Abstract
A potato (Solanum tuberosum) cDNA encoding an isoform of disproportionating enzyme (stDPE2) was identified in a functional screen in Escherichia coli. The stDPE2 protein was demonstrated to be present in chloroplasts and to accumulate at times of active starch degradation in potato leaves and tubers. Transgenic potato plants were made in which its presence was almost completely eliminated. It could be demonstrated that starch degradation was repressed in leaves of the transgenic plants but that cold-induced sweetening was not affected in tubers stored at 4°C. No evidence could be found for an effect of repression of stDPE2 on starch synthesis. The malto-oligosaccharide content of leaves from the transgenic plants was assessed. It was found that the amounts of malto-oligosaccharides increased in all plants during the dark period and that the transgenic lines accumulated up to 10-fold more than the control. Separation of these malto-oligosaccharides by high-performance anion-exchange chromatography with pulsed-amperometric detection showed that the only one that accumulated in the transgenic plants in comparison with the control was maltose. stDPE2 was purified to apparent homogeneity from potato tuber extracts and could be demonstrated to transfer glucose from maltose to oyster glycogen.
Disproportionating enzyme (D-enzyme; 4-α-glucanotransferase, EC 2.4.1.25) catalyzes the transfer (disproportionation) of α-1,4 bonds between different glucans. It is located in plastids and has always been presumed to be involved in starch metabolism. Mutant and transgenic plants, which lack its activity almost entirely, have been isolated (Takaha et al., 1998; Colleoni et al., 1999a, 1999b; Critchley et al., 2001; Wattebled et al., 2003), and based on studies on these, two contradictory proposals have been made as to its role. Following analysis of the Chlamydomonas reinhardtii sta11 mutant, it was suggested that D-enzyme is involved directly in the synthesis of the amylopectin fraction of starch (Colleoni et al., 1999a, 1999b; Wattebled et al., 2003); however, no evidence for this could be found in an Arabidopsis D-enzyme mutant (dpe1; Critchley et al., 2001). It was demonstrated in this mutant that starch degradation was impaired, and it was proposed that the main role of D-enzyme is in malto-oligosaccharide metabolism during mobilization of starch in the dark period. In addition, transgenic potato (Solanum tuberosum) plants have been produced in which 99% of D-enzyme activity was repressed, but no influence on either amylopectin synthesis or starch degradation was reported (Takaha et al., 1998).
In all of the above studies, only one isoform of D-enzyme was repressed. Analysis of the Arabidopsis genome, however, has revealed that it contains two genes that appear to code for different D-enzyme isoforms. The mutation studied by Critchley et al. (2001) was in a gene lying on chromosome 5 (At5g64860), with the other putative isoform being on chromosome 2 (At2g40840). Recently two studies have identified Arabidopsis plants mutated in the gene coding for this second isoform (Lu and Sharkey, 2004; Chia et al., 2004). In this article, we describe the analysis of the role of this isoform in starch metabolism in potato.
RESULTS AND DISCUSSION
Identification of cDNAs Coding for Proteins Capable of Metabolizing Maltose
In an effort to identify plant proteins capable of catabolizing maltose, the Escherichia coli TSM90 mutant was obtained from the CGSC: E. coli Genetic Stock Center. This strain cannot metabolize maltose, but it is not clear which gene is mutated that causes this phenotype. According to information provided by the CGSC stock center, the lesion lies in either the MalP (coding for maltodextrin phosphorylase), or the MalQ (coding for amylomaltase) gene. The TSM90 strain demonstrates a negative phenotype when grown on MacConkey agar supplemented with maltose, unless expressing a protein that gives it the ability to metabolize maltose (Ehrmann and Vogel, 1998). TSM90 cells were transformed with a plasmid cDNA library constructed using RNA isolated from potato leaves during the light-to-dark transition (Scheidig et al., 2002), and plasmid DNA was isolated from colonies demonstrating a positive phenotype. Digestion of the resulting plasmids with restriction enzymes indicated that they could be separated into two different classes, and DNA sequence information was obtained from the inserts of a representative of both. One class demonstrated 100% identity to a previous cloned D-enzyme isoform of potato (National Center for Biotechnology Information [NCBI] no. X68664; Takaha et al., 1993), which we will refer to as the stDPE1 (S. tuberosum disproportionating enzyme isoform 1) isoform, while the other was more similar to a predicted cDNA revealed by the Arabidopsis genomic sequencing project (At2g40840) and to 4-α-glucanotransferases from bacteria. We will refer to this isoform as stDPE2 (S. tuberosum disproportionating enzyme isoform 2). We will refer to the homologous Arabidopsis proteins as atDPE1 and atDPE2, respectively. One of the cDNAs isolated coding for stDPE2 was full length, as indicated by the presence of a stop codon just upstream of the start codon. The cDNA codes for a peptide containing 948 amino acids with a predicted molecular mass of approximately 109 kD. The sequence information is present in the NCBI database with the accession number AY510449.
