Abstract
Isoprenoid biosynthesis in plant plastids occurs via the 1-deoxy-d-xylulose 5-phosphate (DXP) pathway. We used tobacco rattle virus (TRV) to posttranscriptionally silence the expression of the last two enzymes of this pathway, the IspG-encoded (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase (HDS) and the IspH-encoded isopentenyl/dimethylallyl diphosphate synthase (IDDS), as well as isopentenyl/dimethylallyl diphosphate isomerase (IDI), the enzyme that interconverts IPP and DMAPP. TRV-IspG and TRV-IspH infected Nicotiana benthamiana plants had albino leaves that contained less than 4% of the chlorophyll and carotenoid pigments of control leaves. We applied [13C]DXP and [14C]DXP to silenced leaves and found that 2-C-methyl-d-erythritol 2,4-cyclodiphosphate accumulated in plants blocked at HDS while DXP, (E)-4-hydroxy-3-methylbut-2-enyl phosphate and (E)-2-methylbut-2-ene-1,4-diol accumulated in IDDS-blocked plants. Albino leaves from IspG- and IspH-silenced plants displayed a disorganized palisade mesophyll, reduced cuticle, fewer plastids, and disrupted thylakoid membranes. These findings demonstrate the participation of HDS and IDDS in the DXP pathway in plants, and support the view that plastid isoprenoid biosynthesis is metabolically and physically segregated from the mevalonate pathway. IDI-silenced plants had mottled white-pale green leaves with disrupted tissue and plastid structure, and showed an 80% reduction in pigments compared to controls. IPP pyrophosphatase activity was higher in chloroplasts isolated from IDI-silenced plants than in control plant chloroplasts. We suggest that a low level of isoprenoid biosynthesis via the DXP pathway can occur without IDI but that this enzyme is required for full function of the DXP pathway.
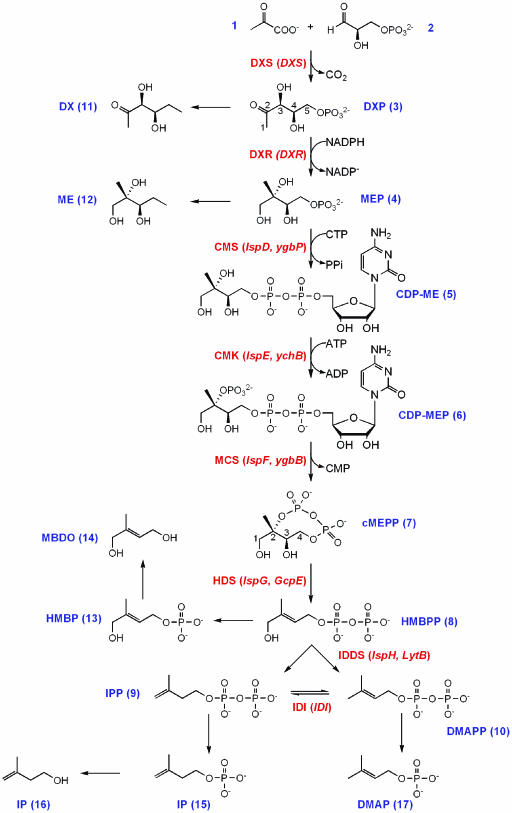
Isoprenoids (also called terpenoids or terpenes) form one of the largest and most structurally diverse groups of metabolites in nature. Members of the plant kingdom alone are estimated to produce >30,000 isoprenoid compounds (Gershenzon, personal communication), many of them secondary metabolites with defensive or signaling properties (Croteau et al., 2000). Plants synthesize all isoprenoids from two C5 precursors, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), with DMAPP serving as a starter unit for chain elongation reactions that yield C10, C15, C20, C30, and C40 diphosphate esters. Such prenyl diphosphates are further metabolized to give monoterpenes (C10), sesquiterpenes (C15), diterpenes, and the phytol side chain of chlorophylls (C20), sterols (C30), and carotenoids (C40). In plants, IPP and DMAPP are synthesized by independent metabolic pathways, which are active in different cellular compartments. The mevalonate (MVA) pathway, which provides C5 precursors for the synthesis of some sesquiterpenes, sterols and the side chain of ubiquinone (in higher plants), is localized in the cytoplasm while the plastid localized 1-deoxy-d-xylulose 5-phosphate (DXP) pathway (Fig. 1) produces the IPP and DMAPP used to synthesize monoterpenes, some sesquiterpenes and diterpenes. The DXP pathway is alternatively known as the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway. Perhaps most significantly for plant function, the two major classes of photosynthetic pigments, chlorophylls and carotenoids, are formed in the plastid with IPP and DMAPP from the DXP pathway (Arigoni et al., 1997; Eisenreich et al., 2001).
Figure 1.
The DXP pathway to isoprenoids. Enzymes of the pathway are labeled in bold with the corresponding genes in parentheses: DXS, DXP synthase; DXR, DXP reductoisomerase; CMS, CDP-ME synthase; CMK, CDP-ME kinase; MCS, cMEPP synthase; HDS, HMBPP synthase; IDDS, IPP/DMAPP synthase; IDI, IPP/DMAPP isomerase. Precursors, intermediates, and products (compounds 1–10) of the DXP pathway, and metabolites derived from them (compounds 11–17), are numbered in bold: 1, pyruvate; 2, d-glyceraldehyde 3-phosphate; 3, 1-deoxy-d-xylulose 5-phosphate (DXP); 4, 2-C-methyl-d-erythritol 4-phosphate (MEP); 5, 4-diphosphocytidyl 2-C-methyl-d-erythritol (CDP-ME); 6, 4-diphosphocytidyl 2-C-methyl-d-erythritol 2-phosphate (CDP-MEP); 7, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (cMEPP); 8, (E)-4-hydroxy-3-methylbut-2-enyl diphosphate (HMBPP); 9, isopentenyl diphosphate (IPP); 10, dimethylallyl diphosphate (DMAPP); 11, 1-deoxy-d-xylulose (DX); 12, 2-C-methyl-d-erythritol (ME); 13, (E)-4-hydroxy-3-methylbut-2-enyl phosphate (HMBP); 14, (E)-2-methylbut-2-ene-1,4-diol (MBDO) 15, isopentenyl phosphate (IP); 16, isopentenol; 17, dimethylallyl phosphate (DMAP).
The enzymatic steps of the DXP pathway and the genes encoding them have been completely elucidated in Escherichia coli and for most steps, also in plants (for reviews, see Rodríguez-Concepción and Boronat, 2002; Rohdich et al., 2003; Rohmer, 2003). The penultimate enzyme of the DXP pathway is (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase (HDS), encoded in E. coli by the IspG/GcpE gene (Cunningham et al., 2000; Altincicek et al., 2001), which has been shown to catalyze the formation of (E)-4-hydroxy-3-methylbut-2-enyl diphosphate (HMBPP) from 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (cMEPP; Hecht et al., 2001). An Arabidopsis IspG-like cDNA complements an IspG-defective E. coli strain (Querol et al., 2002), providing evidence that the plant HDS has a similar function to its bacterial homolog. The final step in the DXP pathway was recently shown to be catalyzed by isopentenyl/dimethylallyl diphosphate synthase (IDDS, encoded by the E. coli IspH/LytB gene and its homologs in other organisms), which converts HMBPP to IPP and DMAPP (Rohdich et al., 2002). While clarified both genetically (Cunningham et al., 2000; McAteer et al., 2001) and biochemically in bacteria, the role of IspH has yet to be functionally analyzed in plants. IPP and DMAPP are interconverted by isopentenyl/dimethylallyl diphosphate isomerase (IDI; EC 5.3.3.2), the protein product of the IDI gene. IDI plays a central role in terpene biosynthesis, apparently regulating the ratio of C5 precursors from the DXP and mevalonate pathways in the plastid and cytosol, respectively. IDI is a nonessential gene in E. coli (Hahn et al., 1999) and it is not clear if it is absolutely required in plants.
