Abstract
Endogenous retroviruses (ERVs) are present in the genome of all vertebrates and originated from infections of the germline of the host by exogenous retroviruses. ERVs have coevolved with their hosts for millions of years and are recognized to contribute to genome plasticity, protect the host against infection of related pathogenic and exogenous retroviruses, and play a vital role in development of the placenta. Consequently, some ERVs have been positively selected and maintained in the host genome throughout evolution. This review will focus on the critical role of ERVs in development of the mammalian placenta and specifically highlight the biological role of sheep JSRV-related endogenous betaretroviruses in conceptus (embryo and associated extraembryonic membranes) development.
Keywords: Endogenous retrovirus, placenta, trophoblast
Introduction
Endogenous retroviruses (ERVs) are present in the genome of all vertebrates and are vertically transmitted as stable, inherited Mendelian genes.1 ERVs are thought to arise from ancient infections of the germline of the host by exogenous retroviruses. The obligatory integration step of the retroviral replication cycle allowed, during evolution, the incorporation of the viral genome (provirus) into the host genome. Retrotransposition or re-infection of the germline can generate further insertions augmenting the number of ERVs loci in the genome.2 ERVs have heavily colonized the genome of all animal species; for example, they account for approximately 8–10% of the human genome.3
A complete ERV ‘provirus’ (i.e. the retroviral genome integrated into the host cell genome) shares the same genomic structure of an exogenous retrovirus, which is four viral genes (gag, pro, pol, and env) flanked by two long terminal repeats (LTRs) (Fig. 1). The gag gene encodes for the major viral structural protein, while pro and pol encode for the viral enzymatic machinery necessary for the viral replication cycle. The env gene encodes for the envelope glycoprotein (Env) that is inserted into the lipid bilayer of the exterior membrane to form the viral envelope and mediates entry of the virus into susceptible cells.
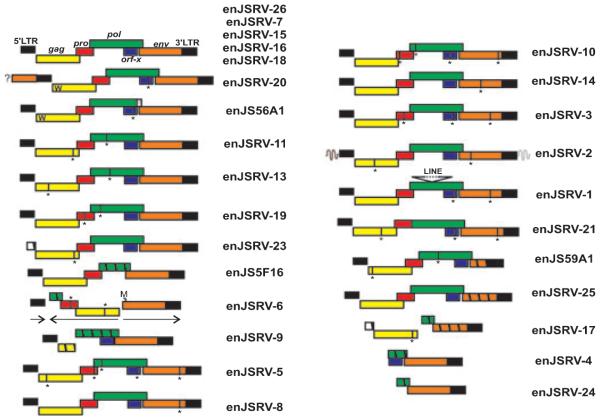
Fig. 1.
Representative enJSRVs proviruses present within the sheep genome. Five enJSRVs display an intact genomic organization typical of replication competent proviruses (top). The ‘W’ present in the Gag protein of the two transdominant proviruses, enJS56A1 and enJSRV-20, indicates the R21W substitution. The 5′ flanking region of enJSRV-20 contains an env gene indicated by a box and a question mark (?). Vertical lines and an asterisk (*) represent stop codons, while hatched boxes indicate deletions. enJSRV-6 harbors a recombined structure with internal sequence in the opposite direction compared to the 5′ and 3′ LTRs of the provirus. The first methionine (indicated by the letter M) of the env gene of enJSRV-6 is present after the usual start codon. Figure reproduced from Arnaud et al.6
The LTRs contain enhancer and promoter elements that direct expression of the viral genes. Most ERVs are destined to extinction if their expression brings deleterious consequences for the host. Thus, their persistence in the host genome is the result of a fine balance reached throughout evolution which usually renders them replication defective because of the accumulation of mutations, deletions, rearrangements, and methylation.1
ERVs are widespread throughout vertebrate genomes.4 Some ERVs are highly related to exogenous retroviruses, including Jaagsiekte sheep retrovirus (JSRV), mouse mammary tumor virus, feline leukemia virus, and avian leukemia virus, which are currently active and infect sheep, mice, cats, and chickens, respectively.1 These ERVs are generally referred to as ‘modern’ ERVs, because they integrated into the host genome after speciation and are closely related to exogenous viruses that are still infectious, while most ERVs do not have an exogenous counterpart. Some modern ERVs are still able to produce infectious virus because of the lack of inactivating mutations. Modern ERVs can also have insertionally polymorphic loci, because they are not completely fixed in a particular population and are still undergoing endogenization. For instance, both koalas and sheep are currently being invaded by the koala retrovirus5 and endogenous JSRVs (enJSRVs),6,7 respectively. In contrast, ‘ancient’ ERVs invaded the genomes before speciation and, consequently, are present in every individual at the same genomic location of phylogenetically related species.8
The biological significance of ERVs has been debated for several decades, and in the past they were generally thought to be ‘junk DNA’.9 However, recent studies suggest that ERVs have a variety of beneficial roles to their host.10–12 At the very least, the abundance of these elements in the host genome suggests that they contribute to genome plasticity. Moreover, the presence of transcriptionally active ERVs with intact open reading frames conserved million of years after integration supports the idea that some ERVs were exapted by the host for specific biological roles.