It should be stressed that this screen was not established specifically to identify D-enzyme isoforms, although that was the result. Indeed, in principle it should not have been possible to identify the stDPE1 isoform in this system, as it is thought not to be able to utilize maltose as a donor substrate (Jones and Whelan, 1969). The most likely explanation for the identification of stDPE1 is that longer malto-oligosaccharides will also be present in trace amounts in the E. coli cell. This will occur either because the E. coli will take up malto-triose (which is a minor contaminant of almost all commercially available maltose) from the growth media or because it will be produced within the E. coli cell by isoamylase action on glycogen (Boos and Schuman, 1998). Because D-enzyme acts as a transferase, the sum of all glycosidic linkages remains constant following its action. For example, disproportionation of malto-triose to maltose can lead to the production of malto-tetraose and glucose. Glucose may also act as an acceptor substrate for D-enzyme (Jones and Whelan, 1969), but it will become metabolized further to glucose-6-phosphate by the action of glucokinase, thus removing it from the equilibrium. This allows the further degradation of maltose and the production of longer malto-oligosaccharides. It is known that the presence of these longer malto-oligosaccharides is necessary for the catabolism of maltose by E. coli's own amylomaltase, an enzyme that shows a similar activity to stDPE1 (for review of maltose metabolism in E. coli, see Boos and Schuman, 1998). The reason for the identification of stDPE2 in this expression system will be discussed later.
Analysis of the predicted stDPE2 primary protein structure by the NCBI conserved domain search (Marchler-Bauer et al., 2002) indicated that it was composed of two domains: the first 300 amino acids at the N terminus contain one motif similar to starch-binding domains, while the final 600 amino acids before the C terminus contained a 4-α-glucanotransferase domain.
The Presence of stDPE2 Increases during Periods of Starch Degradation
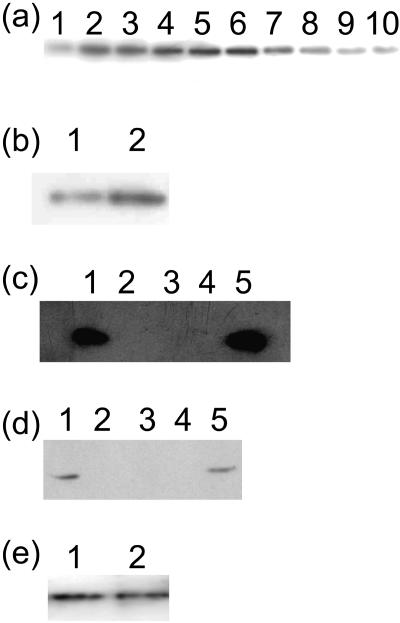
An antibody was produced that recognizes specifically the presence of the protein in plants. It was raised against two peptides, both of which are present within the putative catalytic domain. To ensure the specificity of the antibody for stDPE2, it was affinity purified using the peptides. Immunoblot analysis indicated the presence of one protein of approximately 100 kD in molecular mass in both leaves and tubers of potato plants. A time course was performed to study how the amount of protein changed in leaf tissue during times of starch degradation. It is clear that in leaves, although the protein is present continuously, it increases in amount a just prior to sunset and reduces in amount just before sunrise (Fig. 1a). It was also detected in tubers stored for 8 weeks either at room temperature or at 4°C. The amount of stDPE2 was greater in those stored at 4°C than those stored at room temperature (Fig. 1b).
Figure 1.
Immunoblots of stDPE2. a, Amount of stDPE2 protein in leaves of wild-type potato plants taken at different times of day. Samples were taken every 2 h from 3 pm (lane 1) to 9 am (lane 10). Sunset was at 9 pm and sunrise at 6 am. Thirty micrograms of total protein were loaded in each lane. b, stDPE2 in wild-type tubers stored for 8 weeks at either room temperature (lane 1) or 4°C (lane 2). Twenty micrograms of protein were loaded in each lane. c, stDPE2 protein in leaves of control (lanes 1 and 5) and three transgenic lines, line 8 (lane 2), line 26 (lane 3), and line 28 (lane 4). Sixty micrograms of protein were loaded in each lane. d, Amount of stDPE2 protein in growing tubers of control (lanes 1 and 5) and three transgenic lines, line 8 (lane 2), line 26 (lane 3), and line 28 (lane 4). Thirty micrograms of protein were loaded in each lane. e, stDPE2 in homogenate (lane 1) and chloroplast enriched (lane 2) fraction. Volume equivalent to equal amount of plastidial marker enzyme (NADP-dependant glyceraldehyde 3-phosphate dehydrogenase) was loaded in each lane.
These data would indicate that stDPE2 most probably has a role in starch degradation rather than starch synthesis. This is because the amount of stDPE2 increases during periods of starch degradation, in this case in leaves just before and after sunset and in tubers stored at 4°C. It is also present, however, at times of starch synthesis, such as in leaves during the light period, so based on these data a role in starch synthesis cannot be ruled out.
Transgenic Potato Plants Lacking the Protein Demonstrate a Starch-Excess Phenotype in Their Leaves
To study the role of stDPE2 in plant metabolism, an RNA interference (RNAi) construct was manufactured designed to repress the presence of the protein in potato. Thirty transgenic plants were screened by immunoblots, and three lines (8, 26, and 28) were identified that lacked the protein in their leaves (Fig. 1c). Two of these (8 and 28) were chosen for further study. It was also found that the protein was not present in tubers of the transgenic lines either when growing (Fig. 1d) or when stored for 8 weeks at 4°C (data not shown).
