Information Figure SF1). The AuNP–aptamer construct is stored at 4 °C in the dark until use. Gel electrophoresis (2% agarose) on the hairpin aptamer appended to the AuNP exhibits a single band. TEM analysis shows a spherical, 3 ± 0.5 nm AuNP core, and absorption features consistent with the AuNP (520 nm) and duplex DNA (260 nm). The FAM feature is obscured by the AuNP LSPR by comparison to the DNA aptamer absorption. The UV–vis confirms a stochastic, ∼1:1 coupling ratio for aptamer to AuNP. The aptamer loading is measured by UV–vis absorption spectra by analyzing the ratio of the DNA absorption at 260 nm to the AuNP absorption at 520 nm using the Beer–Lambert law (Supporting Information).
Photoluminescence Energy Transfer Experiments
Photoluminescence (PL) was measured in 0.3 cm quartz cuvettes on a Varian Eclipse fluorimeter in a thermostated optical cell holder (20 °C) by exciting the FAM at 494 nm and collecting the PL spectra from 505 to 650 nm. The aptamer beacon and ATP dilutions were dissolved in TRIS buffer (20 nM TRIS, 300 mM NaCl, and 5 mM MgCl2). The AuNP–aptamer concentration was ∼1 nM and ATP concentrations were varied systematically from 5 nM to 97 mM. UTP, GTP, and AMP binding studies were carried out for selected aptamers identically to the ATP studies to analyze the impact on specificity and selectivity.
Acknowledgments
Support for this work was provided by the Florida State University through an FSU-planning grant and the National Institute of Health through NIH-R01-EB00832.
Supporting Information Available
Detailed synthesis and analysis of the AuNPs, theoretical NSET values for AuNP–fluorescein conjugates; experimental data for the single-stranded control sequence; UTP, GTP and AMP binding studies; raw PL data for the five stem-loop constructs; AuNP controls; statistical analysis of the saturation curves; and Hill plots. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Yun C. S.; Javier C.; Jennings T.; Fisher M.; Hira S.; Peterson S.; Hopkins B.; Reich N. O.; Strouse G. F. (2005) Nanometal surface energy transfer in optical rulers, breaking the FRET barrier. J. Am. Chem. Soc. 127, 3115–3119. [DOI] [PubMed] [Google Scholar]
- Jennings T. L.; Schlatterer J. C.; Singh M. P.; Greenbaum N. L.; Strouse G. F. (2006) NSET molecular beacon analysis of hammerhead RNA substrate binding and catalysis. Nano Lett. 6, 1318–1324. [DOI] [PubMed] [Google Scholar]
- Haller A.; Rieder U.; Aigner M.; Blanchard S. C.; Micura R. (2007) Conformational capture of the saM-II riboswitch. Nat. Chem. Biol. 7, 393–400. [DOI] [PubMed] [Google Scholar]
- Ellington A. D.; Szostak J. W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822. [DOI] [PubMed] [Google Scholar]
- Crameri A.; Stemmer P. C. (1993) 1020-Fold aptamer library amplification without gel purification. Nucleic Acids Res. 21, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton B. E.; Gold L.; Hicke B. J.; Nebojša J.; Jucker F. M.; Sebesta D. P.; Tarasow T. M.; Willis M. C.; Zichi D. A. (1997) Post-SELEX combinatorial optimization of aptamers. Bioorg. Med. Chem. 5, 1087–1096. [DOI] [PubMed] [Google Scholar]
- Famulok M.; Jenne A. (1998) Oligonucleotide libraries - variatio delectat. Curr. Opin. Chem. Biol. 2, 320–327. [DOI] [PubMed] [Google Scholar]
- Wilson D. S.; Szostak J. W. (1999) Selection of functional nucleic acids. Annu. Rev. Biochem. 68, 611–647. [DOI] [PubMed] [Google Scholar]
- Bock L. C.; Griffin L. C.; Latham J. A.; Vermaas E. H.; Toole J. J. (1992) Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 346, 564–566. [DOI] [PubMed] [Google Scholar]
- Huizenga D. E.; Szostak J. W. (1995) A DNA aptamer that binds adenosine and ATP. Biochemistry 34, 656–665. [DOI] [PubMed] [Google Scholar]
- Stojanovic M. N.; de Prada P.; Landry D. W. (2001) Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 123, 4928–4931. [DOI] [PubMed] [Google Scholar]
- Giver L.; Bartel D.; Zapp M.; Pawul A.; Green M.; Ellington A. D. (1993) Selective optimization of the Rev-binding element of HIV-1. Nucleic Acids Res. 21, 5509–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. M.; Ting R.; Perrin D. M. (2004) High affinity DNAzyme-based ligands for transition metal cations—a prototype sensor for Hg2+. Org. Biomol. Chem. 2, 307–312. [DOI] [PubMed] [Google Scholar]
- Lupold S. E.; Hicke B. J.; Lin Y. (2002) Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 62, 4029–4033. [PubMed] [Google Scholar]
- Yamamoto R.; Kumar P. K. R. (2000) Molecular beacon aptamer fluoresces in the presence of Tat protein of HIV-1. Genes Cells 5, 389–396. [DOI] [PubMed] [Google Scholar]
- Hamaguchi N.; Ellington A.; Stanton M. (2001) Aptamer beacons for the direct detection of proteins. Anal. Biochem. 294, 126–131. [DOI] [PubMed] [Google Scholar]
- Zheng D.; Seferos D. S.; Giljohann D. A.; Patel P. C.; Mirkin C. A. (2009) Aptamer Nano-flares for molecular detection in living cells. Nano Lett. 9, 3258–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.; Lubin A. A.; Heeger A. J.; Plaxco K. W. (2005) Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew. Chem., Int. Ed. 44, 5456–5459. [DOI] [PubMed] [Google Scholar]