We used virus-induced gene silencing (VIGS) to analyze the function of IspG, IspH, and IDI in plants. VIGS exploits the RNA silencing pathway to posttranscriptionally block expression of host plant genes (Baulcombe, 1999; Voinnet, 2001). Plant mRNAs targeted in this manner are degraded before translation can occur, leading to a knockout phenotype for the gene of interest. A well-characterized VIGS system is the use of tobacco rattle virus (TRV) to silence genes in Nicotiana benthamiana Domin (Solanaceae). TRV-mediated silencing of phytoene desaturase (PDS), an essential gene of carotenoid biosynthesis, has been demonstrated in N. benthamiana (Ratcliff et al., 2001; Liu et al., 2002). Silencing of PDS leads to photobleaching of the photosynthetic apparatus, and therefore albino leaves. We reasoned that the DXP pathway would be amenable to dissection using VIGS since knockouts would block the formation of photosynthetic pigments, as has been demonstrated for the Arabidopsis cla1-1 mutant in which the DXS gene is disrupted (Mandel et al., 1996). As well, blocking the DXP pathway by mutagenesis or loss-of-function transformation approaches typically produces an embryo lethal phenotype (see Budziszewski et al., 2001). The transient nature of VIGS allowed us to overcome this problem by silencing only in virus-infected leaves.
In this paper we show that the VIGS knockout of IspG and IspH produces plants with albino leaves due to the impairment of the ability to synthesize chlorophylls and carotenoids via the DXP pathway. Silencing IDI produced plants with a mottled white-pale green leaf phenotype. Microscopic examination of silenced tissue found abnormalities in leaf ultrastructure, including degraded chloroplasts, in IspG-, IspH-, and IDI-silenced plants. We characterized the metabolic blocks in IspG- and IspH-silenced plants by feeding labeled DXP and profiling the intermediates of the DXP pathway. In the case of IspG, albino leaves accumulated cMEPP, the substrate of the IspG-encoded HDS enzyme, and leaves of IspH silenced plants accumulated HMBP and MBDO, which are dephosphorylated congeners of HMBPP, the substrate of the IspH-encoded IDDS enzyme. These results provide the first evidence that selective knockout of IspG and IspH prevents the formation of plant isoprenoids. Furthermore, it confirms the fundamental metabolic and physical separation of the MVA and DXP pathways. Silencing of IDI also led to decreased chlorophyll and carotenoid levels, although somewhat less than that observed with IspG and IspH silencing, demonstrating that IDI, unlike its E. coli homolog, is required for normal functioning of isoprenoid biosynthesis in plant plastids.
RESULTS
Cloning of TRV-IspG, -IspH, and -IDI Silencing Constructs
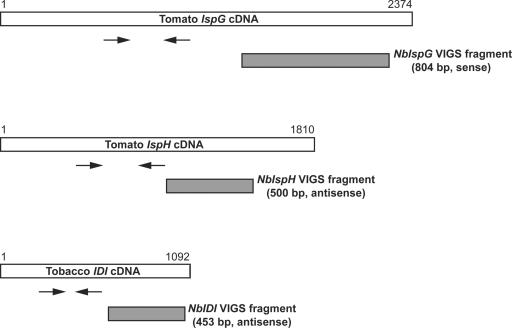
Full-length sequences for N. benthamiana IspG, IspH, and IDI cDNAs have not been published and we used sequences from tobacco (Nicotiana tabacum) and other Solanaceae to design primers to amplify cDNA fragments. PCR with IspG primers gave an 804-bp product, NbIspG, which showed 90% identity with tomato (Lycopersicon esculentum) IspG cDNA. In the same fashion, a 500-bp NbIspH and a 453-bp NbIDI cDNA fragment were amplified. NbIspH was most closely related (98% identity) to a partial cDNA clone of the tobacco IspH gene (AF159699). NbIDI showed 94% nucleotide identity with the IPP1 gene encoding the plastidic isoform of tobacco IDI, compared with 87% identity with the IPP2 gene encoding the cytosolic isoform (Nakamura et al., 2001). The position of N. benthamiana cDNA fragments relative to their tomato (IspG, IspH) and tobacco (IDI) full-length homologs is illustrated in Figure 2.
Figure 2.
Schematic representation of IspG, IspH, and IDI cDNAs and PCR-amplified fragments used for gene silencing. The locations of the cDNA fragments used for silencing (shaded bars) and the primer pairs used for RT-PCR analysis (arrows) are indicated. Since the complete sequences of IspG, IspH, and IDI cDNAs from N. benthamiana are not known, tomato IspG (GenBank accession no. AF435086), tomato IspH (TIGR TC124188), and tobacco IDI (GenBank accession no. AB049815) cDNAs were used for comparison.
Our strategy for constructing TRV plasmids was to ligate PCR amplified cDNAs into the cloning vector, pGEM-Teasy. cDNA fragments were excised by restriction enzyme digestion and ligated into pTV00, a binary vector that expresses a modified TRV-RNA2 under the control of the 35S promoter (Ratcliff et al., 2001). This strategy introduced 4 bp and 46 bp of the pGEM-Teasy multiple cloning site at the 5′ and 3′ end of the inserted cDNAs, respectively. The IspG fragment was in the sense orientation relative to the TRV coat protein while the IspH and IDI fragments were in the antisense orientation. TRV-IspG, -IspH, and -IDI constructs were coinfiltrated with Agrobacterium tumefaciens containing pBINTRA6, a binary vector construct of TRV-RNA1 (Ratcliff et al., 2001), into N. benthamiana leaves. Mock infected plants infiltrated with the A. tumefaciens resuspension buffer and plants infected with pTV00 plasmid without a cDNA insert (empty TRV) served as controls.
Phenotypes of TRV Silenced Plants
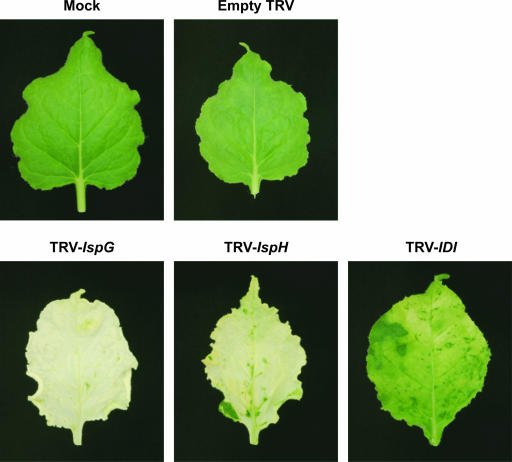
As reported by Ratcliff et al. (2001), infection with TRV gave N. benthamiana plants with a slightly stunted appearance and some crinkled leaves, the latter occurring mainly within the first 10 d after infiltration. Emerging leaves of TRV-IspG and TRV-IspH infected plants showed chlorotic patches 8 to 10 d after infiltration and a distinct leaf phenotype was apparent for all three silencing constructs after 4 weeks (Fig. 3). Most leaves of IspG-silenced plants were white or very pale yellow with wavy leaf margins, although some had islands of green tissue within areas of white tissue. TRV-IspH gave a more variable VIGS phenotype with limited silencing near leaf veins in some plants while others had fully white or pale yellow leaves. IspH-silenced leaves also showed a wavy leaf margin. Leaves from plants infected with TRV-IDI had a mottled white-pale green appearance, often with green tips, and lacked the wavy margin of IspG- and IspH-silenced plants. No changes in pigmentation were observed in mock infected and empty TRV infected plants.
Figure 3.
Leaves of N. benthamiana plants with TRV silenced IspG, IspH, and IDI genes. N. benthamiana plants were infected with TRV viruses containing IspG, IspH, and IDI cDNA fragments. Mock and empty TRV infected plants exhibited no white tissue while silencing of IspG and IspH produced an albino phenotype of white or pale yellow leaf tissue. VIGS of IDI led to mottled pale green leaves but no fully white tissue. Photographed leaves represent typical phenotype for the mock, empty TRV, and TRV-VIGS constructs 4 weeks after infiltration.
