Abstract
Despite major advances in early revascularization techniques, cardiovascular diseases are still the leading cause of death worldwide, and myocardial infarctions contribute heavily to this. Over the past decades, it has become apparent that reperfusion of blood to a previously ischemic area of the heart causes damage in and of itself, and that this ischemia reperfusion induced injury can be reduced by up to 50% by mechanical manipulation of the blood flow to the heart. The recent discovery of remote ischemic preconditioning (RIPC) provides a non-invasive approach of inducing this cardioprotection at a distance. Finding its endogenous mediators and their operative mode is an important step toward increasing the ischemic tolerance. The release of humoral factor(s) upon RIPC was recently demonstrated and several candidate proteins were published as possible mediators of the cardioprotection. Before clinical applicability, these potential biomarkers and their efficiency must be validated, a task made challenging by the large heterogeneity in reported data and results. Here, in an attempt to reproduce and provide more experimental data on these mediators, we conducted an unbiased in-depth analysis of the human plasma proteome before and after RIPC. From the 68 protein markers reported in the literature, only 28 could be mapped to manually reviewed (Swiss-Prot) protein sequences. 23 of them were monitored in our untargeted experiment. However, their significant regulation could not be reproducibly estimated. In fact, among the 394 plasma proteins we accurately quantified, no significant regulation could be confidently and reproducibly assessed. This indicates that it is difficult to both monitor and reproduce published data from experiments exploring for RIPC induced plasma proteomic regulations, and suggests that further work should be directed towards small humoral factors. To simplify this task, we made our proteomic dataset available via ProteomeXchange, where scientists can mine for novel potential targets.
Introduction
Remote ischemic preconditioning (RIPC) is an emerging treatment for reducing ischemia reperfusion injury (IRI) in the heart. Several proof-of-concept studies and small randomized controlled trials have demonstrated that the human heart is amenable to RIPC [1]–[5]. Recently, it was also reported that 3 cycles of 5 min upper arm ischemia substantially reduced myocardial injury after coronary artery bypass graft in large pools of patients. This was demonstrated by a significant decrease in cardiac troponin I (cTnI) release, significantly improving prognosis with a reduction in mortality in the group receiving RIPC compared to controls [6].
The signaling pathway of RIPC in the human heart is just starting to be uncovered [7], but how the RIPC stimulus is transferred from the arm to the heart remains unclear. Compelling preclinical evidence suggests communication via one or more unknown humoral factors: First, it seems that a period of reperfusion after the RIPC stimulus is required for protection, suggesting that wash-out of blood borne factors and transport to the site of protection is involved [8]–[10]. Secondly, it was demonstrated that effluent from preconditioned hearts could transfer the protection to naïve recipient hearts and that the protection is mediated via small, unknown hydrophobic factors of protein nature between 3.5 and 15–30 kDa [11]–[16]. Moreover, the fact that this humoral factor is effective at a remote location after dilution in blood or perfusion fluid, hints at a large concentration change which should be detectable by modern proteomic techniques.
In fact, several proteins were recently found to be regulated after RIPC, paving the way for potential use of these cardioprotective compounds in the clinic. Notably, Hepponstall et al. conducted an ambitious study where 806 differentially expressed peptides were identified after RIPC. Among them, 133 could be mapped to 48 protein sequences [17]. In addition, 2D gel experiments reported 33 regulated spots with 6 identifiable proteins. Surprisingly, only one of the given protein accession numbers could be found in both the mass spectrometry and the 2D gel result sets. Subsequently, Pang et al. found 14 proteins to be differentially regulated by RIPC and validated these findings by Western blotting [18]. Notably, only one of the accessions reported, Gelsolin (UniProt accession number P06396), could be mapped to those published by Hepponstall et al. Additionally, albeit with different sequences, both studies underlined the regulation of Apolipoprotein A-I. However, Pang et al. found this protein to be down-regulated, while Hepponstall et al. found it down-regulated using gels and up-regulated when applying a gel free technique in the same study. The latter result is in concordance with the study of Hibert et al. showing a 30% up-regulation of Apolipoprotein A-I, postulating it to be the principal factor behind RIPC mediated cardioprotection [19]. Later on, however, Hilbert et al. published another report where this protein appeared not to be regulated, but seven other related proteins presented a Mann-Whitney test p-value <0.05 [20]. Notably, no protein passed a more stringent 0.01 threshold and all proteins showed moderate regulations (between 0.58∶1 and 1.2∶1), consistently below the two fold change regulation level generally used in biology. The results of Davidson et al. suggested SDF-1α to be the main mediator of RIPC, presumably communicating the cardioprotection via SDF-1α/CXCR4 signaling [21]. Importantly, Przyklenk [22] criticized the latter study for its limitations, as the plasma levels of SDF-1α should have been monitored both before and after the RIPC stimulus, thereby failing to validate the factor as a mediator of RIPC.
Discovery studies “often significantly overestimate their findings” as claimed by Gosho et al. [23]. Consequently, prior to clinical testing, protein markers must be validated, preferably using targeted mass spectrometry based proteomics [24]. For that, quantitative assays are built in order to target and quantify the compounds of interest. Setting up such an assay requires experimental data on the digestion, peptide elution, ionization, and fragmentation profile of the targeted protein. This information is, however, not available from any of the mentioned mass spectrometry datasets. Notably, they are not available in public repositories, despite this being standard publication guidelines in proteomics [25],[26]. In fact, the peptide information is not available and in the case of Hepponstall et al., not even the estimated protein ratios are reported.
The disagreement in the literature, even in studies from the same group or within the same study, together with the lack of provided information, show the striking need for a transparent proteomic dataset stringently monitoring the proteomic changes induced by RIPC. Here, we present a quantitative in depth sequencing of the plasma proteome before and after RIPC. Six healthy donors underwent RIPC according to the protocol used in consensus with the literature and whose efficiency has been long established. In contrast to previously reported studies, the digested plasma samples were multiplexed using isobaric tags and fractionated, thereby limiting inter-sample artifacts and dramatically increasing sample coverage. The protein identification and quantification was achieved using open source software applying stringent quality criteria to avoid reporting artifact regulations. The acquired raw data was deposited to the ProteomeXchange consortium [27] together with identification results, and can thereby freely be accessed, inspected and even reprocessed to better plan further experiments [28].