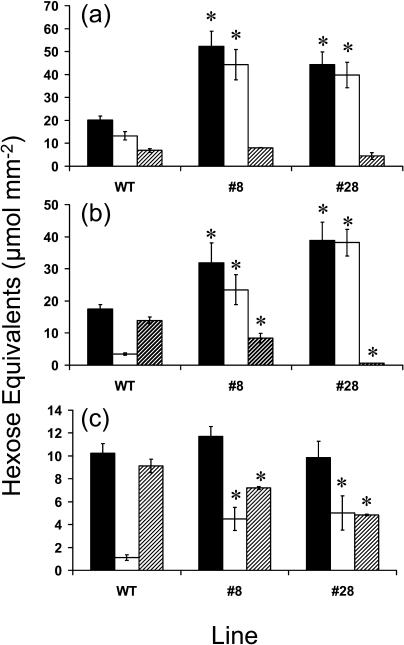
To examine whether starch degradation was impaired in leaves of the transgenic lines, they were wrapped in aluminum foil for 24 h before being bleached with ethanol and stained with Lugols solution to visualize starch. It was found that leaves from both transgenic lines contained starch, in contrast to the control, which did not (data not shown). The amount of starch present in leaves at the beginning and end of the dark period was also determined for potato plants of three different ages (5, 10, and 15 weeks old; Fig. 2). The data are somewhat different between the differently aged plants, either because of changes in climate at the various sampling dates or because of the different ages of the plants. Nevertheless, for all ages the leaf starch contents of the plants were greater at the end of the night period in the transgenic plants in comparison with the control (Fig. 2). This indicates that stDPE2 is involved in starch degradation.
Figure 2.
Amount of starch in leaves from differently aged plants. Starch contents were determined in source leaves of 5- (a), 10- (b), and 15-week-old (c) plants, both at the beginning (black bars) and end (white bars) of the dark period. The difference between these values (amount of starch degraded) is shown as hatched bars. Results are means ± se of at least eight individual probes. Asterisk denotes significant difference in the transgenic plants in comparison with the corresponding wild-type (WT) value at the 5% level (Student's t test).
The Starch-Excess Phenotype Is Not Due to an Increased Rate of Starch Synthesis
Although unlikely, it is still possible that the increase in starch seen in the leaves of the transgenic lines is because starch is synthesized faster there. To examine this, we kept plants without any light for 3 d to ensure that they contained no starch in their leaves. We then illuminated them and measured the amount of starch produced over a 10-h period. Plants from both the control and transgenic lines manufactured starch at a linear rate over this period, and no differences in amounts were found between the lines (data not shown). This demonstrates that the starch-excess phenotype found in the transgenic lines is caused by a lesion in the pathway of starch breakdown.
Tubers of the Transgenic Lines Accumulate Soluble Sugars to the Same Extent as the Control When Stored at 4°C
Potato tubers degrade starch and accumulate soluble sugars in their tubers when stored at low temperatures in a process known as cold sweetening (Müller-Thurgau, 1882). To examine whether this process was inhibited in the transgenic plants, amounts of sucrose, glucose, and fructose were determined in tubers of the transgenic and control plants that had been stored at 4°C for 8 weeks. No differences could be found in the amount of total soluble sugars accumulating in the tubers of the transgenic lines in comparison with the control (data not shown). This demonstrates that stDPE2 on its own does not influence cold sweetening, but there are no reports in the literature of transgenic plants in which starch-degrading enzymes have been repressed and which are inhibited in starch degradation at low temperatures. It is likely, therefore, that there is more redundancy in the pathway of starch degradation in cold-stored tubers than in leaves during the dark period.
The Structure of Starch Is Not Altered in Either Leaves or Tubers
Analysis of an algal D-enzyme mutant indicated that the enzyme was involved directly in amylopectin biosynthesis (Colleoni et al., 1999a, 1999b; Wattebled et al., 2003). One way to study this in the plants generated in this study is to analyze the size distribution of the constituent chains of starch following digestion with the debranching enzyme isoamylase. There is a polymodal distribution of chains within amylopectin, which can be analyzed by high-performance anion-exchange chromatography with pulsed-amperometric detection (HPAEC-PAD; Hizukuri, 1986). Alterations in this distribution would indicate that the amylopectin was altered. We tested this in starches from both leaves and tubers of the transgenic plants but could find no evidence of differences between them and the controls (data not shown). In addition, we measured the amylose contents of the starches using an iodine binding assay. Again, no differences were found between starches isolated from the control and transgenic plants (data not shown).
Leaves of the Transgenic Lines Accumulate Maltose
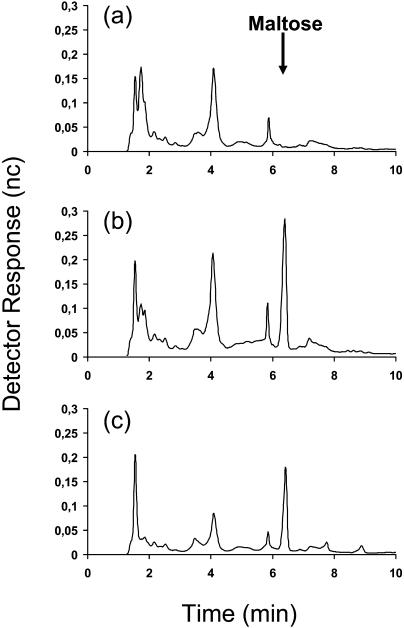
To try and identify the possible substrate for the stDPE2 isoform, malto-oligosaccharides were isolated from the control and the transgenic plants. This is because analysis of an Arabidopsis dpe1 mutant demonstrated that it accumulated malto-oligosaccharides, especially malto-triose, which is its preferred substrate (Critchley et al., 2001). The amounts (Table I) and types (Fig. 3) of malto-oligosaccharides present in leaves of the transgenic plants were determined in samples harvested either during the middle of the light or the middle of the dark period. It was found that leaves of the transgenic lines contained significantly (P ≤ 0.05) more malto-oligosaccharides than the control at both time points (Table I). During the light period, the increase in the transgenics was approximately 2-fold in comparison with the control, and during the dark period it was approximately 10-fold.
