Abstract
Crop plants are regularly challenged by a range of environmental stresses which typically retard their growth and ultimately compromise economic yield. The stress response involves the reprogramming of approximately 4% of the transcriptome. Here, the behavior of AtRD22 and AtUSPL1, both members of the Arabidopsis thaliana BURP (BNM2, USP, RD22 and polygalacturonase isozyme) domain-containing gene family, has been characterized. Both genes are up-regulated as part of the abscisic acid (ABA) mediated moisture stress response. While AtRD22 transcript was largely restricted to the leaf, that of AtUSPL1 was more prevalent in the root. As the loss of function of either gene increased the plant's moisture stress tolerance, the implication was that their products act to suppress the drought stress response. In addition to the known involvement of AtUSPL1 in seed development, a further role in stress tolerance was demonstrated. Based on transcriptomic data and phenotype we concluded that the enhanced moisture stress tolerance of the two loss-of-function mutants is a consequence of an enhanced basal defense response.
Introduction
Abiotic stress factors such as moisture stress, salinity, extreme temperature and variable light intensity can disturb plant metabolism and growth. It has been estimated that crop yield losses due to such stresses lie in the order of 50% [1], so increasing the resilience of crop plants will be an important contributor to yield stability. Abiotic stress affects both photosynthesis and photorespiration, as well as having an impact on the energy and redox status of the plant cell. One of the most damaging products of stress is the group of compounds referred to as reactive oxygen species (ROS). The plant response also includes the induced synthesis of certain enzymes and low molecular weight compounds associated with antioxidant activity, redox regulators, chaperones such as heat shock proteins and late embryogenesis abundant proteins, water and ion transporters, the production of compatible osmolytes to maintain cellular water content and the fine tuning of proteolysis involved in programmed cell death. Numerous attempts, with varying levels of success, have been made to genetically engineer the production of some of these components with a view to enhancing abiotic stress tolerance [2].
The phytohormone abscisic acid (ABA) is involved in the regulation of expression of many stress-responsive genes, although other stress responsive genes are known to be regulated in an ABA-independent manner [3]–[5]. The regulation of several stress-inducible genes is mediated by the interaction of bZIP transcription factors (ABFs) with ABA-response elements (ABRE) in target gene promoters [6].
The Arabidopsis thaliana gene RESPONSIVE TO DEHYDRATION22 (AtRD22), originally identified as a gene which responded to dehydration [7], [8], encodes a member of the BURP protein family, members of which share a highly conserved BURP domain at their C terminus [9]), sometimes also referred to as a U domain [10], [11]. BURP proteins appear to be plant-specific; some examples are the Vicia faba unknown seed protein (USP) [10], the Brassica napus microsporogenesis-specific protein BNM2 [12]–[14], the Panicum maximum apomixis-specific protein [15] and the wheat pollen protein RAFFTIN [16]. The soybean genome harbors 23 BURP protein encoding genes, of which 17 are responsive to stress [17]–[20]. The 18 Populus trichocarpa BURP family members fall into five recognizable sub-families [21]. There are 15 related genes in maize, one of which is specifically expressed in the root cortex parenchyma [22] while the sorghum genome harbors 11 homologs [23]. The ectopic expression of the A. thaliana gene AtUSPL1 has been shown to distort seed development and to alter the morphology of seed lipid vesicles [24].
AtRD22 is up-regulated by moisture stress, salinity stress and exogenously supplied ABA [8] and its induction has been used as a marker for abiotic stress [25]–[29]. The heterologous expression in both A. thaliana and rice of the soybean gene GmRD22 enhances salinity stress tolerance [30]. Members of the BURP family are up-regulated by stress in rice [31], soybean [30] and maize [23]. Members of the BURP family have been described in relation to stress conditions. In cotton, an RD22-like protein interacts with an α-expansin and the over expression of both proteins simultaneously promotes growth and fruit weight [32].Remarkably, the expression programmes active during stress response partially overlap with gene expression during early embryogenesis and seed desiccation [24]. Most likely ancient stress response genes have been recruited to protect seed tissue from dehydration stress in drying seeds [33], [34]. Here, a combination of genetic, molecular and physiological approaches has been applied to isolate and characterize T-DNA insertion mutants of AtRD22 and AtUSPL1.
Materials and Methods
Plant material and growth conditions
A. thaliana seeds (ecotype Col-0 and the three T-DNA insertion lines SALK_146066 (rd22-1), WiscDsLox481-484P12 (rd22-2) and SALK_022325 (uspl1)) obtained from the European Arabidopsis Stock Center (NASC) were stratified before imbibing them for three days at 4°C in the dark. The resulting germinated seedlings were grown in soil for four weeks under 60% relative humidity, a constant temperature of 22°C and under a 16 h photoperiod (light intensity of 120 µmol m− 2 s−1). Drought stress was applied by a. active dehydration under low humidity for 1–5 days and b. withholding water for 1–5 days; the relative soil water content was monitored using an HH2 moisture meter (delta-T devices, Cambridge, UK). Moisture stress was also mimicked in two week old seedlings by transferring them for three days on a MS basal medium containing one of 150 mM or 300 mM NaCl, 100 µM ABA, 4% w/v trehalose, 4% w/v sorbitol, 4% w/v glucose, 4% w/v fructose, 4% w/v sucrose, 300 mM mannitol, 15% w/v PEG 6000 or 4% w/v PEG 20000.
Detailed information is provided in Figure 1/Figure S4. Primers used for genotyping are listed in Table S1. Drought stress was applied by withdrawal of water and relative soil water content [%] was controlled with HH2 moisture meter (delta-T devices Ltd, Cambridge, UK).
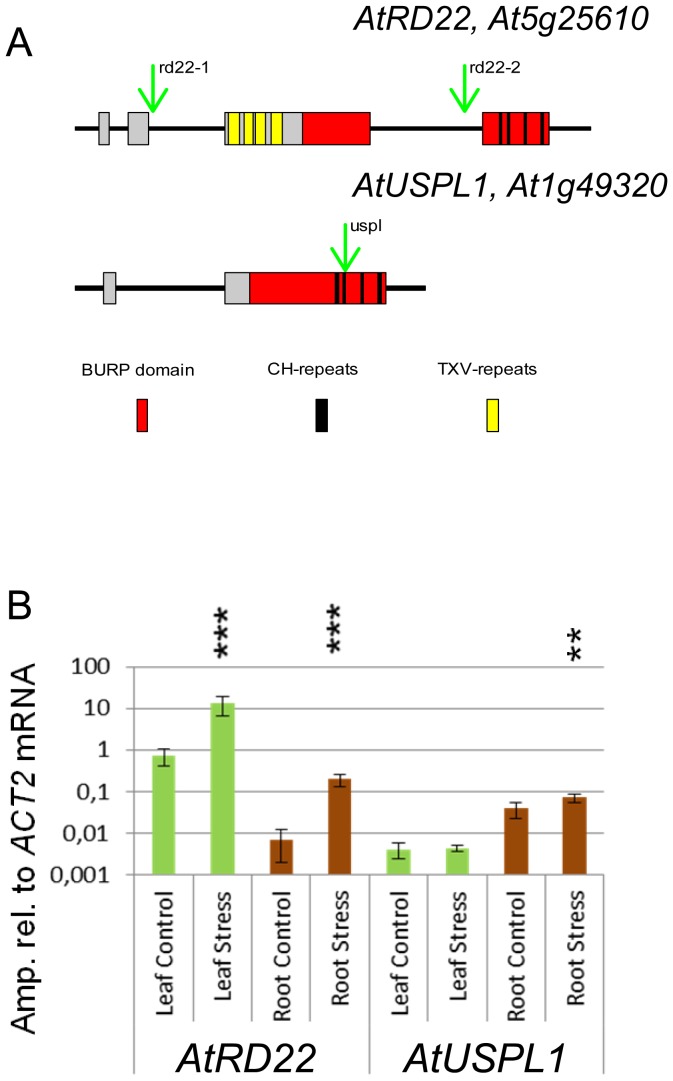
Figure 1. AtRD22 and AtUSPL1, members of the BURP gene family.
A. Scheme of BURP-domain containing proteins AtRD22 and AtUSPL1. BURP (named after BNM2, USP, RD22 and Polygalacturonase isozyme) proteins are identified by their C-terminal BURP-domain (red). The BURP domains contain 4 CH-repeats (black). In comparison to AtUSPL1, AtRD22 contains an additional motif, the TXV repeats (yellow), in AtUSPL1 no repetitive domain structure can be found. AGI ID is given in brackets. Position of T-DNA insertions for the used loss-of function mutants is indicated by a green arrow. B. Quantification of AtRD22 and AtUSPL1 mRNA in leaves and roots. The bar diagram indicates the amplification of AtRD22 (left) and AtUSPL1 mRNA relative to ACT2 mRNA (amp. rel. to ACT2 mRNA). Leaf (green) and root (brown) tissue of well watered and drought stressed (2% RWC) plants (N = 5) was analyzed. Asterisks indicate significant differences (T-test: *** p<0.01, ** = p<0.05).
Plasmid construction and plant transformation
Standard protocols [35] were used to prepare plasmids for Gateway Cloning Technology (Invitrogen GmbH, Karlsruhe, Germany). To achieve its ectopic expression, the AtUSPL1 coding sequence was cloned into the pBENDER GATEWAY vector (a gift from B. Weisshaar, MPI, Cologne).
A 962 bp fragment harboring the AtUSPL1 promoter was amplified, re-sequenced and inserted into the SphI/HindIII cloning site of pBIN101 (Clontech) in front of the uidA GUS reporter gene. Stable transformation of A. thaliana was performed following [36] and subsequent GUS staining to detect transgene expression following [37].
Automated plant phenotyping and image analysis
After 14 days of pre-cultivation pots (diameter 10 cm) were transferred to an automated phenotyping system composed of a conveyor belt transport system and three imaging chambers for plant imaging in the visible (390–750 nm) and NIR (1400–1510 nm with detection peak at 1452 nm) wavelength ranges as well as capturing fluorescence emission (520–750 nm) as well as a watering and weighing station (LemnaTec 3D Scanalyzer; LemnaTec, GmbH, Aachen, Germany) installed in a climate controlled plant cultivation chamber (environmental conditions: 8 h/16 h night/day, 20/22°C (night/day), relative air humidity of 60% and 240 µmol m− 2 s−1 light intensity). Ten blocks (corresponding to ten biological replicates) with each 15 plants (2×5 genotypes under stress conditions, 1×5 under control conditions) were used. Half of the blocks were placed on opposite sides of the cultivation chamber and pots within one block were randomized in order to account for lane or block effects. For reasons of acclimation, plants were grown under control conditions for another week (till day 21 after sowing) before stress application was started. Automated watering was performed every 2nd day to reach defined target values of the pot weight (70% field capacity for control, no watering for soil drought stress). Relative soil water content [%] was controlled in parallel to pot weight with HH2 moisture meter (delta-T devices, Cambridge, UK). The growth rate was calculated as Relative Growth Rate (RGR) = (log(LA[tn+1]) - log(LA[tn]))/((tn+1) - tn) following [38]; Leaf area (LA), timepoint n (tn). Statistical significant growth differences were estimated by Welch t-test from values for plant area determined from top view (visual [39]). Detailed information on calculation of NIR intensities and Cameras used are provided in Table S3.