Materials and Methods
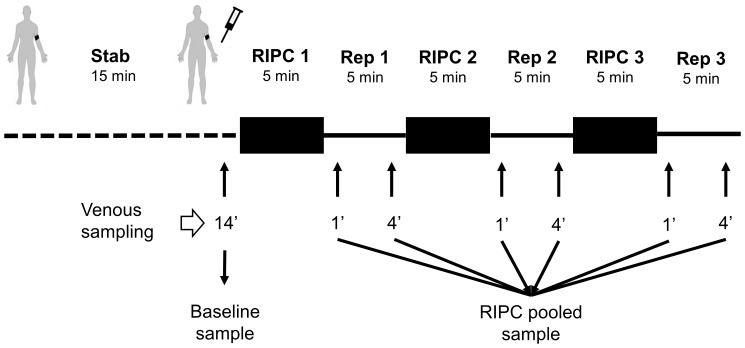
This study was approved by the regional ethics committee for medical and health research in Western Norway (REK 2010/1642-1). Written informed consent was obtained from all participants and the study conformed to the principles in the declaration of Helsinki. Six healthy adult male donors aged 29±2 years old (mean ± SD), not on any medication, underwent the RIPC protocol which consisted of 3 cycles of 5 min upper arm ischemia alternating with 5 min reperfusion (Figure 1). All subjects rested upright on a bench for 15 min before ischemia was induced by inflating a blood pressure cuff to 200 mmHg. Venous blood samples were collected from the ipsilateral arm after 14 min of rest in the beginning, and at 1 and 4 min into each reperfusion period. Blood samples were collected in K2 EDTA-coated tubes (Vacutainer, BD). Samples were centrifuged at 1,500 rcf at 4°C for 15 min within 30 min after sampling. 1.5 ml plasma was transferred to Eppendorf tubes before a second centrifugation at 15,000 rcf at 4°C for 15 min to remove any cell contaminants or cell fragments. ∼90% of the clear supernatant was removed using a 1 ml pipette and stored at −80°C. The whole process from sampling to freezing of each individual sample took less than 75 min. Samples from 1 and 4 min of each of the reperfusion periods were pooled, resulting in 12 samples: a sample before (control) and after RIPC for each of the six individuals.
Figure 1. Experimental protocol.
A peripheral venous catheter was inserted in the cubital fossa of subject for blood sampling. The subject rested for 14 min reclined on a bench before the baseline sample was drawn. The blood pressure cuff was inflated to 200 mmHg for 5 min before being released. Blood samples were drawn at 1 and 4 min into reperfusion from the ipsilateral arm. Blood samples were centrifuged to collect plasma which was stored at −80°C. Before analysis, all six reperfusion samples were pooled for each subject.
Chemicals
Trypsin was purchased from Promega. N-octyl-β-D-glycopyranoside (NOG), acetonitrile (ACN), formic acid (FA), ammonium formate and water were purchased from Sigma-Aldrich. Water and ACN were of HPLC quality.
Abundant protein depletion and concentration
20 µl of plasma from each sample was depleted using a human Multiple Affinity Removal System (MARS Hu-14) 4.6 mm×50 mm LC column (Agilent Technologies) according to the protocol provided by the supplier, using a Dionex 3000-series LC system. The MARS column depletes the plasma of albumin, IgG, antitrypsin, IgA, transferrin, haptoglobin, fibrinogen, alpha-2-macroglobulin, alpha-1-acid glycoprotein, IgM, apolipoprotein AI, apolipoprotein AII, complement C3 and transthyretin. The protein depleted plasma samples were concentrated using 3 kDa ultracentrifugation filters (Amicon Ultra-4, Millipore, Bedford, MA) pre-treated with 0.1% NOG.
Protein digestion and iTRAQ labeling
The entire depleted protein sample was reduced, cysteine blocked, trypsin digested (1∶20, trypsin∶protein, w/w), iTRAQ labeled (114, 115, 116 and 117) and combined according to the protocol using the chemicals provided (AB Sciex). The iTRAQ 4-Plex kit allowed us to multiplex two conditions from two donors per kit, resulting in three parallel experiments as detailed in Table 1. Both conditions (before and after RIPC) followed the exact same downstream workflow for every individual.
Table 1. Repartition on every iTRAQ channel of the samples at baseline and after RIPC for the six donors.
| iTRAQ Channel | Experiment 1 | Experiment 2 | Experiment 3 |
| 114 | Donor 1 baseline | Donor 3 RIPC | Donor 5 baseline |
| 115 | Donor 1 RIPC | Donor 3 baseline | Donor 5 RIPC |
| 116 | Donor 2 baseline | Donor 4 RIPC | Donor 6 baseline |
| 117 | Donor 2 RIPC | Donor 4 baseline | Donor 6 RIPC |
Mix-mode fractionation
iTRAQ labeled peptides were fractionated into 60 fractions using a mixed-mode (MM) reversed phase anion exchange (RP-AX) Sielc Promix column as described by Philips et al. [29] (MP-10.250.0530, 1.0×250 mm, 5 µm, 300 Å, Sielc Technologies, Prospect Heights, Illinois) coupled to an Agilent 1260 series LC system (Agilent Technologies, Palo Alto, CA). The iTRAQ labeled peptides were reconstituted in 20 mM ammonium formate, 3% ACN (buffer A) and loaded on the column in 85% buffer A for 10 minutes at a flowrate of 50 µl/min. The peptides were eluted from the column increasing the contents of buffer B (2 mM ammonium formate, 80% ACN, pH 3.0), from 15% to 60% in 35 minutes and further to 100% buffer over 10 minutes. Buffer B was held constant for 5 minutes before the column was equilibrated for 10 minutes in 85% buffer A. The fractions from the first 10 minutes of the gradient were discarded.
LC-MS/MS analyses
Fifty fractions from the MM RP-AX separation from each sample were analyzed on an LTQ-Orbitrap Velos Pro (Thermo Scientific) coupled to a Dionex Ultimate NCR-3000RS LC system. The fractions were dissolved in 1% FA and trapped on the pre-column (Dionex, Acclaim PepMap 100, 2 cm×75 µm i.d, 3 µm C18 beads) in buffer A (2% ACN, 0.1% FA) at a flowrate of 5 µl/min for 5 minutes before separation by reverse phase chromatography (Dionex, Acclaim PepMap 100, 15 cm×75 µm i.d., 3 µm C18 beads) at a flow of 280 nL/min. The fractions were run on three nano LC gradients: The first fifteen fractions were run on a LC gradient consisting of a gradient starting at 5% buffer B (90%ACN, 0.1% FA) ramping to 12% buffer B over 55 minutes (5–60 min), the gradient was subsequently ramped to 30% buffer B in 30 minutes (60–90 min), increased to 90% B in 10 minutes (90–100 min), held for 5 minutes (100–105 min) followed by ramping to 5% buffer B for 3 minutes (105–108) and equilibration of the column in 12 minutes (108–120); fractions 16–35 were separated on the following LC gradient: 0–5.0 minutes 5% buffer B, 5.0–5.5 minutes 8% buffer B, 5.5–60 minutes 20% buffer, 60–90 minutes 35% buffer B; the last fractions (36–50) were separated using the following gradient: 0–5.0 minutes 5% buffer B, 5.0–5.5 minutes 8% buffer B, 5.5–90 minutes 40% buffer. The last part of the nano LC gradient is similar for all three gradients.
The mass spectrometer was operated in data-dependent-acquisition (DDA) mode to automatically switch between full scan MS and MS/MS acquisition. The instrument was controlled by Tune 2.6.0 and Xcalibur 2.1. Survey full scan MS spectra (from m/z 300 to 2,000) were acquired in the Orbitrap with resolution R = 60,000 at m/z 400 (after accumulation to a target value of 1E6 in the linear ion trap with maximum allowed ion accumulation time of 500 ms). The 7 most intense eluting peptides above an ion threshold of 1,000 counts and charge states 2 or higher, were sequentially isolated in the high-pressure linear ion trap to a target value of 5E5 at a maximum allowed accumulation time of 1,000 ms, and isolation width maintained at 2 Da. Fragmentation in the Higher-Energy Collision Dissociation (HCD) cell was performed with a normalized collision energy of 40%, and activation time of 0.1 ms. Fragments were detected in the Orbitrap at a resolution of 7,500 with first mass fixed at m/z 100.
