Abstract
l-Rhamnose is a component of plant cell wall pectic polysaccharides, diverse secondary metabolites, and some glycoproteins. The biosynthesis of the activated nucleotide-sugar form(s) of rhamnose utilized by the various rhamnosyltransferases is still elusive, and no plant enzymes involved in their synthesis have been purified. In contrast, two genes (rmlC and rmlD) have been identified in bacteria and shown to encode a 3,5-epimerase and a 4-keto reductase that together convert dTDP-4-keto-6-deoxy-Glc to dTDP-β-l-rhamnose. We have identified an Arabidopsis cDNA that contains domains that share similarity to both reductase and epimerase. The Arabidopsis gene encodes a protein with a predicated molecular mass of approximately 33.5 kD that is transcribed in all tissue examined. The Arabidopsis protein expressed in, and purified from, Escherichia coli converts dTDP-4-keto-6-deoxy-Glc to dTDP-β-l-rhamnose in the presence of NADPH. These results suggest that a single plant enzyme has both the 3,5-epimerase and 4-keto reductase activities. The enzyme has maximum activity between pH 5.5 and 7.5 at 30°C. The apparent Km for NADPH is 90 μm and 16.9 μm for dTDP-4-keto-6-deoxy-Glc. The Arabidopsis enzyme can also form UDP-β-l-rhamnose. To our knowledge, this is the first example of a bifunctional plant enzyme involved in sugar nucleotide synthesis where a single polypeptide exhibits the same activities as two separate prokaryotic enzymes.
l-Rhamnose is a component of the plant cell wall pectic polysaccharides rhamnogalacturonan I (RG-I) and rhamnogalacturonan II (RG-II; Ridley et al., 2001) and is also present in diverse secondary metabolites including anthocyanins, flavonoids, and triterpenoids (Das et al., 1987; Bar-Peled et al., 1991; van Setten et al., 1995; Shinozaki et al., 1996; Markham et al., 2000), in certain types of plant glycoproteins (Haruko and Haruko, 1999), and in arabinogalactan proteins (Pellerin et al., 1995). The specific enzymes that attach rhamnose to each molecule are known as rhamnosyltransferases (RhaTs). To date, only a small number of RhaTs have been studied, and those were involved in flavonoid rhamnosylation. The characterized RhaTs utilize UDP-β-l-rhamnose (UDP-β-l-Rha) as the donor substrate (Kamsteeg et al., 1978; Feingold, 1982; Bar-Peled et al., 1991), although in mung bean (Vigna radiata), both dTDP-β-l-rhamnose (dTDP-β-l-Rha) and UDP-β-l-Rha were reported to act as sugar donors for the rhamnosylation of flavonoids (Barber and Neufeld, 1961).
We are studying the enzymes involved in the synthesis of the nucleotide-rhamnose as part of our effort to understand the synthesis of pectic polysaccharides. To date, the rhamnosylation of plant polysaccharides and glycoproteins has not been studied. Thus, the identity of the activated form(s) of rhamnose needed for the synthesis of these macromolecules is not known with certainty. The enzymes required for the synthesis of the activated form(s) of rhamnose in plants have also not been purified.
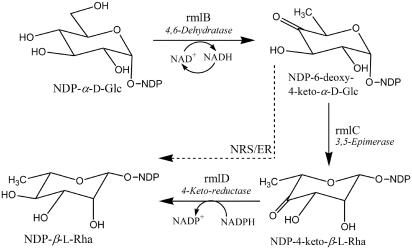
In contrast, much more is known about the synthesis of rhamnose in microorganisms. Gram-negative bacteria are known to form dTDP-β-l-Rha, which is utilized for the synthesis of lipopolysaccharides (for review, see Reeves et al., 1996), and at least two strains of Streptococcus pneumoniae were reported to make UDP-β-l-Rha as well. Some gram-positive bacteria (Kneidinger et al., 2001) utilize both GDP-d-Rha and dTDP-β-l-Rha, while the Paramecium bursaria Chlorella virus (Tonetti et al., 2003) forms GDP-d-Rha, which is utilized for synthesis of the viral capsid glycoprotein. Synthesis of dTDP-β-l-Rha is well characterized in bacteria and is initiated from dTDP-α-d-glucose in a series of reactions catalyzed by three enzymes: dTDP-Glc 4,6-dehydratase (known as rmlB; Allard et al., 2001; Hegeman et al., 2002); dTDP-4-keto-6-deoxy-glucose 3,5-epimerase (known as rmlC; Graninger et al., 1999; Giraud et al., 2000); and dTDP-4-keto-rhamnose reductase (known as rmlD; Blankenfeldt et al., 2002; see Fig. 1). The genes (rmlB, rmlC, and rmlD) that encode each of these enzymes have been identified and cloned from numerous bacteria (Giraud and Naismith, 2000; Li and Reeves, 2000).
Figure 1.
The biosynthetic pathway for the formation of nucleoside diphospho-l-rhamnose in bacteria and plants. Bacteria form dTDP-β-l-Rha from dTDP-α-d-Glc in three reactions (solid arrows) catalyzed by three enzymes: 1) dTDP-Glc 4,6-dehydratase (rmlB); 2) dTDP-6-deoxy-4-keto-d-Glc 3,5-epimerase (rmlC); and 3) dTDP-4-keto-l-Rha NADPH-dependent 4-keto reductase (rmlD). We propose that Arabidopsis forms UDP-β-l-Rha or dTDP-β-l-Rha from UDP-α-d-Glc or dTDP-α-d-Glc in a series of reactions catalyzed by two enzymes: 1) UDP-Glc (or dTDP-Glc) 4,6-dehydratase; and 2) a bifunctional 3,5-epimerase and NADPH-dependent 4-keto reductase (NRS/ER; broken arrow).
Biosynthesis of rhamnose-nucleotides in plants has only been reported in a few studies in which UDP-α-d-Glc was used as the starting material for synthesis of UDP-Rha. As in prokaryotes, UDP-α-d-Glc is converted to UDP-4-keto-6-deoxy-Glc by an enzyme activity similar to the bacterial rmlB (Fig. 1). Since at least some plants generate dTDP-α-d-Glc (Milner and Avigad, 1965; Delmer and Albersheim, 1970) in addition to UDP-α-d-Glc, formation of dTDP-β-l-Rha from dTDP-α-d-Glc was reported as well (for review, see Feingold, 1982). Less is known about the subsequent reactions in the pathway, and it was suggested that the 3,5-epimerization and 4-reduction is mediated by a single enzyme (Kamsteeg et al., 1978). Which nucleotide-activated form(s) of rhamnose plants utilize for various rhamnosylation pathways (glycoproteins, polysaccharides, secondary metabolites, and arabinogalactan proteins) is still unknown.
Here we report that an Arabidopsis gene encodes a protein that contains amino acid motifs that are present in both nucleotide-sugar epimerases and reductases. The recombinant Arabidopsis protein converts dTDP-4-keto-6-deoxy-Glc to a product identified as dTDP-β-l-Rha. Thus, the Arabidopsis protein is a dTDP-rhamnose synthase that possesses 3,5-epimerase and 4-keto reductase activities that we named NRS/ER for nucleotide-rhamnose synthase/epimerase-reductase.