Anatomy of Silenced Leaves
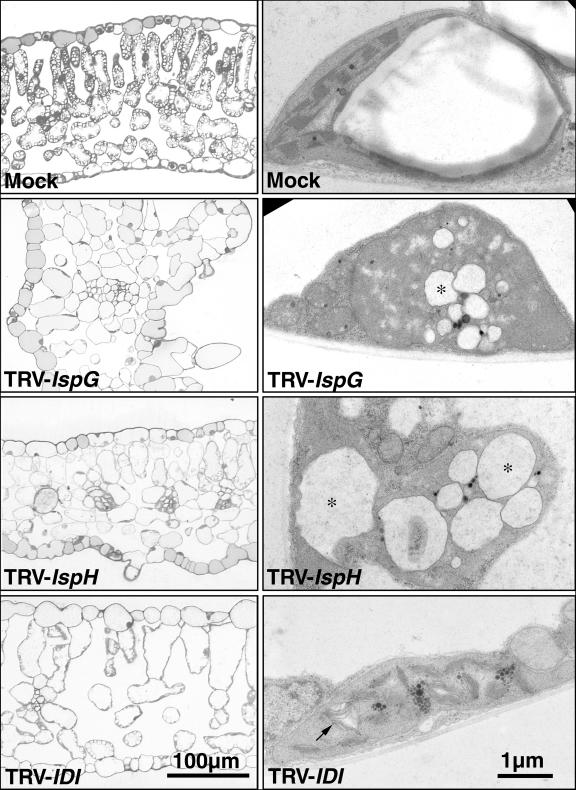
To further characterize the IspG-, IspH-, and IDI-silencing phenotypes, we examined leaf ultrastructure using light and transmission electron microscopy (Fig. 4). Leaf tissue from mock and empty TRV infected plants was indistinguishable, indicating that TRV infection itself had little effect on tissue or plastid structure. A thick cuticle, organized palisade mesophyll, and numerous large starch granules within chloroplasts of mesophyll cells were characteristic features of control leaves. As well, electron microscopy showed that chloroplasts in such plants had robust grana with intact thylakoid membranes. Albino tissue of TRV-IspG infected plants was characterized by a number of unusual features including epidermal protuberances, a cuticle of reduced thickness, and disorganized palisade mesophyll. Chloroplasts were reduced in number and size in all cell types and starch granules were not visible. In chloroplasts that were present, thylakoid membranes were disrupted and globular lytic vesicles were observed. IspH-silenced tissues also had epidermal protuberances, reduced cuticle, and disorganized palisade mesophyll structure, although these features were less pronounced than in IspG-silenced plants. Although chloroplasts were also reduced in number and had severely disrupted thylakoid membranes in most cell types, chloroplasts in vascular tissue and companion cells of IspH-silenced plants appeared normal. The features that marked the albino tissue of IspG and IspH plants were not as distinct in TRV-IDI infected plants. Such leaves contained a normal number of chloroplasts although starch granules were still absent. A notable feature of IDI-silenced leaf tissue was that chloroplasts contained thylakoid membranes with an unusual fluted structure (Fig. 4).
Figure 4.
Microscopic analysis of leaf mesophyll and plastids of mock and TRV-VIGS infected N. benthamiana plants. Light microscopic images (left column) of green tissue from a mock control plant leaf, and silenced tissue of TRV-IspG, -IspH, and -IDI infected leaves. Transmission electron microscopic (right column) images of plastids from the same leaf tissues. Lytic vesicles in TRV-IspG and TRV-IspH plants are marked with an asterisk (*). The unusual structure of thylakoid membranes TRV-IDI plants are indicated with an arrow.
Semiquantitative RT-PCR Analysis of Gene Expression in Silenced Tissue
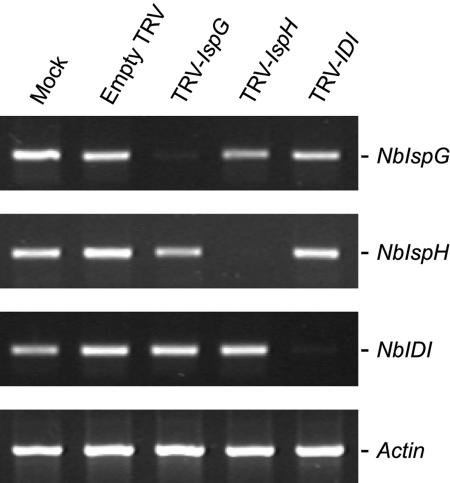
We measured transcript levels of NbIspG, NbIspH, and NbIDI in silenced tissue using RT-PCR with primers annealing at the 5′ end of the silenced genes, outside of the region used for the TRV constructs (Fig. 2). Actin was amplified as a control transcript. The accumulation of each of the three transcripts was reduced by more than 90% in silenced tissue (Fig. 5).
Figure 5.
RT-PCR analysis of IspG, IspH, and IDI gene expression in N. benthamiana plants infected with TRV-IspG, -IspH, and -IDI constructs. Ethidium bromide stained agarose gels showing typical amplification products from 30 cycles of PCR with IspG-, IspH-, IDI-, and Actin-specific primers. First-strand cDNA templates were synthesized from total RNA isolated from silenced leaf tissue from TRV-IspG, TRV-IspH, and TRV-IDI infected plants or equivalent tissue from mock and empty TRV infected plants. An N. benthamiana Actin gene was amplified to determine equal amounts of cDNA template.
Analysis of Photosynthetic Pigments in Silenced Leaf Tissue
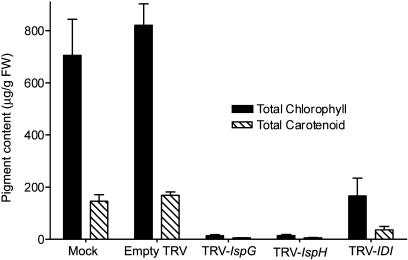
The effect of knocking out IspG, IspH, and IDI expression on isoprenoid biosynthesis was measured by quantifying total chlorophylls and carotenoids in silenced leaves (Fig. 6). Silenced tissue was separated from green tissue and extracted with organic solvent. For mock and empty TRV infected plants, tissue from an equivalent location and leaf age to that used for silenced samples was used. The content of total chlorophylls and carotenoids was slightly higher in leaves of empty TRV infected plants compared to mock infected, an observation that was consistent throughout our experiments. Silenced tissue in TRV-IspG infected plants had total chlorophyll and carotenoid levels that were reduced by 98% and 96% of mock-infected plants, respectively. In TRV-IspH infected leaves pigment levels were reduced by 98% of mock infected for total chlorophylls and 96% for total carotenoids. IDI silencing produced a less dramatic reduction in photosynthetic pigment levels compared to the other TRV constructs; a result in agreement with mottled green appearance of TRV-IDI infected leaves. Total chlorophylls in TRV-IDI leaves were reduced 80%, and total carotenoids by 78%, compared to mock infected leaves.
Figure 6.
Concentrations of photosynthetic pigments in leaf tissues of VIGS silenced N. benthamiana plants compared to mock infected and empty TRV infected plants. The content of total chlorophylls and carotenoids in TRV silenced leaf tissue or equivalent tissue from mock and empty TRV leaves was determined spectrophotometrically. Values represent mean ± sd for three plants.
Metabolic Profiles After [13C]/[14C]DXP Feeding to TRV-IspG and TRV-IspH Silenced Plants
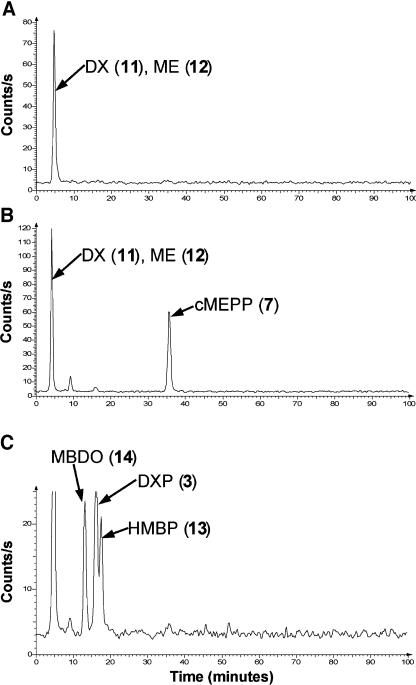
To examine the effect that silencing IspG and IspH has on the intermediates of the DXP pathway, we fed [14C]DXP to detached leaves of mock and empty TRV infected control plants and to albino leaves of TRV-IspG and TRV-IspH silenced plants. This feeding study assumed that DXP is dephosphorylated and then rephosphorylated after entry into cells by d-xylulokinase (Wungsintaweekul et al., 2001). Leaves were extracted, partitioned into organic and aqueous phases, and the amount of radioactivity present in intermediates of the DXP pathway analyzed using a combination of radioHPLC and TLC. The results of this analysis are shown in Table I and Figure 7. As expected for leaves unable to synthesize carotenoids and other isoprenoids, IspG- and IspH-silenced plants incorporated significantly less [14C]DXP into organic soluble metabolites than control plants. Using ion-pair radioHPLC to analyze the aqueous fraction of control plants, we found that more than 90% of the radioactivity was incorporated into the dephosphorylated compounds, DX and ME, which are not separable using our HPLC system (Fig. 7A). In IspG-silenced plants, only about 50% of the radioactivity was present in DX/ME and more than 30% was found in a peak, having the same retention time as authentic cMEPP (Rt, approximately 36 min), that was not present in control plants (Fig. 7B). Treatment by alkaline phosphatase did not alter the retention time of this metabolite. To identify this peak, we fed a mixture of [3,4,5-13C3]- and [3,4,5-14C3]DXP to detached leaves of IspG-silenced plants and purified the corresponding peak using DEAE Sephadex chromatography with detection of radioactive fractions by scintillation counting. Mass spectroscopic and 13C-NMR analysis showed the purified compound was unequivocally [1,3,4-13C3]cMEPP. cMEPP is the substrate for HDS, and therefore a block at this step would be expected to lead to a build up of cMEPP. Metabolites that occur later in the pathway (i.e. HMBPP) were not detected in TRV-IspG infected leaves.