In this review, we will focus on the biological roles of ERVs in development of the placenta and then highlight the biological role of sheep JSRV-related endogenous betaretroviruses (enJSRVs) in conceptus (embryo and associated extraembryonic membranes) development.
ERVs in the human, mouse, and rabbit placenta
ERVs have been speculated to play a physiological role in placenta morphogenesis for almost three decades, considering that retroviral particles have been frequently observed in the reproductive tract.13–18 In fact, ERVs are abundant in the genital tract and placenta of various animal species.17,19 The presence of intact env genes that are expressed in the multinucleated syncytiotrophoblasts of the placenta and preserved over thousands of years, together with the observation that they elicit fusion of cells in vitro, led to the speculation that ERVs play an essential role in placental development and were positively selected for a fundamental role in the evolution of placental mammals and development of viviparity.20–24
Human
HERV-W (ERVWE1), HERV-FRD, and ERV-3 are three human ERVs (HERV) whose intact env genes are expressed in the human placenta.25–27 HERV-W is not present in the human genome as a complete provirus; however, its env gene (ERVWE1), encoding a protein termed syncytin 1, is preferentially expressed in the syncytiotrophoblast. The syncytiotrophoblast is a multinucleated cell that lines the outer surface of the placenta, is derived by intercellular fusion of trophoblast cells, and is responsible for the transport of oxygen, nutrients, and waste products, production of hormones, and immune tolerance.28,29 Syncytin 1 is a glycosylated protein and possesses characteristic features of a retroviral Env protein, such as the presence of a leader peptide, a potential furin cleavage site, a fusion peptide-like sequence, and a putative immunosuppressive region (Fig. 2). It also contains a hydrophobic membrane-spanning domain, suggesting it could be inserted into the plasma membrane.27 There is considerable in vitro information suggesting that syncytin 1 is involved in the fusion of mononuclear cytotrophoblasts to form syncytiotrophoblast in the human placenta. Transfection of a variety of cell lines with HERV-W env induced cellular fusion that was reduced when the cell cultures were treated with an antibody against the HERV-W Env protein.21,26 In addition, induction of fusion of BeWo cells (a human trophoblastic choriocarcinoma cell line) by forskolin was associated with increased expression of syncytin.21 Moreover, inhibition of syncytin 1 expression in primary trophoblast cells reduced the number and size of syncytia formed during culture.30
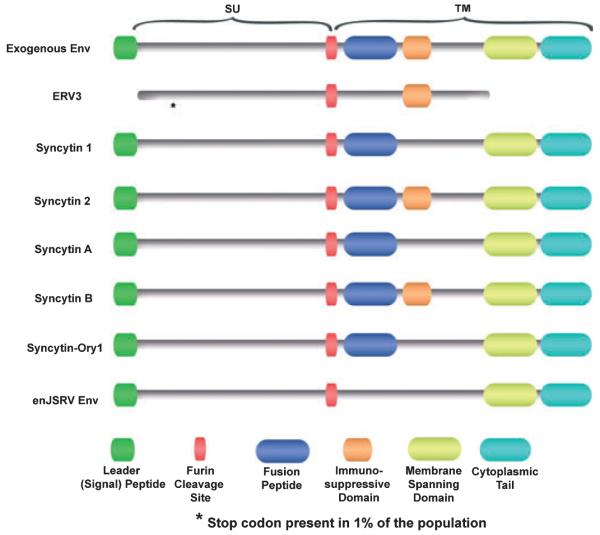
Fig. 2.
Schematic diagram of the functional domains of Env glycoproteins proposed to be involved in placental morphogenesis compared to a typical exogenous betaretroviral Env. ERV3 codes for a truncated Env in humans, with 1% of the population having a stop codon within the amino terminus of the SU region. Syncytin 2 (human) and syncytin-B (mouse) contain all the hallmarks of intact and functional Env proteins, including the leader peptide, furin cleavage site, fusion peptide, immunosuppressive domain, membrane-spanning domain, and a cytoplasmic tail.
The Env glycoprotein of HERV-FRD, termed syncytin 2, is structurally similar to syncytin 1 (see Fig. 2); however, it entered the primate genome before the split of the New World and the Old World Monkeys more than 40 million years ago, while syncytin 1 entered the primate genome approximately 25 millions years ago and is not present in Old World Monkeys.31 Syncytin 2 also elicits cell fusion when transiently transfected into several different cell lines.32 Interestingly, the two syncytins display different properties as both are fusogenic, but syncytin 2 has immunosuppressive properties unlike syncytin 1.33
The Env protein of ERV3 is also present in syncytiotrophoblasts and was the first ERV Env for which a potential physiological function was described.34 Although it has a long open reading frame, the protein is prematurely terminated by the presence of a stop codon in the transmembrane region (Fig. 2), which truncates the hydrophobic domain that is required for anchoring to the cell membrane.35 It also lacks a leader and a fusion peptide and, although it harbors a region with the characteristics of an immunosuppressive domain, its function is likely diminished by the lack of membrane anchorage.36 ERV3 Env does not elicit cell fusion, although its expression increases in BeWo cells treated with forskolin. When ERV3 Env is stably expressed in undifferentiated BeWo cells, it induces changes characteristic of trophoblast differentiation, such as increased levels of chorionic gonadotropin, growth inhibition, and altered morphology.37 Considering that the ERV3 Env is expressed in a variety of normal tissues and particularly in hormone-producing organs, including adrenal and sebaceous glands and testis, it may play a general role in hormone production.36 However, 1% of 150 healthy Caucasian individuals were found to be homozygous for a premature stop codon that would theoretically result in a severely truncated non-functional protein;38 thus, it is debatable whether the ERV3 Env has a critical biological function.