RESULTS
Isolation and Cloning of NRS/ER from Arabidopsis
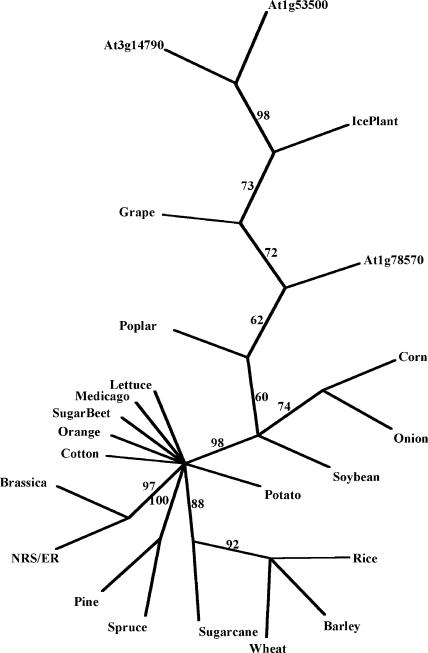
The biosynthetic genes involved in the synthesis of dTDP-β-l-Rha in human pathogens such as Salmonella enterica (Marumo et al., 1992), Yersinia enterocolitica (Zhang et al., 1993), Escherichia coli (Moralejo et al., 1993; Marolda and Valvano, 1995), Shigella flexneri (Macpherson et al., 1994), and in plant-associated bacteria including Xanthomonas campestris pv campestris (Koplin et al., 1993) and Rhizobium meliloti (Becker et al., 1997) were discovered and characterized a decade ago. Concomitantly, we identified some Arabidopsis cDNA clones (T44775, R92767, T88368, and Z26952) obtained from the Expressed Sequence Tag (EST) projects at Michigan State University (Newman et al., 1994) and at Institut National de la Recherche Agronomique (INRA), France, that showed sequence similarity to some of these bacterial genes. This finding provided us with valuable initial information to identify biosynthetic genes involved in formation of activated nucleotide-rhamnose in plants. We subsequently isolated a full-length cDNA corresponding to the Arabidopsis gene that encodes a protein (GenBank accession no. AAF75813). This protein (301 amino acids long, which we named NRS/ER, see below) contains structural and functional motifs that are conserved in a large family of NAD-dependent epimerases, reductase, and dehydratases. The Arabidopsis NRS/ER shares approximately 40% amino acid sequence similarity to the functional rmlD from S. enterica and E. coli (Reeves et al., 1996). Multiple sequence alignments revealed several conserved amino acid regions and motifs in NRS/ER and RmlD from several bacteria (Fig. 2A). Two distinctive features of this family are a conserved catalytic triad composed of a Thr/Ser and a YXXXK (where X is any amino acid), and a modified Rossman motif GXXGXXG (X = any amino acid) involved in the coenzyme-binding domain (Wierenga et al., 1986). The YXXXK motif, likely located on NRS/ER between amino acid 144 and 148, is within a larger conserved domain (EXDX7YXKTKX3EX2L) that appears to be conserved in NRS/ER and rmlD-like proteins from various bacteria (Fig. 2A). The GXXGXXG motif [that is predicted to bind NAD(P)H] is located between amino acids 19 and 26 at the N-terminal region of NRS/ER and is contained within a larger conserved domain: OLOXGXXGXXGXXL (where O = hydrophobic amino acid and X = any amino acid; Fig. 2A). Searches of the database of Expressed Sequence Tags (dbEST) revealed putative NRS/ER orthologs with amino acid sequence identities of approximately 80% over 280 amino acids in cotton, wheat, soybean, rice, maize, potato, tomato, sugar beet, and barley. NRS/ER orthologs with at least 80% amino acid sequence identity were also identified in grape, orange, pine, poplar, spruce, lettuce, and ice plant (Fig. 3). Thus NRS/ER is highly conserved in plants and is likely to carry out important functions in plants. A BLAST search (Altschul et al., 1997) of the Arabidopsis genomic database using the amino acid sequences corresponding to NRS/ER identified three additional coding regions with sequence identities of between 78% and 81% to NRS/ER over 290 amino acids (Fig. 2B). In addition, a BLAST search identified a fungal (Podospora anserina; NCBI accession no. CAD60580) protein with greater than 70% sequence similarity (greater than 55% amino acid identity) to NRS/ER over 275 amino acids. This is interesting since, to the best of our knowledge, rhamnose biosynthesis in fungi has not been reported. However, the biochemical function of this fungal protein has not been determined.
Figure 2.
Amino acid sequence alignment of Arabidopsis NRS/ER. A, Comparing the conserved amino acid sequences between NRS/ER and functional bacterial rmlD proteins. Protein sequence alignment was performed using ClustalX (version 1.83; Jeanmougin et al., 1998). E. coli K-12 (GenBank accession no. AAB88399), S. enterica serovar Typhimurium (accession no. 21730455), Mycobacterium tuberculosis H37Rv (accession no. CAB07093), Fusobacterium nucleatum (accession no. NC_003454), and Arabidopsis NRS/ER (accession no. AAR99502). The putative conserved GX2GX2G motif and the putative catalytic triad YX3K (Giraud et al., 2000) are marked. B, Comparison of the conserved amino acid sequences between NRS/ER and three Arabidopsis proteins predicted from the genomic database (At3g14790, At1g53500, and At1g78570). Protein sequences were aligned with ClustalX software.
Figure 3.
Phylogenetic analysis of NRS/ER proteins. NRS/ER-like protein sequences, translated from dbEST, were aligned with ClustalX software. Alignments were analyzed with the PAUP software (Swofford, 1998) to generate an unrooted tree. Percentage bootstrap values of 1,000 replicates are given at each branch point. Branch lengths are to scale. Accession numbers are in parentheses: cotton (Gossypium arboreum, BE052407); wheat (Triticum aestivum, CD875945); loblolly pine (Pinus taeda, CF386165); sweet potato (Ipomea batatas, CB330308); soybean (Glycine max, CA784305); orange (Citrus sinensis, CB292447); lettuce (Lactuca sativa, BQ871492); grape (Vitis vinifera, CF512199); barley (Hordeum vulgare, BG369398); medicago (Medicago truncatula, BG587952); brassica (Brassica napus, CD839832); sugarcane (Saccharum officinarum, CA258761); onion (Allium cepa, CF443605); ice plant (Mesembryanthemum crystallinum, BF479490); spruce (Picea mariana, AF051236); sugar beet (Beta vulgaris, BF011071); poplar (Populus tremuloides, CF119362); rice (Oryza sativa, AK071766); corn (Zea mays, AY108467); and the conserved amino acid NRS/ER sequence domain within three Arabidopsis genes annotated At3g14790, At1g53500, and At1g78570.
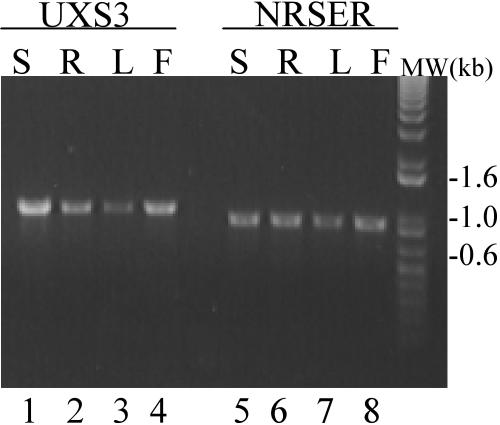
In Arabidopsis, NRS/ER (AY513232; also annotated At1g63000) appears to be transcribed in leaves, roots, and flowers as judged by reverse transcription (RT)-PCR (Fig. 4). These expression data are consistent with information in the EST database, since multiple NRS/ER transcript entries are in cDNA libraries prepared from Arabidopsis flower buds, green siliques, shoots, leaves, and roots (http://www.ncbi.nlm.nih.gov/dbEST/), as well as callus (massively parallel signature sequencing database; http://dbixs001.dbi.udel.edu/MPSS4).
