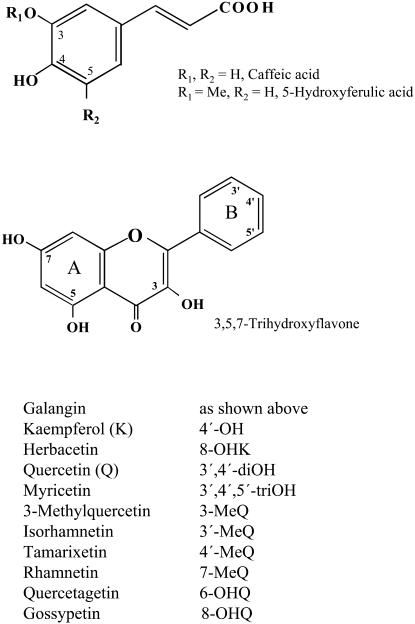
Abstract
Serratula tinctoria (Asteraceae) accumulates mainly 3,3′-dimethylquercetin and small amounts of 3-methylquercetin as an intermediate. The fact that 3-methylquercetin rarely accumulates in plants in significant amounts, and given its important role as an antiviral and antiinflammatory agent that accumulates in response to stress conditions, prompted us to purify and characterize the enzyme involved in its methylation. The flavonol 3-O-methyltransferase (3-OMT) was partially purified by ammonium sulfate precipitation and successive chromatography on Superose-12, Mono-Q, and adenosine-agarose affinity columns, resulting in a 194-fold increase of its specific activity. The enzyme protein exhibited an expressed specificity for the methylation of position 3 of the flavonol, quercetin, although it also utilized kaempferol, myricetin, and some monomethyl flavonols as substrates. It exhibited a pH optimum of 7.6, a pI of 6.0, and an apparent molecular mass of 31 kD. Its Km values for quercetin as the substrate and S-adenosyl-l-Met (AdoMet) as the cosubstrate were 12 and 45 μm, respectively. The 3-OMT had no requirement for Mg2+, but was severely inhibited by p-chloromercuribenzoate, suggesting the requirement for SH groups for catalytic activity. Quercetin methylation was competitively inhibited by S-adenosyl-l-homo-Cys with respect to the cosubstrate AdoMet, and followed a sequential bi-bi reaction mechanism, where AdoMet was the first to bind and S-adenosyl-l-homo-Cys was released last. In-gel trypsin digestion of the purified protein yielded several peptides, two of which exhibited strong amino acid sequence homology, upon protein identification, to a number of previously identified Group II plant OMTs. The availability of peptide sequences will allow the design of specific nucleotide probes for future cloning of the gene encoding this novel enzyme for its use in metabolic engineering.
Flavonoid compounds constitute one of the most ubiquitous groups of natural plant products. They exhibit a wide range of functions and play important roles in the biochemistry, physiology, and ecology of plants. These include their contribution to flower color, protection against UV radiation and pathogenic organisms, promotion of pollen germination and pollen fertility, and activation of Rhizobium nodulation genes. They also act as growth regulators, enzyme inhibitors, insect antifeedants, and antioxidants, and are of potential benefit to human health (Bohm, 1998, and references therein). Flavonoids owe their structural biodiversity to a number of enzyme-catalyzed substitution reactions (Ibrahim and Anzellotti, 2003). Of these, enzymatic O-methylation, which is catalyzed by a family of S-adenosyl-l-Met (AdoMet)-dependent O-methyltransferases (OMTs; Ibrahim and Muzac, 2000), involves the transfer of the methyl group of AdoMet to the hydroxyl groups of an acceptor molecule, with the concomitant formation of the corresponding methyl ether derivative and S-adenosyl-l-homo-Cys (AdoHcy) as products. O-Methylation of flavonoids neutralizes the reactivity of their hydroxyl groups and alters their solubility and, hence, their intracellular compartmentation.
Flavonoid OMTs are substrate-specific, position-oriented enzymes, as was shown with a number of distinct enzymes catalyzing the stepwise O-methylation in Chrysosplenium americanum of the pentahydroxyflavone, quercetin (Q) → 3-methylquercetin (3-MeQ) → 3,7-diMeQ → 3,7,4′-triMeQ. After hydroxylation of the latter intermediate at positions 6 and/or 2′, 3,7,4′-triMeQ is further methylated to 3,7,4′,5′-tetraMeQ or to 3,6,7,2′,4′-pentaMe quercetagetin (Ibrahim et al., 1987), both of which are among the major flavonoid metabolites that accumulate in this plant (Collins et al., 1981). Stepwise O-methylation of Q by distinct OMTs has also been reported in apple (Malus domestica) cell cultures (Macheix and Ibrahim, 1984) and in spinach (Spinacia oleracea) leaves (Thresh and Ibrahim, 1985). In all these examples, O-methylation at position 3 constitutes the first committed step of the methylation sequence. This may explain the reason why 3-O-methyl flavonols rarely accumulate in plants, since they serve as intermediates in the biosynthetic pathway of partially/highly methylated flavonoids. However, 3-MeQ has been detected in small amounts in a number of plant species including Begonia, Centauria, Greyia, Nicotiana, and Serratula (for review, see Gottlieb, 1975).
3-O-Methylation of Q confers some distinct properties to this compound. In addition to being an antiinflammatory and antiviral agent (Malhotra et al., 1996; Middleton and Kandaswami, 1993), 3-MeQ selectively inhibits poliovirus RNA replication (Castrillo and Carrasco, 1987) and more selectively, phosphodiesterase Subtype 3 (Ko et al., 2003). Furthermore, 3-MeQ has recently been reported to accumulate in tobacco (Nicotiana tabacum) leaf trichomes, together with other methylated flavonols, in response to wounding stress and herbivory (Roda et al., 2003). With the exception of the cDNA encoding the 3-O-methylation of flavonols, several flavonoid OMT cDNA clones have been isolated and characterized, including two for chalcones, one for a flavone, two for isoflavones, and three for flavonols (Ibrahim and Muzac, 2000, and references therein).
Serratula tinctoria accumulates mainly 3,3′-dimethylquercetin (3,3′-diMeQ) and small amounts of 3-MeQ as an intermediate (Fig. 1), suggesting the existence in this plant of Q 3-OMT and 3-MeQ 3′-OMT enzyme proteins. Besides its high content of ecdysteroids, these methylated flavonols contribute to the yellow color of the root sap that is used as a folkloric dye, for which it became known as Dyer's savory (Corio-Costet et al., 1991).
Figure 1.
Chemical structures of the substrates used in this study.
We describe in this paper the characterization of the methylated flavonols of S. tinctoria, the partial purification of the flavonol 3-OMT and its physico-chemical properties, as well as the acquisition of internal amino acid sequence information for future cloning of its gene. In spite of the ubiquitous occurrence of flavonols in plants, especially Q (Wollenweber and Dietz, 1981), 3-MeQ rarely accumulates in significant amounts. However, given its role as an antiviral and antiinflammatory agent, as well as being a phytoanticipin (Roda et al., 2003), it would be desirable to bioengineer its constitutive expression in transgenic plants.