Murine
Two murine ERV env genes, syncytin-A (Gm52) and syncytin-B (D930020E02Rik), were identified and found to be expressed in the syncytiotrophoblast component of the labyrinthine zone of the mouse placenta.20 Both are highly fusogenic in transfection assays. Genes orthologous to syncytin-A and -B and disclosing a striking conservation of their coding status are found in all Muridae tested (mouse, rat, gerbil, vole, and hamster), dating their entry into the rodent lineage approximately 20 million years ago. In both humans and mice (Fig. 2), one of the two syncytins (human syncytin 2 and mouse syncytin-B) is immunosuppressive and, rather unexpectedly, the other (human syncytin 1 and mouse syncytin-A) is not although both are able to induce cell–cell fusion.33 Syncytin-A plays an important biological role in syncytiotrophoblast development, because syncytin-A null mice die in utero because of the failure of trophoblast cells to fuse and form one of the two syncytiotrophoblast layers present in the mouse placenta39 that play a key role in transport of nutrients for the developing conceptus.29 Given that two syncytins are immunosuppressive, they may play a role in maternofetal tolerance, although this concept has not been mechanistically tested in vivo.33
Rabbit
Recently, Heidmann et al.24 identified an env gene of retroviral origin in the rabbit Oryctolagus cuniculus, termed syncytin-Ory1, with the characteristic features of human syncytin (Fig. 2). An in silico search for full-length env genes with an uninterrupted open reading frame within the rabbit genome resulted in the identification of an env gene with placenta-specific expression and belonging to a family of endogenous retroelements present at a limited copy number in the rabbit genome. The placenta-expressed env gene demonstrated fusogenic activity in an ex vivo cell–cell fusion assay. Interestingly, the receptor for the rabbit syncytin-Ory1 was found to be the same as that for human syncytin 1, i.e. the previously identified sodium-dependent neutral amino acid transporter type 2 (SLC1A5). Syncytin-Ory1 mRNA was specifically present at the level of the junctional zone of the placenta, where the invading syncytial fetal tissue contacts the maternal decidua to form the labyrinth, consistent with a role in the formation of the syncytiotrophoblast. The identification of a novel syncytin gene within a third order of mammals displaying syncytiotrophoblast formation during placentation strongly supports the notion that on several occasions, retroviral infections have resulted in the independent capture of genes that were positively selected for a convergent physiological role in development of the placenta.24
Endogenous betaretroviruses of sheep
Domestic sheep have at least 27 copies of ERVs in their genome, termed enJSRVs (Fig. 1), because they are highly related to the exogenous and pathogenic JSRV.6,40 JSRV is the causative agent of ovine pulmonary adenocarcinoma, a transmissible lung cancer of sheep.41 A unique feature of JSRV among oncogenic retroviruses is that its Env glycoprotein is the main determinant of cell transformation both in vitro and in vivo.42–48 Expression of the JSRV Env alone is able to transform a variety of cell lines in vitro, including mouse, rat, and chicken fibroblasts as well as human bronchial, canine, and rat epithelial cells.42,44–46,49,50 More importantly, the JSRV Env is able to induce lung adenocarcinomas in immunocompetent sheep when expressed by a JSRV-based vector under the control of the JSRV LTR.47 Thus, JSRV Env is a dominant oncoprotein; however the mechanisms of cell transformation induced by the JSRV Env are not completely understood. Although the mitogen-activated protein kinase (Ras-MEK-MAPK), Rac1, and phosphoinositide 3-kinase (PI3K-AKT-mTOR) pathways are implicated in JSRV-induced cell transformation, it still remains to be determined how the cytoplasmic tail engages the cell signaling network to activate these pathways.50–54
The majority of the 27 enJSRV proviruses are defective as a result of deletions, nonsense mutations, and recombinations; however, five enJSRV proviruses contain intact genomes with uninterrupted open reading frames for all the retroviral genes (Fig. 1).6 These enJSRV loci are insertionally polymorphic in the domestic sheep population. JSRV and enJSRVs have an overall high degree of similarity (approximately 85–89% identity at the nucleotide level). The evolutionary history of these proviruses together with ruminants suggests that integration of enJSRVs began before the split between the genus Ovis and the genus Capra, approximately 5–7 million years ago, and continued after sheep domestication (approximately 10,000 years ago).6,7 Interestingly, one enJSRV provirus, enJSRV-26, is thought to have integrated in the host <200 years ago and may be a unique integration event occurred in a single animal.6 Thus, the enJSRVs are most likely still invading the sheep genome.