Table I.
Incorporation of [14C]DXP into the intermediates of the DXP pathway
| Mock | Empty TRV | TRV-IspG | TRV-IspH | |
|---|---|---|---|---|
| Incorporation in organic phasea | 49 | 43 | 6 | 12 |
| Incorporation in water phasea | 54 | 55 | 93 | 86 |
| cMEPP (7)b | nd | nd | 40 | nd |
| HMBP (13)b | nd | nd | nd | 10 |
| HMBPP (8)b | nd | nd | nd | nd |
| MBDO (14)b | nd | nd | nd | 11 |
Detached leaves of mock and empty TRV infected control plants, and plants infected with TRV-IspG and TRV-IspH, were fed an aqueous solution of 0.5 μCi [3,4,5–14C3]DXP. After incorporation of the labeled precursor over approximately 24 h, the leaves were extracted and the amount of radioactivity present in intermediates of the DXP pathway was determined using scintillation counting and ion-pair radioHPLC.
Values represent percentage compared to the total amount of radioactivity present in ethanolic extract.
Determined by HPLC analysis of the aqueous phase. Values represent percentage compared to the total amount of radioactivity detected by radioHPLC. nd, Not detectable.
Figure 7.
Accumulation of DXP pathway metabolites in green leaf tissue of mock control plants and albino leaf tissue of IspG- and IspH-silenced plants. Detached leaves of TRV-IspG and TRV-IspH infected plants or equivalent green leaves of mock control plants were fed [3,4,5-14C3]DXP (0.5 μCi) over approximately 24 h. The leaves were extracted with boiling 80% (v/v) ethanol, the solvent removed, and the residue partitioned between water and ethyl acetate and the aqueous phase was analyzed by radioHPLC. The peaks indicated were identified by comparison with authentic standards or by purification and structural elucidation. A, Chromatogram of mock infected control plant. Results from mock infected and empty virus plants were equal. B, Chromatogram of TRV-IspG silenced leaf tissue. C, Chromatogram of TRV-IspH silenced leaf tissue. Unlabelled peak at approximately 5 min is a mixture of DX and ME.
Incorporation of [14C]DXP by IspH-silenced leaves led to the accumulation of two new 14C-labeled compounds (Rt approximately 13 and 18 min) not found in either control or TRV-IspG silenced plants, together with a substantial amount of [14C]DXP (Rt, approximately 16 min; Fig. 7C). Based on the comparison of retention times with reference compounds, we tentatively identified the 13 min peak as MBDO. For conclusive identification, we fed detached leaves with [3,4,5-13C3]DXP mixed with a small amount of [3,4,5-14C3]DXP. The leaves were worked up as described above, the aqueous fraction treated with alkaline phosphatase, and the putative MBDO peak purified using TLC. Subsequent mass spectrometric analysis unequivocally demonstrated it to be MBDO, as evidenced by the three 13C-atoms from the applied [3,4,5-13C3]DXP. The 18-min peak shared exact retention time characteristics with an authentic standard of HMBP, a compound not previously reported. Its identification as such was further supported by its conversion to MBDO upon alkaline phosphatase treatment (data not shown). MBDO and HMBP are both dephosporylated congeners of HMBPP, the substrate of IDDS.
Dephosphorylation of IPP by Isolated Chloroplasts from Mock and TRV-IDI Plants
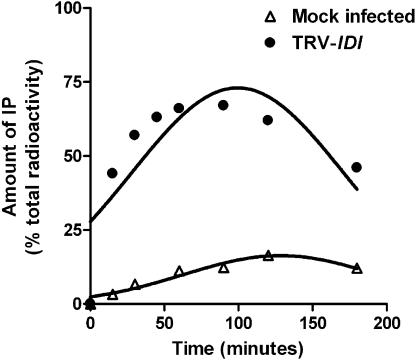
To examine the effect of silencing IDI on the metabolism of IPP, chloroplasts isolated from green leaf tissue from mock infected and white-pale green tissue from TRV-IDI infected plants were incubated with [1-14C]IPP. RadioHPLC analysis found that IPP was dephosphorylated to isopentenyl monophosphate (IP) by chloroplasts of both plants. This conversion was significantly higher in TRV-IDI leaf tissue compared to mock infected tissue. Kinetic analysis (Fig. 8) showed that after a 180-min incubation, less than 20% of the applied [1-14C]IPP was converted to IP by chloroplasts from mock infected plants while more than 50% was converted by chloroplasts from tissue in which IDI was silenced. We also detected significant conversion of IP further to isopentenol in IDI-silenced plants (data not shown).
Figure 8.
Dephosphorylation of [1-14C]IPP to [1-14C]IP by isolated chloroplast from mock infected and TRV-IDI infected plants. Chloroplasts were isolated from green tissue of mock infected plants and from white-pale green tissue of TRV-IDI infected plants and incubated with [1-14C]IPP (0.1 μCi) at 30°C. At 15, 30, 45, 60, 90, 120, and 180 min, the reactions were stopped and the amounts of [1-14C]IPP and [1-14C]IP determined by radioHPLC.
DISCUSSION
We analyzed the function of IspG, IspH, and IDI cDNAs from N. benthamiana using a VIGS approach. The albino leaves of IspG- and IspH-silenced plants, which contained less than 4% of the isoprenoid-derived chlorophyll and carotenoid pigments of control plants, provide evidence for the participation of the IspG-encoded HDS and IspH-encoded IDDS in the DXP pathway. Neither HDS nor IDDS have been previously characterized by knockout in plants; indeed, this is the first report to our knowledge to prove the role of a plant IDDS in the DXP pathway. We analyzed the metabolic blocks caused by silencing using feeding studies with [13C]DXP/[14C]DXP. Leaves in which HDS expression was silenced converted labeled DXP to DX, ME, and cMEPP, the latter of which was purified and identified spectroscopically. cMEPP is the substrate of HDS and has been shown to accumulate in E. coli cells overexpressing a synthetic operon containing the genes (DXS-DXR-IspD-IspE-IspF) in the DXP pathway up to the HDS catalyzed step (Hecht et al., 2001). This compound also builds up in gram-negative bacteria under “oxidative stress” conditions (Ostrovsky et al., 1992; Ostrovsky et al., 1998) and therefore serves as a stable metabolic sink in the DXP pathway. Metabolites that occur after the HDS-catalyzed step in the pathway were not detected in IspG-silenced plants despite the sensitivity afforded by the incorporation of [14C]DXP. In addition to the desphorylated intermediates DX and ME detected in IspG-silenced plants, leaves blocked at the IspH-encoded IDDS step showed incorporation of [14C]DXP into MBDO and HMBP, as identified by comparison of their retention times with standards and mass spectroscopic analysis. A possible explanation for the accumulation of MBDO and HMBP in IspH-silenced plants is that HMBPP is metabolically unstable, and is dephosphorylated first to HMBP (Fig. 1, 13) and finally to the metabolic inert MBDO (Fig. 1, 14).