Estimation of chlorophyll content
Chlorophyll and pheophytin (a chlorophyll degradation product were extracted from the aerial tissue of 6–7 seedlings using a 1∶1 acetone:DMSO mixture, and the absorbance of the extract was recorded at 663, 645 and 553 nm [40] using the UVIKON XS 60/99-90286 spectrophotometer. The experiments comprised three independent replicates. Since the chlorophyll content in the mutant plants grown under non-stressed conditions differed from that present in wild type plants, a relative value was calculated to derive the effect of the stress treatment (Figure S6C). Chlorophyll and pheophytin contents were estimated according to [41], and subjected to a one way ANOVA.
RNA isolation qRT-PCR and Northern analysis
RNA was isolated using an RNeasy kit (Qiagen, Hilden, Germany). For the purposes of Northern blotting, 10 µg RNA was loaded into each lane of a 1.2% w/v agarose, 15% v/v formaldehyde gel, electrophoresed, then transferred passively onto a Hybond N+ membrane (Amersham, GE Healthcare, Waukesha, USA) using 10× SSC as the transfer buffer. The RNA was cross-linked to the membrane by UV irradiation. An AtUSPL1 probe was amplified from A.thaliana genomic DNA using primer pair USPa/b (sequences given in Table S1) and labeled with α-32P dCTP via random priming (Ready Prime Labeling, Pharmacia, GE Healthcare, Waukesha, USA). Membrane/probe hybridizations were carried out in Church hybridization solution [42] at 65°C with pre-hybridization for 6 h and hybridization for 16 h. The membrane was washed for 15 min twice each in 2xSSC, 0.1% SDS, 0.5xSSC, 0.1% SDS and 0.1xSSC, 0.1% SDS at 65°C. Signals were detected and quantified with a Bio-Imaging analyser BAS2000 or X-ray film (Fuji Photo Film Co. Ltd., Tokyo, J). Quantitative real time PCRs (qRT-PCR) were run following [43], in order to assess transcript abundances in leaf and root tissue of six week old plants (five replicate RNA extractions per biological sample, and three technical replications per RNA extract). All primer sequences are given in Table S1.
RNA Isolation, target synthesis and microarray hybridization
Total RNA was isolated from the aerial tissue of two week old seedlings using the TRIzol reagent (Invitrogen) and RNAeasy columns (Qiagen) (see Figure S7 for detailed information). The integrity of the RNA was monitored by agarose gel electrophoresis, and its concentration estimated using a NanoDrop device (Peqlab, Erlangen, Germany), following the manufacturer's protocol. The integrity of the RNA was further confirmed using an Agilent 2100 Bioanalyzer in conjunction with the RNA 6000 Nano assay (Agilent Technologies, Böblingen, Germany). RNA was processed for use on an Affymetrix Arabidopsis ATH1 Genome Array (ATLAS Niolabs GmbH, Berlin, Germany).
Microarray data analysis
The ATH1 chip, which assays 22,392 unique genes, was used to contrast the transcriptomes of wild type Col-0, rd22 and uspl1 plants grown under both control and moisture stressed conditions. Probe set to target gene mappings were taken from the TAIR Web site: ftp://ftp.arabidopsis.org/home/tair/Microarrays/Affymetrix/affy_ATH1 _array_elements2010-12–20.txt. The microarray experiment and basic data interpretation was performed by ATLAS biolabs GmbH (Berlin, Germany) by GeneChip Operating System (GCOS) 1.4. To ensure reliability of the analyses, each GeneChip experiment was performed with two biological replicates. After logarithmic transformation of the data, the average expression for all experimental samples for this probe set was subtracted from each individual expression value, thus leading to a positive value in case of above-average expression levels and a negative value in case of below-average expression levels. GeneSpring GX software (Agilent) was used to gene-wise normalize the expression data. To identify potentially differentially expressed genes, the fold changes >2 and <2 of expression values were identified. This was done separately for the Col-0 and mutant series at different conditions. For the Col-0 and respective mutant sample, a simple moderated t-test was performed and P values were corrected using the Benjamini and Hochberg [44] false discovery rate control, applying standard limma procedures. Differentially expressed genes, between Col-0 and mutant samples were identified for both conditions using the limma nestedF procedure, applying a significance threshold of 0.5 in combination with Benjamini-Hochberg false-discovery rate control and a minimal log2-fold change value of 2. A functional categorization of the differentially transcribed genes was derived using Mapman software [45].
Results
The BURP gene family
The A. thaliana genome contains five BURP family genes; these include both AtUSPL1 (At1g49320) and AtRD22 (At5g25610) (Figure 1A), but also three genes encoding proteins sharing similarity with the tomato non-catalytic β-subunit of polygalacturonase. The latter have been proposed to be designated as AtPG1 (At1g60390), AtPG2 (At1g70370) and AtPG3 (At1g23760). Based on an alignment of related sequences extracted from various species, the family can be subdivided into eight sub-families [23], according to which AtRD22 belongs to the ATRD22-like subgroup, AtUSPL1 to the BNM2-like sub-family, and AtPG1-3 to the PG1β-like subfamily. The A. thaliana genome has no representative of either the V-VIII or the VfUSP like sub-groups. The N-terminal regions of AtPG1-3 each include sequences encoding 21 FXXY–N9–11 repeats (of unknown function) [24]. AtRD22 contains four TXV-repeats, while AtUSPL1 has no repetitive features. Based on the domain structure and phenotype this study is restricted to the functional analysis of AtRD22 and AtUSPL1.
Transcription of AtRD22 and AtUSPL1 during development and in response to stress
Some Arabidopsis BURP genes were described to be preferentially expressed in early embryogenesis [24], their involvement in stress response was less obvious. An exception of this is AtRD22, which was shown previously to be induced under drought treatments [8].
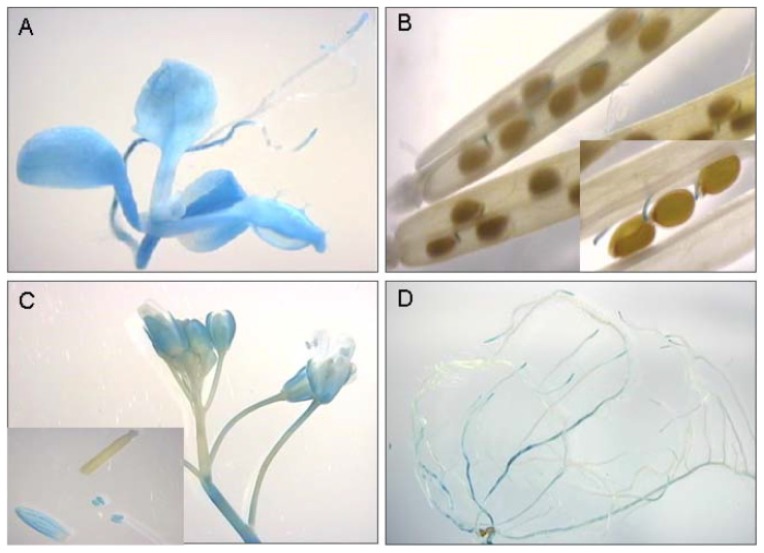
The transcription profiles of AtRD22 and AtUSPL1 were recovered from the Genevestigator database ([46] Figure S1A) and verified using qRT-PCR and Northern blotting. Archival microarray data suggested that while AtRD22 transcript is abundant throughout plant development in the aerial part of the plant, that of AtUSPL1 is low and is restricted to the root. In non-stressed plants, AtRD22 transcription is highest in the leaf, and particularly so in the guard cells [46]. AtUSPL1 transcript is most abundant in the root (Figure 1B, Figure S1B), but is also detectable in the aerial part of the plant early in development. In the root, AtUSPL1 transcription is stimulated by exposure to either mannitol or NaCl, as is that of AtRD22 in the leaf [47]. Here, moisture stress up-regulated AtRD22 transcription was detected, particularly in the leaf. AtUSPL1 transcript was detectable in the root, and its level was enhanced by the imposition of moisture stress (Figure 1B). AtUSPL1 expression was also assayed by tracking GUS expression produced by the pAtUSPL1::GUS transgene. GUS activity was detected in the young leaf, in the hypocotyl and in the stem (Figure 2A). In the silique, its expression was only detectable in the mature seed funiculum (Figure 2B), while in the developing flower and stem, a low level of expression was observed (Figure 2C). GUS activity was especially strong in the root tip (Figure 2D). Compared to the strong AtRD22 promoter activity in aerial parts of the plant [26], ProAtUSPL1 shows strong transcriptional activity in the root tissue.
Figure 2. Histochemical ProAtUSPL1::GUS activity in transgenic Arabidopsis plants.
The AtUSPL1 promoter activity was determined by histochemical localisation of GUS activity derived from the transgenic ProATUSPL1::GUS reporter gene. Activity indicated by blue colour can be seen in A) seedling; B) in funiculus of mature seeds; C) in flowers and stems; and D) in roots.
According to in silico data, treatment with ABA strongly induces AtRD22 in the leaf, and AtUSPL1 responds similarly in the root. AtRD22 is also inducible by exposure to mannitol, glucose, nitrate, high levels of illumination, high temperature and salinity (Figure S2). The gene is down-regulated when the plant is treated with paclobutrazol, an inhibitor of gibberellin synthesis, as well as with cycloheximide, a general inhibitor of protein synthesis, and with syringolin, an inhibitor of cell proliferation.