Data analysis
All RAW data were transformed into mgf peak lists using the ProteoWizard software [30] package version 2.2.2954. The obtained peak lists were searched with OMSSA [31] version 2.1.9 and X!Tandem [32] Cyclone 2013.2.01.1 using SearchGUI [33] version 1.12.2. Peak lists were searched against a concatenated target/decoy [34] version of the human complement of the UniProtKB/Swiss-Prot database [35] (downloaded on September 2012). The decoy sequences were created by reversing the target sequences in SearchGUI. Search settings were as follows: Trypsin with a maximum of 2 missed cleavages; 10 ppm as MS, 0.6 Da as MS/MS tolerances, respectively; fixed modifications: methylthio of Cys (+45.987721 Da) and iTRAQ on Lys and peptide N-term (+144.105918 Da); and variable modifications: oxidation of Met (+15.994915 Da) and iTRAQ on Tyr (+144.105918 Da). All other OMSSA or X!Tandem settings were kept at the default values set in SearchGUI. Peptides and proteins were inferred from the search engine results using PeptideShaker (http://peptide-shaker.googlecode.com) [36]. Peptide to Spectrum Matches (PSMs), peptides and proteins were validated at a stringent 1% FDR estimated using the decoy hits. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium(http://proteomecentral.proteomexchange.org) [27] via the PRIDE partner repository [37],[38] with the dataset identifier PXD000605 and DOI 10.6019/PXD000605.
For every validated protein, the iTRAQ reporter ions were extracted from spectra of validated PSMs and deisotoped using the isotope abundance matrix [39]. Intensities were normalized using the median intensity in order to limit the ratio deviation [40] and peptide and protein ratios were estimated using maximum likelihood estimators [41]. Ratios were log2 converted and normalized to the median to avoid inter-sample bias. Only those proteins presenting two or more validated and quantified peptides were retained for the quantitative analysis. Standard contaminants as well as all proteins with known affinity to the antibodies in the MARS column were excluded from downstream statistical analysis. A paired two-sided t-test was conducted on protein ratios using a p-value threshold of 0.01. Subsequently, in order to distinguish RIPC specific regulations from random biological variability, an asymmetric normal distribution was drawn from the background ratios (calibrated on the median and ±34.1 percentiles) and only those proteins having a probability <1% to be derived from the background were considered confidently regulated. Finally, protein abundance indexes were estimated using spectral counting where the spectral counting index is simply the number of validated PSMs divided by the protein molecular weight [42].
Results
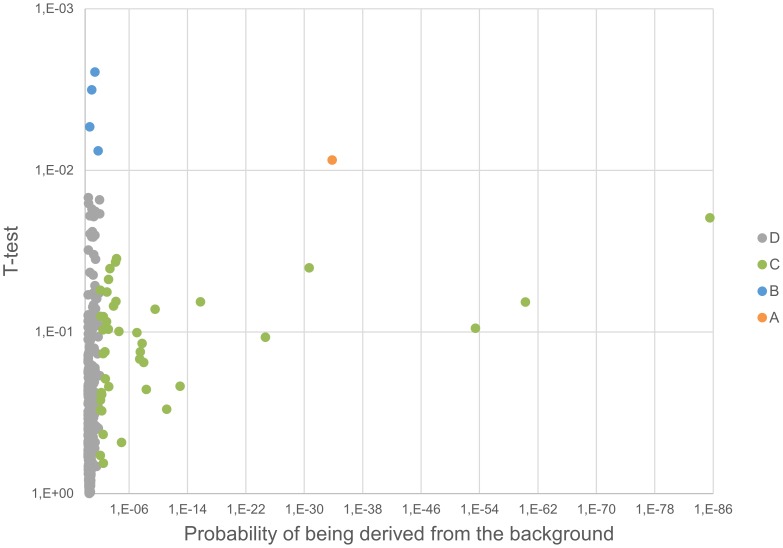
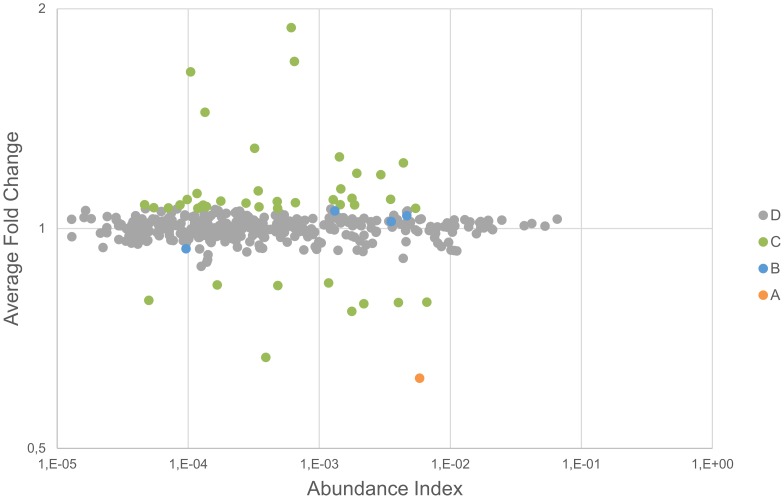
The plasma proteome from six healthy donors was compared at baseline and after RIPC as illustrated in Figure 1 and 2. In total, 727 proteins were confidently identified (<1% FDR). Among them, 409 proteins could be accurately quantified by two or more unique peptides. Of these, 393 remained after exclusion of known contaminant and depletion-affected proteins, and are listed in Table S1. These 393 accurately quantified plasma proteins showed accurate and reproducible stability: 388 (98.7%) presented a fold change between 0.91∶1 and 1.1∶1, which is in accordance with the known technical variability of iTRAQ quantification reported in the literature [43]. In order to assess the relevance of the regulations, the quantified proteins were classified into four categories (as detailed in Table S1): (A) proteins with low variability among donors (t-test passed) and confident regulation to the background (no protein); (B) low donor variability but no confident regulation to the background (4 proteins); (C) high donor variability (t-test failed) but confident regulation to the background (42 proteins); and (D) high donor variability (t-test failed) and no confident regulation to the background (347 proteins). These proteins are plotted in Figure 3 and 4. Notably, no protein was validated by both statistical tests (category A).