Table I.
Comparison of concentrations of malto-oligosaccharides in leaves of the control and transgenic plants taken at two time points
| Concentration (pmol/mm2)
|
||
|---|---|---|
| Middle of Light Period | Middle of Dark Period | |
| Control | 5.5 ± 0.6 | 7.9 ± 2.8 |
| Line 8 | 20.1 ± 0.2* | 97 ± 26.2* |
| Line 28 | 10.8 ± 1.0* | 150.7 ± 19.1* |
Different from control at 5% level, Student's t test (n = 8).
Figure 3.
HPAEC-PAD detection of malto-oligosaccharides in leaves. Samples were taken in the middle of the dark period. a, Untransformed control. b, Transgenic line 8. c, Transgenic line 28.
The malto-oligosaccharides within the samples were also analyzed by HPAEC-PAD to determine which accumulated in the transgenic lines (Fig. 3). It was found that maltose was present in both transgenic lines (Fig. 3, b and c) but was absent from the control (Fig. 3a). The amounts of no other malto-oligosaccharide were consistently altered in the transgenic lines. Maltose must be produced in plastids during starch breakdown, as it is the only product of β-amylase activity, and a β-amylase isoform has recently been demonstrated to be involved in starch degradation in potato leaves (Scheidig et al., 2002). It has always been presumed that maltose would be degraded by α-glucosidase isoforms; however, in the only study in which the activity of an α-glucosidase was repressed in plants, it was demonstrated that it was most likely involved in processing glycoproteins (Taylor et al., 2000). This does not mean, of course, that no α-glucosidases are involved in degrading maltose, but this remains to be proven.
stDPE2 Is Capable of Transferring Glucose from Maltose to Glycogen
It has been demonstrated recently in the dpe2 mutant of Arabidopsis that extracts made from these mutant plants lacked an activity catalyzing the transfer of glucose from maltose to glycogen, and it was assumed that this was the reaction catalyzed by atDPE2 (Chia et al., 2004). We wanted to test if stDPE2 was capable of this reaction and also to examine its activity with other substrates. To do so, we purified it from potato tubers to apparent homogeneity. We then incubated the purified protein with maltose and, separately, with various different potential donor molecules. These were malto-oligosaccharides with a degree of polymerization of 3, 4, 5, 6, or 7, potato amylopectin, or oyster muscle glycogen. After incubation, we separated any low molecular weight glucans by HPAEC. When we incubated stDPE2 with both maltose and glycogen, we found consistently that glucose was produced (data not shown). The incubations with other substrates did not lead to large consistent differences.
To quantify this activity, we incubated the purified protein with maltose and glycogen and measured glucose production enzymatically. We found that samples containing the protein released significant amounts of glucose, and this release was not detected when incubated with either maltose or glycogen alone. In addition, the amounts of glucose released increased linearly with time (data not shown). Based on this data, we calculated the specific activity of the protein, which is shown in Table II. We also tested whether the same activity could be found when we incubated stDPE2 with maltose and amylopectin. In this case, glucose was produced, but the activity was approximately 20-fold less than when glycogen was used as a substrate (Table II). As stDPE1 uses malto-oligosaccharides of many different degrees of polymerization as substrates, we decided also to test if stDPE2 could also metabolize malto-triose. When we incubated stDPE2 with malto-triose and glycogen, however, no glucose production was detected (Table II), indicating that stDPE2 uses exclusively maltose as a donor substrate.
Table II.
Activities of stDPE2 with various substrates, either as amount of glucose released or as amount incorporated into glycogen
| Substrates | Activity (nmol glucose released min−1 mg protein−1) |
|---|---|
| Maltose | 1.0 ± 0.5 |
| Amylopectin | 0.5 ± 0.1 |
| Glycogen | 1.5 ± 0.2 |
| Maltose and amylopectin | 5.5 ± 1.0 |
| Maltose and glycogen | 100.2 ± 1.0 |
| Malto-triose and glycogen | 2.5 ± 1.5 |
| Activity (glucose incorporated min−1 mg protein−1)
|
|
| Maltose and glycogen | 148 ± 40 |
To demonstrate that stDPE2 actually transfers glucose from maltose to glycogen, we repeated the experiment, but included [U-14C]maltose as a substrate. Following incubation, we precipitated the glycogen, washed it, and determined incorporation of radioactivity into it. It could be demonstrated that radioactivity became incorporated into the glycogen and that by using this data a similar specific activity for stDPE2 could be calculated, as was found when production of glucose was measured (Table II). This would be expected, as every transferase event should lead to the incorporation and liberation of equivalent amounts of glucose.
Glycogen itself may not be the acceptor substrate used by stDPE2 in the plant. Although glycogen is thought to be an intermediate of starch biosynthesis (Kossmann and Lloyd, 2000), it is not clear whether it is present during periods of starch degradation. Clearly starch itself could act as a substrate, but there may well be other glucans present that we have not tested that might act as the acceptor in vivo.
These data also help to explain how stDPE2 was identified in the E. coli screening system that we used. E. coli manufacture glycogen, and stDPE2 could have used that as the acceptor substrate for maltose, allowing the production of glucose.