The sheep placenta and conceptus development
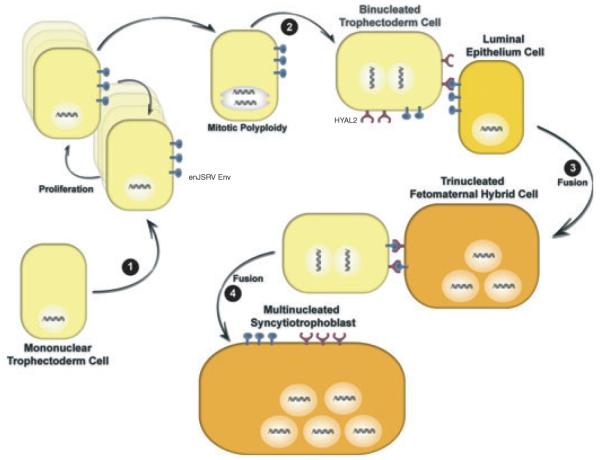
In sheep, the morula-stage embryo enters the uterus by day 5 after mating and forms a blastocyst by day 6 that contains a blastocoele surrounded by a monolayer of trophectoderm.55,56 By day 9, the blastocyst hatches from the zona pellucida, develops into an ovoid conceptus by day 12, and then begins to elongate (reaching 25 cm or more by day 17). Elongation of the conceptus is critical for the production of interferon tau (IFNT), which is the pregnancy recognition signal needed to maintain progesterone production by the corpus luteum, and also for the onset of implantation.57 Implantation of the conceptus involves the apposition, attachment, and adhesion of the conceptus trophectoderm to the endometrial luminal epithelium (LE) of the uterus. Within the outer layer of the conceptus termed the chorion, binucleated trophectoderm cells, termed trophoblast giant binucleate cells (BNC), begin to appear as early as day 14.58 The BNC are thought to be derived from the mononuclear trophectoderm cells by a process referred to as mitotic polyploidy, which involves consecutive nuclear divisions without cytokinesis.59 BNC then fuse with uterine LE to form trinucleate fetomaternal hybrid cells.58 Other BNCs fuse with the trinucleate cells (and likely each other) to form plaques of multinucleated syncytiotrophoblast that have 20–25 nuclei. Trophoblast BNC of the sheep placenta are analogous in many ways to the giant cells of the syncytiotrophoblast of the human placenta.60 The syncytial plaques and BNC form specialized structures on the placenta termed cotyledons that interdigitate with the endometrial caruncles of the maternal uterus to form a structure termed a placentome.61 Blood flow to the uterus and from the fetus is predominantly routed to the placentomes, which provides hematrophic nutrition from the mother to the fetus. Other functions of BNC and multinucleated syncytia include production and synthesis of proteins and hormones, like placental lactogen, pregnancy-associated glycoproteins, and progesterone, that are involved in the growth of uterus and mammary gland and other maternal functions.59
In sheep, enJSRVs are abundantly expressed in the epithelia lining the different tissues of the female reproductive tract (vagina, cervix, uterus, and oviduct).62,63 In the uterus, both RNA and protein of enJSRVs are detected specifically in the endometrial LE and in the glandular epithelia.63–65 In addition, enJSRVs are expressed in the trophectoderm cells of the placenta in a temporal fashion that is coincident with key events in conceptus elongation and onset of trophoblast giant BNC differentiation.62 Within the placenta, enJSRVs are most abundant in the trophoblast giant BNC and multinucleated plaques of syncytiotrophoblast within the placentomes throughout pregnancy. The RNA of enJSRVs is first detected in the conceptus on day 12.62 Interestingly, hyaluronoglucosaminidase 2 (HYAL2), a cellular receptor for both JSRV and enJSRVs Env,6,44 is detected exclusively in the BNC and the multinucleated syncytial plaques of the placenta.62 These observations led to the hypothesis that enJSRVs and HYAL2 are important for placental growth and differentiation in sheep.57 Indeed, injection of morpholinos that inhibit enJSRV Env production into the uteri of pregnant sheep on day 8 of pregnancy compromised conceptus elongation, resulting in reduced mononuclear trophoblast cell outgrowth and loss of trophoblast giant BNC differentiation.66 The biological role of HYAL2 in sheep conceptus development and differentiation has not been determined. Fig. 3 presents a current hypothesis on the biological roles of enJSRVs Env and HYAL2 in trophoblast development and differentiation in the sheep conceptus during early pregnancy.
Fig. 3.