The choice of Serratula as the experimental plant material was dictated by the availability of its seeds and the fact that it contains only two flavonol OMTs, in addition to the lignin monomer OMT, as compared with C. americanum, a semi-aquatic weed that contains at least five flavonol OMTs with similar physico-chemical properties (Ibrahim et al., 1987) rendering the purification of any of them an extremely difficult task. Because of their abundance in Serratula roots, the methylated flavonols were isolated and identified from these tissues. On the other hand, the low growth rate of root relative to leaf tissues, and the fact that both purification and characterization of the flavonol 3-OMT require significant amounts of tissue, prompted us to use the leaves for enzyme work, after verifying the similarity of the flavonoid methylation pattern in both organs.
RESULTS
Characterization of Serratula Flavonoids
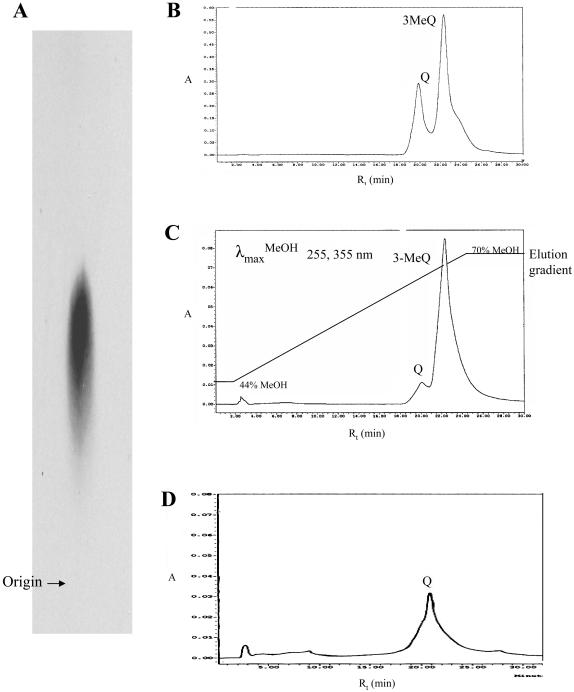
Chromatography of root methanolic extracts on preparative cellulose thin-layer plates, followed by gradient elution from HPLC, revealed two UV-absorbing (dark purple) compounds, I and II, with Rf values (on thin-layer chromatography [TLC]) of 0.72 and 0.76 and Rt values (on HPLC) of 23.0 and 28.0 min, respectively. The mobility of both compounds was not altered after acid hydrolysis, indicating that they were not conjugated.
Compound I exhibited the following UV absorption maxima after the addition of spectral shift reagents: λmaxMeOH (nm) 255, 355; BdII/BdI 0.77; λmaxMeOH+HCl255, 360; λmaxMeOH+NaOH 270, 330,412; λmaxMeOH+NaOAc 275, 390; λmaxMeOH+NaOAc+H3BO3 265, 380; λmaxMeOH+AlCl3 268, 415; λmaxMeOH+AlCl3+HCl 265, 345, 360, 400. It comigrated on TLC and coeluted from HPLC with an authentic sample of 3-MeQ. It also exhibited the same UV fluorescence and UV absorption maxima as those of the reference compound, both in the absence and presence of spectral shift reagents. These characteristics putatively identify compound I as 3-MeQ.
Both the Rf and Rt values of compound II were indicative of a dimethylated flavonol when compared with an authentic sample of 3,3′-diMeQ as a reference compound. The fact that it appeared as a dark purple spot under UV light, which turned yellow upon exposure to ammonia vapors, indicates that position 3 is substituted. It exhibited the following UV maxima: λmaxMeOH (nm) 265, 350; BdII/BdI 0.85; λmaxMeOH+HCl265, 355; λmaxMeOH+NaOH 275,325, 400; λmaxMeOH+NaOAc 275, 390; λmaxMeOH+NaOAc+H3BO3 265, 355; λmaxMeOH+AlCl3 270, 355; λmaxMeOH+AlCl3+HCl 275, 345 355, 395. These spectral properties indicate the presence of free hydroxyl groups at positions 5, 7, and 4′, whereas the AlCl3 shift suggests substitution at either the 3′- or 4′-positions (Harborne, 1967; Markham, 1982). Since the 4′-OH is free, then the second methyl substitution must be at position 3′, which putatively identifies compound II as 3,3′-diMeQ. In addition, it cochromatographed on TLC and coeluted from HPLC with a reference sample of 3,3′-diMeQ. The latter compound exhibits distinct fluoresence characteristics from the 3,4′-diMe isomer after spraying with the diphenylborinate reagent. In addition, 3,3′-diMeQ has been reported to occur in a number of plant species (Wollenweber and Dietz, 1981).
Flavonol 3-OMT Activity of Serratula Leaves
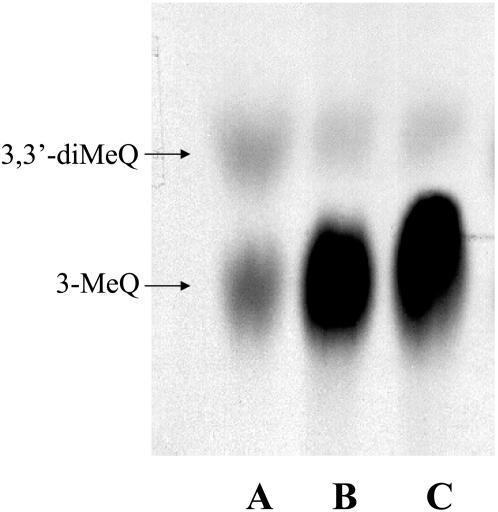
Using Q as substrate, preliminary investigation of the flavonol 3-OMT activity of both root and leaf crude extracts resulted in specific activity values of 0.884 ± 0.15 and 0.187 ± 0.02 pkat g−1, respectively. However, both organs exhibited the same pattern of [14C]labeled enzyme reaction product formation when chromatographed on TLC (Fig. 2). As expected, Q gave rise to only two labeled methylated enzyme reaction products, 3-MeQ and 3,3′-diMeQ, which cochromatographed on TLC with nonlabeled reference samples, thus indicating the existence in both leaves and roots of Serratula of the Q 3-OMT and 3-MeQ 3′-OMT enzyme proteins.
Figure 2.
Autoradiograph of the chromatographed enzyme reaction products of Serratula leaf and root cell-free extracts assayed with Q as substrate and [14C]AdoMet as cosubstrate. Lane A, Control assay of leaf extract without added Q, indicating the presence of endogenous substrates. Lanes B and C, Complete assays of leaf and root extracts, respectively, showing labeled 3-MeQ and 3,3′-diMeQ as reaction products. Label intensity of the leaf reaction products (lane B) does not reflect the leaf enzyme activity relative to roots, since several assay products were applied to ensure visibility of radioactivity.
Purification of the Flavonol 3-O-Methyltransferase
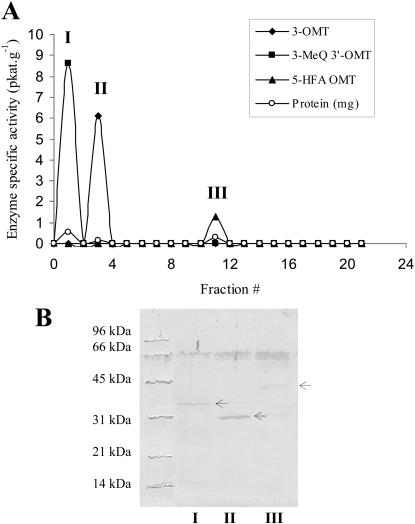
The flavonol 3-OMT was partially purified from Serratula leaves by ammonium sulfate precipitation and successive chromatography on Superose-12, Mono Q, and adenosine-agarose affinity columns. This resulted in a 194-fold purification with a specific activity of 13.8 pkat g−1 and 0.21% recovery (Table I). Chromatography of the enzymatically active Superose-12 fraction on a Mono-Q column allowed the separation of three distinct OMT activities: the flavonol 3-OMT (Fig. 3A, II), 3-MeQ 3′-OMT (Fig. 3A, I), and 5-hydroxyferulic acid (5-HFA) OMT (Fig. 3A, III), with preference for their respective substrates, Q, 3-MeQ, and 5-HFA (Table II). These enzyme activities were immunorecognized by an anti-flavonol 3′-OMT antibody and exhibited distinct Mr values (Fig. 3B, I–III).