Figure 4.
NRS/ER transcript is expressed in all Arabidopsis tissues. Total RNA isolated from stems (S), roots (R), rosette leaves, of 3-week-old plants (L), and flowers (F) was used to amplify a NRS/ER specific 912-bp transcript (lanes 5–8). The AtUXS3 gene, encoding a UDP-GlcA decarboxylase whose RNA is expressed in all Arabidopsis tissues that have been examined (Harper and Bar-Peled, 2002), was used as an internal RT-PCR control and resulted in the predicted 1,046-bp transcript (lanes 1–4). The data are representative of at least three independent RT-PCR reactions.
Characterization of the Enzymatic Properties of Arabidopsis NRS/ER
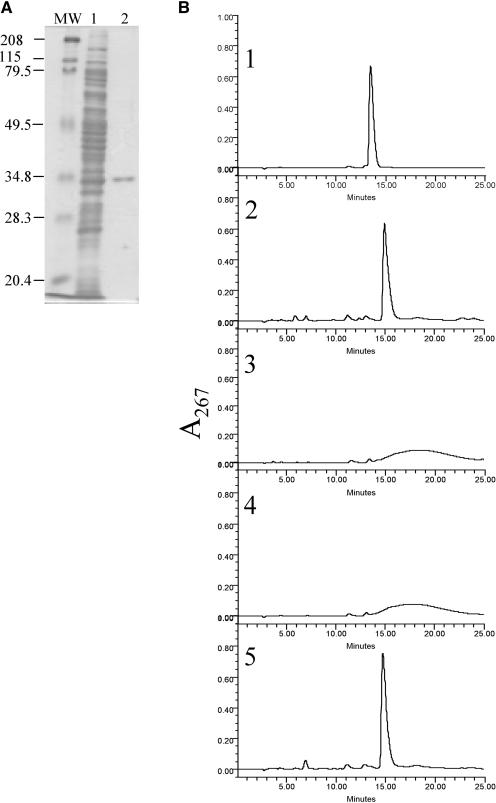
The coding region of NRS/ER, containing a tag at the N-terminal region with six His, was expressed in E. coli, and the recombinant protein was purified by nickel-affinity column chromatography. The purified protein migrated as a single band on SDS-PAGE with an apparent mass of approximately 35 kD (Fig. 5A). Such a value is consistent with the molecular weight of 35,759 predicted from the amino acid sequence of the recombinant protein. Based on the similarity of the NRS/ER to the reductase, epimerase, and dehydratase (RED) family we predicted that this plant protein is an enzyme with dual function (epimerase/reductase), as suggested by Kamsteeg et al. (1978) and by our own work (Bar-Peled et al., 1991).
Figure 5.
Purification and determination of the enzymatic activity of NRS/ER. A, SDS-PAGE analysis of recombinant NRS/ER purified from E. coli: Lane MW, molecular weight marker proteins. Lane 1, total soluble protein from E. coli expressing NRS/ER. Lane 2, NRS/ER purified by Ni-affinity column chromatography. B, NRS/ER encodes a bifunctional epimerase/reductase that converts dTDP-4-keto-6-deoxy-Glc to dTDP-β-l-Rha. 1, The elution profile of dTDP-α-d-Glc on a ODS2-HPLC column. 2, dTDP-β-l-Rha was generated by incubating dTDP-α-d-Glc with the recombinant rmlB, rmlC, and rmlD proteins in the presence of NADPH. 3, dTDP-4-keto-6-deoxy-Glc was generated by incubating dTDP-α-d-Glc with the recombinant rmlB protein alone. The dTDP-4-keto-6-deoxy-Glc eluted as a broad peak from ODS2 column as previously reported (Nakano et al., 2000). 4, Purified NRS/ER was incubated with dTDP-4-keto-6-deoxy-Glc in the absence of NADPH. 5, Purified NRS/ER incubated with dTDP-4-keto-6-deoxy-Glc in the presence of NADPH to yield dTDP-β-l-Rha.
Since the potential substrate for NRS/ER (dTDP-4-keto-6-deoxy-Glc) is not available, we generated it using recombinant rmlB purified from E. coli (Graninger et al., 1999). The bacterial rmlB preferentially converts dTDP-α-d-Glc to dTDP-4-keto-6-deoxy-Glc (Graninger et al., 1999). After incubating recombinant rmlB with dTDP-α-d-Glc, the nucleotide-4-keto-6-deoxy-Glc eluted from a reverse-phase ODS2 HPLC column as a broad peak (Fig. 5B, 3), which is consistent with the results obtained by Nakano et al. (2000). The reason for this peak broadening has not been determined, although the presence of a 4-keto group may increase the hydrophobicity of the sugar nucleotide and thereby promote interaction with the column. 4-Keto sugars are likely to exist on the column as different keto-enol isomers that may also lead to peak broadening on the ODS column. Incubating dTDP-4-keto-6-deoxy-Glc with recombinant NRS/ER in the presence of NADPH resulted in the formation of a product that co-eluted with dTDP-β-l-Rha (Fig. 5B, 5). This major peak, which eluted from the HPLC column at 15.3 min, was collected and analyzed by 1H-NMR spectroscopy. The 1H-NMR spectrum of the product contained a doublet-of-doublet signal at 5.21 ppm (Table I) with coupling constants, 3J1″,P of 8.8 Hz and 3J1″,2″ of less than 2 Hz, diagnostic for proton H-1 of β-l-Rha. The signal at 1.30 ppm is diagnostic for the H-6 methyl protons of rhamnose. All other signals in the 1H-NMR spectra of the enzymatically synthesized compound had chemical shifts and coupling constants that are similar to dTDP-β-l-Rha reported by Nakano et al. (2000). For example, the coupling constant between protons 3″ and 4″, and protons 4″ and 5″ are 9.3 and 9.8 Hz, respectively, indicating the trans configuration that is expected for rhamnose (see Table I). We observed only one point of chemical shift discrepancy for proton 5′, on the deoxy-ribose, from the published report (Nakano et al., 2000). Our data showed a chemical shift of 4.16 ppm (Table I), whereas Nakano et al. (2000) reported a value of 4.03 ppm for H-5′ on deoxy-ribose. However, our assignment for this proton is in complete agreement with the assignment reported for other dTDP sugars (Amann et al., 2001) that indicated a chemical shift of 4.15 for proton 5′ on deoxy-ribose. Thus, our data provide evidence that dTDP-β-l-Rha is formed when the recombinant NRS/ER is incubated with dTDP-4-keto-6-deoxy-Glc.
Table I.