Hypothesis on the biological role of enJSRVs Env and hyaluronoglucosaminidase 2 (HYAL2) in trophoblast differentiation in sheep. During pregnancy, trophoblast giant binucleate cells (BNC) begin to differentiate from mononuclear trophoblast cells (MTC) on day 14. First, MTC begin to express enJSRVs envelope (Env) in the conceptus on day 12 (Step 1). Second, results from microscopy studies support the idea that binucleated trophectoderm cells or trophoblast giant BNC are derived from karyokinesis without cytokinesis (endoreduplication) or mitotic polyploidy (Step 2). Next, the newly formed BNC that are co-expressing enJSRVs env and HYAL2 initially fuse with enJSRVs env-expressing endometrial luminal epithelial (LE) cells, forming a trinucleated fetomaternal hybrid cell (Step 3). During this period, the BNC and LE cells express enJSRV env RNA, whereas only the BNC express HYAL2. In fact, HYAL2 mRNA is not detectable in uterine cells. By days 20–25, virtually all of the endometrial LE cells are fused with the BNC. Fourth, other newly formed BNC fuse with trinucleate cells to form a multinucleated syncytial plaque (Step 4). During most of gestation, the BNC continue to differentiate from the MTC and then fuse with each other and existing multinucleated syncytia to form multinucleated syncytial plaques with 20–25 nuclei. The multinucleated syncytial plaques and BNC form the basis of the cotyledons of the placenta that interdigitate with caruncles of the endometrium to develop and form placentomes.
Interestingly, the enJSRVs Env have a high degree of similarity with the oncogenic exogenous JSRV Env; thus, it is tempting to speculate that both endogenous and exogenous JSRV Env share similar mechanisms to induce trophoblast proliferation/differentiation and cell transformation, respectively, because placental morphogenesis has features similar to tumorigenesis and metastasis.67,68 Although many of these parallels come from comparisons made with the human placenta, trophoblast cells in general have a high proliferation rate, are migratory and invasive, and have the capacity to evade the immune system, which are also characteristics of cancer cells. Thus, it is likely that enJSRV and JSRV Env mediate their effects through the activation of similar albeit not identical pathways.69 Indeed, the Ras-MEK-MAPK, Rac1, and PI3K-Akt-mTOR signaling pathways involved in JSRV-induced cell transformation are important regulators of trophoblast growth and differentiation in human and rodent placentae.69
Conclusions
ERVs are present in the genomes of all vertebrates2 and can be used as DNA fossils to unravel virus– host coevolution over millions of years.8 The domestic sheep constitutes a powerful model to study the biological significance of ERVs given the contemporary presence in this animal species of a pathogenic exogenous retrovirus (JSRV) and the biologically active enJSRVs. Indeed, the study of enJSRVs provided the first in vivo evidence of a physiological role for ERVs in conceptus and placental development.66 Collective evidence from studies of primates, rodents, rabbits, and sheep supports the idea that independent ERVs influenced mammalian evolution and were positively selected for a convergent physiological role in placental morphogenesis. Finally, it is likely that ERVs have other biological roles in reproduction including protection of the host reproductive tract from infectious and pathogenic exogenous retroviruses as well as fetomaternal tolerance.
Acknowledgments
We are grateful to the members of the Laboratory for Uterine Biology and Pregnancy of Texas A&M University and the Laboratory of Viral Pathogenesis of the University of Glasgow Faculty of Veterinary Medicine for stimulating discussions. Work in the laboratory of the authors is supported by NIH grant HD052745, a program grant of the Wellcome Trust and by a Strategic Research Developmental Grant by the Scottish Funding Council.
References
- 1.Boeke JD, Stoye JP. Retrotransposons, endogenous retroviruses and the evolution of retroelements. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. pp. 343–436. [PubMed] [Google Scholar]
- 2.Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, Szustakowki J, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Herniou E, Martin J, Miller K, Cook J, Wilkinson M, Tristem M. Retroviral diversity and distribution in vertebrates. J Virol. 1998;72:5955–5966. doi: 10.1128/jvi.72.7.5955-5966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarlinton RE, Meers J, Young PR. Retroviral invasion of the koala genome. Nature. 2006;442:79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