The Activity of stDPE2, But Not Other Starch Degradative Enzymes, Is Decreased in Leaves of the Transgenic Lines
The activities of α-amylase, β-amylase, α-glucosidase, starch phosphorylase, stDPE1, and stDPE2 were determined in leaf samples of the control and transgenic lines (Table III). The only enzyme reduced in activity in the transgenic lines was stDPE2, the activity of which was undetectable. No significant difference was found in starch phosphorylase activity between leaves of the transgenic and control plants; however, the activities of all the other enzymes were significantly (P ≤ 0.05) increased in the transgenic lines in comparison with the control. It is not unusual for the activities of enzymes involved in starch degradation to increase in plants demonstrating a starch-excess phenotype (Zeeman et al., 1998; Critchley et al., 2001; Scheidig et al., 2002), and these data demonstrate that the decreased starch degradation found in the transgenic plants of this study is probably not due to some pleiotropic effect on another starch degradative enzyme.
Table III.
Comparison of maximum catalytic activities of starch-degrading enzymes in leaves of untransformed control and transgenic potato lines
| Activity (nmol p-nitrophenol released min−1 mg protein−1)
|
|||
|---|---|---|---|
| Enzyme | Control | Line 8 | Line 28 |
| α-amylase | 1.9 ± 0.6 | 7.7 ± 2.9* | 6.3 ± 1.4* |
| β-amylase | 40.7 ± 2.5 | 76.8 ± 19.6* | 66.2 ± 8.4* |
| Activity (nmol min−1 mg protein−1)
|
|||
| Starch phosphorylase | 11.3 ± 0.6 | 10.7 ± 0.9 | 14.4 ± 2.3 |
| stDPE1 | 30.4 ± 3.1 | 63.7 ± 6.8* | 61.1 ± 5.8* |
| α-glucosidase | 0.5 ± 0.1 | 1.2 ± 0.4* | 1.8 ± 0.2* |
| stDPE2 | 1.9 ± 0.8 | 0 ± 0* | 0 ± 0* |
Different from control at 5% level, Student's t test (n = 8).
The Protein Is Present in Potato Chloroplasts
Plants often contain multiple isoforms of starch degradative enzymes, some of which are extraplastidial. We decided, therefore, to examine the subcellular localization of stDPE2. To do this, we isolated chloroplasts from potato leaves using the method of Stitt et al. (1989). We measured the activities of the marker enzymes UDP-glucose pyrophosphorylase (cytoplasm) and NADP-dependent glyceraldehyde 3-phosphate dehydrogenase (plastid). It was found that the cytosolic marker enzyme activity was reduced approximately 40-fold in comparison with the plastid marker enzyme in the plastid enriched fraction. Amounts of both the homogenate and plastid fractions, equivalent to equal activities of the plastid marker enzyme, were separated by SDS-PAGE. The gel was blotted onto a nylon membrane and probed with the antiserum that recognizes stDPE2. As can be seen in Figure 1e, stDPE2 is present in both fractions in equal amounts, demonstrating the presence of stDPE2 in the plastid.
Many of the data presented in this study in potato are extremely similar to those found in the studies of the Arabidopsis dpe2 mutants (Lu and Sharkey, 2004; Chia et al., 2004). Those mutants showed both a maltose- and a starch-excess phenotype, something that we also found. The main difference between those studies and this one is that the Arabidopsis DPE2 isoform was demonstrated to be localized in the cytosol (Chia et al., 2004), while we identified the potato one as being present in the plastid. Irrespective of its subcellular localization, DPE2 is clearly involved in maltose metabolism, leading to an effect on starch degradation. It was argued in the paper of Chia et al. (2004) that an extraplastidial localization of atDPE2 could be postulated because of the recently discovered plastidial maltose transporter (Niittylä et al., 2004). Indeed, the data from an Arabidopsis mutant lacking this transporter supports the localization of atDPE2 in the cytosol, as those mutant plants accumulated very large amounts of maltose (Niittylä et al., 2004). Equally though, it is possible that stDPE2 is involved in starch degradation while being localized in the plastid. This is because the product of DPE2 that would be exported to the cytosol is glucose, and a plastidial glucose transporter has been identified previously (Weber et al., 2000). It is likely that potato also contains a maltose transporter, as there are potato expressed sequence tags that demonstrate similarity to the sequence of the Arabidopsis cDNA. Based on our data, however, we would propose that maltose catabolism proceeds in potato leaves for the most part through stDPE2 and the glucose transporter. Any maltose that is exported by the maltose transporter into the cytosol could be catabolized by, for example, an as yet unidentified, extraplastidial α-glucosidase isoform. It may be that this route via the maltose transporter plays a more important role in starch degradation in tubers. This might help to explain why we could find no difference in cold-induced starch degradation in the transgenic lines lacking stDPE2.
MATERIALS AND METHODS
Materials
Escherichia coli strains were obtained from the CGSC E. coli genetic stock center. Unless otherwise stated, all chemicals were purchased from Sigma (St. Louis). Columns used for protein purification were purchased from Amersham Biosciences (Hillerød, Denmark).
Isolation of the cDNA Showing a Maltose Positive Phenotype in the E. coli Strain TSM90
A cDNA library was transformed in the E. coli strain TSM90 (CGSC no. 6145) and grown overnight at 37°C on MacConkey agar supplemented with 2% (w/v) maltose and the appropriate antibiotic. Plasmid DNA was isolated from 20 colonies demonstrating a maltose positive phenotype using a commercially available kit (Qiagen USA, Valencia, CA). These plasmids were named pMDE1 to pMDE20.
DNA Sequencing
Sequencing of the inserts from the plasmids pMDE2, pMDE11, and pMDE18 was performed by a commercial company (AGOWA GmbH, Berlin).