Proton chemical shifts and coupling constants of dTDP-β-l-Rha synthesized from dTDP-α-d-Glc using rmlB and NRS/ER
| Proton | Thymine | Deoxyribose Chemical shifts, δ | dTDP-β-l-Rha Rhamnose | Rha coupling constants | |
|---|---|---|---|---|---|
| ppma | Hz | ||||
| 1 | — | 6.33 (6.37) | 5.21 (5.21) | J1″,P 8.8 | J1″,2″ < 2 |
| 2 | — | 2.39 (2.38) | 4.08 (4.08) | J2″,3″ 3.4 | |
| 3 | — | 4.62 (4.61) | 3.62 (3.63) | J3″,4″ 9.3 | |
| 4 | — | 4.16 (4.19) | 3.35 (3.36) | J4″,5″ 9.8 | |
| 5 | (CH3) 1.92 (1.95) | 4.16 (4.03b) | 3.43 (3.43) | J5″,6″ 6.3 | |
| 6 | 7.74 (7.74) | — | 1.30 (1.30) | ||
Chemical shifts are in ppm relative to acetone signal. Rhamnose proton-proton coupling constants in Hz are indicated as well as the 3J1″,P coupling values between phosphate and the H-1 proton of Rha. Values in parentheses are chemical shifts of published data (Nakano et al., 2000).
The chemical shift for this proton is 4.15 ppm in other dTDP-sugars derivatives (Amann et al., 2001).
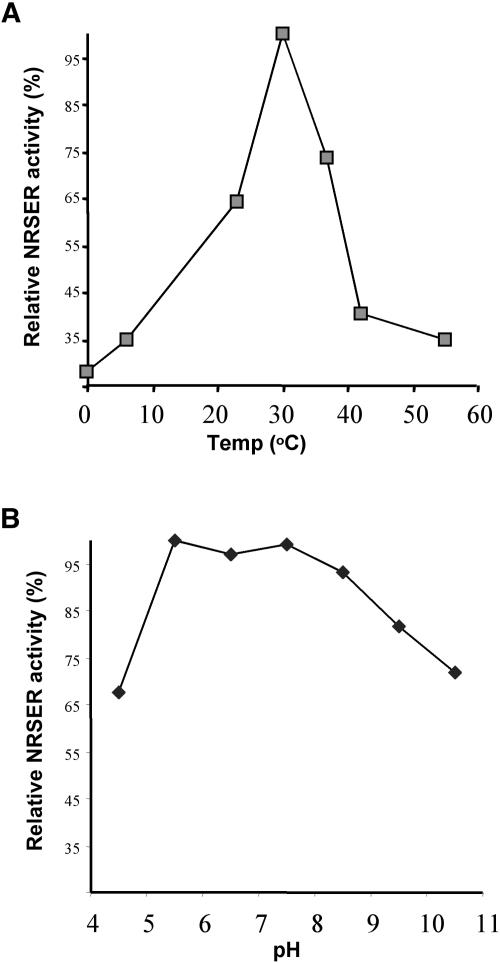
We further characterize NRS/ER properties using dTDP-4-keto-6-deoxy-Glc as substrate. The cofactor NADPH is required for enzymatic activity of the recombinant protein, and without it no conversion to dTDP-β-l-Rha is observed (Fig. 5B, 4). We found that NADH is also a hydride ion donor for the reduction of the 4-keto derivative, but only 40% of dTDP-4-keto-6-deoxy-Glc was converted to dTDP-β-l-Rha by NRS/ER when 1 mm NADH was used compared to 1 mm NADPH. The addition of up to 10 mm MgCl2 or CaCl2 had no discernible effect on the activity nor was NRS/ER activity inhibited by millimolar concentrations of the divalent cation chelator EDTA. In contrast to the plant epimerase/reductase, EDTA strongly inhibits the prokaryote 3,5-epimerase and 4-reductase activities, and the magnesium ion is essential for their activities (Graninger et al., 1999). The NRS/ER is stable for at least 30 months when stored as a crude extract at −20°C. However, more than 60% of NRS/ER activity is lost when the purified enzyme is subjected to repeated freezing and thawing. The recombinant NRS/ER is active between pH 4.5 and 10.5 with maximum activity between pH 5.5 and 7.5 (Fig. 6B). Maximum enzyme activity is obtained at 30°C, and approximately 35% of the activity is retained at 45°C (Fig. 6A). Under optimal conditions enzyme activity is linear for 30 min, and, thus, kinetic analyses were performed using 20-min reactions. The apparent Km for NADPH is 90 μm and 16.9 ± 0.7 μm for dTDP-4-keto-6-deoxy-Glc.
Figure 6.
Effect of temperature and pH on the relative activity of NRS/ER. A, NRS/ER was incubated with dTDP-6-deoxy-4-keto-Glc and NADPH in reaction buffer for 5 min at different temperatures. B, NRS/ER was incubated for 5 min at 30°C with dTDP-6-deoxy-4-keto-Glc and NADPH in reaction buffer of different 0.1-m sodium phosphate pH values. The data are the average of duplicate assays, and the data are presented as the relative activity of NRS/ER.
We tested the specificity of the NRS/ER toward other nucleotide-4-keto-6-deoxy-sugars. To determine if NRS/ER can utilize GDP-4-keto-6-deoxy-Man as a substrate we first purified recombinant GDP-Man 4,6-dehydratase (GMD) as described (Sturla et al., 1997) and incubated GMD with GDP-α-d-Man. HPLC analysis was used to confirm full conversion of GDP-α-d-Man to GDP-4-keto-6-deoxy-Man. The product was then incubated, in control experiments, with recombinant human GDP-Man 3,5-epimerase/4-keto-reductase (Tonetti et al., 1996) that readily converted GDP-4-keto-6-deoxy-Man to GDP-Fuc as determined by HPLC. However, no conversion to GDP-Fuc was evident when NRS/ER was incubated withGDP-4-keto-6-deoxy-Man (with or without NADPH). We also tested the activity of NRS/ER toward UDP-4-keto-6-deoxy-Glc. For these experiments we used a recombinant purified rmlB (Hegeman et al., 2002). Although the recombinant rmlB has preference for dTDP-α-d-Glc, Hegeman et al. (2002) reported that partial conversion of UDP-α-d-Glc to UDP-4-keto-6-deoxy-Glc was achieved using the rmlB. Our initial trials to convert UDP-α-d-Glc to UDP-4-keto-6-deoxy-Glc using recombinant rmlB failed to produce a stable major product, and only trace amounts of putative UDP-4-keto-6-deoxy-Glc were produced after 60 min. Incubation of rmlB with UDP-α-d-Glc for longer periods of time (up to 20 h) in an attempt to achieve more conversion to UDP-4-keto-6-deoxy-Glc resulted in partial degradation of the product, preventing quantitation and characterization of UDP-4-keto-6-deoxy-Glc. However, co-incubation of UDP-α-d-Glc and rmlB with NRS/ER and NADPH resulted in an HPLC peak at retention time 9.3 min, which was collected. This conversion was NADPH dependent. Since the amount of product converted was low (presumably due to the low activity of rmlB toward UDP-α-d-Glc), we pooled the peaks from several reactions and analyzed the product by NMR. The 1H-NMR spectrum of the enzymatic product contained a doublet-of-doublet signal at 5.21 ppm (Table II) with coupling constants, 3J1″,P of 8.8 Hz and 3J1″,2″ of less than 2 Hz, diagnostic for the H-1 of β-l-Rha. The signal at 1.30 ppm is diagnostic of the H-6 methyl protons of rhamnose. Other rhamnose signals in the 1H-NMR spectra of the NRS/ER-derived compound had chemical shifts and coupling constants similar to those for dTDP-β-l-rhamnose (Table I). The uridine proton assignments in the 1H-NMR spectra are characteristic as those uridines of other UDP-sugars (for example, UDP-Xyl; Harper and Bar-Peled, 2002). Thus, our data provide evidence that UDP-β-l-Rha is formed when the recombinant NRS/ER protein is incubated with UDP-4-keto-6-deoxy-Glc. We therefore named the protein NRS/ER for nucleotide rhamnose synthase/3,5-epimerase;4-reductase. Unfortunately, due to low conversion of UDP-α-d-Glc to UDP-4-keto-6-deoxy-Glc by rmlB, complete enzymatic characterization of NRS/ER was not possible.