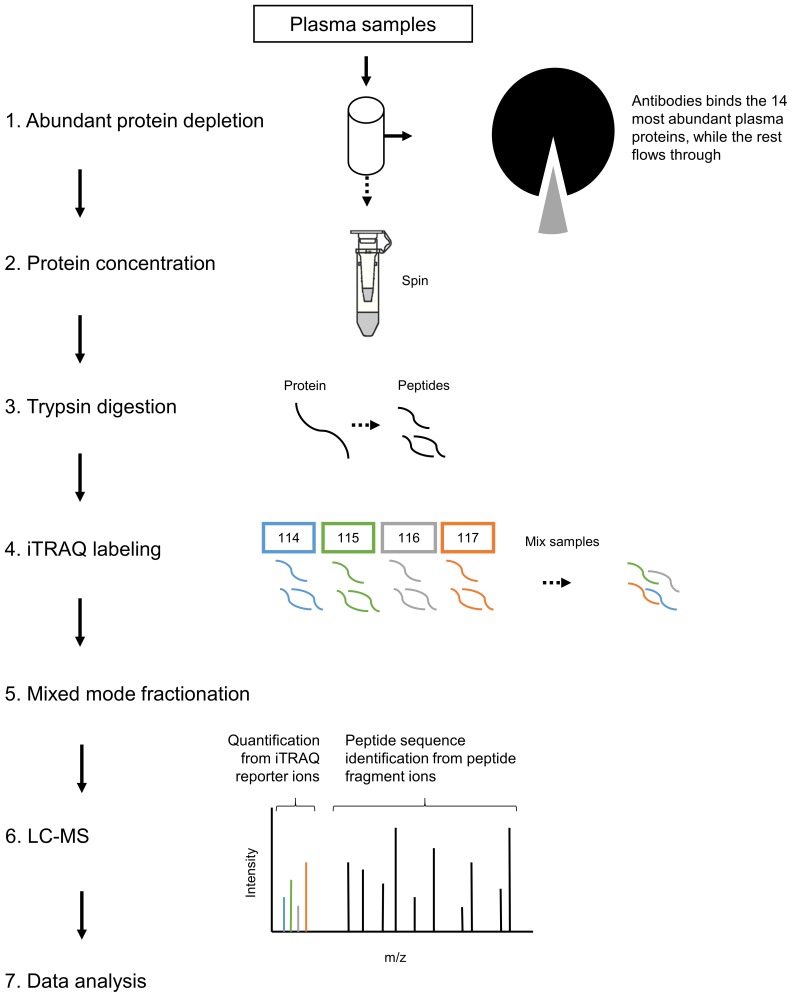
Figure 2. Analysis workflow.
Plasma samples were depleted by a MARS Hu-14 column and subsequently concentrated by 3 kDa ultracentrifugation filters. Next, samples were reduced, cysteine blocked and trypsin digested before iTRAQ labeling. The iTRAQ labeled peptides were fractioned into 60 fractions using a mixed-mode reverse phase anion exchanger. Finally, fractions were analyzed on an LTQ-Orbitrap Velos Pro connected to a Dionex Ultimate NCR-3000RS LC system.
Figure 3. Volcano plot.
The significance of the relative regulation of 394 proteins was inspected using (1) a paired two-sided t-test (y axis) and (2) by estimating the probability for the regulation derived from the background (x axis). Proteins are clustered into four categories based on the statistical test passing the threshold (see text for details).
Figure 4. Protein regulation vs. abundance index.
The protein regulation is plotted against the abundance index and every protein is classified according to the result of the statistical tests.
Four proteins had a significant p-value (<0.01) between the baseline and RIPC samples for the six subjects examined (category B; Kallistatin, Complement C2, Lactoylglutathione lyase (GLO1), and Cysteine-rich secretory protein 3 (CRISP-3))(Table S1). The fold changes for these, however, were very low (<10% change). We considered changes below 20% to be within what can be explained by the technical variability of the analytical approach. This was corroborated by the statistical analysis, showing no confident regulation to the background. Interestingly, all highly regulated proteins were detected in a much lower abundance (see Figure 4) and failed the t-test (p-value>0.01), suggesting that these cannot be trusted for clinical application.
In contrast, high ratios were systematically found for contaminants and proteins affected by depletion. Remarkably, the remaining amount of proteins targeted by the MARS hu-14 depletion column was consistently less abundant in RIPC samples, hinting at a systematic artifact in the depletion procedure. Among them, fibronectin, previously reported to be regulated after RIPC has been identified in the bound fraction after MARS hu-14 depletion [44]. Fibronectin was confidently regulated to the background (ratios of 0.62∶1) in close agreement with Apolipoprotein A-I (ratio of 0.61∶1) and Haptoglobin (ratio of 0.63∶1), both also targeted by the MARS column. In conclusion, we cannot rule out that the difference in relative abundance measured for these proteins are not simply protocol related. On the other hand, our ability to quantify these potential experimental artifacts demonstrates the reliability of the quantification procedure and rules out the hypothesis that regulations were systematically lost along the workflow.
Discussion
RIPC of the upper arm is an easy, practical and non-invasive way of inducing protection from ischemia reperfusion induced injury (IRI) in the heart, and the remote ischemic conditioning stimulus can even be applied during or immediately after an ischemic insult (as reviewed in [45]–[47]). The protocol used in the present study was chosen for its proven clinical efficiency: It was demonstrated to reduce the release of troponins (marker for cardiac damage) [3]–[5] and improve survival [6] after coronary artery bypass graft surgery. Shimizu et al. used a human RIPC protocol of 4×5 min, sampled venous blood before and after the RIPC intervention, and demonstrated that both whole plasma and plasma dialysate <15 kDa could reduce IRI in the isolated rabbit heart [15]. This protocol is comparable, but not identical, to the one used for the proteomic studies mentioned above, ensuring comparability between the results.
Table S2 gives a generated list of all biomarkers found in the literature, in addition to the ones identified in this study, including the measured ratios and t-test results. As detailed in the introduction, Hepponstall et al. [17] report 53 regulated proteins after RIPC. The provided accession numbers were mapped to UniProt entries using the Picr service of the European Bioinformatics Institute [48], leading to mapping of only 14 Swiss-Prot sequences – all other sequences having low or no evidence. 11 of them were confidently quantified in our study (see Table S2). Only Fibronectin (t-test passed) and Haptoglobin (t-test failed) were found to be regulated. However, the regulation of these factors may be experimental artifacts as discussed earlier.
From the 14 proteins reported as regulated by Pang et al. [18], 13 could be mapped to Swiss-Prot accessions, and 12 were accurately quantified in our experiment. Among these, only Apolipoprotein A-I was found to be regulated (0.61∶1 ratio). However, as detailed already, it is one of the targets of the depletion procedure. As a result, it is impossible to validate whether this controversial protein is actually regulated by RIPC or whether the up- and down-regulations reported in the literature may be due to experimental artifacts.
Furthermore, we did not quantify SDF-1α, a purported signaling molecule of RIPC [21],[22]. Nor did we identify a single of its peptides. This protein is a good example of the challenges posed when identifying RIPC mediators, as peptide based protein quantification heavily relies on the ability to detect at least two peptides, which can become defying for small proteins. It is important to note that most of the reported potential mediators of RIPC mediated cardioprotection do not meet the mass expectation (<30 kDa). More experimental data are, thus, necessary to assess the role of SDF-1α in RIPC.