Production of an RNAi Construct for Transformation of Potato Plants
An intron-less RNAi construct was produced in the plant transformation vector pBinARHyg by the following method. A SacI fragment of the cDNA pMDE11 was isolated from an agarose gel following electrophoresis, then blunted and ligated into the XhoI site of pMDE11, which also had been blunted. The orientation of the fragment was determined by restriction digests, and the new plasmid was named pMDE11RNAi. This plasmid was digested with the restriction enzyme XbaI, yielding a fragment of approximately 2,500 bp, which was ligated into the XbaI site of the binary vector pBinARHyg (Abel et al., 1996), producing the vector pBinMDE11RNAiHyg.
Production of Transgenic Plants and Growth Conditions
Potato (Solanum tuberosum) L. cv Désirée was obtained from Saatzucht Fritz Lange KG (Bad Schwartau, Germany). Plants in tissue culture were kept under a 16-h-light/8-h-dark regime on Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 2% (w/v) sucrose at 22°C. Potato plants were transformed with the plasmid pBinMDE11RNAiHyg by Agrobacterium tumefaciens-mediated gene transfer (Rocha-Sosa et al., 1989). Following transfer to soil, plants were kept in a glasshouse without artificial lighting.
Production of Antibody and Immunoblots
Antibodies were manufactured by a commercial organization (Eurogentec, Seraing, Belgium) using their double XP protocol. In outline, rabbits were injected with two peptides present in the stDPE2 primary protein sequence (TGAPPDYFDKNGQNW and VRGSGRFYPQKDLESG). Following bleeding, immunoglobulins that recognize the peptides were affinity purified by the company.
Protein extracts were denatured by heating at 95°C for 5 min in sample buffer (Laemmli, 1970) and separated by SDS-PAGE before being blotted onto nylon membranes (Hybond-P; Amersham Biosciences) using a semidry system. Membranes were incubated with the affinity-purified primary antibody followed by horseradish (Armoracia lapathifolia) peroxidase-conjugated goat anti-rabbit antiserum (DakoCytomation, Glostrup, Denmark). The blots were developed in a chemiluminescent system using 5-amino-2,3-dihydro-1,4-phthalazinedione, p-coumaric acid (both from ICN, Montreal), and hydrogen peroxide (Merck, Rahway, NJ) as substrates.
Isolation and Analysis of Malto-Oligosaccharides in Leaves of the Transgenic Plants
Malto-oligosaccharides were isolated from leaves of potato plants by the method of Critchley et al. (2001). The method was modified slightly to prepare samples for HPAEC analysis in that they were passed through 1 mL of a mixed-bed resin (Bio-Rad Laboratories, Hercules, CA) rather than Dowex before being eluted with water. Oligosaccharides were separated by HPAEC (Dionex, Sunnyvale, CA) and detected using a pulsed amperometric detector as described by Blennow et al. (1998).
Analysis of Starch and Soluble Sugars
Starch, sucrose, glucose, and fructose were determined according to the method of Müller-Röber et al. (1992).
Determination of Hydrolytic Activities in Leaves
For the determination of enzyme activities in plant material, frozen leaf discs were ground in extraction buffer containing 50 mm HEPES-KOH, pH 7.5, 5 mm MgCl2, 2 mm EDTA, 2 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, and 8.7% (v/v) glycerol. The samples were centrifuged for 10 min at 4°C and 20,000g, and the supernatant was immediately frozen in liquid nitrogen before being stored at −80°C. The protein content of the supernatant was determined by the method of Bradford (1976).
Activities of starch degradative enzymes were measured in these samples. α-amylase and β-amylase were determined by monitoring the degradation of malto-oligosaccharides linked to a p-nitrophenyl group by a glucosidic bond at the reducing end, using assay kits from Megazyme (Wicklow, Ireland) according to the method of Scheidig et al. (2002). Fifty microliters of plant extract were mixed with 225 μL of buffer containing 100 mm MES-KOH, pH 6.2, 1 mm EDTA, and 0.1% (v/v) β-mercaptoethanol. Assays were started by adding 25 μL of substrate and coupling enzyme (final concentration 0.4 mm oligosaccharide and 2.5 units of α-glucosidase) and stopped after 10 min at 40°C by adding 2.5 volumes of 1% (w/v) Trizma base. The activity was determined as liberated p-nitrophenolate detected spectrophotometrically at 410 nm. Starch phosphorylase activity was assayed in the direction of amylopectin degradation according to Zeeman et al. (1998). The activities of α-glucosidase and stDPE1 were determined in stopped assays based on the liberation of glucose from malto-triose or maltose, respectively, as described by Zeeman et al. (1998). StDPE2 activity was measured using the method of Chia et al. (2004).
Analysis of Starch
Starch was isolated from tubers by the method of Lloyd et al. (1999) and from leaves by the method of Ritte et al. (2000). Starch was dissolved at a concentration of 5 mg/mL in water by heating at 95°C. Ten units isoamylase (Megazyme) were added, and the samples were allowed to digest for at least 2 h at 37°C. The obtained free amylopectin unit chains were separated by HPAEC-PAD as described by Blennow et al. (1998).
Isolation of Chloroplasts
Chloroplasts were isolated from leaves of 4-week-old potato plants by the method of Stitt et al. (1989). Marker enzymes in the homogenate and plastid fractions were measured by the methods of Zrenner et al. (1995; UDP-glucose pyrophosphorylase) or Stitt et al. (1989; NADP-dependent glyceraldehyde 3-phosphate dehydrogenase).