Table II.
Proton chemical shifts and coupling constants of UDP-β-l-Rha synthesized from UDP-α-d-Glc using rmlB and NRS/ER
| Proton | Uracil | Ribose Chemical shifts, δ | UDP-β-l-Rha Rhamnose | Rha coupling constants | |
|---|---|---|---|---|---|
| ppma | Hz | ||||
| 1 | — | 5.96 | 5.21 (5.21) | J1″,P 8.8 | J1″,2″ < 2 |
| 2 | — | 4.36 | 4.08 (4.08) | J2″,3″ 3.4 | |
| 3 | — | 4.28 | 3.62 (3.63) | J3″,4″ 9.3 | |
| 4 | — | 4.34 | 3.35 (3.36) | J4″,5″ 9.8 | |
| 5 | 5.96 | 4.19 | 3.43 (3.43) | J5″,6″ 6.3 | |
| 6 | 7.93 | — | 1.30 (1.30) | ||
Chemical shifts are in ppm relative to acetone signal. Rhamnose proton-proton coupling constants in Hz are indicated as well as the 3J1″,P coupling values between phosphate and the H-1 proton of Rha. Values in parentheses are chemical shifts of published data for dTDP-Rha (Nakano et al., 2000); chemical shifts for uridine protons are the same as for UDP-sugars.
UDP-Glc, dTDP-Glc, UDP-GlcA, UDP-Xyl, GDP-Man, or GDP-Fuc were not substrates for NRS/ER either in the presence or absence of NADPH.
DISCUSSION
The Arabidopsis gene NRS/ER (also annotated At1g6300) encodes an approximately 33.5-kD enzyme (NRS/ER) which, in the presence of NADPH, converts dTDP-4-keto-6-deoxy-Glc and UDP-4-keto-6-deoxy-Glc to dTDP-β-l-Rha and UDP-β-l-Rha, respectively (Tables I and II). NRS/ER is similar to other plant enzymes involved in the formation of UDP-β-l-Rha (Barber and Chang, 1967; Kamsteeg et al., 1978;Bar-Peled et al., 1991) since all these enzymes have an absolute requirement and preference for NADPH. Our data also provide support for the previous suggestion that the 3,5-epimerase and 4-reductase activities in plants may be due to a single enzyme (Kamsteeg et al., 1978; see Fig. 1).
The combined 3,5-epimerase/4-reductase activities of NRS/ER toward dTDP-4-keto-6-deoxy-Glc and UDP-4-keto-6-deoxy-Glc are similar to the single bifunctional 3,5-epimerase/4-reductase enzyme involved in the formation of GDP-β-l-fucose from GDP-d-4-keto-6-deoxy-Man (Bonin and Reiter, 2000). In the synthesis of GDP-l-Fuc, GDP-α-d-Man is first converted to the 4-keto-6-deoxy-derivative by a GMD (Bonin et al., 1997). The 4-keto-6-deoxy-Man is then converted to GDP-Fuc by a single bifunctional enzyme (GDP-4-keto-6-deoxy-mannose 3,5-epimerase/4-reductase) that is referred to as either GDP-Man epimerase-reductase (GER; Bonin and Reiter, 2000), GDP-fucose synthase (GFS; Menon et al., 1999), FX (Tonetti et al., 1996), or GDP-Man epimerase/reductase (GMER; Rizzi et al., 1998). The GDP-Fuc synthesizing enzymes have been cloned from humans (Tonetti et al., 1996; Sullivan et al., 1998), plants (Bonin et al., 1997; Bonin and Reiter, 2000), and bacteria (Sturla et al., 1997; Menon et al., 1999; Mattila et al., 2000). There is no primary amino acid sequence similarity between the bifunctional NRS/ER and the bifunctional GFS (GDP-Fuc Synthase). Furthermore GDP-4-keto-6-deoxy-mannose is not a substrate for NRS/ER. This suggests that GFS and NRS/ER recognize the different base moieties in the sugar nucleotides and that NRS/ER is a unique member of the RED family. This report also suggests the NRS/ER is a flexible enzyme and can accommodate either dTDP- or UDP-linked 4-keto-6-deoxy-Glc.
The formation of dTDP-β-l-Rha or UDP-β-l-Rha from the corresponding 4-keto-6-deoxy derivative is catalyzed by a single protein in Arabidopsis, whereas the products of two genes (rmlC and rmlD) are required to perform the corresponding epimerization and reduction reaction in bacteria. Prokaryotes may require a separate epimerase and reductase to allow them to convert a common intermediate into two 6-deoxy sugars that differ in their stereochemistry at C-4. In bacteria, dTDP-4-keto-6-deoxy-Glc can be epimerized and reduced to yield dTDP-β-l-Rha, which has an equatorial hydroxyl at C-4, or to yield dTDP-6-deoxy-l-talose, which has an axial hydroxyl at C-4. Indeed, a gene from Actinobacillus actinomycetemcomitans has been identified and shown to encode a dTDP-4-keto-rhamnose reductase that converts dTDP-4-keto-l-Rha to dTDP-6-deoxy-l-Tal (Nakano et al., 2000); 6-deoxy-l-talose is a component of the capsular polysaccharide synthesized by this bacterium. To our knowledge, no6-deoxy-l-talose-containing nucleotides and polysaccharides have been identified in plants.
The Arabidopsis genome contains three additional genes (At1g53000, At1g78570, and At3g14790) that each encode large proteins (approximately 670 amino acids) having two domains: an N-terminal domain (approximately 330 amino acids long) with amino acid sequence similarity to 4,6-dehydratase followed by a C-terminal domain (greater than 320 amino acids) that shares over 80% amino acid sequence identity to the functional NRS/ER described in this report. It is likely that these three Arabidopsis genes are also involved in the synthesis of activated Rha. Indeed, we have generated recombinant At1g78570 C-terminal domain in E. coli and found that the recombinant protein is able to convert dTDP-4-keto-6-deoxy-Glc to dTDP-β-l-Rha (data not shown). Additional studies are required to determine the catalytic activity of the full products of At1g78570, At1g53000, and At3g14790 and to establish if Arabidopsis has several genes encoding the same catalytic activities (i.e. isoforms) for the synthesis of activated nucleotide-rhamnose.
In this report we identified a gene product, NRS/ER, and showed that in vitro it forms both UDP-β-l-Rha and dTDP-β-l-Rha. While it is assumed that UDP-β-l-Rha is the activated sugar used for the synthesis of flavonoids, we cannot exclude the possibility that plants convert dTDP-α-d-Glc to dTDP-β-l-Rha for synthesis of other macromolecules. Isolation of the various RhaTs involved in the synthesis of pectic polysaccharides and glycoproteins, and characterization of their nucleotide-rhamnose preference, is thus essential. The identification of all the genes and the characterization of the proteins involved in the formation of rhamnose in plants are required to determine how many activated forms of rhamnose are used in plants.