Table I.
Partial purification of the flavonol 3-OMT from Serratula leavesa
| Purification Step | Total Protein | Specific Activity | Total Activity | Purification | Recovery |
|---|---|---|---|---|---|
| mg | pkat g−1 | pkat | -fold | % | |
| (NH4)2SO4 | 56 | 0.071 | 0.004 | - | 100 |
| Superose-12 | 20 | 0.81 | 0.016 | 11.5 | 35.7 |
| Mono-Q | 4 | 3.26 | 0.013 | 46 | 7.2 |
| Adenosine-agarose | 0.12 | 13.8 | 0.0017 | 194 | 0.21 |
Leaves (approximately 20 g) were extracted with Pi buffer, the homogenate filtered and centrifuged, and the protein that precipitated between 30% and 70% ammonium sulfate saturation was desalted on PD-10 column before successive chromatography on Superose-12, Mono-Q, and adenosine-agarose columns.
Enzyme assays were carried out using Q as the substrate, as described in the “Materials and Methods” section.
Figure 3.
A, Elution profile of the different OMT activity peaks separated on a Mono-Q column. Peaks: I, 3-MeQ 3′-OMT activity; II, 3-OMT activity; III, 5-HFA OMT activity (see Table II for the substrates used). B, Western-blot analysis of the protein extracts corresponding to OMT activities in A (I, II, and III), showing their apparent Mr values, as revealed by the alkaline phosphatase immuno-staining.
Table II.
Characteristics of the OMT activities separated on Mono-Q chromatography
The partially purified protein was applied to a Mono-Q column and eluted with a linear (50–500 mm) NaCl gradient in buffer C, and 2-mL fractions were collected and assayed for 3-OMT, 3-MeQ 3′-OMT, and 5-HFA OMT activities using Q, 3-MeQ, and 5-HFA as substrates, respectively, as described in the “Materials and Methods” section.
As eluted from the Mono-Q column (Fig. 3A).
Represents the only accepted substrate for each designated fraction (Fig. 1).
See Figure 3B.
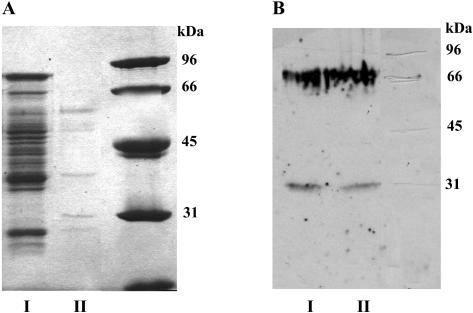
The 3-OMT activity was further purified by affinity chromatography on an adenosine-agarose column. SDS-PAGE of the original Mono-Q (Fig. 4A, I) and the adenosine-agarose (Fig. 4A, II) eluates showed several Coomassie Brilliant Blue-stained protein bands, one of which was immunorecognized by the anti-flavonol 3′-OMT antibody and migrated at an Mr of 31,000 (Fig. 4B, I and II).
Figure 4.
SDS-PAGE of the flavonol 3-OMT before and after elution from the adenosine-agarose affinity column. A, Coomassie Brilliant Blue-stained gels: I, Mono-Q eluate (see Fig. 3A, II) and II, Adenosine-agarose eluate. B, Western blot of A, showing an immunoreactive band that migrates at 31 kD, as revealed by the horseradish peroxidaseimmuno-staining. The bands observed at 66 kD are artifacts arising from irreversible binding of β-mercaptoethanol with an extraneous protein.
Identification of the Enzyme Reaction Product
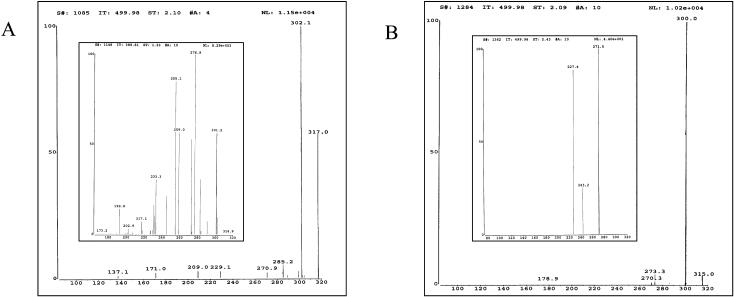
The enzymatic methylation of Q as the substrate and [14CH3]AdoMet as the cosubstrate gave rise to a single labeled reaction product (Fig. 5A) that was identified as 3-MeQ by cochromatography on TLC, comparison of its Rt value on HPLC, and the UV spectral maxima of the nonlabeled product with an authentic sample of 3-MeQ (Fig. 5, B–D).
Figure 5.
Identification of the flavonol 3-OMT enzyme reaction product using Q as the substrate, [14CH3]AdoMet as the cosubstrate, and the Mono-Q fraction II (Fig. 3A) as the enzyme source. A, Autoradiogram of the enzyme reaction product cochromatographed with a reference sample of 3-MeQ on cellulose TLC as desribed in the “Materials and Methods” section. B, HPLC elution profile of reference samples of Q and 3-MeQ. C, HPLC elution profile of the enzyme reaction product showing the residual substrate Q and the product 3-MeQ. The latter exhibits the same retention time and spectral values as the reference compound. D, HPLC elution profile of the reaction products of an enzyme assay lacking the cosubstrate, AdoMet. Notice the lack of substrate utilization or product formation.
Mass spectrometry (MS) analysis of the nonlabeled enzyme reaction product, using the positive and negative atmospheric pressure chemical ionization (gave molecular ions of m/z 317.0 [M+H]+ and 315 [M−H]−, which correspond to the molecular mass of 3-MeQ (Fig. 6, A and B, respectively). The reference compound exhibited similar molecular ions m/z 317 (+) and 315.2 (−), respectively (profiles not shown). However, it was not possible to determine the position of methylation on the flavonoid ring, since the methyl group lost upon ionization. This is demonstrated by the presence of the molecular ions m/z 302.1 [M+H−CH3]+ (Fig. 6A) and 300.0 [M−H−CH3]− (Fig. 6B) for the reaction product, and m/z 302.1 [M+H−CH3]+ and 300.1 [M−H−CH3]− for the reference compound, respectively (profiles not shown). Several other molecular ions in both samples represent fragments resulting from the loss of one or more H, CO, HCO, or CO2.
Figure 6.
Finnigan LCQ-APCI MS analysis of the enzyme reaction product, 3-MeQ in the positive (A) and negative (B) modes, showing molecular ions m/z of 317.2 (+) and 315 (−), respectively. Inset in A and B represents the molecular ions of fragments resulting from the loss of one or more H, CO, CHO, or CO2.