- Ikebukuro K.; Kiyohara C.; Sode K. (2005) Novel electrochemical sensor system for protein using the aptamers in sandwich manner. Biosens. Bioelectron. 20, 2168–2172. [DOI] [PubMed] [Google Scholar]
- Lu W.; Arumugam S. R.; Senapati D.; Singh A. K.; Arbneshi T.; Khan S. A.; Yu H.; Ray P. C. (2010) Multifunctional oval-shaped gold-nanoparticle-based selective detection of breast cancer cells using simple colorimetric and highly sensitive two-photon scattering assay. ACS Nano 4, 1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov V.; Xiao Y.; Shlyahovsky B.; Willner I. (2004) Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J. Am. Chem. Soc. 126, 11768–11769. [DOI] [PubMed] [Google Scholar]
- Ray P. C.; Fan Z.; Crouch R. A.; Sinha S. S.; Pramanik A. (2014) Nanoscopic optical rulers beyond the FRET distance limit: fundamentals and applications. Chem. Soc. Rev. 43, 6370–6404. [DOI] [PubMed] [Google Scholar]
- Yamanaa K.; Ohtania Y.; Nakanoa H.; Saitob I. (2003) Bis-pyrene labeled DNA aptamer as an intelligent fluorescent biosensor. Bioorg. Med. Chem. Lett. 13, 3429–3431. [DOI] [PubMed] [Google Scholar]
- Porchetta A.; Vallée-Bélisle A.; Plaxco K. W.; Ricci F. (2012) Using distal site mutations and allosteric inhibition to tune, extend and narrow the useful dynamic range of aptamer-based sensors. J. Am. Chem. Soc. 134, 20601–20604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtab R.; Rogers J. P.; Singleton C. P.; Murphy C. J. (1996) Preferential adsorption of a ’kinked’ DNA to a neutral curved surface: comparisons to and implications for nonspecific DNA-protein interactions. J. Am. Chem. Soc. 118, 7028–7032. [Google Scholar]
- Kang D.; Vallée-Bélisle A.; Porchetta A.; Plaxco K. W.; Ricci F. (2012) Re-engineering electrochemical biosensors to narrow or extend their useful dynamic range. Angew. Chem., Int. Ed. 51, 6717–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci F.; Vallée-Bélisle A.; Porchetta A.; Plaxco K. W. (2012) The rational design of allosteric inhibitors and activators using the population-shift model: in vitro validation and application to an artificial biosensor. J. Am. Chem. Soc. 134, 15177–15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. H.; Patel D. J. (1997) Structural basis of DNA folding and recognition in an AMP-DNA aptamer complex: distinct architectures but common recognition motifs for DNA and RNA aptamers complexed to AMP. Chem. Biol. 4, 817–832. [DOI] [PubMed] [Google Scholar]
- Breshike C. J.; Riskowski R. A.; Strouse G. F. (2013) Leaving Förster resonance energy transfer behind: Nanometal Surface Energy Transfer predicts the size-enhanced energy coupling between a metal nanoparticle and an emitting dipole. J. Phys. Chem. C 117, 23942–23949. [Google Scholar]
- Oh E.; Kimihiro S.; Goswami R.; Mattoussi H. (2010) One-phase synthesis of water-soluble gold nanoparticles with control over size and surface functionalities. Langmuir 226, 7604–7613. [DOI] [PubMed] [Google Scholar]
- Nicewarner Peña S. R.; Raina S.; Goodrich G. P.; Federoff N. V.; Keating C. D. (2002) Hybridization and enzymatic extension of Au nanoparticle-bound oligonucleotides. J. Am. Chem. Soc. 124, 7314–7323. [DOI] [PubMed] [Google Scholar]
- Bin X.; Sargent E. H.; Kelley S. O. (2010) Nanostructuring of sensors determines the efficiency of biomolecular capture. Anal. Chem. 82, 5928–5931. [DOI] [PubMed] [Google Scholar]
- Mahtab R.; Rogers J. P.; Murphy C. J. (1995) Protein-sized quantum dot luminescence can distinguish between ″straight″, ″bent″, and ″kinked″ oligonucleotides. J. Am. Chem. Soc. 117, 9099–9100. [Google Scholar]
- Gearhart L. A.; Ploehn H. J.; Murphy C. J. (2001) Oligonucleotide adsorption to gold nanoparticles: A surface-enhanced raman spectroscopy study of intrinsically bent DNA. J. Phys. Chem. B 105, 12609–12615. [Google Scholar]
- Achenbach J. C.; Nutiu R.; Li Y. (2005) Structure-switching allosteric deoxyribozymes. Anal. Chim. Acta 234, 41–51. [Google Scholar]
- Arnaut V.; Langecker M.; Simmel F. (2013) Nanopore force spectroscopy of aptamer-ligand complexes. Biophys. J. 105, 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura I.; Shi A.; Nutiu R.; Yu J. M. Y.; Li Y. (2009) Kinetic modeling of designed signaling DNA aptamers. Phys. Rev. E 79, 031906. [DOI] [PubMed] [Google Scholar]
- Porchetta A.; Vallée-Bélisle A.; Plaxco K. W.; Ricci F. (2013) Allosterically-tunable, DNA-based switches triggered by heavy metals. J. Am. Chem. Soc. 135, 13238–13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui-Jain A.; Grand C. L.; Bearss D. J.; Hurley L. H. (2003) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A. 99, 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y.; Kim D. S. (2009) Dramatic effect of single-base mutation on the conformational dynamics of human telomeric G-quadruplex. Nucleic Acids Res. 37, 3625–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.