- 6.Arnaud F, Caporale M, Varela M, Biek R, Chessa B, Alberti A, Golder M, Mura M, Zhang YP, Yu L, Pereira F, Demartini JC, Leymaster K, Spencer TE, Palmarini M. A paradigm for virus-host coevolution: sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog. 2007;3:e170. doi: 10.1371/journal.ppat.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chessa B, Pereira F, Arnaud F, Amorim A, Goyache F, Mainland I, Kao RR, Pemberton JM, Beraldi D, Stear MJ, Alberti A, Pittau M, Iannuzzi L, Banabazi MH, Kazwala RR, Zhang YP, Arranz JJ, Ali BA, Wang Z, Uzun M, Dione MM, Olsaker I, Holm LE, Saarma U, Ahmad S, Marzanov N, Eythorsdottir E, Holland MJ, Ajmone-Marsan P, Bruford MW, Kantanen J, Spencer TE, Palmarini M. Revealing the history of sheep domestication using retrovirus integrations. Science. 2009;324:532–536. doi: 10.1126/science.1170587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin JM. Evolution of retroviruses: fossils in our DNA. Proc Am Philos Soc. 2004;148:264–280. [PubMed] [Google Scholar]
- 9.Bock M, Stoye JP. Endogenous retroviruses and the human germline. Curr Opin Genet Dev. 2000;10:651–655. doi: 10.1016/s0959-437x(00)00138-6. [DOI] [PubMed] [Google Scholar]
- 10.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 11.Varela M, Spencer TE, Palmarini M, Arnaud F. Friendly viruses: the special relationship between endogenous retroviruses and their host. Ann N Y Acad Sci. 2009;1178:157–172. doi: 10.1111/j.1749-6632.2009.05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurth R, Bannert N. Beneficial and detrimental effects of human endogenous retroviruses. Int J Cancer. 2010;126:306–314. doi: 10.1002/ijc.24902. [DOI] [PubMed] [Google Scholar]
- 13.Kalter SS, Helmke RJ, Heberling RL, Panigel M, Fowler AK, Strickland JE, Hellman A. Brief communication: C-type particles in normal human placentas. J Natl Cancer Inst. 1973;50:1081–1084. doi: 10.1093/jnci/50.4.1081. [DOI] [PubMed] [Google Scholar]
- 14.Kalter SS, Heberling RL, Helmke RJ, Panigel M, Smith GC, Kraemer DC, Hellman A, Fowler AK, Strickland JE. A comparative study on the presence of C-type viral particles in placentas from primates and other animals. Bibl Haematol. 1975;40:391–401. doi: 10.1159/000397557. [DOI] [PubMed] [Google Scholar]
- 15.Vernon ML, McMahon JM, Hackett JJ. Additional evidence of type-C particles in human placentas. J Natl Cancer Inst. 1974;52:987–989. doi: 10.1093/jnci/52.3.987. [DOI] [PubMed] [Google Scholar]
- 16.Smith CA, Moore HD. Expression of C-type viral particles at implantation in the marmoset monkey. Hum Reprod. 1988;3:395–398. doi: 10.1093/oxfordjournals.humrep.a136714. [DOI] [PubMed] [Google Scholar]
- 17.Harris JR. The evolution of placental mammals. FEBS Lett. 1991;295:3–4. doi: 10.1016/0014-5793(91)81370-n. [DOI] [PubMed] [Google Scholar]
- 18.DeHaven JE, Schwartz DA, Dahm MW, Hazard ES, 3rd, Trifiletti R, Lacy ER, Norris JS. Novel retroviral sequences are expressed in the epididymis and uterus of Syrian hamsters. J Gen Virol. 1998;79:2687–2694. doi: 10.1099/0022-1317-79-11-2687. [DOI] [PubMed] [Google Scholar]
- 19.Harris JR. Placental endogenous retrovirus (ERV): structural, functional, and evolutionary significance. Bioessays. 1998;20:307–316. doi: 10.1002/(SICI)1521-1878(199804)20:4<307::AID-BIES7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 20.Dupressoir A, Marceau G, Vernochet C, Benit L, Kanellopoulos C, Sapin V, Heidmann T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC, Jr, McCoy JM. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 22.Voisset C, Bouton O, Bedin F, Duret L, Mandrand B, Mallet F, Paranhos-Baccala G. Chromosomal distribution and coding capacity of the human endogenous retrovirus HERV-W family. AIDS Res Hum Retroviruses. 2000;16:731–740. doi: 10.1089/088922200308738. [DOI] [PubMed] [Google Scholar]
- 23.Villarreal LP. On viruses, sex, and motherhood. J Virol. 1997;71:859–865. doi: 10.1128/jvi.71.2.859-865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heidmann O, Vernochet C, Dupressoir A, Heidmann T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new ‘‘syncytin’’ in a third order of mammals. Retrovirology. 2009;6:107. doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venables PJ, Brookes SM, Griffiths D, Weiss RA, Boyd MT. Abundance of an endogenous retroviral envelope protein in placental trophoblasts suggests a biological function. Virology. 1995;211:589–592. doi: 10.1006/viro.1995.1442. [DOI] [PubMed] [Google Scholar]
- 26.Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benirschke K, Kaufmann P. Pathology of the human placenta. 4th Springer Verlag; New York: 1999. [Google Scholar]
- 29.Watson ED, Cross JC. Development of structures and transport functions in the mouse placenta. Physiology (Bethesda) 2005;20:180–193. doi: 10.1152/physiol.00001.2005. [DOI] [PubMed] [Google Scholar]