Substrate Specificity of Serratula OMT
The substrate specificity of the flavonol 3-OMT was studied using the enzyme fraction II of the Mono Q column (Fig. 3A, II). It was tested against 11 flavonol aglycones with different substitution patterns, as well as two flavanones (naringenin and eriodyctiol), two flavones (apigenin and luteolin), and three phenylpropanoids (caffeic acid, caffeoyl CoA, and 5-HFA). The enzyme exhibited an expressed specificity for position 3 of Q as confirmed by cochromatography of the enzyme reaction product on TLC and coelution from HPLC with a reference sample of 3-MeQ (Fig. 5, A–D), as well as by MS analysis (Fig. 6, A and B ). Such position specificity is further confirmed by the fact that 3-MeQ was not accepted as substrate for further methylation, since position 3 is already substituted. The enzyme also accepted other flavonols as substrates, especially kaempferol > isorhamnetin ≈ tamarixetin > galangin, with enzyme activities ranging from 60% to 90% relative to that of Q (Table III). The products of isorhamnetin and tamarixetin assays cochromatographed on TLC with reference samples of 3,3′- and 3,4′-diMeQ, respectively, indicating their O-methylation at position 3. On the other hand, quercetagetin, gossypetin, and herbacetin were methylated to a much lower extent, whereas rhamnetin was a very poor methyl acceptor. The relatively low methyl acceptor ability of the latter substrates may be due to their modified A-ring substitution beyond the usual 5,7-dihydroxy substitution. The fact that none of the flavanones, flavones, or phenylpropanoids tested exhibited any methylating activity (Table III) indicates the specificity of this enzyme for flavonols as methyl acceptors. Furthermore, caffeoyl CoA was not accepted as substrate when tested at concentrations between 10 and 100 μm, thus confirming that this enzyme is not a Group I OMT, in spite of its relatively small molecular size.
Table III.
| Substrate | Relative Activity | Km value | Vmax | Vmax/Km |
|---|---|---|---|---|
| %c | μm | pkat mg−1 | ||
| Q | 100 | 12 | 61 | 5.08 |
| Kaempferol | 93 | 13 | 169 | 13 |
| Myricetin | 46 | 7 | 15 | 2.14 |
The enzyme protein fraction II eluted from the Mono-Q column (Fig. 3A) was assayed against the indicated substrates at a concentration of 50 μm as described in “Materials and Methods”.
See Figure 1 for structures of the substrates used.
No OMT activity was detected with either catechol, caffeic acid, caffeoyl CoA, or 3-MeQ, nor with naringenin, eriodyctiol, apigenin, or luteolin when used as substrates. Other substrates were accepted to varying degrees (% of control); these are: isorhamnetin (88%), tamarixetin (85%), herbacetin (33%), rhamnetin (13%), galangin (60%), quercetagetin (39%), and gossypetin (38%).
Equivalent to a specific activity of 3.26 pkat g−1.
Due to the unavailability of reference compounds other than the methylated Q derivatives, it was not possible to confirm the position of enzymatic methylation of the other flavonols tested, which can only be achieved by NMR analysis, which requires important amounts of the products. However, the fact that (1) enzymatic methylation of Q gave only one reaction product that was rigorously identified (Fig. 5), (2) 3-MeQ was not accepted as substrate for further methylation as did isorhamnetin and tamarixetin, and (3) none of the flavanones or flavones tested acted as methyl acceptors possible due to the lack of a 3-OH group, indicate the specificity of the Serratula OMT for position 3 of flavonols. Nevertheless, the 3-OMT activity is more susceptible to any modification of A-ring, than to B-ring, substitution of the flavonol substrate, as indicated by the low enzyme activity obtained with rhamnetin, quercetagetin, gossypetin, or herbacetin, as substrates (Table III).
Physicochemical Properties
The pH optimum of Q methyation was 7.6 in Tris-HCl buffer. An increase or a decrease of 1.0 pH unit resulted in approximately 60% reduction in catalytic activity. The pI value of the 3-OMT was 6.0 as determined by chromatography on a Mono P chromatofocusing column (data not shown).
Q 3-OMT was completely (100%) inhibited in the presence of 1 mm p-chloromercuribenzoate, as compared with the controls, and by 90% in presence of 0.1 mm of p-chloromercuribenzoate. Only 16% of the enzymatic activity was restored after the addition of 140 mm of β-mercaptoethanol, indicating the requirement of SH groups for catalytic activity.
The apparent molecular mass of the 3-OMT was estimated to be 35 kD ±2.1 after elution from a calibrated Superose-12 column, and an apparent Mr of 31,000 on SDS-PAGE (Fig. 4B), indicating that the enzyme is a monomeric protein.
Kinetic Analysis
The kinetic profiles of the flavonol 3-OMT were determined using Q as the substrate and the Mono Q-purified enzyme fraction II (Fig. 3A). The formation of 3-MeQ was linear with time and the amount of protein added. The replots of the kinetic data gave Km values of 12 μm and 45 μm for Q and AdoMet, respectively (Table IV) and a Vmax value of 61 pkat mg−1 for the conversion of Q to 3-MeQ by the partially purified enzyme.
Table IV.
Kinetic parameters for Serratula flavonol 3-O-methyltransferase
| Substrate
|
||||
|---|---|---|---|---|
| a | b | Parameter Valuea | Standard Deviation | |
| AdoMet | Q | Ka | 45 μm | 0.0001 |
| Kia | 2.2 μm | - | ||
| Ka/Kia | 20 | - | ||
| Kb | 12 μm | 0.259 | ||
| Kip | 21 μm | - | ||
| Kiq | 3.3 μm | - | ||
| Vmax(b) | 61 pkat mg−1 | 0.008 | ||
| Kcat/Km | 15.8 μm−1 s−1 | - | ||
Kip and Kiq represent the inhibition constants for the reaction products, 3-MeQ and AdoHcy, respectively.
Initial Velocity Patterns
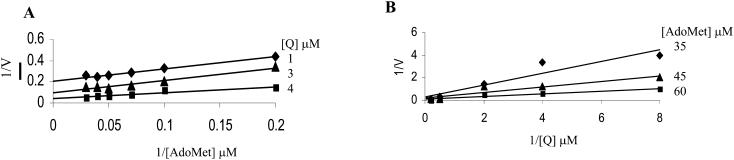
These were obtained by varying the concentration of one substrate in the presence of different fixed concentrations of the second substrate. Accordingly, both Lineweaver-Burk plots of various concentrations of AdoMet at different fixed concentrations of Q (Fig. 7A), and of various concentrations of Q at different fixed concentrations of AdoMet (Fig. 7B) both resulted in intersecting patterns.
Figure 7.
Initial velocity patterns of the flavonol 3-OMT. A, 1/v versus 1/AdoMet at various fixed concentrations of Q. B, 1/v versus 1/Q at various fixed concentrations of AdoMet.