In our study, we identified 4 proteins (category B) to be significantly regulated according to the t-test, but showed no confident regulation to the background; Kallistatin, Complement C2, Lactoylglutathione lyase (GLO1), and Cysteine-rich secretory protein 3 (CRISP-3) (Table S1). Despite claiming an overall negative study outcome, as no factors were released in abundance and confidently regulated to the background, it is perhaps timely to consider whether RIPC might be mediated by a consortium of slightly regulated humoral factors, acting on one or several signalling networks, which in concert induce protection. Interestingly, we identify 3 proteins (kallistatin, kallikrein, and kininogen-1) that might inter-relate in the kallikrein-kinin signalling pathway. Of further interest, and adding to the complexity, Complement 2 as part of the complement system may also exerts cross-talk with proteins of the kinin-generating systems. The 4 category B proteins will be discussed further below.
Kallistatin, in particular, proves to be a very interesting candidate in terms of cytoprotective capabilities (UniProt accession P29622, 48 kDa, p-value 0.002, fold-change 1.04). Chao and co-workers identified kallistatin as a tissue kallikrein binding protein (KBP) and a unique serine proteinase inhibitor (serpin) [49]. Later, kallistatin has been ascribed many other functions unrelated to its interaction with tissue kallikrein, including lowering blood pressure, vasodilatation, preventing cardiac remodelling and offering protection against cardiovascular injuries by preventing apoptosis, oxidative stress, and inflammation [50]–[54]. Moreover, the effects of kallistatin seems to be mediated via pro-survival PI3K/Akt/NO dependent signalling, and is postulated to be activated by a yet unidentified kallistatin specific cell surface receptor or binding protein [52]. Kallistatin may also act as an inhibitor of kallikrein [49], and we found kallikrein to be slightly down-regulated in our data (UniProt accession P03952, category D, 71 kDa, p-value 0.014, fold change 0.96). Kallikrein may be activated by lowered plasma pH [55] due to flow restriction imposed by the ischemic conditioning cycles, which in turn can reduce kinin breakdown, enhance bradykinin (BK) formation [56]–[58], and inhibit BK degradation [59]. We also observed that the MK-RPPGFSPFR-SS peptide located at amino-acids 381 to 389 of Kininogen-1 (UniProt accession P01042, catgory D, 72 kDa) was recorded with a 10 times increase in the number of spectra when compared to the median of the peptides for this protein. The peptide may be a product of Kininogen-1 degradation. Plasma and tissue kallikreins converts kiniogens to produce vasoactive kinin peptides, such as bradykinin and lys-bradykinin. BK is known to exert anti-ischemic effects and for being a possible mediator of ischemic preconditioning, although the peptide presence could not directly be related to RIPC in our experiment (ratio of 1.02∶1, p-value of 0.43). But kinin receptors were previously shown to be influenced by RIPC [60], so monitoring kininogen degradation products might be a promising approach elucidating the RIPC mechanism.
The complement system, which is part of the innate humoral immune system, was slightly up-regulated (UniProt accession P06681, 83 kDa, p-value 0.003, fold-change 1.02). Weissman et al. [61] demonstrated that complement components are deposited in ischemia reperfused myocardium. In addition, animal models of IR in other organ systems like the gut, kidney, and skeletal muscle indicate that the complement system is a key mediator of IRI [62]–[67]. However, the precise mechanism of complement activation in ischemic tissue has not been clearly elucidated due to the lack of appropriate experimental models, restricted knowledge of the molecular processes causing complement activation during hypoxia in cells, and how it exerts cross-talk between different complement activation pathways [68]. Despite the fact that complement activation during IR is associated with cellular injuries; it is intriguing that the complement system exerts cross-talk with proteins of the kinin generating systems [69].
Lactoylglutathione lyase (also known as glyoxalase I or GLO1) was slightly down-regulated after RIPC (UniProt accession Q04760, 23 kDa, p-value 0.005, fold-change 0.92). Glyoxalase 1 (GLO1) in combination with glyoxalase 2 and the co-factor gluthatione constitute the glyoxalase system, which is responsible for the detoxification of methylglyoxal (MG) [70]. MGs are highly reactive metabolites of glucose degradation pathway, protein and fatty acid metabolism. MG itself is cytotoxic and pro-apoptotic. GLO1 might be a key factor for detoxifying MG and protecting organs against IR injury [71], and may also prevent hyperglycemia-induced diabetic complications [72],[73]. Despite a potential protective role of GLO1, it appears down regulated in our study, and could, thus, be unrelated to the cardioprotective properties of RIPC mediating humoral factors.
Our data also identified glycoprotein human cysteine-rich secretory protein 3 (CRISP-3) as slightly up-regulated (UniProt accession P54108, 28 kDa, p-value 0.007, fold-change 1.06). CRISP-3 is believed to play a role in innate immunity. High levels of human CRISP-3 was found in plasma bound to α1-1B-glycoprotein (A1BG-like) (a plasma protein of unknown function) [74]. The A1BG–CRISP-3 complex is thought to inhibit the toxic effect of snake venom metalloproteinases or myotoxins. Udby et al. suggests that the A1BG–CRISP-3 complex displays a similar function in protecting the circulation from a potentially harmful effect of free CRISP-3, although the overall function of CRIPS-3 is unclear [74]. Cardiac related effects of CRISP-3 has not been described in the literature as of yet. We also identified the proposed A1BG binding partner in our data (P04217; category D; 54 kDa; p-value 0.16; fold change 1.02).
It is crucial to consider that even with the best analytical approaches available today, there is a limit to how many proteins that can be identified and quantified in plasma. Notably, isoforms, posttranslational modifications and degraded proteins are a vast field of investigation and future experiments might, thus, be directed at other targets in blood. Moreover, we decided to pool all the reperfusion samples for each individual, making sure all potential relevant proteins were present in the sample. However, pooling of the samples may have masked possible time-dependent RIPC induced protein/mediator alterations, in addition to averaging the relative abundance of potential candidate proteins. It also reduces the ability to monitor changes occurring after a single conditioning cycle only. Future work might, thus, improve the time resolution of the experiment.
In addition to technical analytical factors as described here, other aspects of the RIPC procedure such as the number of cycles and stimulus site should be considered when mining for blood borne humoral factors. Loukogeogakis et al. elegantly demonstrated a dose-response effect with regards to both site and number of cycles, when exploring for the protective effect of RIPC on endothelial IRI of the arm. The maximum protective effect was obtained with 3 cycles of IR of the arm and at least 2 cycles of the leg [75]. This study is complemented by the study of Hong et al., suggesting that the protective effects of RIPC by lower limb ischemia are greater than those induced by upper limb ischemia [76]. The reason that fewer cycles of the lower limb induced protection may possibly be due to a larger lower limb mass leading to greater release of humoral factors. However, according to a very recent systematic review and meta-analysis [77], it is clear that the optimal RIPC stimulus has not been demonstrated, and it is unknown whether upper or lower limb ischemia is superior. Furthermore, the most favorable timing/duration of the stimulus is unclear. Once the optimal stimulus algorithm, site and timing for the RIPC procedure and more certain indications of the RIPC mediated humoral factor(s) mediating protection are established, it will be timely to compare the healthy plasma RIPC proteome to that of diseased patient populations. Furthermore, comparison of age-matched young and old, male and females might also delineate possible and important age and sex differences.