- 30.Frendo JL, Olivier D, Cheynet V, Blond J-L, Bouton O, Vidaud M, Rabreau M, Evain-Brion D, Mallet F. Direct involvment of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol Cell Biol. 2003;23:3566–3574. doi: 10.1128/MCB.23.10.3566-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Parseval N, Heidmann T. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet Genome Res. 2005;110:318–332. doi: 10.1159/000084964. [DOI] [PubMed] [Google Scholar]
- 32.Blaise S, de Parseval N, Benit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangeney M, Renard M, Schlecht-Louf G, Bouallaga I, Heidmann O, Letzelter C, Richaud A, Ducos B, Heidmann T. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci USA. 2007;104:20534–20539. doi: 10.1073/pnas.0707873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyd MT, Bax CM, Bax BE, Bloxam DL, Weiss RA. The human endogenous retrovirus ERV-3 is upregulated in differentiating placental trophoblast cells. Virology. 1993;196:905–909. doi: 10.1006/viro.1993.1556. [DOI] [PubMed] [Google Scholar]
- 35.Cohen M, Powers M, O’Connell C, Kato N. The nucleotide sequence of the env gene from the human provirus ERV3 and isolation and characterization of an ERV3-specific cDNA. Virology. 1985;147:449–458. doi: 10.1016/0042-6822(85)90147-3. [DOI] [PubMed] [Google Scholar]
- 36.Rote NS, Chakrabarti S, Stetzer BP. The role of human endogenous retroviruses in trophoblast differentiation and placental development. Placenta. 2004;25:673–683. doi: 10.1016/j.placenta.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Lin L, Xu B, Rote NS. Expression of endogenous retrovirus ERV-3 induces differentiation in BeWo, a choriocarcinoma model of human placental trophoblast. Placenta. 1999;20:109–118. doi: 10.1053/plac.1998.0337. [DOI] [PubMed] [Google Scholar]
- 38.de Parseval N, Heidmann T. Physiological knockout of the envelope gene of the single-copy ERV-3 human endogenous retrovirus in a fraction of the Caucasian population. J Virol. 1998;72:3442–3445. doi: 10.1128/jvi.72.4.3442-3445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci USA. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.York DF, Vigne R, Verwoerd DW, Querat G. Nucleotide sequence of the Jaaksiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J Virol. 1992;66:4930–4939. doi: 10.1128/jvi.66.8.4930-4939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmarini M, Sharp JM, De las Heras M, Fan H. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J Virol. 1999;73:6964–6972. doi: 10.1128/jvi.73.8.6964-6972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allen TE, Sherrill KJ, Crispell SM, Perrott MR, Carlson JO, DeMartini JC. The jaagsiekte sheep retrovirus envelope gene induces transformation of the avian fibroblast cell line DF-1 but does not require a conserved SH2 binding domain. J Gen Virol. 2002;83:2733–2742. doi: 10.1099/0022-1317-83-11-2733. [DOI] [PubMed] [Google Scholar]
- 43.Maeda N, Palmarini M, Murgia C, Fan H. Direct transformation of rodent fibroblasts by jaagsiekte sheep retrovirus DNA. Proc Natl Acad Sci USA. 2001;98:4449–4454. doi: 10.1073/pnas.071547598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rai SK, Duh FM, Vigdorovich V, Danilkovitch-Miagkova A, Lerman MI, Miller AD. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc Natl Acad Sci USA. 2001;98:4443–4448. doi: 10.1073/pnas.071572898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu SL, Miller AD. Transformation of madin-darby canine kidney epithelial cells by sheep retrovirus envelope proteins. J Virol. 2005;79:927–933. doi: 10.1128/JVI.79.2.927-933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zavala G, Pretto C, Chow Y-HJ, Jones L, Alberti A, Grego E, De las Heras M, Palmarini M. Relevance of Akt phosphorylation in cell transformation induced by jaagsiekte sheep retrovirus. Virology. 2003;312:95–105. doi: 10.1016/s0042-6822(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 47.Caporale M, Cousens C, Centorame P, Pinoni C, De las Heras M, Palmarini M. Expression of the jaagsiekte sheep retrovirus envelope glycoprotein is sufficient to induce lung tumors in sheep. J Virol. 2006;80:8030–8037. doi: 10.1128/JVI.00474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wootton SK, Halbert CL, Miller AD. Sheep retrovirus structural protein induces lung tumours. Nature. 2005;434:904–907. doi: 10.1038/nature03492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danilkovitch-Miagkova A, Duh FM, Kuzmin I, Angeloni D, Liu SL, Miller AD, Lerman MI. Hyaluronidase 2 negatively regulates RON receptor tyrosine kinase and mediates transformation of epithelial cells by jaagsiekte sheep retrovirus. Proc Natl Acad Sci USA. 2003;100:4580–4585. doi: 10.1073/pnas.0837136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varela M, Chow YH, Sturkie C, Murcia P, Palmarini M. Association of RON tyrosine kinase with the Jaagsiekte sheep retrovirus envelope glycoprotein. Virology. 2006;350:347–357. doi: 10.1016/j.virol.