Product Inhibition Patterns
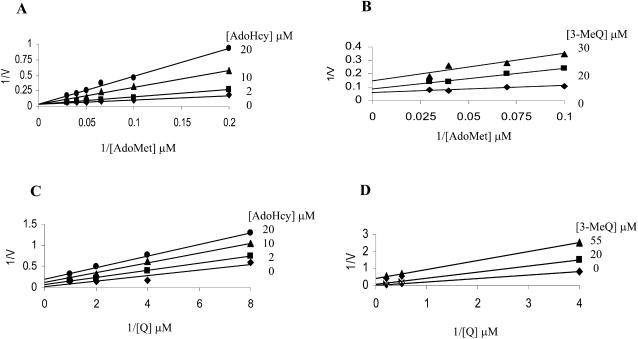
The order of substrate binding and product release was determined from product inhibition studies. AdoHcy competitively inhibited the methylation reaction with respect to AdoMet (Fig. 8A), but gave a mixed inhibition pattern with respect to Q (Fig. 8B). In addition, 3-MeQ produced mixed inhibition patterns with respect to both the flavonol substrate, Q (Fig. 8C) and the cosubstrate, AdoMet (Fig. 8D). Their kinetic values are given in Table IV. These kinetic patterns, where one of the four is competitive, are consistent with an ordered sequential bi-bi reaction mechanism (Segel, 1975) in which AdoMet binds first, followed by Q, and product release in the reverse order. The kinetic patterns obtained, therefore, are inconsistent with a random binding mechanism with dead-end complexes, in which case four competitive inhibition patterns would be expected.
Figure 8.
Product inhibition patterns of the flavonol 3-OMT. A, 1/v versus 1/AdoMet at different fixed concentrations of AdoHcy and a constant concentration (5 μm) of Q. B, 1/v versus 1/Q at different fixed concentrations of AdoHcy and a constant concentration (60 μm) of AdoMet. C, 1/v versus 1/Q at different fixed concentrations of 3-MeQ and a constant concentration (60 μm) of AdoMet. D, 1/v versus 1/AdoMet at different fixed concentrations of 3-MeQ and a constant concentration (5 μm) of Q.
MS/MS Protein Identifcation
Analysis of the 31-kD SDS PAGE-purified protein band (Fig. 4A, II) by Matrix-Assisted Laser Desorption Ionization mass spectrometry yielded several polypeptides, six of which were assigned the highest MASCOT score. These are (with % sequence identity) 1, TTMMHRLK (38%); 2, ICRLLER (28%); 3, VLMESWYHLK (100%); 4, GMSDHSTMSMKK (83%); 5, GVIILAALPK (27%); and 6, VIALIHK (43%). These peptides exhibited high homology to a number of Group II OMTs (Joshi and Chiang, 1998), including the C. americanum Q 3′-OMT (accession no. Q42653), four Thalictrum tuberosum multifunctional OMTs (AF064693.1 to AF064696.1), and several caffeic acid OMTs from tobacco (S36403), poplar (Populus spp. Q43047, Q41086), and Arabidopsis (NM_124796.2), among others. Peptides 3 and 4, in particular, exhibited 100% and 83% identity to the analogous regions of the C. americanum Q 3′-OMT. Surprisingly, however, none of these peptides was aligned within any of the previously reported five OMT conserved motifs (Ibrahim et al., 1998). Rather, they are mostly distribued within the N-terminal half of the protein sequence, except for peptide 6 which aligned near the C-terminal.
DISCUSSION
3-MeQ is known for its roles as an anti-inflammatory and antiviral agent (Middleton and Kandaswami, 1993; Malhotra et al., 1996), as a specific inhibitor of viral RNA replication (Castrillo and Carrasco, 1987), and as a phytoanticipin that accumulates in response to wounding and insect herbivory (Roda et al., 2003). However, it rarely accumulates in plants grown under normal conditions, since it is an intermediate in the pathway of biosynthesis of partially/highly methylated flavonols (Ibrahim et al., 1987; Macheix and Ibrahim, 1984; Thresh and Ibrahim, 1985). This prompted us to purify and characterize the OMT involved in its biosynthesis, the flavonol 3-OMT, with the ultimate goal of cloning its gene and expressing the gene product in transgenic plants.
The flavonol 3-OMT activity was partially purified from S. tinctoria leaves and was enriched to a 194-fold increase in its specific activity (13.8 pkat g−1) with 0.21% recovery. Such a low level of recovery indicates its low abundance in Serratula. This is not unexpected for an enzyme that catalyzes the methylation of an intermediate metabolite that accumulates in small amounts in this plant. Chromatography of the partially purified protein on a Mono Q column allowed the separation of the flavonol 3-OMT from two other methylating activities, the 3-MeQ 3′-OMT and the lignin monomer OMT (Fig. 3A), each of which exhibited distinctly different substrate specificity and apparent Mr value (Table II), as revealed by western-blot analysis (Fig. 3B).
Substrate specificity studies indicate the preference of Serratula OMT for position 3 of Q, whose reaction product was rigorously identified (Fig. 5). The chromatographic and UV spectral properties of the latter are characteristic of a 3-methylated Q to the exclusion of other positions (Harborne, 1967; Markham, 1982). In addition, the fact that 3-MeQ was not accepted as a substrate for further methylation, as was the 3′- or 4′-isomers (Table III), indicates that enzymatic methylation takes place at that position. The higher Vmax/Km of the enzyme with kaempferol than Q, as substrates, is difficult to explain in view of the Q-based methylated products that accumulate in vivo. Susbtitution of ring A of flavonols, other than the usual 5,7-dihydroxylation pattern, seems to impair the methylating activity as shown with the extra methyl group in rhamnetin and the methoxyl group in herbacetin, as well as the presence of an additional hydroxyl group at positions 6 or 8 in quercetagetin and gossypetin, respectively. On the other hand, B-ring hydroxylation and/or methylation seems to affect enzymatic methylation only slightly, except in the case of myricetin where the three vicinal OH groups may result in steric hinderance, and galangin, which lacks B-ring substitution; both resulted in significantly low enzyme activities. Nevertheless, the apparent versatility of the binding site of Serratula 3-OMT to accommodate two ubiquitous flavonols and its high affinity for both substrates, allows the 3-O-methylation of relatively small endogenous levels of common flavonols. These attributes would render the 3-OMT gene amenable to metabolic engineering for the constitutive expression of a variety of functionally important 3-O-methylated flavonols in plants.
Whereas the general physico-chemical properties of Serratula 3-OMT are similar to other flavonol OMTs (Forkmann and Heller, 1999), they differ from those of the corrresponding enzyme in C. americanum (De Luca and Ibrahim, 1985a), in having a relatively high pI value (6.0 instead of 4.8) and a much lower molecular mass (approximately 31 kD instead of 57 kD). The high pI value of Serratula 3-OMT indicates a lower abundance of acidic residues than in the case of the C. americanum enzyme. The high Mr value reported for the Chrysosplenium 3-OMT was probably due to its binding of other proteins during gel filtration chromatography, which could not be verified by SDS-PAGE due to its extremely low abundance, or by western-blot analysis due to unavailability of a suitable antibody.
The relatively low Mr of Serratula 3-OMT resembles those of Group I OMTs that utilize caffeoyl CoA as the preferred substrate, as compared with the 40 to 45 kD enzyme proteins characteristic of Group II OMTs that utilize a variety of o-dihydroxy substrates including phenylpropanoids, alkaloids, and flavonoids (Joshi and Chiang, 1998). However, the fact that the flavonol 3-OMT does not utilize caffeoyl CoA as a substrate excludes the possibility of being a Group I OMT. Rather, it may be considered a distinct member of Group II enzymes with certain physico-chemical attributes that allow the methylation of a quasi aliphatic hydroxyl group instead of the truly aromatic hydroxyls of the flavonol ring system. This is corroborated by the fact that of the various hydroxyl groups of galangin, kaempferol, and Q, the most acidic 3-hydroxyl group exhibits the highest negative electron density, as determined by the complete neglect of differential overlap method (Pople and Beveridge, 1970), thus enhancing its reactivity and specific methylation (Brunet and Ibrahim, 1980). However, further confirmation of the actual molecular mass of this novel enzyme will have to await the cloning of its gene and the characterization of its recombinant protein.