In conclusion, using shotgun quantification techniques, the detectable portion of the plasma proteomes of six healthy adult males was stable after remote ischemic preconditioning, and in contrast to the literature, no significant changes were identified, other than those that appeared to be related to contaminants or the analytical process itself. Four proteins did, however, show low donor variability (passed the t-test) but was not confidently regulated to the background. One way of moving forward could be to increase the workflow sensitivity, with regards to time or biological targets, e.g. by selecting on hydrophobicity and size.
Supporting Information
The complete table of experimental data from all three iTRAQ experiments. All proteins we identified were classified into four categories based on two statistical tests: (A) proteins with low variability among donors (t-test passed) and confident regulation to the background (no protein); (B) low donor variability but no confident regulation to the background (4 proteins); (C) high donor variability (t-test failed) but confident regulation to the background (42 proteins); and (D) high donor variability (t-test failed) and no confident regulation to the background (347 proteins).
(XLSX)
A list of all biomarkers found in the literature, in addition to the ones identified in this study. This includes their measured ratios and t-test results.
(XLSX)
Acknowledgments
The EBI is acknowledged for the quality of the products they make available to the community, notably Picr and PRIDE which were central for this study. The PRIDE team is acknowledged for efficient handling of our data.
Contributions
Conceived and designed the experiments: AKJ LB EH JEN FSB. Performed the experiments: AKJ LB EH MV ØSS HKG. Analyzed the data: EH MV HKG FSB. Contributed reagents/materials/analysis tools: AKJ LB EH MV ØSS FSB JEN. Wrote the paper: AKJ EH LB MV FSB.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Most of the data are within the MS or supporting files, while the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) [27] via the PRIDE partner repository [37], [38] with the dataset identifier PXD000605 and DOI 10.6019/PXD000605.
Funding Statement
This study was funded by the F. Mohn foundation, the Grieg Foundation, Bergen Heart Foundation, Lærdal Foundation for Acute Medicine and Bergen Medical Research Foundation. Erik Helgeland was supported by the University of Bergen. Lars Breivik was supported by the Norwegian Council on Cardiovascular disease. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Botker HE, Kharbanda R, Schmidt MR, Bottcher M, Kaltoft AK, et al. (2010) Remote ischaemic conditioning before hospital admission, as a complement to angioplasty, and effect on myocardial salvage in patients with acute myocardial infarction: a randomised trial. Lancet 375: 727–734. [DOI] [PubMed] [Google Scholar]
- 2. Cheung MM, Kharbanda RK, Konstantinov IE, Shimizu M, Frndova H, et al. (2006) Randomized controlled trial of the effects of remote ischemic preconditioning on children undergoing cardiac surgery: first clinical application in humans. J Am Coll Cardiol 47: 2277–2282. [DOI] [PubMed] [Google Scholar]
- 3. Hausenloy DJ, Mwamure PK, Venugopal V, Harris J, Barnard M, et al. (2007) Effect of remote ischaemic preconditioning on myocardial injury in patients undergoing coronary artery bypass graft surgery: a randomised controlled trial. Lancet 370: 575–579. [DOI] [PubMed] [Google Scholar]
- 4. Thielmann M, Kottenberg E, Boengler K, Raffelsieper C, Neuhaeuser M, et al. (2010) Remote ischemic preconditioning reduces myocardial injury after coronary artery bypass surgery with crystalloid cardioplegic arrest. Basic Res Cardiol 105: 657–664. [DOI] [PubMed] [Google Scholar]
- 5. Venugopal V, Hausenloy DJ, Ludman A, Di Salvo C, Kolvekar S, et al. (2009) Remote ischaemic preconditioning reduces myocardial injury in patients undergoing cardiac surgery with cold-blood cardioplegia: a randomised controlled trial. Heart 95: 1567–1571. [DOI] [PubMed] [Google Scholar]
- 6. Thielmann M, Kottenberg E, Kleinbongard P, Wendt D, Gedik N, et al. (2013) Cardioprotective and prognostic effects of remote ischaemic preconditioning in patients undergoing coronary artery bypass surgery: a single-centre randomised, double-blind, controlled trial. Lancet 382: 597–604. [DOI] [PubMed] [Google Scholar]
- 7. Heusch G, Musiolik J, Kottenberg E, Peters J, Jakob H, et al. (2012) STAT5 activation and cardioprotection by remote ischemic preconditioning in humans: short communication. Circ Res 110: 111–115. [DOI] [PubMed] [Google Scholar]
- 8. Gho BC, Schoemaker RG, van den Doel MA, Duncker DJ, Verdouw PD (1996) Myocardial protection by brief ischemia in noncardiac tissue. Circulation 94: 2193–2200. [DOI] [PubMed] [Google Scholar]
- 9. McClanahan TB, Nao BS, Wolke LJ, Martin BJ, T E, Mertz aKPG (1993) Brief renal occlusion and reperfusion induces myocardial infarct size in rabbits. FASEB J 7. [Google Scholar]
- 10. Weinbrenner C, Nelles M, Herzog N, Sarvary L, Strasser RH (2002) Remote preconditioning by infrarenal occlusion of the aorta protects the heart from infarction: a newly identified non-neuronal but PKC-dependent pathway. Cardiovasc Res 55: 590–601. [DOI] [PubMed] [Google Scholar]
- 11. Breivik L, Helgeland E, Aarnes EK, Mrdalj J, Jonassen AK (2010) Remote postconditioning by humoral factors in effluent from ischemic preconditioned rat hearts is mediated via PI3K/Akt-dependent cell-survival signaling at reperfusion. Basic Res Cardiol 106: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dickson EW, Lorbar M, Porcaro WA, Fenton RA, Reinhardt CP, et al. (1999) Rabbit heart can be “preconditioned” via transfer of coronary effluent. Am J Physiol 277: H2451–2457. [DOI] [PubMed] [Google Scholar]
- 13. Serejo FC, Rodrigues LF Jr, da Silva Tavares KC, de Carvalho AC, Nascimento JH (2007) Cardioprotective properties of humoral factors released from rat hearts subject to ischemic preconditioning. J Cardiovasc Pharmacol 49: 214–220. [DOI] [PubMed] [Google Scholar]
- 14. Dickson EW, Blehar DJ, Carraway RE, Heard SO, Steinberg G, et al. (2001) Naloxone blocks transferred preconditioning in isolated rabbit hearts. J Mol Cell Cardiol 33: 1751–1756. [DOI] [PubMed] [Google Scholar]
- 15. Shimizu M, Tropak M, Diaz RJ, Suto F, Surendra H, et al. (2009) Transient limb ischaemia remotely preconditions through a humoral mechanism acting directly on the myocardium: evidence suggesting cross-species protection. Clin Sci (Lond) 117: 191–200. [DOI] [PubMed] [Google Scholar]
- 16. Lim SY, Yellon DM, Hausenloy DJ (2010) The neural and humoral pathways in remote limb ischemic preconditioning. Basic Res Cardiol 105: 651–655. [DOI] [PubMed] [Google Scholar]