MATERIALS AND METHODS
Cloning and RT-PCR Analysis of Arabidopsis NRS/ER
Arabidopsis expressed tag cDNA databases (dbEST) were searched to identify cDNA with amino acid sequence similarity to bacterial rmlB, rmlC, and rmlD. Several ESTs (T44775, R92767, T88368, and Z26952) that showed similarity to these bacterial gene products were identified and used to design primers to obtain the corresponding Arabidopsis genes by RT-PCR. Briefly, total RNA from Arabidopsis ecotype Columbia plants was isolated using Trizol reagent (Chomczynski, 1993). RNA was reverse transcribed into cDNA at 42°C for 60 min using 1 μm oligo(dT) primer (5′TTCTAGAATTCAGCGGCCGCTT15-TTV) in a 20-μL total reaction consisting of 50 mm Tris-HCl (pH 8.4), 75 mm KCl, 3 mm MgCl2, 0.2 mm of each deoxynucleotide triphosphates (dNTPs), 10 mm dithiothreitol, and 200 units SuperScript II RNase H− reverse transcriptase (Gibco-BRL, Gaithersburg, MD). Following the reaction two units of RNase H (Gibco-BRL) were added. An aliquot of the resulting reverse-transcribed products was used as a template for PCR using 1 unit of high fidelity Platinum Taq DNA polymerase mixed with GB-D proofreading DNA polymerase and Platinum antibody (Gibco-BRL), 0.2 μm of the sense primer [80AAF75813#1S/1-27 5′CATATGGTTGCAGACGCAAACGGTTCATC], 0.2 μm of the antisense primer [80AAF75813#2AS/883-917 5′GCGGCCGCTCAAGCTTTAACTTCAGTCTTCTTGTT], and 0.2 mm of each dNTPs (Roche, Indianapolis, IN) in buffer containing 60 mm Tris-SO4 (pH 8.9), 18 mm ammonium sulfate, and 1 mm MgSO4. RT-PCR reaction product was separated by agarose-gel electrophoresis, purified, and cloned into pCR2.1-TOPO plasmid (Invitrogen, Carlsbad, CA). The cloned RT-PCR product was sequenced and the nucleotide sequence submitted to GenBank (accession no. AY513232, NRS/ER). The Nde I-Not I DNA fragment from pCR2.1:81.5, consisting of the coding region for NRS/ER, was subcloned into a pET28b E. coli expression vector (Novagen, Madison, WI). The resulting clone, pET28b:81.5.3, was constructed to give an in-frame N-terminal 6His tag-NRS/ER gene fusion. The mass of the recombinant NRS/ER (approximately 35 kD) is larger than the native protein (approximately 33 kD) due to the presence of the 6His amino acid tag.
For NRS/ER expression studies in Arabidopsis Columbia, total RNA from flowers, fully expanded rosette leaves and stems of 6-week-old plants, rosette leaves of 3-week-old plants, or roots of 4-week-old plants grown in liquid media as described (Bar-Peled and Raikhel, 1997) was reverse transcribed into cDNA in 20-μL reactions using 200 units of SuperScript II reverse transcriptase (Invitrogen) and 1 μm oligo(dT) primer using the manufacturer's recommended buffer. One-twentieth of each of the reverse-transcribed products was used as a template for PCR reactions using 0.5 units Taq DNA polymerase (Roche, Basel, Switzerland), the manufacturer's buffer, 0.2 mm dNTPs, 1.5 mm MgCl2, and 0.2 μm NRS/ER gene-specific sense and antisense primers (see above). As internal RT-PCR controls, we used AtUxs3 (UDP-GlcA decarboxylase) gene primer [101-1S 5′AGAATTCCCATGGCAGCTACAAGTGAGAAACAG, and 101-2AS 5′GCGGCCGCTTAGTTTCTTGGGACGTTAAGCCTTAG] as described (Harper and Bar-Peled, 2002). PCR conditions were one cycle at 95°C for 2 min, 30 cycles (95°C, 30 s; 54°C, 30 s; 70°C, 1 min), and a final extension at 70°C for 5 min. One-tenth of each sample and the DNA Mr marker (1KB plus, Invitrogen) were resolved on a TAE-1% agarose gel and visualized by staining with ethidium bromide.
Protein Expression and Purification
Fifteen milliliters of an overnight culture of E. coli strain BL21(DE3)pLysS (Novagen), carrying the pET28b:81.5 (NRS/ER) or control pET28b vector alone were used to inoculate 0.5 L Luria-Bertani broth supplemented with 50 μg/mL kanamycin and 30 μg/mL chloramphenicol. Cells were grown at 37°C while shaking (200 rpm) until a cell density of A600 = approximately 0.6, and then induced by addition of isopropylthio-β-galactoside (final concentration, 1 mm). After approximately 3 h at approximately 25°C, cells were collected by centrifugation (10 min at 6,000g at 4°C). Cells were washed with cold water, resuspended in 20 mL extraction buffer (50 mm MOPS-NaOH, pH 7.6, 0.5 mm EDTA) supplemented with fresh 1 mm dithiothreitol and 0.5 mm phenylmethylsulfonyl fluoride, and ruptured by 10 sonication intervals (10-s pulse followed by 10-s rest) on ice, using a microtip probe and a Fisher model 550 sonicator (Fisher Scientific, Pittsburgh, PA) set at power of 4.5. The suspension was centrifuged (20,000g, approximately 30 min, 4°C), and the supernatant was collected and loaded onto a Sepharose CL-6B-nickel column (2 mL resin volume [NTA-Ni, Qiagen USA, Valencia, CA] packed in a 0.6-cm [i.d.] × 5-cm polypropylene column [Bio-Rad, Hercules, CA]) that was pre-equilibrated with buffer A (50 mm sodium phosphate, 0.3 m NaCl, pH 8). The recombinant protein was purified as described (Harper and Bar-Peled, 2002) and stored at −20°C until analysis. All chromatography steps were performed at 4°C. Protein concentration was determined using the Bradford dye-binding assay (Bio-Rad), using bovine serum albumin as a standard. Proteins were separated by 0.1% SDS-12% PAGE as previously described (Bar-Peled et al., 1991) alongside Mr markers (Bio-Rad) and visualized by staining with SimplyBlue (Invitrogen).
Enzyme Assays
The substrate dTDP-4-keto-6-deoxy-glucose was generated from dTDP-α-d-Glc using recombinant dTDP-Glc 4,6-dehydratase (the product of the rmlB gene; a gift from Paul Messner [University of Vienna, Austria] and Perry Frey [University of Wisconsin, Madison, WI]) as described (Graninger et al., 1999; Hegeman et al., 2002).