The fact that Serratula 3-OMT exhibits low Km values for Q suggests its low concentration in the cell. This is in contrast with the relatively high Km value for AdoMet that is utilized in several other metabolic processes. The relatively low Ki of the enzyme for the flavonoid product 3-MeQ suggests that it is an inhibitor of the enzyme reaction, and allows accumulation of the latter compound in catalytic amounts, which is compatible with its role in Serratula as an intermediate metabolite in the methylation sequence: Q → 3MeQ → 3,3′-diMeQ. These results, together with the extremely low Ki values for both AdoMet and AdoHcy (Table IV), suggest a tight control of methylated flavonol biosynthesis in this plant. The kinetic patterns obtained suggest that Serratula 3-OMT follows the same kinetic mechanism reported for Chrysosplenium 3-OMT, with a similar Ka/Kia value (De Luca and Ibrahim, 1985b), except for a 2.5-fold lower Km for AdoMet and a 2.5-fold higher Vmax for Q, indicating a higher efficiency of the Serratula enzyme as shown by its relatively high Kcat/Km value (Table IV).
The peptide sequence information obtained from the purified flavonol 3-OMT shows strong homology with a number of Group II OMTs. This flavonol-specific, position-oriented enzyme derives its novelty from being structurally and functionally a member of Group II OMTs, in spite of its low molecular mass that is reminescent of Group I enzymes. Future studies will be directed toward the cloning of the gene encoding this enzyme, characterization of the gene product, and the regulation of its expression under different conditions of stress.
MATERIALS AND METHODS
Plant Material
Seeds of Serratula tinctoria (Asteracea) were obtained from the Botanical Garden of the University of Göttingen, Germany. They were cultivated under greenhouse conditions and supplemented with a 250-mg L−1 solution of N-P-K (20:20:20) as required.
Flavonoid Extraction and Analysis
Because of their abundance in roots, flavonoids were isolated and characterized from these tissues. Fresh roots were extracted twice at room temprature with 100% and 80% aqueous MeOH and the combined extracts were evaporated in vacuo to an aqueous residue that was hydrolyzed with 2 n HCl. The acid hydrolyzate was extracted twice with dry ethyl acetate, and the organic layer evaporated to dryness, then dissolved in MeOH. The latter was applied onto preparative cellulose TLC plates and chromatographed using EtOAc-HOAc-H2O (1:3:7, v/v/v) as solvent. The dark UV-absorbing (366 nm) bands were eluted with MeOH for further purification on HPLC. The latter was performed with a Symmetry RP C18 silica colmn (5 μm particle size, 4.6 × 250 mm, Waters), using a 25-min gradient of 45% to 99.5% MeOH containing 0.5% HOAc. The major peaks were collected for UV spectral analysis using spectral shift reagents (Harborne, 1967; Markham, 1982).
Chemicals
Most flavonoid compounds were from our laboratory collection; both 3,3′-diMeQ and 3,4′-diMeQ were a generous gift from Prof. E. Wollenweber, caffeoyl CoA from Dr. M. Abou-Zaid, and 3-MeQ from Dr. Y. Fukushi. S-Adenosyl-l-[14CH3] Met (55 mCi mmol−1) was purchased from American Radiolabeled Chemicals (St. Louis) and both the unlabeled AdoMet and and the adenosine-agarose ligand chemicals were obtained from Sigma (Oakville, Canada). AdoHcy and Polybuffer PB-74 were purchased from Amersham Biosciences (Baie d'Urfé, Quebec).
Buffers
The following buffers were used: A, 0.2 m sodium phosphate, pH 7.6, 14 mm β-mercaptoethanol, 10% glycerol (v/v), 5 mm EDTA, 10% polyvinylpyrrolidone (w/v), 0.2% diethylaminodithiocarbamate (w/v), 1 mm phenylmethanesulfonyl fluoride; B, 50 mm Tris-HCl, pH 7.6, 14 mm β-mercaptoethanol, 10% glycerol (v/v), 0.3 m NaCl; C, 50 mm Tris-HCl, pH 7.6, 14 mm β-mercaptoethanol, 10% glycerol (v/v), 50 mm NaCl; D, 0.1 m Tris-HCl, pH 7.5, 0.1 m MgCl2; E, 20 mm Tris-HCl, pH 7.6, 14 mm β-mercaptoethanol, 10% glycerol (v/v), 1 mm EDTA; F, 50 mm Tris-HCl, pH 7.1, 14 mm β-mercaptoethanol, 10% glycerol (v/v); G, Pollybuffer 74 (1:15, v/v), pH 4.0, 14 mm β-mercaptoethanol, 10% glycerol (v/v), 5% betaine (w/v), 10 mm KCl. All FPLC buffers were filtered, degassed, and stored at 4°C.
Preparation of Adenosine-Agarose Affinity Gel
Preparation of the ligand was carried out according to the protocol of Rakwal et al. (2000) with some modification in the buffer used to equilibrate the adenosine-agarose column (buffer E), and the calf intestinal alkaline phosphatase reaction buffer D (pH 7.5).
Protein Extraction and Purification of the 3-OMT
The lack of sufficient amounts of root tissue, coupled with the requirement for several extractions for protein purification and characterization, prompted us to use leaf tissue after having verified the existence of the flavonol 3-OMT activity in both organs (see the “Results” section). All steps were carried out at 4°C unless otherwise stated. Leaf tissues (approximately 20 g), harvested from the tips of 2- to 3-month-old plants, were ground to a fine powder in liquid nitrogen before being homogenized with buffer A (1:5, w/v). The homogenate was filtered through one layer of Miracloth and the filtrate centrifuged at 12,000g for 20 min. The supernatant was stirred for one h with Dowex 1×2 resin (10%, w/v), which had previously been equilibrated with the extraction buffer, in order to remove contaminating phenolics, then filtered. Solid ammonium sulfate was then added to the filtrate and the protein which precipitated between 30% and 70% salt saturation was collected by centrifugation at 12,000g for 15 min. The protein pellet was resuspended in buffer B and used immediately for enzyme purification.
The Amersham Biosciences FPLC system was used for enzyme purification. The protein pellet resulting from ammonium sulfate precipitation was first desalted on a PD-10 column before being applied onto a Superose-12 (prep grade) gel filtration column (16 mm × 500 mm) that was previously equilibrated with buffer B. The column was run at a flow rate of 1 mL min−1 (0.5 MPa) and 2-mL fractions were collected and assayed for 3-OMT activity. The active fractions were pooled, and buffer B was exchanged to buffer C on a PD-10 column before being applied to a Mono-Q HR 5/5 column (5 mm × 50 mm) preequilibrated with buffer C at a flow rate of 0.5 mL/min (2.5 MPa). The bound proteins were eluted with 50 mL of a linear (50–500 mm) NaCl gradient in buffer C, and 2-mL fractions were collected and assayed for the various OMT activities using Q, 3-methylquercetin, and 5-HFA as substrates. The Mono-Q fraction containing the 3-OMT activity was concentrated in an Amicon stirred cell (model 8050) and the buffer was exchanged with buffer E on a PD-10 column. Finally, the protein fraction containing 3-OMT activity was loaded on an adenosine-agarose affinity column (7 mm × 130 mm) that had previously been equilibrated with buffer E. The column was washed first with 50 mL of the same buffer, then eluted with 15 mL of 4 mm AdoMet in the same buffer containing 0.2 m KCl. Fractions (2.5 mL) were collected and assayed for 3-OMT activity using Q as the substrate. The enzmatically active fractions were pooled and dialyzed overnight in buffer E containing 0.2 m KCl for SDS-PAGE and MS/MS peptide analysis.