The albino phenotype caused by IspG- and IspH-silencing parallels that of loss-of-function mutants of the DXP pathway. T-DNA insertions in DXS were found to be responsible for the albino Arabidopsis cla1-1 mutant (Mandel et al., 1996; Estévez et al., 2000) and a mutant uncovered in a seedling-lethal screen (Budziszewski et al., 2001). The latter study also found albino mutants resulting from disruption of DXR and IspD. Cosuppression of DXR gave peppermint (Mentha piperita) plants with light green leaves and reduced chlorophyll levels (Mahmoud and Croteau, 2001. As well, Arabidopsis plants with a lethal albino phenotype were observed after transformation of an antisense construct of CMS (Okada et al., 2002). It is worth noting that the DXP pathway is also blocked using fosmidomycin, an inhibitor of DXR (Okada et al., 2002; Zeidler et al., 1998).
VIGS seems an ideal method for knockout of these genes as it allowed us to overcome the problem of seedling lethality or dwarfism that characterizes plants blocked in the DXP pathway. N. benthamiana plants infected with TRV silencing constructs of PDS (Ratcliff et al., 2001) or IspG, IspH, and IDI (this study) show albino tissue only in leaves that develop from infected meristems, while the lower green leaves continue to support plant growth. VIGS knockouts also obviate the need to culture DXP pathway blocked mutants such as cla1-1 on medium containing Suc. Kasahara et al. (2002) suggest that Suc may perturb the MVA and DXP pathways, thus complicating experiments in blocked mutants. The VIGS approach to loss-of-function knockouts in the DXP pathway gave a near complete block at the targeted enzymatic steps. This was demonstrated by the albino leaf phenotypes, semiquantitative reverse transcription (RT)-PCR analysis showing reduced transcript levels and the absence of metabolites occurring after the blocked enzyme steps. Okada et al. (2002) found that Arabidopsis CMK antisense plants or those treated with fosmidomycin contained 7% of the chlorophyll and 14% of the carotenoids of wild-type plants. VIGS silencing of IspG and IspH, albeit with different genes and only in infected tissues, produced leaves containing less than 4% of control pigment levels. These findings clearly demonstrate the utility of VIGS for silencing genes that are likely to result in seedling lethality.
Plants infected with TRV-IspG and TRV-IspH plants displayed somewhat different leaf phenotypes. Plants infected with TRV-IspG developed albino leaves earlier than TRV-IspH plants and IspG silencing typically resulted in a greater proportion of fully white leaves than IspH plants, which often had white tissue only along leaf veins. Although some IspH-silenced plants developed fully white leaves after virus infection had proceeded for 2 to 3 weeks, the ability of the TRV-IspH construct to spread was impaired relative to TRV-IspG. Insertion of heterologous sequences into a viral genome may alter replication or infectivity. The observed differences in silencing phenotype may therefore be due to intrinsic properties, such as RNA secondary structure, of the IspG and IspH cDNA fragments used for silencing. It is interesting to note that Ham et al. (1999) found that tobacco IDDS interacts with the 2b movement protein of cucumber mosaic virus and that IspH expression was induced by cucumber mosaic virus infection. Although this observation was not made with TRV, it raises the possibility that IDDS may modify virus spread. Reduction in IDDS expression through VIGS may therefore reduce the ability of TRV to mediate silencing.
A body of evidence from feeding studies shows that precursor exchange (metabolic crosstalk) occurs between the DXP and MVA pathways. Examples of plant metabolites formed from both DXP and MVA precursors include monoterpenes and sesquiterpene volatiles in Phaseolus lunatus (Piel et al., 1998), sesquiterpenes in chamomile (Matricaria recutita) (Adam and Zapp, 1999), gingkolide diterpenes in Gingko biloba (Schwarz and Arigoni, 1999), and gibberellins in Arabidopsis (Kasahara et al., 2002). In most cases the amount of carbon exchanged seems to be small (1%–2% in the case of the gingkolides). Based on the albino leaves of IspG- and IspH-silenced plants, we conclude that MVA-derived DMAPP and IPP are not able to compensate for blocks in the DXP pathway at the HDS- or IDDS-catalyzed step. From these results it seems clear that the DXP pathway is metabolically segregated from the MVA pathway with little movement of precursors from the cytoplasm to the chloroplast. This finding is in agreement with studies showing albino phenotypes resulting from genetic or chemical blocks in the DXP pathway: mutants with disrupted DXS (i.e. cla1-1), DXR and ISPD genes, transgenic plants with silenced DXR and CMK genes, and fosmidomycin-treated plants. These results are in contrast to Nagata et al. (2002) who found that MVA could restore chlorophyll levels and plastid structure in the cla1-1 mutants. In recent papers, Laule et al. (2003) and Bick and Lange (2003) have shown that unidirectional transport of IPP and GPP from the plastid to the cytosol occurs in plants. Our results shed no light on such a transport system, other than to emphasize that transport in the other direction is insignificant. The evolutionary utility of plastid to cytosol export may be that the DXP pathway, situated in the carbon-fixing compartment of the plant cell, is a more robust source of isoprenoid precursors and thus more suited to compensating for deficiencies in the MVA pathway than the other way around.
We observed profound changes in the tissue and plastid structure of IspG, IspH, and IDI-silenced leaves. The most notable perturbations at the tissue level were the disorganization of the palisade mesophyll and reduction in chlorophyll number; characteristic changes in plastids included the disruption (or degradation) of thylakoids and the appearance of lytic vesicles. Such changes in structure were most obvious in IspG-silenced plants, followed by IspH- and IDI-silenced plants. Similar changes in plastid ultrastructure have been reported in Arabidopsis cla1-1, which was first isolated as an altered chloroplast mutant, and immutans mutants (Aluru et al., 2001). IMMUTANS, which encodes a protein involved in carotenoid desaturation, also showed the disorganized palisade mesophyll we observed. Chlorophylls and carotenoids play central roles in photosynthesis but also in the differentiation of chloroplasts (Park et al., 2002). The plastid phenotype observed after silencing of IspG, IspH and IDI adds to evidence that DXP-derived isoprenoid products have a key but as yet unclear function in plastid development.
The mottled white-pale green leaves of IDI-silenced plants were strikingly different from the albino leaves of IspG and IspH-silenced plants. Chlorophyll levels in IDI-silenced leaves were reduced by about 80% compared to empty virus controls, but were 10-fold higher than in TRV-IspG and TRV-IspH infected plants. This difference was probably not due to the ability of TRV-IDI to mediate silencing since RT-PCR analysis showed that VIGS reduced IDI expression to a similar extent as for IspG and IspH transcripts in TRV-IspG and TRV-IspH plants, respectively (Fig. 5). IDI plays a role in both the DXP and MVA pathways with plastid (DXP pathway) and cytosol (MVA pathway) targeted isoforms of IDI reported in tobacco (Nakamura et al., 2001) and Arabidopsis (Campbell et al., 1997). The 453-bp NbIDI sequence used for silencing is likely derived from the cDNA coding for the DXP pathway-specific isoform of IDI, based on its higher similarity to tobacco IPP1 than to IPP2 (Nakamura et al., 2001). However, IPP1 and IPP2 differ little in their coding regions, and VIGS should knockout both the DXP and MVA IDI isoforms to an equal extent (see Yoshioka et al. [2003] for an example of silencing of homologous genes). The silencing of the cytosolic IDI matters little since MVA-derived C5 precursors appear not to be extensively transported to the chloroplast.