The assays for dTDP-4-keto-6-deoxy-Glc epimerase-reductase activity (final reaction volume of 75 μL) were performed in two steps. First, dTDP-4-keto-6-deoxy-Glc was produced by incubating dTDP-α-d-Glc (1.3 mm) with 5 μg recombinant rmlB in 50 μL of sodium phosphate, pH 8.5, for 10 min at 30°C. The recombinant dTDP/UDP-4-keto-6-deoxy-Glc epimerase-reductase (NRS/ER; 5 μg) and NADPH (1.3 mm) were added, and after a 20-min incubation at 30°C, the reactions were terminated by the addition of 20 μL chloroform. The mixture was vortexed and centrifuged at 16,000g for 5 min at room temperature. The aqueous phase was retained, and the organic phase extracted with 80 μL water. The aqueous phases were combined and analyzed by HPLC using a Phenosphere-ODS2 column (250 × 4.6 mm; Phenomenex, Torrance, CA) or a Spheresorb-ODS2 column (250 × 4.6 mm; Waters, Milford, MA) eluted at 1 mL min−1 with 0.5 m KH2PO4 (A) for 15 min, followed by a gradient to 80% A and 20% methanol over 14 min at a flow rate of 0.7 mL min−1. Peaks eluted from the ODS2 column were injected onto a Phenosphere-SAX ion exchange column (250 × 4.6 mm; Phenomenex) and eluted at 1 mL min−1 with a linear 2- to 600-mm ammonium-formate gradient formed over 25 min (Bar-Peled et al., 2001). Nucleotides and nucleotide-sugars were detected by UV absorbance using a Waters photodiode array detector. The λmax for thymidine and uridine were 267 nm and 261 nm, respectively. The columns were calibrated with authentic nucleotide-sugars (Sigma, St. Louis).
1H-NMR Spectroscopic Analysis of the Products Formed by Incubating dTDP- and UDP-4-Keto-6-Deoxy-Glc with NRS/ER
UV-absorbing peaks eluting from the SAX column were collected and lyophilized to remove the ammonium formate. The residues were dissolved in water, relyophilized twice, and then exchanged twice with 99.96% D2O. Proton NMR spectroscopy was performed at 25°C on Varian Inova spectrometers (Palo Alto, CA) operating at 500 MHz and 600 MHz (Bar-Peled et al., 2001). The chemical shifts are in ppm and were normalized using acetone (2.224 ppm) as external standard.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers 21730455, AAB88399, AAR99502, AF051236, AK071766, AY108467, AY513232, BE052407, BF011071, BF479490, BG369398, BG587952, BQ871492, CA258761, CA784305, CAD60580, CAB07093, CB292447, CB330308, CD839832, CD875945, CF119362, CF386165, CF443605, CF512199, and NC_003454.
Acknowledgments
The authors thank Professors Paul Messner (University of Vienna), Perry Frey (Department of Biochemistry, University of Wisconsin, Madison), and Michela Tonetti (University of Genova, Italy) for their generous gifts of the gene products used in this study. We thank Michael Hahn, Debra Mohnen, and Malcolm O'Neill for their comments on the manuscript. We also thank the anonymous reviewers for their valuable comments and Bob Kuzoff for assistance with phylogenetic analyses.
This work was supported in part by the U.S. Department of Agriculture (grant no. 2002–35318–12620 to M.B.-P.) and by the U.S. Department of Energy (center grant no. DE–FG05–93ER20097) for the DOE Center for Plant and Microbial Complex Carbohydrates.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.037192.
References
- Allard STM, Giraud MF, Whitfield C, Graninger M, Messner P, Naismith JH (2001) The crystal structure of dTDP-d-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar typhimurium, the second enzyme in the dTDP-l-rhamnose pathway. J Mol Biol 307: 283–295 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann S, Drager G, Rupprath C, Kirschning A, Elling L (2001) (Chemo)enzymatic synthesis of dTDP-activated 2,6-dideoxysugars as building blocks of polyketide antibiotics. Carbohydr Res 335: 23–32 [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, Griffith CL, Doering TL (2001) Functional cloning and characterization of a UDP-glucuronic acid decarboxylase: the pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc Natl Acad Sci USA 98: 12003–12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M, Lewinsohn E, Fluhr R, Gressel J (1991) UDP-rhamnose-flavanone-7-O-glucoside-2″-O-rhamnosyltransferase - purification and characterization of an enzyme catalyzing the production of bitter compounds in citrus. J Biol Chem 226: 20953–20959 [PubMed] [Google Scholar]
- Bar-Peled M, Raikhel NV (1997) Characterization of AtSEC12 and AtSAR1. Proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiol 114: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GA, Chang MT (1967) Synthesis of uridine diphosphate L-rhamnose by enzymes of Chlorella pyrenoidosa. Arch Biochem Biophys 118: 659–663 [DOI] [PubMed] [Google Scholar]
- Barber GA, Neufeld EF (1961) Rhamnosyl transfer from TDP l-rhamnose catalyzed by plant enzyme. Biochem Biophys Res Commun 6: 44–48 [DOI] [PubMed] [Google Scholar]
- Becker A, Ruberg S, Kuster H, Roxlau AA, Keller M, Ivashina T, Cheng HP, Walker GC, Puhler A (1997) The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J Bacteriol 179: 1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenfeldt W, Kerr ID, Giraud MF, McMiken HJ, Leonard G, Whitfield C, Messner P, Graninger M, Naismith JH (2002) Variation on a theme of SDR: dTDP-6-deoxy-l-lyxo-4-hexulose reductase (Rm1D) shows a new Mg2+-dependent dimerization mode. Structure 10: 773–786 [DOI] [PubMed] [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter W-D (1997) The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-mannose-4,6-dehydratase catalyzing the first step in the de novo synthesis of GDP-l-fucose. Proc Natl Acad Sci USA 94: 2085–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin CP, Reiter W-D (2000) A bifunctional epimerase-reductase acts downstream of the MUR1 gene product and completes the de novo synthesis of GDP-L-fucose in Arabidopsis. Plant J 21: 445–454 [DOI] [PubMed] [Google Scholar]
- Chomczynski P (1993) A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15: 536–537 [PubMed] [Google Scholar]
- Das MC, Bachhawat BK, Mahato SB, Basu MK (1987) Plant glycosides in a liposomal drug-delivery system. Biochem J 247: 359–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP, Albersheim P (1970) The biosynthesis of sucrose and nucleoside diphosphate glucose in Phaseolus aureus. Plant Physiol 45: 782–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold DS (1982) Aldo (and keto) hexoses and uronic acids. In FA Loewus, W Tanner, eds, Plant Carbohydrates I, Intracellular Carbohydrates, Vol 13A. Springer-Verlag, Heidelberg, pp 3–76
- Giraud MF, Leonard GA, Field RA, Berlind C, Naismith JH (2000) RmlC, the third enzyme of dTDP-l-rhamnose pathway, is a new class of epimerase. Nat Struct Biol 7: 398–402 [DOI] [PubMed] [Google Scholar]
- Giraud MF, Naismith JH (2000) The rhamnose pathway. Curr Opin Struct Biol 10: 687–696 [DOI] [PubMed] [Google Scholar]
- Graninger M, Nidetzky B, Heinrichs DE, Whitfield C, Messner P (1999) Characterization of dTDP-4-dehydrorhamnose 3,5-epimerase and dTDP-4-dehydrorhamnose reductase, required for dTDP-l-rhamnose biosynthesis in Salmonella enterica serovar Typhimurium LT2. J Biol Chem 274: 25069–25077 [DOI] [PubMed] [Google Scholar]