OMT Assay
The enzyme assay contained 10 to100 μm of the phenolic substrate (dissolved in 50% dimethyl sulfoxide), 10 μm of AdoMet (containing 25 nCi of [14CH3]AdoMet (55 mCi mmol−1) as the cosubstrate, and 10 to 100 μg of the enzyme protein in a final volume of 100 μL. The assay mixture was incubated for 30 min at 30°C, and the reaction was terminated by the addition of 10 μL of 6 n HCl. The labeled O-methylated products were extracted by shaking for 3 min with 500 μL of a mixture of C6H6-EtOAc (1:1, v/v). Aliquots of the organic phase were counted for radioactivity by liquid scintillation, or used for identification of the reaction products by TLC or HPLC. The enzyme reaction products were cochromatographed with the available reference compounds on cellulose TLC plates in EtOAc-HOAc-H2O (1:3:7, v/v/v). Assays were conducted in triplicates and values that differed by >15% were excluded. Control assays lacking the substrate or cosubstrate were also conducted. Enzyme assays were repeated more than once to ensure for reproducibility of results.
Identification of Enzyme Reaction Products
The identity of the methylated products was verified by cochromatography on TLC with reference compounds, when available, visualization under UV light (366 nm), and autoradiography on x-ray film. Several enzyme assays prepared with unlabeled AdoMet were used for HPLC analysis. The product was lyophilized and dissolved in 150 μL HPLC-grade MeOH and filtered through 0.20 μm Millipore syringe-driven filters prior to injection onto a Symmetry RP C18 silica column (Waters, Milford, MA) using a linear gradient starting with a mixture of MeOH-H2O-HOAc (44:55:1, v/v/v) for 2 min to 70:29:1 (v/v/v) over 25 min, and remained at that plateau for 10 min before gradient descent to the starting conditions. Identity of the product was confirmed by comparison of its Rt value and UV-absorption maxima with those of a reference sample of 3-MeQ. The HPLC-purified product was lyophilized and used for MS analysis.
MS Analysis of the Enzyme Raction Product
Both the enzyme reaction product and the 3-MeQ standard were subjected to MS analysis in the positive and negative mode using Finnigan LCQ atmospheric pressure chemical ionization (APCI) with ion trap mass analyzer. Vaporization temperature 450°C, discharge current 5 μA, and capillary temperature 150°C. Fragmentation occurred within the ion trap and reported as percentages of the available collision energy used to fragment the ions. The latter were the average of ten scans.
Other Physico-Chemical Methods
The molecular mass of 3-OMT was estimated by gel filtration on a calibrated Superose-12 column, using the reference proteins, bovine serum albumin (67.0 kD), carbonic anhydrase (31.0 kD), and chymotrypsinogen A (25.0 kD).
The pI value of the enzyme was determined from its elution pattern on a Mono-P HR 5/20 column (5 mm × 200 mm) preequilibrated with buffer F, using the active Mono-Q fractions after desalting on a PD-10 column. The column was washed with 50 mL of the same buffer, and the bound proteins were eluted with 50 mL of a descending pH gradient (pH 7–4) in buffer G at a flow rate of 0.5 mL min−1, and 1-mL fractions were collected and assayed for 3-OMT activity.
The purity of protein fractions was monitored by SDS-PAGE (Laemmli, 1970) using 12.5% acrylamide gels and Mr markers (Bio-Rad, Hercules, CA) ranging from 14.4 to 97.4 kD for calibration under denaturing conditions. After electrophoresis, proteins were stained with Coomassie Brilliant Blue (R-250). For western blotting, the gels were submitted to semi-dry electrophoretic transfer onto nitrocellulose membranes and probed with 1:500 dilution of IgG-purified Chrysosplenium americanum anti-flavonol 3′-OMT and horseradish peroxidase-linked donkey anti-rabbit IgGs (Amersham Biosciences) as described previously (Séguin et al., 1998). Alternatively, membranes were probed with 1:500 dilution of the IgG-purified Arabidopsis anti-flavonol 3′-OMT antibody (Muzac et al., 2000) and 1:3,000 diluted secondary antibody labeled with alkaline phosphatase. Determination of alkaline phosphatase activity was carried out using 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium as substrates.
Protein content was determined using the Bio-Rad protein assay (Bradford, 1976) and bovine serum albumin (Sigma) as the standard protein.
Kinetic Analysis
Kinetic studies were performed using the Mono-Q fraction II protein (Fig. 3A). Product formation was linear with respect to the assay time and the amount of protein used. Assays of each graph point were repeated five times within the same set of the experiment. Values with differences greater than 15%, compared to the norm in each set, were repeated. Standard deviations of the intercept and the slope of each replot used in the determination of 3-OMT kinetic parameters were calculated according to Zar (1984).
Kinetic analyses of substrate interactions between Q and AdoMet were carried out using the 10 μg of protein and varying concentrations of AdoMet in the presence of fixed, constant concentrations of Q. The inverse was also performed with different concentrations of Q in the presence of fixed, constant concentrations of AdoMet. Results of the substrate interaction kinetics are presented as Lineweaver-Burk plots (Segel, 1975) fitted by linear regression. The replots of the intercept and slope values obtained from the Lineweaver-Burk plot were used for the calculation of substrate Km values, the Ki for AdoMet, Vmax of Q, catalytic efficiency of 3-OMT, and the enzyme turnover rate. The order of substrate binding, and the Kip and Kiq values of 3-MeQ and AdoHcy, respectively, were determined from the Lineweaver-Burk plot patterns of the product inhibition studies.
MS/MS Protein Identification
After SDS-PAGE, the 31-kD affinity-purified protein band (Fig. 4A, II) was excised and subjected to in-gel trypsin digestion. The resulting peptides were extracted with acidified acetonitrile, mixed with matrix (α-cyano-4-hydroxycinnamic acid) and analyzed by Matrix-Assisted Laser Desorption Ionization (Waters-Micromass) mass spectroscopy. The peptide masses obtained were used to search the nonredundant protein database using MASCOT (Matrix Science, London) for preliminary peptide identification based on mass finger printing. Another portion of the extracted peptides was injected into LC-MS/MS system (liquid chromatography-electrospray ionization-quadruple-time of flight) at a flow rate of 0.1 mL min−1 in the positive mode, and MASCOT was used to identify the protein. These steps were carried out by Genome Québec, Montreal.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers Q42653, AF064693.1 to AF064696.1, S36403, Q43047, Q41086, and NM_124796.2.