The behavior of AtRD22 and AtUSPL1 T-DNA insertion mutants
In wild type plants, the expression of the two selected BURP-gene family members was confirmed by quantitative RT real-time PCR (Figure 1B) and microarray analysis of the aerial part and among them AtRD22 transcript is abundant under stress treatments (Figure S3). The loss of function mutants achieved by T-DNA insertions were analysed for characterization of AtRD22 and AtUSPL1 genes in the functional context of drought stress tolerance (Figure S4A). For AtRD22 two independent T-DNA insertion alleles were used: rd22-1 and rd22-2. For AtUSPL1 the mutant line uspl1 was used. To analyse the functional loss of both members of the BURP-gene family the rd22-1/uspl1 double mutant was analysed. In the two analysed T-DNA insertion lines rd22-1 and uspl1 no mRNA of the respective gene was detectable by semi-quantitative PCR and Northern analysis (Figure S4B), which is corroborated by microarray analysis (Figure S4C). Therefore we assume that the used T-DNA mutants represent a loss of function mutation of the respective gene.
Loss of AtRD22 and AtUSPL1 lead to enhanced drought tolerance
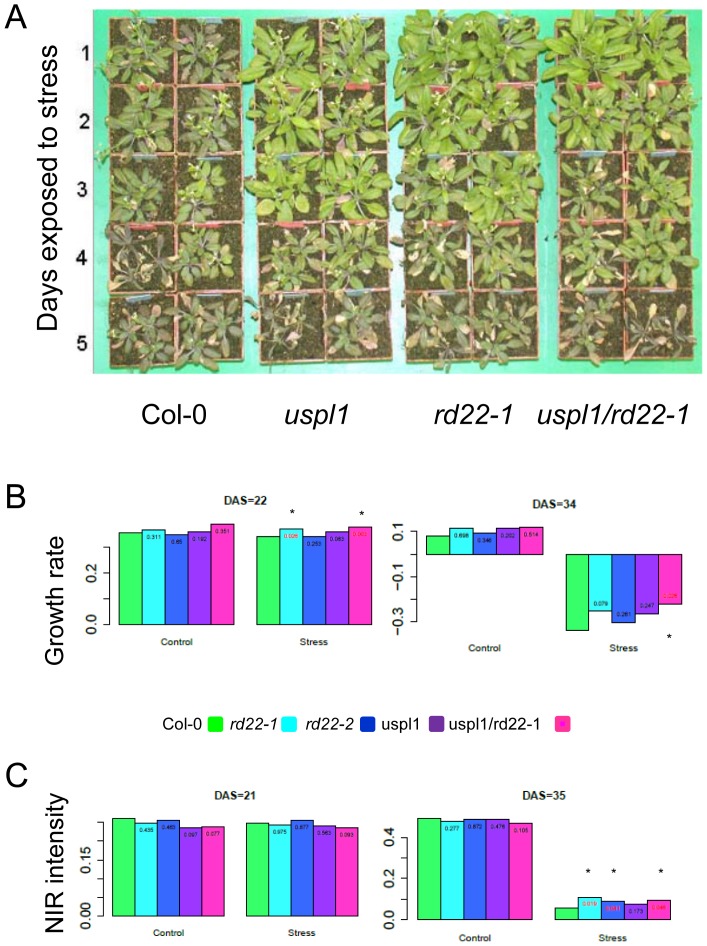
The response of the loss-of-function AtRD22 and AtUSPL1 T-DNA insertion mutants to moisture stress is summarized in Figure 3 and Figure S4/S5. Two independent AtRD22 (rd22-1 and rd22-2) and one AtUSPL1 (uspl1) mutants were analysed, along with the rd22-1/uspl1 double mutant. In both rd22-1 and uspl1, transcript of the mutated gene was not detected based on either semi-quantitative PCR or Northern blotting (Figure S4B), corroborating the prediction of microarray analysis (Figure S4C). When subjected to moisture stress for 2–3 days, wild type plants became discolored as a result of an accumulation of anthocyanin and their growth ceased, whereas both the single and double mutant plants remained green and showed no evidence of any growth retardation (Figure 3A). Exposure to a longer period of moisture stress discriminated between wild type and the mutants in a similar manner (Figure S5A).
Figure 3. Influence of drought stress treatment on single and double loss of function mutants.
A) Four weeks old wild type (Col-0), single and double mutant plants (rd22-1 and uspl1, rd22-1/uspl1) were drought stressed for 1–5 days before they were returned to climate chamber conditions. Time of stress treatment in days is indicated left. B) Growth rates of plants under control and drought stress conditions. Bars indicate the growth rates at 22 days after sowing (DAS) for early phase of drought stress and 34 for the late phase of drought stress. For application of drought stress stop of watering started at 21 DAS. Wild type (Col-0): green bar; rd22-1: bright blue bar; rd22-2: dark blue bar; uspl1: purple bar; rd22-1/uspl1pink bar. Original data: Figure S5 B, Table S2, Asterisks indicate significant differences (p<0.01). C) NIR reflection as a water content-related parameter. Bars indicate the NIR intensity at 21 days after sowing (DAS) for start of experiment and at 35 DAS for the end of experiment. Wild type (Col-0): green bar; rd22-1: bright blue bar; rd22-2: dark blue bar; uspl1: purple bar; rd22-1/uspl1pink bar. Ncontrol = 5, Nstress = 10 plants. Original data in Table S3.
Detection of plant growth via automated phenotyping
To quantify the drought response of the mutant plants an automated phenotyping platform (LemnaTec) was used. The growth of 30 plants of control (Col-0), rd22-1, rd22-2, uspl1 and rd22-1/uspl1 each under defined control and drought stress condition was analysed. Drought stress was applied from 21 DAS by complete withdrawal of water. To monitor the growth of the plants, leaf area was estimated from top view images ([39], [48], Figure S5B, Table S2) and used for calculation of relative growth rates [38]. An elevated growth rate of the rd22-1 single mutant plant as well as for the double mutant plants compared to wild type could be found in the early phase of the experiment (22 DAS, Figure 3B). The first significant drought related difference in growth rate was detectable after 7 days without watering (28 DAS, Figure S5B). A significant difference in the growth rate of the rd22/uspl1 double mutant plants compared to the wild type plants was detectable almost throughout the entire experiment initially from day 24 till day 32 after sowing under stress conditions (Figure 3B, Figure S5B).
Plant senescence as consequence of the drought stress was monitored by quantifying the ratio of yellow to green pixels. Leaves of the mutant plants rd22-1, uspl1 and the double mutant showed a lower accumulation of yellow stained material, indicative for reduced senescence (33 DAS, Figure S5C). In order to analyse the relative water status of the plants the reflected near-infrared (NIR) radiation (1450 nm) from leaves was detected [49]. NIR intensity is calculated as 1 – NIR reflectance and illustrates the relation to the water content of the leaves. The drought stress related decrease of the NIR intensity was detectable at 33 DAS (Table S3). All genotypes analysed showed a similar level of detectable NIR reflectance in the beginning of the experiment and continuously under control conditions. At the end of the drought stress exposure rd22 and rd22/uspl1 double mutant plants showed a higher NIR intensity, indicating higher water content in the leaves (Figure 3C).
Plant growth response to salinity and osmotic stress and exposure to ABA
To further investigate the role of AtRD22 and AtUSPL1 during salinity stress and correlating responses on the transcriptional level, wild type and mutant plants were grown on plates with the respective treatments. Two-weeks-old seedlings were transferred for three days to MS basal medium containing one of the following stress inducing compounds: 150 mM and 300 mM NaCl, 100 µM ABA, 4% trehalose, 4% sorbitol, 4% glucose, 4% fructose, 4% sucrose, 300 mM manitol, 15% PEG 6000 and 4% PEG 20000. Although these conditions are artificially mimicking drought, they were chosen to achieve a uniform plant response to the stimulus. When exposed to either 150 mM NaCl or 100 µM ABA, only minor signs of stress symptoms were apparent, but in the former case, the growth of both the wild type and single mutant plants was retarded (Figure S6A). Wild type and rd22-1 plants exhibited the least extent of leaf bleaching, while the double mutant and particularly the uspl1 single mutant, were more visibly affected. Exposure to the various sugars had only a mild effect on plant growth, but a general tendency was for the mutant plants to be more vigorous than the wild type ones. Therefore NaCl 150 mM conditions were chosen for further transcriptome and chlorophyll analysis.
Chlorophyll and pheophytin content
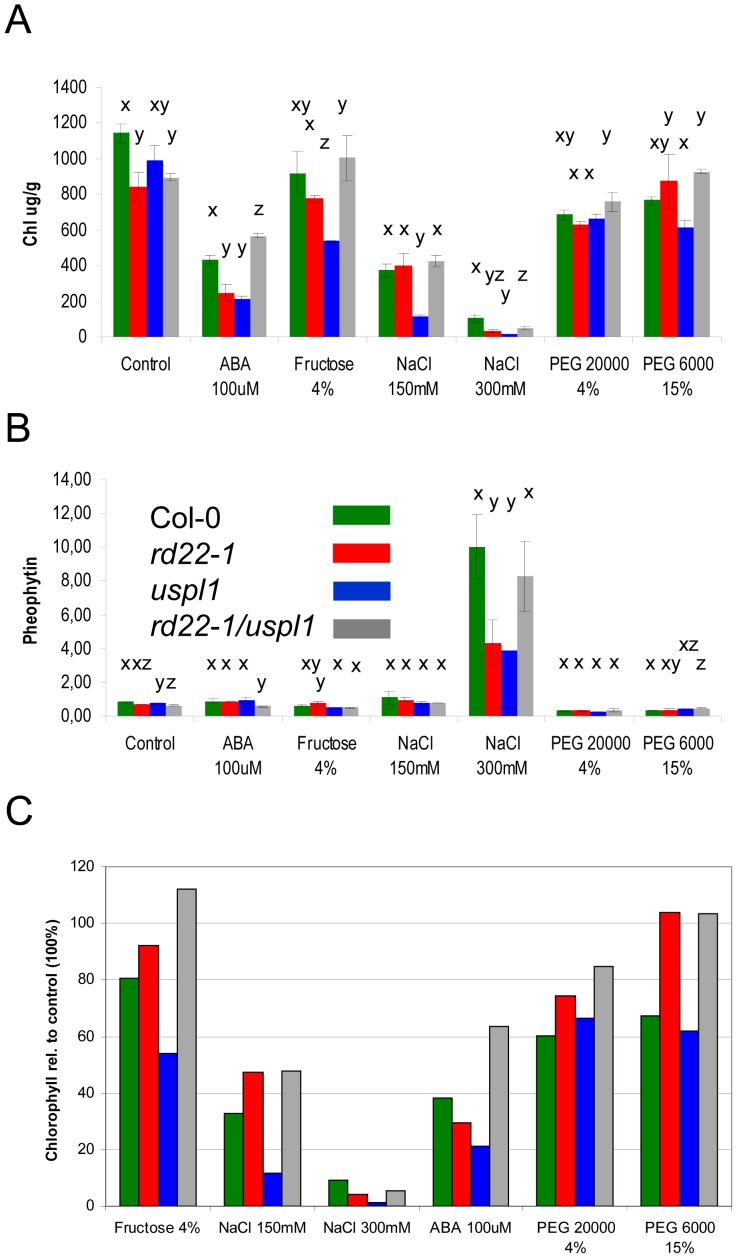
After a four day exposure to moisture stress, the quantity of chlorophyll a and b in the rd22-1 mutant and the double mutant leaves was less than that in non-stressed plants (Figure S6C). Since the contents of chlorophyll a and chlorophyll b were strongly correlated with one another, subsequent measurements considered the total chlorophyll content. Pheophytin is a degradation product of chlorophyll, that accumulates during senescence, dark [50] and salt stress [51]. In plants grown in the presence of 300 mM NaCl, the content of chlorophyll (Figure 4A) fell sharply, while that of pheophytin rose (Figure 4B). However when challenged by a lesser level of stress (100 µM ABA or 150 mM NaCl), while the chlorophyll content was reduced, there was no measurable increase in pheophytin content (Figure 4C). Chlorophyll content in the rd22-1 and uspl1 mutants was more severely reduced in the presence of either 150 mM NaCl or 100 µM ABA. The uspl1 mutant was the most compromised genotype with respect to chlorophyll content when the medium was supplemented with fructose, while the rd22-1 and the double mutant plants out-performed the wild type and uspl1 mutant plants when the stress was imparted by PEG. To clearly indicate this different chlorophyll degradation in the rd22-1 single and double mutant plants the amount [%] of total clorophyll is indicated relative to the amount of clorophyll at control conditions (Figure 4C). Compared to the amount of chlorophyll under control conditions the rd22-1 and uspl1 mutant plants show the strongest reduction upon 150 mM NaCl and 100 µM ABA. In addition, the reduction of chlorophyll in the uspl1 mutant with fructose show a major difference compared to the other genotypes. In the rd22-1 and rd22-1/uspl1 mutant plants the amount of chlorophyll is higher on fructose, 150 mM NaCl and PEG supplemented media compared to the wild type and uspl1 mutant plants. Taken into account that the chlorophyll content in the mutant in the unstressed condition varies from the wild type a relative value is indicated to show the change due to the treatment (Figure S6B).