We suggest that an explanation for the IDI-silenced leaf phenotype lies in the physiological role of IDI in plastid isoprenoid biosynthesis. Loss-of-function IDI mutants have not previously been reported in plants and this study represents the first attempt to knockout IDI activity. Insertional disruption of IDI in E. coli has no effect on the organism's growth rate (Hahn et al., 1999) and the genome of Synechocystis, an organism that relies exclusively on the DXP pathway, contains no IDI gene (Ershov et al., 2000). The DXP pathway bifurcates at the final enzymatic step with IDDS converting HMBPP into DMAPP and IPP at a ratio of 5:1 in E. coli (Adam et al., 2002; Altincicek et al., 2002). As others have noted, a pathway that yields both DMAPP and IPP makes possible the formation of GPP and longer prenyl diphosphates in the absence of IDI. Our results suggest that even without IDI, a rudimentary form of isoprenoid biosynthesis can occur in the plastid from the DMAPP and IPP formed by IDDS, thus accounting for the chlorophylls and carotenoids found in IDI-silenced tissue. However, we argue that while not absolutely required, IDI plays a highly significant role in plastidic isoprenoid biosynthesis in higher plants, as evidenced by the 80% reduction in the pigments when it is knocked out. The importance of IDI is further shown by the disrupted plastids in IDI-silenced plants, which lack starch granules and have thylakoids coalescing into fluted structures or lytic vesicles similar to those that result from IspG- and IspH-silencing. The color complementation assay used by Kajiwara et al. (1997) to identify cDNAs capable of increasing carotenoid formation in an engineered strain of E. coli led to the isolation of an IDI cDNA and the demonstration that increased IDI activity led to more pigment formation. These authors concluded that IDI has an “influential” role in isoprenoid metabolism in E. coli, even though IDI is a nonessential gene. Our conclusions concerning the function of IDI in plants is formally in agreement with its function in E. coli; plastidic isoprenoid biosynthesis can occur to a minor degree when IDI is absent but is necessary for normal plant function. Comparison of IPP metabolism by chloroplasts of mock control and IDI-silenced plants showed that IDI-silenced tissues dephosphorylated IPP to IP at a higher rate than control tissue. This suggests that IDI knockout leads to the induction of a system for metabolically inactivating IPP and possibly other prenyl diphosphates. Recently, Martin et al. (2003) concluded that E. coli engineered to express the MVA pathway suffered growth inhibition due to the toxic buildup of IPP. Perhaps the role of IDI is to adjust the DMAPP to IPP ratio in plastids, preventing an imbalance in C5 diphosphates, and thereby optimizing the flux of C5 precursors into isoprenoids.
MATERIALS AND METHODS
Cloning of IspG, IspH, and IDI Gene Fragments
Total RNA was isolated from Nicotiana benthamiana Domin leaves using guanidium thiocyanate-phenol-chloroform. Five micrograms were reverse transcribed using Superscript II reverse transcriptase and an oligo(dT) primer. PCR for all three genes was performed using 2.5 units Taq polymerase, 200 μm dNTPs, and 200 nm of each primer in a 50-μL volume. PCR conditions were 94°C, 3 min followed by 94°C, 30 s; 50°C, 30 s; 72°C, 1 min for 30 cycles and finally 72°C, 7 min. Primer pairs were 5′-ATGCCATTCAAGGATCTGGC-3′ and 5′-CGTCTTTCCGACATAAAGGTC-3′, 5′-TGTGAAGAACATGGCAGAGG-3′ and 5′-TCTCAACCAACTCACCATGC-3′ and 5′-GGAAAGTGGGGAGAACATGA-3′ and 5′-TCGACAGAAAGCCACAGCTA-3′ designed to amplify fragments of IspG, IspH, and IDI cDNAs, respectively. PCR products were purified, ligated into pGEM-T easy (Promega, Madison, WI), and sequenced. Plasmid DNA was digested with Apa1 and Spe1 and the released DNA fragments purified before ligation into Apa1 and Spe1 digested pTV00. Constructs were confirmed by sequencing before transformation into Agrobacterium tumefaciens GV3101/pSa_RepA (Hellens et al., 2000) via electroporation.
Growth and Infiltration of Nicotiana benthamiana
N. benthamiana plants were grown in a controlled environment chamber with 16 h/22°C days and 8 h/20°C nights. Cultures of A. tumefaciens containing pBINTRA6 or pTV constructs were grown overnight. After centrifugation, bacteria were resuspended in buffer containing 1 mm MES (pH 5), 10 mm MgCl2, and 100 μm acetosyringone to OD600 = 1 and allowed to stand at room temperature for 2 to 4 h. Suspensions of pBINTRA6 and pTV containing cultures were mixed 1:1 and infiltrated into the underside of two or three leaves of 4- to 5-week-old plants using a 2-mL syringe. Mock-infected control plants were infiltrated with Agrobacterium resuspension buffer. Empty TRV control plants were infiltrated with pBINTRA6 and pTV00 without cDNA insert.
Carotenoid and Chlorophyll Analysis
Carotenoids and chlorophylls were extracted from N. benthamiana leaves 3 to 4 weeks after infiltration using a method modified from Fraser et al. (2000). Briefly, silenced leaf tissue from TRV-IspG, TRV-IspH, and TRV-IDI infected plants and equivalent green tissue from mock and empty TRV plants was frozen in liquid nitrogen and ground to a fine powder. One hundred 150-mg portions were extracted with 300 μL of methanol for 5 min at 4°C on a rotary shaker followed by the addition of 300 μL of 50 mm Tris-HCl (pH 7.5) containing 1 m NaCl and further mixing for 5 min at 4°C. Chloroform (800 μL) was added and the solution mixed for 10 min at 4°C. The aqueous and organic fractions were separated by centrifugation at 3,000g for 5 min and 500 μL of the lower phase was transferred to a microcentrifuge tube. The solvent was removed by SpeedVac and the residue stored in the dark at −20°C. For spectrophotometric analysis, pigment residues were taken up in 500 μL of acetone. Aliquots were measured directly or as dilutions at 470, 645, and 662 nm using a UV-vis spectrophotometer (Ultrospec 3000, Amersham Pharmacia Biotech, Uppsala). Carotenoid and chlorophyll concentrations were calculated as described in Lichtenthaler (1987) for 100% acetone.
RT-PCR Analysis
Total RNA was isolated from 100-mg portions of silenced leaf tissue from three plants using guanidium/ammonium thiocyanate-phenol-chloroform. First-strand cDNA was reverse transcribed from 5 μg total RNA using Superscript II reverse transcriptase and random primers. PCR for all genes was performed using 1 unit Taq polymerase, 200 μm dNTPs, and 200 nm of each primer in a 20-μL volume. To control for an equal amount of cDNA in each reaction, the amount of cDNA required to produce Actin amplification products of similar intensity for all samples was titered using 5′-ATGGCAGACGGTGAGGATATTCA-3′ and 5′-GCCTTTGCAATCCACATCTGTTG-3′ designed to amplify a 1.1-kb Actin cDNA fragment (Romeis et al., 2001). PCR conditions for Actin were 94°C, 3 min followed by 94°C, 30 s; 50°C, 30 s; 72°C, 1 min 15 s for 30 cycles and finally 72°C, 7 min. Once cDNA amounts were equalized, PCR for the detection of IspG, IspH, and IDI cDNAs was performed using at 94°C, 3 min followed by 94°C, 30 s; 50°C, 30 s; 72°C, 1 min 15 s for 30 cycles and finally 72°C, 7 min. Primer pairs were 5′-ACTTCGAGTGGCTGAGTGCT-3′ and 5′-CGTATCACCCAAACCATCCT-3′ for IspG, 5′-CTCTTGAGCTGTTGAATCAGGA-3′ and 5′-TATTTCCCAGCAAAGGATGC-3′ for IspH, and 5′-CGCCGTCTCATGTTTGAAG-3′ and 5′-GACCCAATGGGGTGAACTG-3′ for IDI. PCR products were analyzed on a 1.2% agarose gel stained with ethidium bromide and visualized on a GelDoc 1000 (Bio-Rad, Hercules, CA).
Incorporation of [13C]- and [14C]DXP
Fully white leaves of TRV-IspG and TRV-IspH infected plants or equivalent leaves from mock or empty TRV plants were detached at the petiole and placed in an aqueous solution of 0.5 μCi [3,4,5-14C3]DXP (specific activity 155 μCi/μmol) and 2.3 μmol [3,4,5-13C3]DXP. After most of the solution had been taken up, 500 μL of water was added in 100 μL aliquots over 7 to 8 h after which the excised leaf was provided with water ad libitum for 16 h. Leaves were weighed and leaf frozen in liquid nitrogen before extraction twice with 10 mL boiling 80% (v/v) ethanol for 20 min. The extract was concentrated under reduced pressure to give a residue that was partitioned between 1 mL water and 2 × 750 μL ethyl acetate. Total radioactivity in the ethanolic extract, and aqueous and organic phases, was measured by scintillation counting. The aqueous phase was lyophilized, redissolved in 250 μL water, and analyzed by ion-pair radioHPLC performed using a 4.6 mm × 250 mm Luna 5-μm C8 column (Phenomenex, Torrance, CA) with an ion-pair solvent system consisting of solvent A (10 mm tetra-n-butylammonium hydrogen sulfate in water) and solvent B (10 mm tetra-n-butylammonium hydrogen sulfate in 70% [v/v] methanol) and a gradient elution of 0 min (100% A), 20 min (100% A), 80 min (40% A:60% B), 85 min (100% A), and 100 min (100% A). Radioactivity was measured with a Ramona 2000 detector (Raytest, Straubenhardt, Germany). Alkaline phosphatase treatment was performed in a 1-mL reaction containing 50 mm Tris-HCl pH 8, 10 mm MgCl2, and 5 units alkaline phosphatase at 37° for 2 h. MBDO, DXP, HMBP, and cMEPP were identified by comparison with authenticate standards.