- 17. Hepponstall M, Ignjatovic V, Binos S, Monagle P, Jones B, et al. (2012) Remote ischemic preconditioning (RIPC) modifies plasma proteome in humans. PLoS One 7: e48284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pang T, Zhao Y, Zhang NR, Jin SQ, Pan SQ (2013) Transient limb ischemia alters serum protein expression in healthy volunteers: complement C3 and vitronectin may be involved in organ protection induced by remote ischemic preconditioning. Oxid Med Cell Longev 2013: 859056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hibert P, Prunier-Mirebeau D, Beseme O, Chwastyniak M, Tamareille S, et al. (2013) Apolipoprotein a-I is a potential mediator of remote ischemic preconditioning. PLoS One 8: e77211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hibert P, Prunier-Mirebeau D, Beseme O, Chwastyniak M, Tamareille S, et al. (2014) Modifications in rat plasma proteome after remote ischemic preconditioning (RIPC) stimulus: identification by a SELDI-TOF-MS approach. PLoS One 9: e85669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davidson SM, Selvaraj P, He D, Boi-Doku C, Yellon RL, et al. (2013) Remote ischaemic preconditioning involves signalling through the SDF-1alpha/CXCR4 signalling axis. Basic Res Cardiol 108: 377. [DOI] [PubMed] [Google Scholar]
- 22. Przyklenk K (2013) ‘Going out on a limb’: SDF-1alpha/CXCR4 signaling as a mechanism of remote ischemic preconditioning? Basic Res Cardiol 108: 382. [DOI] [PubMed] [Google Scholar]
- 23. Gosho M, Nagashima K, Sato Y (2012) Study designs and statistical analyses for biomarker research. Sensors (Basel) 12: 8966–8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aebersold R, Burlingame AL, Bradshaw RA (2013) Western blots versus selected reaction monitoring assays: time to turn the tables? Mol Cell Proteomics 12: 2381–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kinsinger CR, Apffel J, Baker M, Bian X, Borchers CH, et al. (2012) Recommendations for mass spectrometry data quality metrics for open access data (corollary to the Amsterdam Principles). J Proteome Res 11: 1412–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martens L, Nesvizhskii AI, Hermjakob H, Adamski M, Omenn GS, et al. (2005) Do we want our data raw? Including binary mass spectrometry data in public proteomics data repositories. Proteomics 5: 3501–3505. [DOI] [PubMed] [Google Scholar]
- 27. Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, et al. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotech 32: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barsnes H, Martens L (2013) Crowdsourcing in proteomics: public resources lead to better experiments. Amino Acids 44: 1129–1137. [DOI] [PubMed] [Google Scholar]
- 29. Phillips HL, Williamson JC, van Elburg KA, Snijders AP, Wright PC, et al. (2010) Shotgun proteome analysis utilising mixed mode (reversed phase-anion exchange chromatography) in conjunction with reversed phase liquid chromatography mass spectrometry analysis. Proteomics 10: 2950–2960. [DOI] [PubMed] [Google Scholar]
- 30. Kessner D, Chambers M, Burke R, Agus D, Mallick P (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24: 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, et al. (2004) Open mass spectrometry search algorithm. J Proteome Res 3: 958–964. [DOI] [PubMed] [Google Scholar]
- 32. Craig R, Beavis RC (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467. [DOI] [PubMed] [Google Scholar]
- 33. Vaudel M, Barsnes H, Berven FS, Sickmann A, Martens L (2011) SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics 11: 996–999. [DOI] [PubMed] [Google Scholar]
- 34. Elias JE, Gygi SP (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214. [DOI] [PubMed] [Google Scholar]
- 35. Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, et al. (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32: D115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barsnes H, Vaudel M, Colaert N, Helsens K, Sickmann A, et al. (2011) compomics-utilities: an open-source Java library for computational proteomics. BMC Bioinformatics 12: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, et al. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41: D1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martens L, Hermjakob H, Jones P, Adamski M, Taylor C, et al. (2005) PRIDE: the proteomics identifications database. Proteomics 5: 3537–3545. [DOI] [PubMed] [Google Scholar]
- 39. Vaudel M, Sickmann A, Martens L (2010) Peptide and protein quantification: a map of the minefield. Proteomics 10: 650–670. [DOI] [PubMed] [Google Scholar]
- 40. Vaudel M, Sickmann A, Martens L (2013) Introduction to opportunities and pitfalls in functional mass spectrometry based proteomics. Biochim Biophys Acta [DOI] [PubMed] [Google Scholar]
- 41. Burkhart JM, Vaudel M, Zahedi RP, Martens L, Sickmann A (2011) iTRAQ protein quantification: a quality-controlled workflow. Proteomics 11: 1125–1134. [DOI] [PubMed] [Google Scholar]
- 42. Powell DW, Weaver CM, Jennings JL, McAfee KJ, He Y, et al. (2004) Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol 24: 7249–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burkhart JM, Vaudel M, Gambaryan S, Radau S, Walter U, et al. (2012) The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood 120: e73–82. [DOI] [PubMed] [Google Scholar]
- 44. Yadav AK, Bhardwaj G, Basak T, Kumar D, Ahmad S, et al. (2011) A systematic analysis of eluted fraction of plasma post immunoaffinity depletion: implications in biomarker discovery. PLoS One 6: e24442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hausenloy DJ (2013) Cardioprotection techniques: preconditioning, postconditioning and remote con-ditioning (basic science). Curr Pharm Des 19: 4544–4563. [DOI] [PubMed] [Google Scholar]
- 46. Heusch G (2013) Cardioprotection: chances and challenges of its translation to the clinic. Lancet 381: 166–175. [DOI] [PubMed] [Google Scholar]
- 47. Przyklenk K (2013) Reduction of Myocardial Infarct Size with Ischemic “Conditioning”: Physiologic and Technical Considerations. Anesth Analg [DOI] [PubMed] [Google Scholar]
- 48. Cote RG, Jones P, Martens L, Kerrien S, Reisinger F, et al. (2007) The Protein Identifier Cross-Referencing (PICR) service: reconciling protein identifiers across multiple source databases. BMC Bioinformatics 8: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou GX, Chao L, Chao J (1992) Kallistatin: a novel human tissue kallikrein inhibitor. Purification, characterization, and reactive center sequence. J Biol Chem 267: 25873–25880. [PubMed] [Google Scholar]
- 50. Chao J, Stallone JN, Liang YM, Chen LM, Wang DZ, et al. (1997) Kallistatin is a potent new vasodilator. J Clin Invest 100: 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao L, Yin H, S. Smith R J, Chao L, Chao J (2008) Role of kallistatin in prevention of cardiac remodeling after chronic myocardial infarction. Lab Invest 88: 1157–1166. [DOI] [PubMed] [Google Scholar]