Purification of stDPE2
Liquid chromatography was performed at 20°C using a ÄKTAexplorer 10 system from Amersham Biosciences. All other procedures were performed at 4°C. Columns used for D-enzyme purification were purchased from Amersham Biosciences.
Fifty grams of potato tubers were homogenized in 125 mL of ice-cold extraction buffer (50 mm HEPES-KOH, pH 7.5, 5 mm EDTA, 5 mm dithiothreitol, 5 mm MgCl2, 2 mm ɛ-aminocaproic acid, 0.5 mm phenylmethylsulfonyl fluoride, and 0.1% (w/v) Triton X-100). The slurry was filtered through two layers of nylon mesh and centrifuged at 2,000g for 10 min. Ammonium sulfate was added to the supernatant, and the protein fraction that precipitated between 25% to 50% (w/v) ammonium sulfate was isolated by centrifugation at 20,000g for 60 min. The precipitated proteins were dissolved in 50 mL of the sample buffer described above containing 1 m ammonium sulfate.
The following buffers were used in the enzyme purification: buffer A, 50 mm Tris-HCl, pH 8.0, 1 mm EDTA, 2 mm 2-mercaptoethanol, and 1 m ammonium sulfate; buffer B, same as buffer A but without ammonium sulfate; and buffer C, same as buffer B, but containing 0.5 m KCl.
The proteins were chromatographed by hydrophobic interactions using a RESOURCE ETH column (0.96-mL matrix) equilibrated in buffer A. After loading, a linear gradient (100%–0% in 25 column volumes [CV]) of buffer B was applied at a flow rate of 2 mL min−1. Fractions of 1 mL were collected and subsequently immuno-assayed for presence of stDPE2. Fractions containing stDPE2 were pooled and dialyzed against buffer B. The dialyzed protein was chromatographed on a RESOURCE Q (0.96 mL) column equilibrated in buffer B. The column was washed in 10 CV of buffer B before a linear gradient (0%–100% in 25 CV) of buffer C was applied at a flow rate of 2 mL min−1. Fractions of 1 mL were collected and immuno-assayed for presence of stDPE2. Fractions containing stDPE2 were pooled and dialyzed against buffer B. The dialyzed protein was chromatographed on a Mono Q (0.98 mL) column equilibrated in buffer B. The column was washed in 3 CV of buffer B before a linear gradient (0%–100% in 25 CV) of buffer C was applied at a flow rate of 0.5 mL min−1. Fractions of 0.5 mL were collected and immuno-assayed for presence of stDPE2. These fractions were pooled, aliquoted, and frozen at −80°C until use.
Determination of stDPE2 Activity
stDPE2 protein was incubated in 50 mm MOPS, pH 6.8, 30 mm maltose, and individual potential substrates. These were 5 mm malto-triose, maltoheptaose, maltopentaose maltohexaose, maltoheptaose, or 1.5% (w/v) oyster glycogen or potato amylopectin. Following incubation, the reactions were stopped by heating for 2 min at 90°C.
HPAEC Analysis of the Samples
Three volumes of ethanol were added to the samples containing either glycogen or amylopectin, and the precipitated polysaccharides were removed by centrifugation. The sugars in 100-μL samples of all of these were separated by HPAEC, as described by Blennow et al. (1998), with detection via a PAD.
Detection of Glucose Production
stDPE2 was incubated as described above, although as a control maltose was omitted from some samples. As blanks, stDPE2 protein, which had been heated at 95°C for 5 min, was used. The amount of glucose in 50-μL samples was determined enzymatically by the method of Müller-Röber et al. (1992). Activities were calculated as amount of glucose released in the presence of stDPE2, subtracting the amount released in the presence of the denatured protein.
Incorporation of [14C]maltose into Glycogen
stDPE2 protein was incubated as described above with 1.5% (w/v) oyster glycogen and 92.5 mBq mol−1 [14C]maltose (Amersham). As blanks, stDPE2 protein, which had been heated at 95°C for 5 min, was used. Following incubation, glycogen was precipitated from 50-μL samples by the addition of three volumes of 75% (v/v) methanol and 1% (w/v) KCl. The precipitate was harvested by centrifugation at 20,000g for 10 min and then washed with 1 mL 75% (v/v) methanol and 1% (w/v) KCl, before being dissolved in 500 μL of water. Five milliliters of liquid scintillation fluid was added to this, and the amount of radioactivity determined. Activities were calculated as amount of radioactivity incorporated in the presence of stDPE2, subtracting the amount incorporated in the presence of the denatured protein.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY510449.
Acknowledgments
We thank Per Lassen Nielsen for assistance with the HPAEC-PAD and Romy Baran with the potato transformation.
This work was supported by PlantTec Biotechnology GmbH, Potsdam, Germany (grant to J.R.L.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.038026.