Purification and Structural Elucidation of cMEPP
Detached albino leaves from TRV-IspG infected plants were fed 0.5 μCi [3,4,5-14C3]DXP and 2.3 μmol [3,4,5-13C3]DXP and extracted as described above. The aqueous phase was applied to a DEAE Sephadex (formate form) column (1 × 18 cm), which was eluted with a linear gradient of 0.06 m to 0.56 m ammonium formate (pH 8) at a flow-rate of 1.1 mL/min. Fractions were analyzed by scintillation counting and those containing the cyclic diphosphate were combined and lyophilized in vacuum. HRMS and 13C-NMR data of the residue so obtained showed that it was [1,3,4-13C3]cMEPP as follows. ESI-FT-ICRMS (negative ion mode) m/z: 276.98749 (14.4%, cMEPP, calc. for C5H11O9P2− 276.98838) and 279.99782 (100%, [1,3,4-13C3]cMEPP, calc. for C213C3H11O9P2− 279.99844); 13C-NMR data (500 MHz, H2O): 69.57 (d, J = 42.7 Hz, C-3), 68.14 (d, JCP = 4.1 Hz, C-1), 66.82 (dd, JCC = 42.7 Hz, JCP = 6.1 Hz, C-4).
Purification and Structure Elucidation of MBDO
Detached albino leaves from TRV-IspH infected plants were fed [3,4,5-14C3]DXP and [3,4,5-13C3]DXP and extracted as described for TRV-IspG plants. The resulting aqueous fraction was incubated with 3 units alkaline phosphatase for 2 h at 37°C. After centrifugation, the supernatant was separated on a silica plate (SIL-G/UV254, Macherey-Nagel, Duren, Germany) developed in chloroform-methanol-H2O (4:1:0.1). The band corresponding to [1,3,4-13C3]MBDO, identified by comparison of its Rf with an authenticate standard, was extracted with methanol and analyzed by radioHPLC and gas chromatography-mass spectrometry (GC-MS). The isolated compound and authentic MBDO showed the same behavior by ion-pair radioHPLC (RT: 13 min) and GC-MS. The 70 eV-EI mass spectrum of [1,3,4-13C3]MBDO was RT = 15.76 min, (m/z [rel. int., %]): 235 ([M-CH3]+, 0.4), 192 (45), 191 (13), 161 (5.6), 160 (7.4), 159 (3.6), 156 (2.9), 148 (22), 147 (78), 146 (42), 145 (23), 144 (8.6), 143 (19), 141 (2.2), 117 (5.0), 75 (43), and 73 (100). The isotopic distribution of 13C in the labeled MBDO was calculated using the [M-TMSiOH]+-ion (m/z 156 in nonlabeled MBDO) with values of 20% 13C0, 1% 13C1, 3% 13C2, 21% 13C3, 37% 13C4, and 17% 13C5. The 70 eV-EI mass spectrum of MBDO was RT = 15.68 min, (m/z [rel. int., %]): 231 ([M-CH3]+, 0.4), 191 (32), 156 ([M-TMSiOH]+, 9.6), 148 (10), 147 (66), 144 (8.8), 143 (70), 141 (7.7), 115 (9.1), 75 (27), and 73 (100). The GC-MS of MBDO and [1,3,4-13C3]MBDO were performed with a GC-MS system (Voyager, ThermoQuest, Dreieich, Germany). The following conditions were used: 70 eV EI, source temperature 200°C, column DB-5MS (J&W, 30 m × 0.25 mm, 0.25 μm film thickness), injection temperature 250°C, interface temperature 300°C, carrier gas He, flow rate 1.0 mL/min, constant flow mode, splitless injection. The column temperature program was 40°C for 5 min, rising to 300°C at a rate of 10°C/min and then held at 300°C for 4 min. The trimethylsilylation of the samples was carried out with N-methyl-N-trimethylsilylfluoroacetamide.
IPP Metabolism in Chloroplasts
One and one-half grams of silenced tissue from TRV-IDI infected leaves or equivalent mock infected tissue were homogenized in 60 mL ice-cold buffer A (50 mm HEPES, pH 8.0, 1 mm EDTA, 1 mm DTE, and 0.4 m Suc) with an Ultraturrax at 9500 rpm for 20 s. The homogenate was filtered through four layers of nylon gauze (50 μm) and the filtrate was centrifuged at 3,076g for 10 min at 4°C. The resulting green pellet was carefully resuspended in buffer A and centrifuged as above. The pellet was taken up in 200 μL buffer B (50 mm HEPES, pH 7.6, and 1 mm dithioerythritol) and filtered through one layer of nylon gauze. The resulting chloroplast suspension contained approximately 2.5 mg chlorophyll/ml as determined using the method of Joyard et al. (1987). The IPP conversion assay contained 100 mm HEPES, pH 7.6, 6 mm MnCl2, 10 mm MgCl2, 7 mm NaF, 1.72 nmol [1-14C]IPP (0.1 μCi, specific activity 58 mCi/mmol, Amersham Biosciences), and 50 μg chlorophyll in a total volume of 500 μL. Assays were incubated at 30°C for 15, 30, 60, 90, 120, and 180 min. The assays were extracted, partitioned into organic and aqueous fractions as described in Fellermeier et al. (2001), and the amount of radioactivity in both fractions measured using scintillation counting. RadioHPLC analysis of the aqueous fraction was performed as described above. IPP and IP were identified by comparison with authenticate standards. ESI-FT-ICRMS (negative ion mode) of IP: m/z 165.03202 [M-H]− (calc. for C5H10O4P− 165.03222).
Microscopy
Plant material was prepared for microscopic investigations as described in Hause and Hahn (1998). Semithin sections were observed with a Zeiss (Jena, Germany) Axioskop light microsope. A Zeiss EM900 was used for electron microscopic investigations.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY497303 (NbIspG), AY497304 (NbIspH), and AY497305 (NbIDI).
Acknowledgments
We are grateful to David Baulcombe (Sainsbury Laboratory, Norwich, UK) and Plant Biosciences Limited (Norwich, UK) for making available the TRV vectors. At the IPB-Halle, we thank Verona Dietl for technical assistance, Ursula Schäfer for help with plant cultivation, Christine Kuhnt for performing the GC-MS measurements of the MBDO samples, and Dr. Andrea Porzel for measuring the NMR spectra.
This work was supported in part by grants from the Deutsche Forschungsgemeinschaft (to M.H.Z.) and the Fonds der Chemischen Industrie, Frankfurt (to T.M.K. and M.H.Z.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.038133.