- 52. Chao J, Yin H, Yao YY, Shen B, Smith RS Jr, et al. (2006) Novel role of kallistatin in protection against myocardial ischemia-reperfusion injury by preventing apoptosis and inflammation. Hum Gene Ther 17: 1201–1213. [DOI] [PubMed] [Google Scholar]
- 53. Shen B, Gao L, Hsu YT, Bledsoe G, Hagiwara M, et al. (2010) Kallistatin attenuates endothelial apoptosis through inhibition of oxidative stress and activation of Akt-eNOS signaling. Am J Physiol Heart Circ Physiol 299: H1419–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Y, Bledsoe G, Hagiwara M, Shen B, Chao L, et al. (2012) Depletion of endogenous kallistatin exacerbates renal and cardiovascular oxidative stress, inflammation, and organ remodeling. Am J Physiol Renal Physiol 303: F1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Renaux JL, Thomas M, Crost T, Loughraieb N, Vantard G (1999) Activation of the kallikrein-kinin system in hemodialysis: role of membrane electronegativity, blood dilution, and pH. Kidney Int 55: 1097–1103. [DOI] [PubMed] [Google Scholar]
- 56. Dray A, Perkins M (1993) Bradykinin and inflammatory pain. Trends Neurosci 16: 99–104. [DOI] [PubMed] [Google Scholar]
- 57. Dietze GJ, Wicklmayr M, Rett K, Jacob S, Henriksen EJ (1996) Potential role of bradykinin in forearm muscle metabolism in humans. Diabetes 45 Suppl 1: S110–114. [DOI] [PubMed] [Google Scholar]
- 58. Boix F, Rosenborg L, Hilgenfeldt U, Knardahl S (2002) Contraction-related factors affect the concentration of a kallidin-like peptide in rat muscle tissue. J Physiol 544: 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Edery H, Lewis GP (1962) Inhibition of plasma kininase activity at slightly acid ph. Br J Pharmacol Chemother 19: 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Saxena P, Shaw OM, Misso NL, Naran A, Shehatha J, et al. (2011) Remote ischemic preconditioning stimulus decreases the expression of kinin receptors in human neutrophils. J Surg Res 171: 311–316. [DOI] [PubMed] [Google Scholar]
- 61. Weisman HF, Bartow T, Leppo MK, Marsh HC Jr, Carson GR, et al. (1990) Soluble human complement receptor type 1: in vivo inhibitor of complement suppressing post-ischemic myocardial inflammation and necrosis. Science 249: 146–151. [DOI] [PubMed] [Google Scholar]
- 62. Diepenhorst GM, van Gulik TM, Hack CE (2009) Complement-mediated ischemia-reperfusion injury: lessons learned from animal and clinical studies. Ann Surg 249: 889–899. [DOI] [PubMed] [Google Scholar]
- 63. Pemberton M, Anderson G, Vetvicka V, Justus DE, Ross GD (1993) Microvascular effects of complement blockade with soluble recombinant CR1 on ischemia/reperfusion injury of skeletal muscle. J Immunol 150: 5104–5113. [PubMed] [Google Scholar]
- 64. Wada K, Montalto MC, Stahl GL (2001) Inhibition of complement C5 reduces local and remote organ injury after intestinal ischemia/reperfusion in the rat. Gastroenterology 120: 126–133. [DOI] [PubMed] [Google Scholar]
- 65. Zhao H, Montalto MC, Pfeiffer KJ, Hao L, Stahl GL (2002) Murine model of gastrointestinal ischemia associated with complement-dependent injury. J Appl Physiol (1985) 93: 338–345. [DOI] [PubMed] [Google Scholar]
- 66. Stahl GL, Xu Y, Hao L, Miller M, Buras JA, et al. (2003) Role for the alternative complement pathway in ischemia/reperfusion injury. Am J Pathol 162: 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Karpel-Massler G, Fleming SD, Kirschfink M, Tsokos GC (2003) Human C1 esterase inhibitor attenuates murine mesenteric ischemia/reperfusion induced local organ injury. J Surg Res 115: 247–256. [DOI] [PubMed] [Google Scholar]
- 68. Schwaeble WJ, Lynch NJ, Clark JE, Marber M, Samani NJ, et al. (2011) Targeting of mannan-binding lectin-associated serine protease-2 confers protection from myocardial and gastrointestinal ischemia/reperfusion injury. Proc Natl Acad Sci U S A 108: 7523–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bossi F, Peerschke EI, Ghebrehiwet B, Tedesco F (2011) Cross-talk between the complement and the kinin system in vascular permeability. Immunol Lett 140: 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Engelbrecht B, Stratmann B, Hess C, Tschoepe D, Gawlowski T (2013) Impact of GLO1 knock down on GLUT4 trafficking and glucose uptake in L6 myoblasts. PLoS One 8: e65195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Inagi R, Kumagai T, Fujita T, Nangaku M (2010) The role of glyoxalase system in renal hypoxia. Adv Exp Med Biol 662: 49–55. [DOI] [PubMed] [Google Scholar]
- 72. Ahmed U, Dobler D, Larkin SJ, Rabbani N, Thornalley PJ (2008) Reversal of hyperglycemia-induced angiogenesis deficit of human endothelial cells by overexpression of glyoxalase 1 in vitro. Ann N Y Acad Sci 1126: 262–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wautier JL, Schmidt AM (2004) Protein glycation: a firm link to endothelial cell dysfunction. Circ Res 95: 233–238. [DOI] [PubMed] [Google Scholar]
- 74. Udby L, Sorensen OE, Pass J, Johnsen AH, Behrendt N, et al. (2004) Cysteine-rich secretory protein 3 is a ligand of alpha1B-glycoprotein in human plasma. Biochemistry 43: 12877–12886. [DOI] [PubMed] [Google Scholar]
- 75. Loukogeorgakis SP, Williams R, Panagiotidou AT, Kolvekar SK, Donald A, et al. (2007) Transient limb ischemia induces remote preconditioning and remote postconditioning in humans by a K(ATP)-channel dependent mechanism. Circulation 116: 1386–1395. [DOI] [PubMed] [Google Scholar]
- 76. Hong DM, Jeon Y, Lee CS, Kim HJ, Lee JM, et al. (2012) Effects of remote ischemic preconditioning with postconditioning in patients undergoing off-pump coronary artery bypass surgery–randomized controlled trial. Circ J 76: 884–890. [DOI] [PubMed] [Google Scholar]
- 77. The Remote Preconditioning Trialists G, Healy DA, Khan WA, Wong CS, Moloney MC, et al. (2014) Remote preconditioning and major clinical complications following adult cardiovascular surgery: Systematic review and meta-analysis. Int J Cardiol [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The complete table of experimental data from all three iTRAQ experiments. All proteins we identified were classified into four categories based on two statistical tests: (A) proteins with low variability among donors (t-test passed) and confident regulation to the background (no protein); (B) low donor variability but no confident regulation to the background (4 proteins); (C) high donor variability (t-test failed) but confident regulation to the background (42 proteins); and (D) high donor variability (t-test failed) and no confident regulation to the background (347 proteins).
(XLSX)
A list of all biomarkers found in the literature, in addition to the ones identified in this study. This includes their measured ratios and t-test results.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Most of the data are within the MS or supporting files, while the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) [27] via the PRIDE partner repository [37], [38] with the dataset identifier PXD000605 and DOI 10.6019/PXD000605.