References
- Abel GWR, Springer F, Willmitzer L, Kossmann J (1996) Cloning and functional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato (Solanum tuberosum L.). Plant J 10: 981–991 [DOI] [PubMed] [Google Scholar]
- Blennow A, Bay-Schmidt AM, Wischmann B, Olsen CE, Møller BL (1998) The degree of starch phosphorylation is related to the chain length distribution of the neutral and the phosphorylated chains of amylopectin. Carbohydr Res 307: 45–54 [Google Scholar]
- Boos W, Schuman H (1998) Maltose/maltodextrin system of Escherichia coli: transport, metabolism and regulation. Microbiol Mol Biol Rev 62: 204–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 243–254 [DOI] [PubMed] [Google Scholar]
- Chia T, Thorneycroft D, Chapple A, Messerli G, Chen J, Zeeman S, Smith SM, Smith AM (2004) A cytosolic glycosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J 36: 853–863 [DOI] [PubMed] [Google Scholar]
- Colleoni C, Dauvilée D, Mouille G, Buléon A, Gallant D, Bouchert B, Morell M, Samuel M,, Delrue B, d'Hulst C, Bliard C, Nuzillard J-M, et al (1999. a) Genetic and biochemical evidence for the involvement of α-1,4 glucanotransferase in amylopectin biosynthesis. Plant Physiol 120: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni C, Dauvillée D, Mouille G, Morell M, Samuel M, Slomiany M-C, Liénard L, Wattebled F, d'Hulst C, Ball S (1999. b) Biochemical characterization of the Chlamydomonas reinhardtii α-1,4 glucanotransferase supports a direct function in amylopectin biosynthesis. Plant Physiol 120: 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM (2001) A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J 26: 89–100 [DOI] [PubMed] [Google Scholar]
- Ehrmann MA, Vogel RF (1998) Maltose metabolism of Lactobacillus sanfranciscensis: cloning and heterologous expression of the key enzymes, maltose phosphorylase and phosphoglucomutase. FEMS Microbiol Lett 169: 81–86 [DOI] [PubMed] [Google Scholar]
- Hizukuri S (1986) Polymodal distribution of the chain lengths of amylopectins, and its significance. Carbohydr Res 147: 342–347 [Google Scholar]
- Jones G, Whelan WJ (1969) The action pattern of D-enzyme, a transmaltodextrinylase from potato. Carbohydr Res 9: 483–490 [Google Scholar]
- Kossmann J, Lloyd JR (2000) Understanding and influencing starch biochemistry. CRC Crit Rev Plant Sci 19: 171–226 [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lloyd JR, Springer F, Buléon A, Müller-Röber B, Willmitzer L, Kossmann J (1999) The influence of alterations in ADP glucose pyrophosphorylase activities on starch structure and composition in potato tubers. Planta 209: 230–238 [DOI] [PubMed] [Google Scholar]
- Lu Y, Sharkey TD (2004) The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218: 466–473 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res 30: 281–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Röber B, Sonnewald U, Willmitzer L (1992) Inhibition of the ADP-glucose-pyrophosphorylase in transgenic potato tubers leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. EMBO J 11: 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Thurgau H (1882) Ueber zuckeranhäufung in pflanzentheilen in folge niederer temperatur. Landw Jahrb Schweiz 11: 751–828 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Niittylä T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC (2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 303: 87–89 [DOI] [PubMed] [Google Scholar]
- Ritte G, Lorberth R, Steup M (2000) Reversible binding of the starch-related R1 protein to the surface of transitory starch granules. Plant J 21: 387–391 [DOI] [PubMed] [Google Scholar]
- Rocha-Sosa M, Sonnewald U, Frommer W, Stratmann M, Schell J, Willmitzer L (1989) Both developmental and metabolic signals activate the promoter of the class I patatin gene. EMBO J 66: 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidig A, Fröhlich A, Schulze S, Lloyd JR, Kossmann J (2002) Downregulation of a chloroplast-targeted β-amylase leads to a starch-excess phenotype in leaves. Plant J 30: 581–591 [DOI] [PubMed] [Google Scholar]
- Stitt M, Lilley RM, Gerhardt R, Heldt HW (1989) Metabolite levels in specific cells and subcellular compartments of plant leaves. Methods Enzymol 174: 518–552 [Google Scholar]
- Takaha T, Critchley J, Okada S, Smith SM (1998) Normal starch content and composition in tubers of antisense potato plants lacking D-enzyme (4-α-glucanotransferase). Planta 205: 445–451 [Google Scholar]
- Takaha T, Yanase M, Okada S, Smith SM (1993) Disproportionating enzyme (4-alpha-glucanotransferase; EC 2.4.1.25) of potato. Purification, molecular cloning, and potential role in starch metabolism. J Biol Chem 268: 1391–1396 [PubMed] [Google Scholar]
- Taylor MA, Ross HA, McRea D, Stewart D, Roberts I, Duncan G, Wright F, Millam S, Davies HV (2000) A potato α-glucosidase gene encodes a glycoprotein-processing α-glucosidase II like activity. Demonstration of enzyme activity and effects of down-regulation in transgenic plants. Plant J 24: 305–316 [DOI] [PubMed] [Google Scholar]
- Wattebled F, Ral J-P, Dauvillée D, Myers A, James M, Schlichting R, Giersch C, Ball S, D'Hulst C (2003) STA11, a Chlamydomonas reinhardtii locus required for normal starch granule biogenesis, encodes disproportionating enzyme. Further evidence for a function of α-1,4 glucanotransferases during starch granule biosynthesis in green algae. Plant Physiol 132: 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Servaites JC, Geiger DR, Kofler H, Hille D, Gröner F, Hebbeker U, Flügge U-I (2000) Identification, purification, and molecular cloning of a putative plastidic glucose transporter. Plant Cell 12: 787–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Northrop F, Smith AM, Rees T (1998) A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J 15: 357–365 [DOI] [PubMed] [Google Scholar]
- Zrenner R, Salanoubat M, Willmitzer L, Sonnewald U (1995) Evidence for the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J 7: 97–107 [DOI] [PubMed] [Google Scholar]