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 51.Maeda N, Fu W, Ortin A, de las Heras M, Fan H. Roles of the Ras-MEK-mitogen-activated protein kinase and phosphatidylinositol 3-kinase-Akt-mTOR pathways in Jaagsiekte sheep retrovirus-induced transformation of rodent fibroblast and epithelial cell lines. J Virol. 2005;79:4440–4450. doi: 10.1128/JVI.79.7.4440-4450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Las Heras M, Ortin A, Benito A, Summers C, Ferrer LM, Sharp JM. In-situ demonstration of mitogen-activated protein kinase Erk 1 / 2 signalling pathway in contagious respiratory tumours of sheep and goats. J Comp Pathol. 2006;135:1–10. doi: 10.1016/j.jcpa.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Palmarini M, Maeda N, Murgia C, De-Fraja C, Hofacre A, Fan H. A phosphatidylinositol-3-kinase (PI-3K) docking site in the cytoplasmic tail of the Jaagsiekte sheep retrovirus transmembrane protein is essential for envelope-induced transformation of NIH3T3 cells. J Virol. 2001;75:11002–11009. doi: 10.1128/JVI.75.22.11002-11009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maeda N, Fan H. Signal transduction pathways utilized by enzootic nasal tumor virus (ENTV-1) envelope protein in transformation of rat epithelial cells resemble those used by jaagsiekte sheep retrovirus. Virus Genes. 2008;36:147–155. doi: 10.1007/s11262-007-0193-x. [DOI] [PubMed] [Google Scholar]
- 55.Guillomot M. Cellular interactions during implantation in domestic ruminants. J Reprod Fertil Suppl. 1995;49:39–51. [PubMed] [Google Scholar]
- 56.Spencer TE, Johnson GA, Bazer FW, Burghardt RC. Implantation mechanisms: insights from the sheep. Reproduction. 2004;128:657–668. doi: 10.1530/rep.1.00398. [DOI] [PubMed] [Google Scholar]
- 57.Spencer TE, Johnson GA, Bazer FW, Burghardt RC, Palmarini M. Pregnancy recognition and conceptus implantation in domestic ruminants: roles of progesterone, interferons and endogenous retroviruses. Reprod Fertil Dev. 2007;19:65–78. doi: 10.1071/rd06102. [DOI] [PubMed] [Google Scholar]
- 58.Wooding FB. Role of binucleate cells in fetomaternal cell fusion at implantation in the sheep. Am J Anat. 1984;170:233–250. doi: 10.1002/aja.1001700208. [DOI] [PubMed] [Google Scholar]
- 59.Wooding FB. Current topic: the synepitheliochorial placenta of ruminants: binucleate cell fusions and hormone production. Placenta. 1992;13:101–113. doi: 10.1016/0143-4004(92)90025-o. [DOI] [PubMed] [Google Scholar]
- 60.Hoffman LH, Wooding FB. Giant and binucleate trophoblast cells of mammals. J Exp Zool. 1993;266:559–577. doi: 10.1002/jez.1402660607. [DOI] [PubMed] [Google Scholar]
- 61.Igwebuike UM. Trophoblast cells of ruminant placentas–A minireview. Anim Reprod Sci. 2006;93:185–198. doi: 10.1016/j.anireprosci.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 62.Dunlap KA, Palmarini M, Adelson DL, Spencer TE. Sheep endogenous betaretroviruses (enJSRVs) and the hyaluronidase 2 (HYAL2) receptor in the ovine uterus and conceptus. Biol Reprod. 2005;73:271–279. doi: 10.1095/biolreprod.105.039776. [DOI] [PubMed] [Google Scholar]
- 63.Palmarini M, Hallwirth C, York D, Murgia C, de Oliveira T, Spencer T, Fan H. Molecular cloning and functional analysis of three type D endogenous retroviruses of sheep reveals a different cell tropism from that of the highly related exogenous jaagsiekte sheep retrovirus. J Virol. 2000;74:8065–8076. doi: 10.1128/jvi.74.17.8065-8076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spencer TE, Stagg AG, Joyce MM, Jenster G, Wood CG, Bazer FW, Wiley AA, Bartol FF. Discovery and characterization of endometrial epithelial messenger ribonucleic acids using the ovine uterine gland knockout model. Endocrinology. 1999;140:4070–4080. doi: 10.1210/endo.140.9.6981. [DOI] [PubMed] [Google Scholar]
- 65.Palmarini M, Gray CA, Carpenter K, Fan H, Bazer FW, Spencer T. Expression of endogenous betaretroviruses in the Ovine uterus: effects of neonatal age, estrous cycle, pregnancy and progesterone. J Virol. 2001;75:11319–11327. doi: 10.1128/JVI.75.23.11319-11327.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, Spencer TE. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci USA. 2006;103:14390–14395. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferretti C, Bruni L, Dangles-Marie V, Pecking AP, Bellet D. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum Reprod Update. 2007;13:121–141. doi: 10.1093/humupd/dml048. [DOI] [PubMed] [Google Scholar]
- 68.Soundararajan R, Rao AJ. Trophoblast ‘pseudo-tumorigenesis’: significance and contributory factors. Reprod Biol Endocrinol. 2004;2:15. doi: 10.1186/1477-7827-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pollheimer J, Knofler M. Signalling pathways regulating the invasive differentiation of human trophoblasts: a review. Placenta. 2005;26(Suppl A):S21–S30. doi: 10.1016/j.placenta.2004.11.013. [DOI] [PubMed] [Google Scholar]