References
- Adam P, Hecht S, Eisenreich W, Kaiser J, Grawert T, Arigoni D, Bacher A, Rohdich F (2002) Biosynthesis of terpenes: studies on 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase. Proc Natl Acad Sci USA 99: 12108–12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam KP, Thiel R, Zapp J (1999) Incorporation of 1-[1-(13)C]deoxy-D-xylulose in chamomile sesquiterpenes. Arch Biochem Biophys 369: 127–132 [DOI] [PubMed] [Google Scholar]
- Altincicek B, Kollas A-K, Sanderbrand S, Wiesner J, Hintz M, Beck E, Jomaa H (2001) GcpE is involved in the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli J Bacteriol 183: 2411–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altincicek B, Duin EC, Reichenberg A, Hedderich R, Kollas AK, Hintz M, Wagner S, Wiesner J, Beck E, Jomaa H (2002) LytB protein catalyzes the terminal step of the 2-C-methyl-D-erythritol-4-phosphate pathway of isoprenoid biosynthesis. FEBS Lett 532: 437–440 [DOI] [PubMed] [Google Scholar]
- Aluru MR, Bae H, Wu D, Rodermel SR (2001) The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol 127: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigoni D, Sagner S, Latzel C, Eisenreich W, Bacher A, Zenk MH (1997) Terpenoid biosynthesis from 1-deoxy-D-xylulose in higher plants by intramolecular skeletal rearrangement. Proc Natl Acad Sci USA 94: 10600–10605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe DC (1999) Fast forward genetics based on virus-induced gene silencing. Curr Opin Plant Biol 2: 109–113 [DOI] [PubMed] [Google Scholar]
- Bick JA, Lange BM (2003) Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch Biochem Biophys 415: 146–154 [DOI] [PubMed] [Google Scholar]
- Budziszewski GJ, Lewis SP, Glover LW, Reineke J, Jones G, Ziemnik LS, Lonowski J, Nyfeler B, Aux G, Zhou Q, McElver J, Patton DA, et al (2001) Arabidopsis genes essential for seedling viability: isolation of insertional mutants and molecular cloning. Genetics 159: 1765–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M, Hahn FM, Poulter CD, Leustek T (1997) Analysis of the isopentenyl diphosphate isomerase family from Arabidopsis thaliana. Plant Mol Biol 36: 323–328 [DOI] [PubMed] [Google Scholar]
- Croteau R, Kutchan TM, Lewis NG (2000) Natural products (secondary metabolites). In B. Buchanan, W. Gruissem, R. Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 1250–1268
- Cunningham FX Jr, Lafond TP, Gantt E (2000) Evidence of a role for lytB in the nonmevalonate pathway of isoprenoid biosynthesis. J Bacteriol 182: 5841–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich W, Rohdich F, Bacher A (2001) Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci 6: 78–84 [DOI] [PubMed] [Google Scholar]
- Ershov Y, Gantt RR, Cunningham FX, Gantt E (2000) Isopentenyl diphosphate isomerase deficiency in Synechocystis sp. strain PCC6803. FEBS Lett 473: 337–340 [DOI] [PubMed] [Google Scholar]
- Estévez JM, Cantero A, Romero C, Kawaide H, Jimenez LF, Kuzuyama T, Seto H, Kamiya Y, Leon P (2000) Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol 124: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellermeier M, Raschke M, Sagner S, Wungsintaweekul J, Schuhr CA, Hecht S, Kis K, Radykewicz T, Adam P, Rohdich F, et al (2001) Studies on the nonmevalonate pathway of terpene biosynthesis: the role of 2C-methyl-D-erythritol 2,4-cyclodiphosphate in plants. Eur J Biochem 268: 6302–6310 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Pinto ME, Holloway DE, Bramley PM (2000) Technical advance: application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J 24: 551–558 [DOI] [PubMed] [Google Scholar]
- Hahn FM, Hurlburt AP, Poulter CD (1999) Escherichia coli open reading frame 696 is IDI, a nonessential gene encoding isopentenyl diphosphate isomerase. J Bacteriol 181: 4499–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham BK, Lee TH, You JS, Nam YW, Kim JK, Paek KH (1999) Isolation of a putative tobacco host factor interacting with cucumber mosaic virus-encoded 2b protein by yeast two-hybrid screening. Mol Cells 9: 548–555 [PubMed] [Google Scholar]
- Hause G, Hahn H (1998) Cytological characterization of multicellular structures in embryogenic microspore cultures of Brassica napus L. Bot Acta 111: 204–211 [Google Scholar]
- Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F (2001) Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (ispG) protein. Proc Natl Acad Sci USA 26: 14837–14842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Joyard J, Dorne AJ, Douce R (1987) Use of thermolysin to probe the cytosolic surface of the outer envelope membranes from plastids. In L Packer, R Douce, eds, Plant Cell Membranes. Academic Press, San Diego, pp 195–206
- Kajiwara S, Fraser PD, Kondo K, Misawa N (1997) Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem J 324: 421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H, Hanada A, Kuzuyama T, Takagi M, Kamiya Y, Yamaguchi S (2002) Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. J Biol Chem 277: 45188–45194 [DOI] [PubMed] [Google Scholar]
- Laule O, Furholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange M (2003) Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thialiana. Proc Natl Acad Sci USA 100: 6866–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. In L Packer, R Douce, eds, Plant Cell Membranes. Academic Press, San Diego, pp 350–382
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Mahmoud SS, Croteau RB (2001) Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc Natl Acad Sci USA 98: 8915–8920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAteer S, Coulson A, McLennan N, Masters M (2001) The lytB Gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J Bacteriol 183: 7403–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, Leon P (1996) CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J 9: 649–658 [DOI] [PubMed] [Google Scholar]
- Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21: 796–802 [DOI] [PubMed] [Google Scholar]
- Nagata N, Suzuki M, Yoshida S, Muranaka T (2002) Mevalonic acid partially restores chloroplast and etioplast development in Arabidopsis lacking the non-mevalonate pathway. Planta 216: 345–350 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Shimada H, Masuda T, Ohta H, Takamiya K (2001) Two distinct isopentenyl diphosphate isomerases in cytosol and plastid are differentially induced by environmental stresses in tobacco. FEBS Lett 506: 61–64 [DOI] [PubMed] [Google Scholar]
- Okada K, Kawaide H, Kuzuyama T, Seto H, Curtis IS, Kamiya Y (2002) Antisense and chemical suppression of the nonmevalonate pathway affects ent-kaurene biosynthesis in Arabidopsis. Planta 215: 339–344 [DOI] [PubMed] [Google Scholar]
- Ostrovsky D, Diomina G, Lysak E, Matveeva E, Ogrel O, Trutko S (1998) Effect of oxidative stress on the biosynthesis of 2-C-methyl-D-erythritol-2,4-cyclopyrophosphate and isoprenoids by several bacterial strains. Arch Microbiol 171: 69–72 [DOI] [PubMed] [Google Scholar]
- Ostrovsky D, Shipanova I, Sibeldina L, Shashkov A, Kharatian E, Malyarova I, Tantsyrev G (1992) A new cyclopyrophosphate as a bacterial antistressor? FEBS Lett 298: 159–161 [DOI] [PubMed] [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J, Donath J, Bandemer K, Boland W (1998) Mevalonate-independent biosynthesis of volatiles in plants: induced and constitutive emission of volatiles. Angew Chem Int Ed Engl 37: 2478–2481 [DOI] [PubMed] [Google Scholar]
- Querol J, Campos N, Imperial S, Boronat A, Rodríguez-Concepción M (2002) Functional analysis of the Arabidopsis thaliana GCPE protein involved in plastid isoprenoid biosynthesis. FEBS Lett 514: 343–346 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Rohdich F, Hecht S, Gärtner K, Adam P, Krieger C, Amslinger S, Arigoni D, Bacher A, Eisenreich W (2002) Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc Natl Acad Sci USA 99: 1158–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdich F, Hecht S, Bacher A, Eisenreich W (2003) The deoxyxylulose phosphate pathway of isoprenoid biosynthesis. Discovery and function of the ispDEFGH genes and their cognate enzymes. Pure Appl Chem 75: 393–405 [Google Scholar]
- Rodríguez-Concepción M, Boronat A (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol 130: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M (2003) Mevalonate-independent methylerythritol phosphate pathway for isoprenoid biosynthesis: elucidation and distribution. Pure Appl Chem 75: 375–388 [Google Scholar]
- Romeis T, Ludwig AA, Martin R, Jones JD (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20: 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Arigoni D (1999) Ginkgolide biosynthesis. In D Cane, ed, Comprehensive Natural Product Chemistry, Vol 2. Pergamon, Oxford, pp 367–399
- Voinnet O (2001) RNA silencing as a plant immune system against viruses. Trends Genet 17: 449–459 [DOI] [PubMed] [Google Scholar]
- Wungsintaweekul J, Herz S, Hecht S, Eisenreich W, Feicht R, Rohdich F, Bacher A, Zenk MH (2001) Phosphorylation of 1-deoxy-D-xylulose by xylulokinase of Escherichia coli. Eur J Biochem 268: 310–316 [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Numata N, Nakajima K, Katou S, Kawakita K, Rowland O, Jones JD, Doke N (2003) Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15: 706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler J, Schwender J, Müler C, Wiesner J, Weidemeyer C, Beck E, Jomaa H, Lichtenthaler HK (1998) Inhibition of the non-mevalonate 1-deoxy-D-xylulose 5-phosphate pathway of plant isoprenoid biosynthesis by fosmidomycin. Z Naturforsch C 53: 980–986 [Google Scholar]