Figure 4. Chlorophyll and pheophytin content in single and double BURP mutants.
The bars represent the total A) chlorophyll and B) pheophytin content in leaves from wild type (Col-0, green bar), single and double mutant plants (rd22-1, red bar, uspl1, blue bar and rd22-1/uspl1, grey bar). Chlorophyll a and b was determined separately (Figure S6A) and subsumed as total chlorophyll content. Error bar represents standard error. N = 5–6 plants in duplicate. Statistical analysis was performed by oneway ANOVA at alpa = 0.05 Tukey post hoc test: same letters indicate no difference, different letters indicate significant difference. C) The bars show the total chlorophyll content [%] relative to unstressed control plants.
The conclusion was that AtRD22 acts to suppress chlorophyll degradation under moisture stress.
Transcriptome analysis
The transcriptional responses to exposure to 150 mM NaCl and to 4% w/v trehalose overlapped by about 10% (Figure S6A). The 150 mM NaCl treatment resulted in a changed transcript abundance for 913 of the 22,392 genes represented on the ATH1 chip. Salinity stress and both the trehalose and sorbitol treatments marginally up-regulated AtRD22, while the other BURP family genes were hardly affected (Figure S6D). The four genes showing the greatest change in transcript abundance between wild type and mutant plants grown under non-stressed conditions (Table 1) were At4g33720 (encoding CAP, a PR protein under the control of the DREB2A tanscription factor, [52]: up-regulated by 12 fold in both rd22-1 and uspl1 compared to wild type); At5g49700 (encoding a DNA-binding protein associated with moisture stress [53]: up-regulated by 56 fold in rd22-1 and by 40 fold in uspl1); At2g27550 (encoding ATC, a systemic inhibitor of floral initiation [54]: up-regulated by 23 fold in both rd22-1 and uspl1); and At2g44380 (encoding a DC1-domain containing protein, involved in the abiotic stress response, [55]: up-regulated by 22 fold in both rd22-1 and uspl1). The full set of differentially transcribed genes detected in response to the various treatments is given in Table S4 (see also Figure S7). A total of 77 genes displayed differential transcription between wild type and rd22-1 plants grown under non-stressed conditions; 31 out of these are associated with either biotic or abiotic regulatory pathways (Figure S7A) and included eight encoding a peroxidase putatively involved in H2O2 degradation. The absence of AtUSPL1 only re-programmed 18 genes, of which five (one encoding a peroxidase) are associated with either biotic or abiotic regulatory pathways (Figure S7B). Exposure to 150 mM NaCl resulted in the up-regulation of a number of genes associated with either moisture stress or pathogen defense (Table 2). In rd22-1, out of the 764 differentially transcribed genes, 231 fell into one of these two categories (Figure S7C), while in the uspl1 mutant, only 18 genes were up-regulated, of which five were associated with either the biotic or the abiotic stress response (Figure S7D). In the latter mutant, AtRD22 was strongly up-regulated in the presence of salinity stress. When the stress was applied by the addition of trehalose to the medium, 55 of the 171 differentially transcribed genes identified in the rd22-1 mutant were associated with either the abiotic or the biotic stress response (Figure S7E). Over 50% (111 out of 212) of the genes responding to trehalose treatment reacted similarly to salinity stress (Figure S3A, Table S4). The set of common up- regulated genes included the transcription factor genes AtMYB15 (At3g23250) and WRKY33 (At2g38470), LPT3 and LPT4 (encoding lipid transporters), At1g35910 (a putative trehalose-6-phosphate phosphatase), At1g61120 (terpene synthase 04), At1g16850 (unknown function) and At1g78410 (VQ motif-containing protein). Although fewer genes were induced by the trehalose (212) than by the 150 mM NaCl (913) treatment, the conclusion was that the two stress agents must affect a similar class of gene. The response to both stress agents also included the down-regulation of several photosynthesis-related genes.
Table 1. Top differential expressed genes in A) rd22-1 and B) uspl1 at standard growth conditions in the aerial part of 2 week old seedlings.
| A) rd22 | |||||||||
| Probe Set ID | fold change | Gene | AGI ID | put. Function | Probe Set ID | fold change | Gene | AGI ID | put. Function |
| 257162_s_at | 59 | At3G24290 | amm. transporter | 265364_at | −45 | At2G13330 | Transposon | ||
| 248564_at | 56 | At5G49700 | DNA binding | 246908_at | −40 | RD22 | At5G25610 | Disease Defense | |
| 249052_at | 47 | PDF1.2 | At5G44420 | Disease Defense | 251677_at | −38 | ORG3 | At3G56980 | DNA bind |
| 245276_at | 41 | ATHB-2 | At4G16780 | 257453_at | −30 | At1G65130 | hydrolase | ||
| 265102_at | 39 | At1G30870 | Peroxidase | 261684_at | −29 | At1G47400 | |||
| 259276_at | 38 | At3G01190 | PER27 Peroxidase | 265665_at | −28 | At2G27420 | |||
| 263096_at | 38 | AHB1 | At2G16060 | 249645_at | −26 | THI2.2 | At5G36910 | toxin receptor | |
| 264567_s_at | 37 | At1G05250 | Peroxidase | 266472_at | −26 | ||||
| 254044_at | 35 | XTR9 | At4G25820 | hydrolase | 254170_at | −22 | At4G24430 | lyase | |
| 254200_at | 34 | At4G24110 | 247252_at | −22 | At5G64770 |
Complete list of differential expressed genes in Table S4.
Table 2. Differential expressed genes in A) rd22-1 and B) uspl1 on medium containing 150mM NaCl in the aerial part of 2 week old seedlings.
| A) rd22 on NaCl | ||
| up regulated genes | ||
| Probe Set ID | Fold change | Gene Symbol |
| 247215_at | 40 | PROPEP3 |
| 266017_at | 38 | |
| 252984_at | 34 | ELI3-2 |
| 252487_at | 33 | |
| 256627_at | 33 | |
| 246340_s_at | 32 | FAMT |
| 264005_at | 31 | AGP2 |
| 257206_at | 29 | |
| 267035_at | 26 | AGT3 |
| 259975_at | 25 | |
| 253872_at | 24 | RD26 |
| 254101_at | 24 | AMY1 |
| 266267_at | 23 | ATGSTU4 |
| 256245_at | 22 | HSP70 |
| 255502_at | 21 | |
| 247308_at | 20 | |
| 256436_at | 20 | |
| 266142_at | 11 | |
| 256603_at | 11 | |
| 258791_at | 10 | PR4 |
| 264415_at | 6 | RAP2.6 |
| 257517_at | 4 | |
| 258277_at | 4 | PAD3 |
| 260919_at | 3 | |
| 249481_at | 2 | |
| down regulated | ||
| Probe Set ID | Fold change | Gene Symbol |
| 246366_at | −27 | |
| 256772_at | −22 | BGAL1 |
| 250366_at | −21 | |
| 261118_at | −20 | |
| 261684_at | −20 | |
| 267264_at | −17 | SCPL11 |
| 258497_at | −11 | COL2 |
| 261413_at | −11 | PLL5 |
| 246908_at | −7 | RD22 |
| 247450_at | −2 | |
| 261351_at | −2 | |
| 266363_at | −1 | |
Discussion
RD22 and USPL1 have suppressor function during drought stress
AtRD22 and AtUSPL1 were transcribed predominantly in, respectively, the leaf and the root. The induction of both genes by moisture stress generated a slight growth reduction. While the induction by moisture stress of AtRD22 has been noted previously, this was not the case for AtUSPL1, most likely due to a concentration on the short-term stress response. Microarray-based transcriptomic analyses have shown that exposure to either NaCl or trehalose induces not only the up-regulation of salinity, trehalose and moisture stress responsive genes, but also the down-regulation of photosynthesis-related genes. The latter is implied by both the induction of leaf chlorosis and senescence and the measured fall in the chlorophyll content of the leaves of stressed plants. The present data confirmed that both salinity and trehalose activate the ABA-mediated moisture stress response. [56]–[58]. The sole BURP gene family to be up- regulated in the aerial part of the plant was AtRD22, consistent with observations based on the expression of the transgene pAtRD22::GUS [26]. The root specificity of AtUSPL1 expression was confirmed both by the behavior of the pAtUSPL1::GUS transgene and by Northern hybridization experiments.