Note Added in Proof
After acceptance of this report, four position-specific flavone/flavonol OMT cDNA clones (accession numbers AY337457–AY337461) were reported from mint (Mentha x piperita) that were used for the combinatorial generation of different flavonoid structures based on the in vitro activities of their recombinant proteins [Willits MG, Giovanni M, Prata RTN, Kramer CM, De Luca V, Steffens JC, Graser G (2004) Bio-fermentation of modified flavonoids: an example of in vivo diversification of secondary metabolites. Phytochemistry 65: 31–41].
Acknowledgments
We wish to thank Aka Meyers (Göttingen Botanical Gardens, Germany) for the generous gifts of Serratula seeds, Prof. E. Wollenweber (TU Darmastadt, Germany) for the 3,3′-diMeQ and 3,4′-diMeQ, Dr. M. Abou-Zaid (Natural Resources Canada, Sault Ste Marie, Canada) for caffeoyl CoA, Dr. Y. Fukuski (Hokkaido University, Japan) for the 3-MeQ, Dr. M. Di Falco (Genome Québec, Montreal) for the MS/MS peptide analysis, and Dr. L. Davin (Washington State University, Pullman, WA) for MS analysis of the enzyme reaction product and 3-MeQ reference.
This work was supported by grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada, and by Formation des chercheurs et l'aide á la recherche (FCAR), Department of Higher Education, Québec. D.A. was the recipient of both NSERC and FCAR postgraduate scholarships.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036442.
References
- Bohm BA (1998) Flavonoid functions in nature. In Introduction to Flavonoids. Chemistry and Biochemistry of Organic Natural Products, Vol 2. Harwood Academic Publishers, Amsterdam, pp 339–364
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brunet G, Ibrahim RK (1980) O-Methylation of flavonoids by cell-free extracts of Calamondin orange. Phytochemistry 19: 741–746 [Google Scholar]
- Castrillo JL, Carrasco L (1987) Action of 3-methylquercetin on poliovirus RNA replication. J Virol 61: 3319–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FW, De Luca V, Ibrahim RK, Voirin B, Jay M (1981) Polymethylated flavonols in Chrysosplenium americanum: Identification and enzymatic synthesis. Z Naturforsch 36c: 730–736 [Google Scholar]
- Corio-Costet MF, Chaius L, Malosse C, Scalla R, Delbecque JP (1991) A plant producing ecdysteroids: plants and cell culture. In Kouchkowsky Y, ed, Plant Sciences Today, Vol 59. INRA, Paris, pp 243–247
- De Luca V, Ibrahim RK (1985. a) Enzymatic synthesis of polymethylated flavonols in Chrysosplenium americanum. I. Partial purification and some properties of 3-, 6-, 7- and 4′- O-methyltransferases. Arch Biochem Biophys 238: 496–605 [DOI] [PubMed] [Google Scholar]
- De Luca V, Ibrahim RK (1985. b) Enzymatic synthesis of polymethylated flavonols in Chrysosplenium americanum. II. Substrate interaction and product inhibition studies of flavonol 3-, 6- and 4′-O-methyltransferases. Arch Biochem Biophys 238: 596–605 [DOI] [PubMed] [Google Scholar]
- Forkmann G, Heller W (1999) Biosynthesis of flavonoids. In Sankawa U, ed, Polyketides and Other Secondary Metabolites Including Fatty Acids and Their Derivatives, Vol 1. Elsevier, Amsterdam, pp 713–748
- Gottlieb OR (1975) Flavonols. In JB Harborne, TJ Mabry, H Mabry, eds, The Flavonoids, Vol. 1. Academic Press, New York, pp 296–375
- Harborne JB (1967) Comparative Biochemistry of the Flavonoids. Academic Press, New York
- Ibrahim RK, Anzellotti D (2003) The enzymatic basis of flavonoid biodiversity. In Romeo JT, ed, Interactive Phytochemistry: From Ethnobotany to Molecular Ecology. Elsevier, Amsterdam, pp 1–36
- Ibrahim RK, Bruneau A, Bantignies B (1998) Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol Biol 36: 1–10 [DOI] [PubMed] [Google Scholar]
- Ibrahim RK, De Luca V, Khouri HE, Latchinian L, Brisson L, Charest P-M (1987) Enzymology and compartmentation of polymethylated flavonol glucosides in Chrysosplenium americanum. Phytochemistry 26: 1237–1245 [Google Scholar]
- Ibrahim RK, Muzac I (2000) The methyltransferase gene superfamily: a tree with multiple branches. In Romeo JT, Ibrahim R, Varin L, De Luca V, eds, Evolution of Metabolic Pathways. Elsevier Science, Amsterdam, pp 349–384
- Joshi CP, Chiang VL (1998) Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent O-methyltransferases. Plant Mol Biol 37: 663–674 [DOI] [PubMed] [Google Scholar]
- Ko WC, Chen MC, Wang SH, Lai YH, Chen JH, Lin CN (2003) 3-O-Methylquercetin more selectively inhibits phosphodiesterase Subtype 3. Planta Med 69: 310–315 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Macheix J-J, Ibrahim RK (1984) The O-methyltransferase system of apple fruit cell culture. Biochem Physiol Pflanz 179: 659–664 [Google Scholar]
- Malhotra B, Onnyilagha JC, Bohm BA, Towers GHN, James D, Harborne JB, French CJ (1996) Inhibition of tomato ring-spot virus by flavonoids. Phytochemistry 43: 1271–1276 [Google Scholar]
- Markham KR (1982) Techniques of Flavonoid Identification. Academic Press, New York
- Middleton E, Kandaswami C (1993) The impact of plant flavonoids on mammalian biology: implications for immunity, inflammation and cancer. In JB Harborne, ed, The Flavonoids, Advances in Research Since 1986. Chapman & Hall, London, pp 619–652
- Muzac I, Wang J, Anzellotti D, Zhang H, Ibrahim RK (2000) Functional expression of an Arabidopsis cDNA clone encoding a flavonol 3′-O-methyltransferase and characterization of the gene product. Arch Biochem Biophys 375: 385–388 [DOI] [PubMed] [Google Scholar]
- Pople JA, Beveridge D (1970) Approximate Molecular Orbital Theory. McGraw-Hill, New York
- Rakwal R, Agrawal GK, Yonekura M, Kodama O (2000) Naringenin 7-O-methyltransferase involved in the biosynthesis of the flavanone sakuranetin from rice (Oryza sativa). Plant Sci 155: 213–221 [DOI] [PubMed] [Google Scholar]
- Roda AL, Oldham NJ, Svatos A, Baldwin IT (2003) Allometric analysis of the induced flavonols on the leaf surface of wild tobacco (Nicotiana attenuata). Phytochemistry 62: 527–536 [DOI] [PubMed] [Google Scholar]
- Segel IH (1975) Enzyme Kinetics. Wiley, New York
- Séguin J, Muzac I, Ibrahim RK (1998) Purification and immunological characterization of a recombinant trimethylflavonol 3′-O-methyltransferase. Phytochemistry 49: 319–325 [DOI] [PubMed] [Google Scholar]
- Thresh K, Ibrahim RK (1985) Are spinach chloroplasts involved in flavonoid O-methylation? Z Naturforsch 40c: 331–335 [Google Scholar]
- Wollenweber E, Dietz VH (1981) Occurrence and distribution of free flavonoid aglycones in plants. Phytochemistry 20: 869–932 [Google Scholar]
- Zar J (1984) Biostatistical Analysis, Ed 2. Prentice-Hall, Englewood Cliffs, NJ