- Harper AD, Bar-Peled M (2002) Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol 130: 2188–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruko U, Haruko O (1999) Glycobiology of the plant glycoprotein epitope: structure, immunogenicity and allergenicity of plant glycotopes. Trends Glycosci Glycotech 11: 413–428 [Google Scholar]
- Hegeman AD, Gross JW, Frey PA (2002) Concerted and stepwise dehydration mechanisms observed in wild-type and mutated Escherichia coli dTDP-glucose 4,6-dehydratase. Biochemistry 41: 2797–2804 [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23: 403–405 [DOI] [PubMed] [Google Scholar]
- Kamsteeg J, van Brederode J, van Nigtevecht G (1978) The formation of UDP-l-rhamnose from UDP-D-glucose by an enzyme preparation of red campion (Silene dioica (L) Clairv) leaves. FEBS Lett 91: 281–284 [DOI] [PubMed] [Google Scholar]
- Kneidinger B, Graninger M, Adam G, Puchberger M, Kosma P, Zayni S, Messner P (2001) Identification of two GDP-6-deoxy-D-lyxo-4-hexulose reductases synthesizing GDP-D-rhamnose in Aneurinibacillus thermoaerophilus L420–91T. J Biol Chem 276: 5577–5583 [DOI] [PubMed] [Google Scholar]
- Koplin R, Wang G, Hotte B, Priefer UB, Puhler A (1993) A 3.9-kb DNA region of Xanthomonas campestris pv. campestris that is necessary for lipopolysaccharide production encodes a set of enzymes involved in the synthesis of dTDP-rhamnose. J Bacteriol 175: 7786–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Reeves PR (2000) Genetic variation of dTDP-l-rhamnose pathway genes in Salmonella enterica. Microbiol 146: 2291–2307 [DOI] [PubMed] [Google Scholar]
- Macpherson DF, Manning PA, Morona R (1994) Characterization of the dTDP-rhamnose biosynthetic genes encoded in the rfb locus of Shigella flexneri. Mol Microbiol 11: 281–292 [DOI] [PubMed] [Google Scholar]
- Markham KR, Gould KS, Winefield CS, Mitchell KA, Bloor SJ, Boase MR (2000) Anthocyanic vacuolar inclusions - their nature and significance in flower colouration. Phytochemistry 55: 327–336 [DOI] [PubMed] [Google Scholar]
- Marolda CL, Valvano MA (1995) Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: Identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J Bacteriol 177: 5539–5546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumo K, Lindqvist L, Verma N, Weintraub A, Reeves PR, Lindberg AA (1992) Enzymatic synthesis and isolation of thymidine diphosphate-6-deoxy-D-xylo-4-hexulose and thymidine diphosphate-L-rhamnose. Production using cloned gene products and separation by HPLC. Eur J Biochem 204: 539–545 [DOI] [PubMed] [Google Scholar]
- Mattila P, Rabina J, Hortling S, Helin J, Renkonen R (2000) Functional expression of Escherichia coli enzymes synthesizing GDP-l-fucose from inherent GDP-d-mannose in Saccharomyces cerevisiae. Glycobiology 10: 1041–1047 [DOI] [PubMed] [Google Scholar]
- Milner Y, Avigad G (1965) Thymidine diphosphate nucleotides as substrates in the sucrose synthetase reaction. Nature 206: 825. [DOI] [PubMed] [Google Scholar]
- Menon S, Stahl M, Kumar R, Xu GY, Sullivan F (1999) Stereochemical course and steady state mechanism of the reaction catalysed by the GDP-fucose synthetase from Escherichia coli. J Biol Chem 274: 26743–26750 [DOI] [PubMed] [Google Scholar]
- Moralejo P, Egan SM, Hidalgo E, Aguilar J (1993) Sequencing and characterization of a gene cluster encoding the enzymes for L-rhamnose metabolism in Escherichia coli. J Bacteriol 175: 5585–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Suzuki N, Yoshida Y, Nezu T, Yamashita Y, Koga T (2000) Thymidine diphosphate-6-deoxy-L-lyxo-4-hexulose reductase synthesizing dTDP-6-deoxy-l-talose from Actinobacillus actinomycetemcomitans. J Biol Chem 275: 6806–6812 [DOI] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, Retzel E, Somerville C (1994) Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol 106: 1241–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin P, Vidal S, Williams P, Brillouet JM (1995) Characterization of five type II arabinogalactan-protein fractions from red wine of increasing uronic acid content. Carbohydr Res 277: 135–143 [DOI] [PubMed] [Google Scholar]
- Reeves PR, Hobbs M, Valvano MA, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz CR, Rick PD (1996) Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol 4: 495–503 [DOI] [PubMed] [Google Scholar]
- Ridley BL, O'Neill MA, Mohnen D (2001) Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57: 929–967 [DOI] [PubMed] [Google Scholar]
- Rizzi M, Tonetti M, Vigevani P, Sturla L, Bisso A, Flora AD, Bordo D, Bolognesi M (1998) GDP-4-keto-6-deoxy-D-mannose epimerase/reductase from Escherichia coli, a key enzyme in the biosynthesis of GDP-L-fucose, displays the structural characteristics of the RED protein homology superfamily. Structure 6: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Shinozaki Y, Tobita T, Mizutani M, Matsuzaki T (1996) Isolation and identification of two new diterpene glycosides from Nicotiana tabacum. Biosci Biotechnol Biochem 60: 903–905 [DOI] [PubMed] [Google Scholar]
- Sturla L, Bisso A, Zanardi D, Benatti U, De Flora A, Tonetti M (1997) Expression, purification and characterization of GDP-d-mannose 4,6-dehydratase from Escherichia coli. FEBS Lett 412: 126–130 [DOI] [PubMed] [Google Scholar]
- Sullivan K, Kumar R, Kriz R, Stahl M, Xu G-Y, Rouse J, Chang X-J, Boodhoo A, Potvin B, Cumming DA (1998) Molecular cloning of Human GDP-mannose 4,6-dehydratase and reconstructions of GDP-fucose biosynthesis in vitro. J Biol Chem 273: 8193–8202 [DOI] [PubMed] [Google Scholar]
- Swofford DL (1998) PAUP. Phylogenetic Analysis Using Parsimony and Other Methods. Version 4. Sinauer Associates, Sunderland, MA
- Tonetti M, Sturla L, Bisso A, Benatti U, De Flora A (1996) Synthesis of GDP-fucose by the Human FX protein. J Biol Chem 271: 27274–27279 [DOI] [PubMed] [Google Scholar]
- Tonetti M, Zanardi D, Gurnon JR, Fruscione F, Armirotti A, Damonte G, Sturla L, De Flora A, Van Etten JL (2003) Paramecium bursaria Chlorella virus 1 encodes two enzymes involved in the biosynthesis of GDP-L-fucose and GDP-D-rhamnose. J Biol Chem 278: 21559–21565 [DOI] [PubMed] [Google Scholar]
- van Setten DC, van de Werken G, Zomer G, Kersten GF (1995) Glycosyl compositions and structural characteristics of the potential immuno-adjuvant active saponins in the Quillaja saponaria Molina extract quil A. Rapid Commun Mass Spectrom 9: 660–666 [DOI] [PubMed] [Google Scholar]
- Wierenga RK, Terpstra P, Hol WGJ (1986) Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol 187: 101–107 [DOI] [PubMed] [Google Scholar]
- Zhang L, al-Hendy A, Toivanen P, Skurnik M (1993) Genetic organization and sequence of the rfb gene cluster of Yersinia enterocolitica serotype O:3: similarities to the dTDP-l-rhamnose biosynthesis pathway of Salmonella and to the bacterial polysaccharide transport systems. Mol Microbiol 9: 309–321 [DOI] [PubMed] [Google Scholar]