The T-DNA insertion mutants of both AtRD22 and AtUSPL1 produced no detectable relevant transcript, so each was taken as a genuine loss-of- function allele. In both cases, a reduced photosynthesis phenotype was exhibited, as reflected by a fall in both chlorophyll content under non-stressed growing conditions and photosynthesis-associated gene transcript abundance in stressed plants. Both under prolonged moisture stress mimicking conditions and actual moisture stress, stress tolerance was enhanced in all of the mutants. The mutants' performance (and particularly that of the double mutant), as measured by either the development of leaf area or by the rate of leaf tissue senescence, showed that they were more tolerant of moisture stress than were wild type plants. NIR intensity appeared to reliably reflect relative water content, which was higher in each of the mutants than in wild type.
Given that photosynthesis is clearly compromised by moisture stress, it was not surprising that the transcription of several photosynthesis-associated genes was altered when the plants were exposed to moisture stress. Although the mutant plants' transcriptomes were indistinguishable from that of the wild type with respect to photosynthesis-related genes, nevertheless their leaves contained less chlorophyll than the leaves of wild type plants raised under non-stressed conditions. The reduction in chlorophyll content in the mutants induced by exposure to either NaCl or PEG was less severe than that in wild type plants. Notably, under moisture stress conditions, the leaves of double mutant plants retained more chlorophyll than those of wild type ones. The retention of chlorophyll can be expected to support a higher rate of photosynthesis, so that less transpiration is required to generate a given quantity of assimilate. Since the plant's capacity to retain its water is improved, its water content under moisture stress conditions was greater (Figure 3C).
The observed up-regulation of peroxidase encoding genes in the mutants suggested a secondary effect of the loss-of-function mutations. The transcriptomic data implied that the oxidative state within the mutant plants differed from that within the wild type, with a knock-on effect on gene expression in both ABA-dependent and ABA-independent pathways [59]. Both the phenotype of the mutants as well as the transcriptional response of both genes suggested that under moisture stress conditions, their products exerted a suppressor function. Similar conclusions have been drawn regarding the effect of mutations of genes encoding SnRK2 [60] the effect of which is an almost complete abolition of the ABA response. In particular, AtRD22 is not transcribed in these mutants when the growing medium is supplemented with ABA reflecting moisture stress condition. Under normal growing conditions, the SnRK2 mutants exhibit reduced growth, which (along with plant survival) is improved by the addition of ABA to the growing medium. The inference is that the BURP domain containing proteins AtRD22 and AtUSPL1 act as suppressors of the ABA-mediated moisture stress response.
RD22 act as suppressor predominantly in the leaf, while USPL1 act in the root
Although both AtRD22 and AtUSPL1 exhibited organ-specific transcription, the loss of function of each gene induced a comparable improvement in the plant's tolerance to moisture stress, at least at the level of the vigour of the leaves. The sensitivity of the microarray platform rules out any trans-silencing effects of either T-DNA insertion on other members of the BURP gene family. Since this sort of silencing has been associated with several T-DNA insertion mutations [61], the assumption is that both AtRD22 and AtUSP1 are involved in a holistic moisture stress response. The microarray analysis demonstrated that the two loss-of-function mutations resulted in the induction of a partially overlapping set of genes in the aerial part of the plant, even though AtUSPL1 was transcribed specifically in the root. Not only was the tolerance of moisture stress enhanced in both mutants, but also their chlorophyll content was reduced. As the lack of AtRD22 and AtUSP1 led to an enhanced tolerance to moisture stress, the proposed suppressor function of the BURP domain-containing proteins during an episode of moisture stress includes an organ-specific component. Both proteins are part of an ABA mediated moisture stress response pathway, with AtRD22 functioning mainly in the aerial part of the plant and AtUSPL1 in the root.
Increased drought resistance might be correlated to an increased defence gene response
The transcriptomic analysis of the two mutants revealed an increased transcript abundance compared to the wild type levels with respect to various genes associated with the response to biotic stress (for example, the gene PDF1.2 was up-regulated by nearly 50 fold). This class of genes was differentially transcribed both under non-stressed and moisture stressed conditions. Several peroxidase encoding genes were also up-regulated. Peroxidases are known to represent an integral component of the plant's hypersensitive response [62]. H2O2 is also known as signal molecule during the stress response [63], [64]. A loss of control over the production of H2O2 might result in the described reduction of chlorophyll content [65], [66] and increased water content under drought stress conditions. Such regulation would subsequently lead to better performance of the mutant plants under drought stress.
The up-regulation of peroxidase encoding genes implies a level of linkage between the ABA-mediated moisture stress response and defense against pathogen invasion. Such a connection has been proposed in a suggested model for the function of OCP3, a homeodomain transcription factor. The loss of OCP3 function results in an enhanced tolerance to moisture stress and at the same time an increased sensitivity to ABA. The abundance of AtRD22 transcript in the ocp3 mutant is not different to that in the wild type, and the plant's susceptibility to Botrytis cinerea infection is reduced [67]. The ocp3 mutant was initially identified and named after the phenotype of constitutive over expression of a cationic peroxidase [68]. Here, AtOCP3 was marginally down regulated when the plants were challenged by NaCl, but there was no transcriptional difference between wild type and either of the two mutants. The implication is that the BURP-containing proteins act to enhance the plant's moisture stress response via the up-regulation of peroxidase encoding genes. In soybean, a direct interaction between RD22 and cell wall-localized peroxidases has been described [69]. The up- regulation of H2O2 detoxifying enzymes enhances moisture stress tolerance [70] as well as explaining the link with the pathogen defense response.
A reduced chlorophyll content, in conjunction with an elevated level of transcription of defense response genes under non-stressed growing conditions, suggests that the mutant plants are primed to mount a stronger and/or more rapid set of measures to prevent the accumulation of ROS. A change in the accumulation of H2O2 would not only have an impact on the defense response [71], but also on the response to moisture stress [63], [72]. Due to the overall elevated expression of H2O2-detoxifying enzymes such responses could be limited, leading to an enhanced drought resistance. Our study provides the first functional approach to investigate BURP-domain encoding gene function in addition to the previously published structural comparisons.
Supporting Information
Expression profile of the Arabidopsis thaliana BURP gene family. A) Expression profile of the Arabidopsis thaliana BURP gene family. Data obtained from Genevestigator database (Zimmerman et al., 2004). Relative expression of AtRD22 (red) and AtUSPL1 (blue) is given for the different developmental stages of Arabidopsis life cycle (left to right: germinating seed, seedling, Young rosette, developed rosette, bolting, young flower, developed flower, flowers and siliques, mature siliques, senescence). B) Expression of AtUSPL1 confirmed by Northern Blot analysis. Expression of AtUSPL1 was determined from root, leaf, shoot, young silique and total flower tissue of Arabidopsis thaliana (Col-0) plants. For equal loading of the RNA samples probing of the membrane with specific probe against housekeeping mRNA of AtROC1 (rotamase cyclophilin, renamed in AtCYP1) was performed. C) Expression of AtRD22 obtained from Arabidopsis eFP Browser (Winter et al., 2007). The expression of selected stimuli (Cold: 4°C, Osmotic: 300 mM Mannitol, Salt: 150 mM NaCl and Drought: air steam 15 min) is displayed for the aerial as well as the hypogeic part of the plant. AtRD22 expression is induced in the aerial part of the plant after applying osmotic, salt stress and slightly increased after loss of water due to airstream treatment. D) Expression of AtUSPL1 obtained from Arabidopsis eFP Browser (Winter et al., 2007). The expression of selected stimuli (Cold: 4°C, Osmotic: 300 mM mannitol, Salt: 150 mM NaCl and Drought: air steam 15 min) is displayed for the aerial as well as the hypogeic part of the plant. AtUSPL1 expression is induced in the hypogeic part of the plant after applying osmotic, salt stress and slightly increased after loss of water due to airstream treatment.
(TIF)
Expression of the BURP domain containing gene family in Arabidopsis thaliana . Expression analysis of AtRD22 (At5G25610) and AtUSPL1 (At1G49320) obtained from Genevestigator database (Zimmerman et al., 2004) displaying induced and reduced expression after different conditions and stresses. Displayed are only changes in expression upon stress/treatment above threefold with a statistic significance p<0.001. Red indicates up-regulation; Green indicates down-regulation.
(TIF)
Comparison of gene expression in Arabidopsis wild type plants grown on 150 mM NaCl and 4% trehalose supplemented MS medium. BURP gene family mRNA in Col-0 under selected stress conditions. Bars indicate the expression pattern obtained by microarray analysis using ATH1 chip: AtRD22 (red): 246908_at; AtUSPL1 (blue): 262388_at; AtPG1 (dark green): 265131_at; AtPG2 (green): 264277_at; AtPG3 (bright green): 264315_at. Displayed is the rel. Abundance of mRNA.
(TIF)
Characterization of BURP mutant plants. A) Scheme of the AtRD22 and AtUSPL1 gene model. In blue the exon-intron structure within the coding region of the respective gene is given. The protein structure refers to Figure 1. In the encoded protein parts are given in aminoacids [aa]. And the size of the fulllenght protein is given below the gene description. Position of T-DNA insertions of used mutant plants are indicated by black lines. Mutant alleles for rd22: rd22-1 (SALK_146066) and rd22-2 (WiscDsLox481-484P12). Mutant alleles for uspl1: uspl1: (SALK_022325). The T-DNA insertion line SALK_146066 (rd22-1) is based on pROK2 conferring kanamycin resistance and the WiscDsLox481-484P12 (rd22-2) is based on pWiscDs-Lox conferring phosphinotricin (BASTA) resistance. uspl1 T-DNA insertion lines SALK_022325 (referred to as uspl1, based on pROK2 conferring kanamycin resistance from Nottingham Arabidopsis Stock Centre) was analyzed. The position of the T-DNA insertion in At1G49320 (AtUSPL1) was determined by PCR and subsequent sequencing. The position of the T-DNA insertion are depicted and confirmed by PCR. Double mutant rd22-1/uspl1 line was generated by crossing SALK_146066 and SALK_022325 and identified in the F3 generation by PCR. Kanamycin and phosphinotricin resistance of the plants was tested on germination medium (1 MS salts; 10 g/l sucrose) plates with 40 mg/l kanamycin or 20 mg/l glufosinate-ammonium under long day conditions. B) Analysis of used T-DNA insertion mutants. The absence of AtRD22 and AtUSPL1 mRNA in homozygous rd22 and uspl1 mutant plants was determined by Northern Blot analysis (left) and semi quantitative RT-PCR. C) Analysis of BURP-gene family mRNA in rd22-1 and uspl1 mutants by microarray analysis on MS medium. Bars indicate the rel. expression signal obtained by microarray analysis using ATH1 chip from each single experiment: AtRD22 (red): 246908_at; AtUSPL1 (blue): 262388_at; AtPG1 (dark green): 265131_at; AtPG2 (green): 264277_at; AtPG3 (bright green): 264315_at.
(TIF)
Increased drought stress resistance of the rd22 and uspl1 mutant plants. A) The plants were drought stressed by withdrawal of water. Top: day 0 (80% RWC in the soil); Bottom: appearance of plants after 8 days without watering. B) Top: Projected area of wild type and mutant plants under control conditions and drought stress (dotted line) obtained by lemnatec phenotyping; Drought stress was started 21 days after sawing (DAS). Middle: Growth rates calculated based on Poorter and Lewis 1986 for individual days. Wild type (Col-0): green line; rd22-1: bright blue line; rd22-2: dark blue line; uspl1: purple line; rd22-1/uspl1pink line (+/- s.e.m.). Bottom: Statistical analysis or growth rates at 28 DAS. Asterisks indicate significant differences (p<0.05) between control and stress. C) Estimation of senescence after drought stress. The graph indicates the ration of yellow to green pixels in the plant area of the analysed top view images from day 33. Wild type (Col-0): green bar; rd22-1: bright blue bar; rd22-2: dark blue bar; uspl1: purple bar; rd22-1/uspl1: pink bar. Ncontrol = 5, Nstress = 10 plants. Asterisks indicate significant differences (p<0.05).
(TIF)
A) Differentially expressed genes (Col-0) between 150 mM NaCl containing medium and standard growth conditions sorted by relation to pathway (Mapman). Top: Differentially expressed genes (Col-0) between 150 mM NaCl containing medium and standard growth conditions sorted by relation to pathway (Mapman). The different numbers indicate different categories/pathways and are described in the table. Bottom: Differentially expressed genes (Col-0) between standard growth conditions and 4% trehalose containing medium. The bar diagram indicates the percentage of common (red) and inverse (yellow) regulated genes upon salt (grey) and sugar (blue) treatment. Approximately half of the genes induced by 4% trehalose treatment are also reacting on 150 mM NaCl treatment. B) Influence of ABA and NaCl on single and double loss of function mutants. Growth phenotypes of wild type (Col-0), single and double mutant plants (rd22-1 and uspl1, rd22-1/uspl1) on standard MS-medium and 150 mM NaCl, 300 mM NaCl and 100 µM ABA supplemented MS-Medium. Two week old seedlings were transferred for 3 days to the MS basal and supplemented medium. C) Chlorophyll and pheophytin content of wild type and rd22-1, uspl1 and rd22-1/uspl1 mutant plants on different supplemented media. Two week old seedlings were transferred to the MS basal + one of the following stress treatments for 4 days: 100 µM ABA, 4% fructose, 150 mM NaCl, 300 mM NaCl, 4% PEG 20000 and 15% PEG 6000. From Top to bottom: Chlorophyll a and b content. The error bar represents standard error. Content was estimated from 5-6 plants in duplicate. Statistical analysis was performed by oneway ANOVA at alpa = 0.05 Tukey post hoc test: same letters indicate no difference, different letters indicate significant difference. Chlorophyll a and b and pheophytin content [%] relative to unstressed control plants.
(TIF)
Differentially expressed genes in the rd22-1 and uspl1 plants grown on control, 150 mM NaCl and 4% trehalose containing medium. List of used categories is given in Figure 3 A. Top: Display of top regulated category (Bin 20, biotic and abiotic stress pathways) of differential regulated genes. Bottom: Overview of all differentially regulated genes mapped to categories (Bins) by MAPMAN. For RNA extraction tissue was grinded in liquid nitrogen and the homogenized powder was added to 1 ml TRIZOL and incubated at RT for 5 min. Samples were centrifuged at 10.000 rpm for 10 min and the supernatant was transferred to a new tube. 200 µl of chloroform were added and incubated at room temperature for 2–3 min. Samples were again centrifuged as described above and the aqueous supernatant was transferred to the QIAshredder column and centrifuged for 30 s at 10000 rpm. 350 µl of RLT buffer (plus β-mercaptoethanol) and 250 µl of absolute ethanol were added to the flow-through and passed through an RNAeasy spin column. All the following steps were performed as described in the manufacturer's protocol followed by in-column DNAse digestion. A) Schematic display of differentially expressed genes in rd22-1 by MAPMAN. 31 out of 77 differential regulated genes are mapping to biotic and abiotic stress pathways. B) Schematic display of differentially expressed genes in uspl1 by MAPMAN. 5 out of 18 differential regulated genes are mapping to biotic and abiotic stress pathways. C) Schematic display of differentially expressed genes in rd22-1 on 150 mM NaCl by MAPMAN. 231 out of 764 differentially regulated genes are mapping to biotic and abiotic stress pathways. D) Schematic display of differentially expressed genes in uspl1 on 150 mM NaCl by MAPMAN. 7 out of 12 differentially regulated genes are mapping to biotic and abiotic stress pathways. E) Schematic display of differentially expressed genes in rd22 on 150 mM NaCl by MAPMAN. 55 out of 171 differentially regulated genes are mapping to biotic and abiotic stress pathways.
(TIF)
Primer information.
(XLS)
Average projected plant area (mm2) from top images.
(XLSX)
Average near-infrared intensity as observed from top images, obtained using a Nir 300 camera from VDS Vosskühler (now Allied Vision Technologies). High values indicate relative high water content.
(XLSX)
List of differentially expressed genes.
(XLSX)
Acknowledgments
We thank Elke Liemann and Jana Lorenz for excellent technical assistance, Dijun Chen for support with statistical analysis of lemnatec data, Ingo Mücke for his support using the automated phenotyping system.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was funded by Vietnamese government. The junior Research Group Abiotic Stress Genomics (MK and NS) is funded by IZN (Interdisciplinary Center for Crop Plant Research, Halle (Saale), Germany. VTH and CS are funded by BMBF (GABI-GRAIN: Integrative genomics approach for exploring seed quality and yield under terminal drought; FKZ 0315041A and FKZ 0315041C) and IB-BMBF (Ind09/526). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bray EA, Bailey-Serres J, Weretilnyk E (2000) Responses to abiotic stresses. Biochemistry and Molecular Biology of Plants, Rockville, MD, USA: American Society of Plant Physiologists. pp.1158–1203.
- 2. Sreenivasulu N, Sopory SK, Kishor PBK (2007) Deciphering the regulatory mechanisms of abiotic stress tolerance in plants by genomic approaches. Gene 388: 1–13. [DOI] [PubMed] [Google Scholar]
- 3. Agarwal PK, Agarwal P, Reddy MK, Sopory SK (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25: 1263–1274. [DOI] [PubMed] [Google Scholar]
- 4. Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant Cellular and Molecular Responses to High Salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499. [DOI] [PubMed] [Google Scholar]
- 5. Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803. [DOI] [PubMed] [Google Scholar]
- 6. Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnology 17: 287–291. [DOI] [PubMed] [Google Scholar]
- 7. Yamaguchi-Shinozaki K, Shinozaki K (1992) A novel Arabidopsis DNA binding protein contains the conserved motif of HMG-box proteins. Nucleic Acids Res 20: 6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamaguchi-Shinozaki K, Shinozaki K (1993) The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet 238: 17–25. [DOI] [PubMed] [Google Scholar]
- 9. Hattori J, Boutilier KA, van Lookeren Campagne MM, Miki BL (1998) A conserved BURP domain defines a novel group of plant proteins with unusual primary structures. Mol Gen Genet 259: 424–428. [DOI] [PubMed] [Google Scholar]
- 10. Baumlein H, Boerjan W, Nagy I, Bassuner R, Van Montagu M, et al. (1991) A novel seed protein gene from Vicia faba is developmentally regulated in transgenic tobacco and Arabidopsis plants. Mol Gen Genet 225: 459–467. [DOI] [PubMed] [Google Scholar]
- 11. Wohlfarth T, Braun H, Kirik V, Kolle K, Czihal A, et al. (1998) Regulation and evolution of seed globulin genes. Journal of Plant Physiology 152: 600–606. [Google Scholar]
- 12. Boutilier KA, Gines MJ, DeMoor JM, Huang B, Baszczynski CL, et al. (1994) Expression of the BnmNAP subfamily of napin genes coincides with the induction of Brassica microspore embryogenesis. Plant Mol Biol 26: 1711–1723. [DOI] [PubMed] [Google Scholar]
- 13. Teerawanichpan P, Xia Q, Caldwell SJ, Datla R, Selvaraj G (2009) Protein storage vacuoles of Brassica napus zygotic embryos accumulate a BURP domain protein and perturbation of its production distorts the PSV. Plant Molecular Biology 71: 331–343. [DOI] [PubMed] [Google Scholar]
- 14. Treacy BK, Hattori J, Prudhomme I, Barbour E, Boutilier K, et al. (1997) Bnm1, a Brassica pollen-specific gene. Plant Molecular Biology 34: 603–611. [DOI] [PubMed] [Google Scholar]
- 15. Chen LZ, Guan LM, Seo M, Hoffmann F, Adachi T (2005) Developmental expression of ASG-1 during gametogenesis in apomictic guinea grass (Panicum maximum). Journal of Plant Physiology 162: 1141–1148. [DOI] [PubMed] [Google Scholar]
- 16. Wang AM, Xia Q, Xie WS, Datla R, Selvaraj G (2003) The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proceedings of the National Academy of Sciences of the United States of America 100: 14487–14492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Batchelor AK, Boutilier K, Miller SS, Hattori J, Bowman LA, et al. (2002) SCB1, a BURP-domain protein gene, from developing soybean seed coats. Planta 215: 523–532. [DOI] [PubMed] [Google Scholar]
- 18. Datta N, Lafayette PR, Kroner PA, Nagao RT, Key JL (1993) Isolation and Characterization of 3 Families of Auxin down-Regulated Cdna Clones. Plant Molecular Biology 21: 859–869. [DOI] [PubMed] [Google Scholar]
- 19. Granger C, Coryell V, Khanna A, Keim P, Vodkin L, et al. (2002) Identification, structure, and differential expression of members of a BURP domain containing protein family in soybean. Genome 45: 693–701. [DOI] [PubMed] [Google Scholar]
- 20.Xu HL, Li YX, Yan YM, Wang K, Gao Y, et al.. (2010) Genome-scale identification of Soybean BURP domain-containing genes and their expression under stress treatments. Bmc Plant Biology 10. [DOI] [PMC free article] [PubMed]
- 21. Shao YH, Wei G, Wang L, Dong Q, Zhao Y, et al. (2011) Genome-Wide Analysis of BURP Domain-Containing Genes in Populus trichocarpa. Journal of Integrative Plant Biology 53: 743–755. [DOI] [PubMed] [Google Scholar]
- 22. Held BM, John I, Wang H, Moragoda L, Tirimanne TS, et al. (1997) Zrp2: a novel maize gene whose mRNA accumulates in the root cortex and mature stems. Plant Mol Biol 35: 367–375. [DOI] [PubMed] [Google Scholar]
- 23. Gan D, Jiang H, Zhang J, Zhao Y, Zhu S, et al. (2011) Genome-wide analysis of BURP domain-containing genes in maize and sorghum. Mol Biol Rep 38: 4553–4563. [DOI] [PubMed] [Google Scholar]
- 24. Van Son L, Tiedemann J, Rutten T, Hillmer S, Hinz G, et al. (2009) The BURP domain protein AtUSPL1 of Arabidopsis thaliana is destined to the protein storage vacuoles and overexpression of the cognate gene distorts seed development. Plant Molecular Biology 71: 319–329. [DOI] [PubMed] [Google Scholar]
- 25. Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, et al. (1997) Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iwasaki T, Yamaguchishinozaki K, Shinozaki K (1995) Identification of a Cis-Regulatory Region of a Gene in Arabidopsis-Thaliana Whose Induction by Dehydration Is Mediated by Abscisic-Acid and Requires Protein-Synthesis. Molecular and General Genetics 247: 391–398. [DOI] [PubMed] [Google Scholar]
- 27. Park MY, Chung MS, Koh HS, Lee DJ, Ahn SJ, et al. (2009) Isolation and functional characterization of the Arabidopsis salt-tolerance 32 (AtSAT32) gene associated with salt tolerance and ABA signaling. Physiol Plant 135: 426–435. [DOI] [PubMed] [Google Scholar]
- 28. Sanchez JP, Chua NH (2001) Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 13: 1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Song HM, Zhao RM, Fan PX, Wang XC, Chen XY, et al. (2009) Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta 229: 955–964. [DOI] [PubMed] [Google Scholar]
- 30. Wang HM, Zhou L, Fu YP, Cheung MY, Wong FL, et al. (2012) Expression of an apoplast-localized BURP-domain protein from soybean (GmRD22) enhances tolerance towards abiotic stress. Plant Cell and Environment 35: 1932–1947. [DOI] [PubMed] [Google Scholar]
- 31. Ding Z, Li S, An X, Liu X, Qin H, et al. (2009) Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics 36: 17–29. [DOI] [PubMed] [Google Scholar]
- 32. Xu B, Gou JY, Li FG, Shangguan XX, Zhao B, et al. (2013) A Cotton BURP Domain Protein Interacts With -Expansin and Their Co-Expression Promotes Plant Growth and Fruit Production. Molecular Plant 6: 945–958. [DOI] [PubMed] [Google Scholar]
- 33. Schallau A, Kakhovskaya I, Tewes A, Czihal A, Tiedemann J, et al. (2008) Phylogenetic footprints in fern spore- and seed-specific gene promoters. Plant Journal 53: 414–424. [DOI] [PubMed] [Google Scholar]
- 34. Sreenivasulu N, Radchuk V, Strickert M, Miersch O, Weschke W, et al. (2006) Gene expression patterns reveal tissue-specific signalling networks controlling programmed cell death and ABA-regulated maturation in developing barley seeds (vol 47, pg 310, 2006). Plant Journal 47: 987–987. [DOI] [PubMed] [Google Scholar]
- 35. Orkin S (1990) Molecular-Cloning - a Laboratory Manual, 2nd Edition - Sambrook, J, Fritsch, Ef, Maniatis, T. Nature. 343: 604–605. [Google Scholar]
- 36. Bechthold N, Elli J, Pelletier G (1993) In planta Agrobacterium-mediated transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Paris 316: 871–883.8076216 [Google Scholar]
- 37. Jefferson RA, Kavanagh TA, Bevan MW (1987) Gus Fusions - Beta-Glucuronidase as a Sensitive and Versatile Gene Fusion Marker in Higher-Plants. Embo Journal 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poorter H, Lewis C (1986) Testing Differences in Relative Growth-Rate - a Method Avoiding Curve Fitting and Pairing. Physiologia Plantarum 67: 223–226. [Google Scholar]
- 39.Klukas C, Chen D, Pape JM (2014) IAP: an open-source information system for high-throughput plant phenotyping. Plant Physiol. [DOI] [PMC free article] [PubMed]
- 40. Govind G, Harshavardhan VT, Patricia JK, Dhanalakshmi R, Senthil Kumar M, et al. (2009) Identification and functional validation of a unique set of drought induced genes preferentially expressed in response to gradual water stress in peanut. Mol Genet Genomics 281: 591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hiscox JD, Israelstam GF (1979) Method for the Extraction of Chlorophyll from Leaf Tissue without Maceration. Canadian Journal of Botany-Revue Canadienne De Botanique 57: 1332–1334. [Google Scholar]
- 42. Church GM, Gilbert W (1984) Genomic sequencing. Proceedings of the National Academy of Sciences of the United States of America 81: 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seiler C, Harshavardhan VT, Reddy PS, Hensel G, Kumlehn J, et al. (2014) Abscisic acid flux alterations result in differential abscisic acid signaling responses and impact assimilation efficiency in barley under terminal drought stress. Plant Physiol 164: 1677–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 57: 289–300. [Google Scholar]
- 45. Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, et al. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant Journal 37: 914–939. [DOI] [PubMed] [Google Scholar]
- 46. Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, et al.. (2007) An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large-Scale Biological Data Sets. Plos One 2. [DOI] [PMC free article] [PubMed]
- 48. Klukas C, Pape JM, Entzian A (2012) Analysis of high-throughput plant image data with the information system IAP. J Integr Bioinform 9: 191. [DOI] [PubMed] [Google Scholar]
- 49. Munns R, James RA, Sirault XR, Furbank RT, Jones HG (2010) New phenotyping methods for screening wheat and barley for beneficial responses to water deficit. J Exp Bot 61: 3499–3507. [DOI] [PubMed] [Google Scholar]
- 50. Schelbert S, Aubry S, Burla B, Agne B, Kessler F, et al. (2009) Pheophytin Pheophorbide Hydrolase (Pheophytinase) Is Involved in Chlorophyll Breakdown during Leaf Senescence in Arabidopsis. Plant Cell 21: 767–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Santos CLV, Campos A, Azevedo H, Caldeira G (2001) In situ and in vitro senescence induced by KCl stress: nutritional imbalance, lipid peroxidation and antioxidant metabolism. Journal of Experimental Botany 52: 351–360. [PubMed] [Google Scholar]
- 52. Qin F, Sakuma Y, Tran LSP, Maruyama K, Kidokoro S, et al. (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, et al. (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang NC, Jane WN, Chen J, Yu TS (2012) Arabidopsis thaliana CENTRORADIALIS homologue (ATC) acts systemically to inhibit floral initiation in Arabidopsis. Plant Journal 72: 175–184. [DOI] [PubMed] [Google Scholar]
- 55. Li C, Lv J, Zhao X, Ai X, Zhu X, et al. (2010) TaCHP: a wheat zinc finger protein gene down-regulated by abscisic acid and salinity stress plays a positive role in stress tolerance. Plant Physiol 154: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barrero JM, Rodriguez PL, Quesada V, Piqueras P, Ponce MR, et al. (2006) Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ 29: 2000–2008. [DOI] [PubMed] [Google Scholar]
- 57. Hwang IW, Goodman HM (1995) An Arabidopsis-Thaliana Root-Specific Kinase Homolog Is Induced by Dehydration, Aba, and Nacl. Plant Journal 8: 37–43. [DOI] [PubMed] [Google Scholar]
- 58. Rosado A, Schapire AL, Bressan RA, Harfouche AL, Hasegawa PM, et al. (2006) The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for osmotic stress responses and abscisic acid sensitivity. Plant Physiology 142: 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shinozaki K, Yamaguchi-Shinozaki K (1992) [Plant genes induced by drought stress and ABA]. Tanpakushitsu Kakusan Koso 37: 1190–1199. [PubMed] [Google Scholar]
- 60. Fujii H, Verslues PE, Zhu JK (2011) Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci U S A 108: 1717–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Daxinger L, Hunter B, Sheik M, Jauvion V, Gasciolli V, et al. (2008) Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends in Plant Science 13: 4–6. [DOI] [PubMed] [Google Scholar]
- 62. Almagro L, Gomez Ros LV, Belchi-Navarro S, Bru R, Ros Barcelo A, et al. (2009) Class III peroxidases in plant defence reactions. J Exp Bot 60: 377–390. [DOI] [PubMed] [Google Scholar]
- 63. Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, et al. (2004) ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. Journal of Experimental Botany 55: 205–212. [DOI] [PubMed] [Google Scholar]
- 64. Hancock JT, Desikan R, Neill SJ (2001) Role of reactive oxygen species in cell signalling pathways. Biochem Soc Trans 29: 345–350. [DOI] [PubMed] [Google Scholar]
- 65. Kar RK, Choudhuri MA (1987) Possible Mechanisms of Light-Induced Chlorophyll Degradation in Senescing Leaves of Hydrilla-Verticillata. Physiologia Plantarum 70: 729–734. [Google Scholar]
- 66. Hynninen PH, Kaartinen V, Kolehmainen E (2010) Horseradish peroxidase-catalyzed oxidation of chlorophyll a with hydrogen peroxide Characterization of the products and mechanism of the reaction. Biochimica Et Biophysica Acta-Bioenergetics 1797: 531–542. [DOI] [PubMed] [Google Scholar]
- 67. Ramirez V, Coego A, Lopez A, Agorio A, Flors V, et al. (2009) Drought tolerance in Arabidopsis is controlled by the OCP3 disease resistance regulator. Plant Journal 58: 578–591. [DOI] [PubMed] [Google Scholar]
- 68. Coego A, Ramirez V, Gil MJ, Flors V, Mauch-Mani B, et al. (2005) An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OFCATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell 17: 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang YN, Liu C, Li KX, Sun FF, Hu HZ, et al. (2007) Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Molecular Biology 64: 633–644. [DOI] [PubMed] [Google Scholar]
- 70. Miao Y, Lv D, Wang P, Wang XC, Chen J, et al. (2006) An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mullineaux P, Ball L, Escobar C, Karpinska B, Creissen G, et al. (2000) Are diverse signalling pathways integrated in the regulation of arabidopsis antioxidant defence gene expression in response to excess excitation energy? Philos Trans R Soc Lond B Biol Sci 355: 1531–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Neill S, Barros R, Bright J, Desikan R, Hancock J, et al. (2008) Nitric oxide, stomatal closure, and abiotic stress. J Exp Bot 59: 165–176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression profile of the Arabidopsis thaliana BURP gene family. A) Expression profile of the Arabidopsis thaliana BURP gene family. Data obtained from Genevestigator database (Zimmerman et al., 2004). Relative expression of AtRD22 (red) and AtUSPL1 (blue) is given for the different developmental stages of Arabidopsis life cycle (left to right: germinating seed, seedling, Young rosette, developed rosette, bolting, young flower, developed flower, flowers and siliques, mature siliques, senescence). B) Expression of AtUSPL1 confirmed by Northern Blot analysis. Expression of AtUSPL1 was determined from root, leaf, shoot, young silique and total flower tissue of Arabidopsis thaliana (Col-0) plants. For equal loading of the RNA samples probing of the membrane with specific probe against housekeeping mRNA of AtROC1 (rotamase cyclophilin, renamed in AtCYP1) was performed. C) Expression of AtRD22 obtained from Arabidopsis eFP Browser (Winter et al., 2007). The expression of selected stimuli (Cold: 4°C, Osmotic: 300 mM Mannitol, Salt: 150 mM NaCl and Drought: air steam 15 min) is displayed for the aerial as well as the hypogeic part of the plant. AtRD22 expression is induced in the aerial part of the plant after applying osmotic, salt stress and slightly increased after loss of water due to airstream treatment. D) Expression of AtUSPL1 obtained from Arabidopsis eFP Browser (Winter et al., 2007). The expression of selected stimuli (Cold: 4°C, Osmotic: 300 mM mannitol, Salt: 150 mM NaCl and Drought: air steam 15 min) is displayed for the aerial as well as the hypogeic part of the plant. AtUSPL1 expression is induced in the hypogeic part of the plant after applying osmotic, salt stress and slightly increased after loss of water due to airstream treatment.
(TIF)
Expression of the BURP domain containing gene family in Arabidopsis thaliana . Expression analysis of AtRD22 (At5G25610) and AtUSPL1 (At1G49320) obtained from Genevestigator database (Zimmerman et al., 2004) displaying induced and reduced expression after different conditions and stresses. Displayed are only changes in expression upon stress/treatment above threefold with a statistic significance p<0.001. Red indicates up-regulation; Green indicates down-regulation.
(TIF)
Comparison of gene expression in Arabidopsis wild type plants grown on 150 mM NaCl and 4% trehalose supplemented MS medium. BURP gene family mRNA in Col-0 under selected stress conditions. Bars indicate the expression pattern obtained by microarray analysis using ATH1 chip: AtRD22 (red): 246908_at; AtUSPL1 (blue): 262388_at; AtPG1 (dark green): 265131_at; AtPG2 (green): 264277_at; AtPG3 (bright green): 264315_at. Displayed is the rel. Abundance of mRNA.
(TIF)
Characterization of BURP mutant plants. A) Scheme of the AtRD22 and AtUSPL1 gene model. In blue the exon-intron structure within the coding region of the respective gene is given. The protein structure refers to Figure 1. In the encoded protein parts are given in aminoacids [aa]. And the size of the fulllenght protein is given below the gene description. Position of T-DNA insertions of used mutant plants are indicated by black lines. Mutant alleles for rd22: rd22-1 (SALK_146066) and rd22-2 (WiscDsLox481-484P12). Mutant alleles for uspl1: uspl1: (SALK_022325). The T-DNA insertion line SALK_146066 (rd22-1) is based on pROK2 conferring kanamycin resistance and the WiscDsLox481-484P12 (rd22-2) is based on pWiscDs-Lox conferring phosphinotricin (BASTA) resistance. uspl1 T-DNA insertion lines SALK_022325 (referred to as uspl1, based on pROK2 conferring kanamycin resistance from Nottingham Arabidopsis Stock Centre) was analyzed. The position of the T-DNA insertion in At1G49320 (AtUSPL1) was determined by PCR and subsequent sequencing. The position of the T-DNA insertion are depicted and confirmed by PCR. Double mutant rd22-1/uspl1 line was generated by crossing SALK_146066 and SALK_022325 and identified in the F3 generation by PCR. Kanamycin and phosphinotricin resistance of the plants was tested on germination medium (1 MS salts; 10 g/l sucrose) plates with 40 mg/l kanamycin or 20 mg/l glufosinate-ammonium under long day conditions. B) Analysis of used T-DNA insertion mutants. The absence of AtRD22 and AtUSPL1 mRNA in homozygous rd22 and uspl1 mutant plants was determined by Northern Blot analysis (left) and semi quantitative RT-PCR. C) Analysis of BURP-gene family mRNA in rd22-1 and uspl1 mutants by microarray analysis on MS medium. Bars indicate the rel. expression signal obtained by microarray analysis using ATH1 chip from each single experiment: AtRD22 (red): 246908_at; AtUSPL1 (blue): 262388_at; AtPG1 (dark green): 265131_at; AtPG2 (green): 264277_at; AtPG3 (bright green): 264315_at.
(TIF)
Increased drought stress resistance of the rd22 and uspl1 mutant plants. A) The plants were drought stressed by withdrawal of water. Top: day 0 (80% RWC in the soil); Bottom: appearance of plants after 8 days without watering. B) Top: Projected area of wild type and mutant plants under control conditions and drought stress (dotted line) obtained by lemnatec phenotyping; Drought stress was started 21 days after sawing (DAS). Middle: Growth rates calculated based on Poorter and Lewis 1986 for individual days. Wild type (Col-0): green line; rd22-1: bright blue line; rd22-2: dark blue line; uspl1: purple line; rd22-1/uspl1pink line (+/- s.e.m.). Bottom: Statistical analysis or growth rates at 28 DAS. Asterisks indicate significant differences (p<0.05) between control and stress. C) Estimation of senescence after drought stress. The graph indicates the ration of yellow to green pixels in the plant area of the analysed top view images from day 33. Wild type (Col-0): green bar; rd22-1: bright blue bar; rd22-2: dark blue bar; uspl1: purple bar; rd22-1/uspl1: pink bar. Ncontrol = 5, Nstress = 10 plants. Asterisks indicate significant differences (p<0.05).
(TIF)
A) Differentially expressed genes (Col-0) between 150 mM NaCl containing medium and standard growth conditions sorted by relation to pathway (Mapman). Top: Differentially expressed genes (Col-0) between 150 mM NaCl containing medium and standard growth conditions sorted by relation to pathway (Mapman). The different numbers indicate different categories/pathways and are described in the table. Bottom: Differentially expressed genes (Col-0) between standard growth conditions and 4% trehalose containing medium. The bar diagram indicates the percentage of common (red) and inverse (yellow) regulated genes upon salt (grey) and sugar (blue) treatment. Approximately half of the genes induced by 4% trehalose treatment are also reacting on 150 mM NaCl treatment. B) Influence of ABA and NaCl on single and double loss of function mutants. Growth phenotypes of wild type (Col-0), single and double mutant plants (rd22-1 and uspl1, rd22-1/uspl1) on standard MS-medium and 150 mM NaCl, 300 mM NaCl and 100 µM ABA supplemented MS-Medium. Two week old seedlings were transferred for 3 days to the MS basal and supplemented medium. C) Chlorophyll and pheophytin content of wild type and rd22-1, uspl1 and rd22-1/uspl1 mutant plants on different supplemented media. Two week old seedlings were transferred to the MS basal + one of the following stress treatments for 4 days: 100 µM ABA, 4% fructose, 150 mM NaCl, 300 mM NaCl, 4% PEG 20000 and 15% PEG 6000. From Top to bottom: Chlorophyll a and b content. The error bar represents standard error. Content was estimated from 5-6 plants in duplicate. Statistical analysis was performed by oneway ANOVA at alpa = 0.05 Tukey post hoc test: same letters indicate no difference, different letters indicate significant difference. Chlorophyll a and b and pheophytin content [%] relative to unstressed control plants.
(TIF)
Differentially expressed genes in the rd22-1 and uspl1 plants grown on control, 150 mM NaCl and 4% trehalose containing medium. List of used categories is given in Figure 3 A. Top: Display of top regulated category (Bin 20, biotic and abiotic stress pathways) of differential regulated genes. Bottom: Overview of all differentially regulated genes mapped to categories (Bins) by MAPMAN. For RNA extraction tissue was grinded in liquid nitrogen and the homogenized powder was added to 1 ml TRIZOL and incubated at RT for 5 min. Samples were centrifuged at 10.000 rpm for 10 min and the supernatant was transferred to a new tube. 200 µl of chloroform were added and incubated at room temperature for 2–3 min. Samples were again centrifuged as described above and the aqueous supernatant was transferred to the QIAshredder column and centrifuged for 30 s at 10000 rpm. 350 µl of RLT buffer (plus β-mercaptoethanol) and 250 µl of absolute ethanol were added to the flow-through and passed through an RNAeasy spin column. All the following steps were performed as described in the manufacturer's protocol followed by in-column DNAse digestion. A) Schematic display of differentially expressed genes in rd22-1 by MAPMAN. 31 out of 77 differential regulated genes are mapping to biotic and abiotic stress pathways. B) Schematic display of differentially expressed genes in uspl1 by MAPMAN. 5 out of 18 differential regulated genes are mapping to biotic and abiotic stress pathways. C) Schematic display of differentially expressed genes in rd22-1 on 150 mM NaCl by MAPMAN. 231 out of 764 differentially regulated genes are mapping to biotic and abiotic stress pathways. D) Schematic display of differentially expressed genes in uspl1 on 150 mM NaCl by MAPMAN. 7 out of 12 differentially regulated genes are mapping to biotic and abiotic stress pathways. E) Schematic display of differentially expressed genes in rd22 on 150 mM NaCl by MAPMAN. 55 out of 171 differentially regulated genes are mapping to biotic and abiotic stress pathways.
(TIF)
Primer information.
(XLS)
Average projected plant area (mm2) from top images.
(XLSX)
Average near-infrared intensity as observed from top images, obtained using a Nir 300 camera from VDS Vosskühler (now Allied Vision Technologies). High values indicate relative high water content.
(XLSX)
List of differentially expressed genes.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.