Abstract
The TOUSLED (TSL)-like nuclear protein kinase family is highly conserved in plants and animals. tsl loss of function mutations cause pleiotropic defects in both leaf and flower development, and growth and initiation of floral organ primordia is abnormal, suggesting that basic cellular processes are affected. TSL is more highly expressed in exponentially growing Arabidopsis culture cells than in stationary, nondividing cells. While its expression remains constant throughout the cell cycle in dividing cells, TSL kinase activity is higher in enriched late G2/M-phase and G1-phase populations of Arabidopsis suspension culture cells compared to those in S-phase. tsl mutants also display an aberrant pattern and increased expression levels of the mitotic cyclin gene CycB1;1, suggesting that TSL represses CycB1;1 expression at certain times during development or that cells are delayed in mitosis. TSL interacts with and phosphorylates one of two Arabidopsis homologs of the nucleosome assembly/silencing protein Asf1 and histone H3, as in humans, and a novel plant SANT/myb-domain protein, TKI1, suggesting that TSL plays a role in chromatin metabolism.
TOUSLED (TSL) is a widely expressed gene in Arabidopsis required for proper development (Roe et al., 1993, 1997b). TSL is a nuclear Ser/Thr protein kinase composed of a C-terminal catalytic domain and an N-terminal regulatory domain (Roe et al., 1997a). TOUSLED-like kinases (TLKs) are highly conserved and are found in both plants and animals. Humans and mice both have two highly related TLK genes, TLK1 and TLK2 (Yamakawa et al., 1997; Shalom and Don, 1999; Sillje et al., 1999; Zhang et al., 1999; Li et al., 2001). Human TLK expression is constitutive throughout the cell cycle, while TLK protein kinase activity (both TLK1 and TLK2) oscillates during the cell cycle, with a peak of activity in S-phase (Sillje et al., 1999). Inhibition of DNA polymerase α causes a decrease in TLK activity, suggesting that activation of TLKs is linked to DNA replication (Sillje et al., 1999). In addition, recent evidence indicates that TLKs are targets of DNA damage checkpoints (Groth et al., 2003). Mutations in TLK genes cause severe defects in both plants and animals; tsl loss-of-function mutations in Arabidopsis cause flower and leaf abnormalities (Roe et al., 1993), whereas mutations in or suppression of TLK homologs in Drosophila melanogaster and Caenorhabditis elegans cause an early arrest in the embryo (Carrera et al., 2003; Han et al., 2003; T. Ratliff, M. Herman, and J. Roe, unpublished data). Overexpression of TLK1 in mouse cells conferred enhanced resistance to ionizing radiation (Li et al., 2001). A dominant negative form caused missegregation of chromosomes, whereas siRNA-mediated suppression of TLK1 blocked cell division in mouse cells (Sunavala-Dossabhoy et al., 2003).
The target(s) of TSL and the TLKs have remained elusive until recently. Sillje and Nigg (2001) identified the human homolog of Asf1 (also known as CIA) as a candidate TLK target. Asf1 is a silencing protein first identified in yeast (Saccharomyces cerevisiae) that recruits histone H3 and H4 during nucleosome assembly (Le et al., 1997; Tyler et al., 1999; Munakata et al., 2000). It functions redundantly with chromatin assembly factor, CAF-1, and can interact directly with a CAF-1 subunit (Tyler et al., 2001; Mello et al., 2002). Asf1 also interacts with protein complexes involved in chromatin remodeling and transcription initiation, such as the SWI/SNF Brahma remodeling complex in D. melanogaster, and with transcription initiation factor IID (TFIID) in humans and yeast (Chimura et al., 2002; Moshkin et al., 2002). Asf1 likely participates in several silencing pathways in yeast (Singer et al., 1998; Tyler et al., 1999; Sharp et al., 2001). It also acts during DNA repair, where it reassembles nucleosomes following repair (Emili et al., 2001; Hu et al., 2001). The two human Asf1 proteins, Asf1a and Asf1b, bind to and are substrates in vitro of both human TLK1 and TLK2 (Sillje and Nigg, 2001). Phosphorylation of the Asf1s is cell cycle regulated and parallels the cell cycle-regulated activity of TLK1 and TLK2, suggesting that Asf1 is a TLK target in vivo (Sillje and Nigg, 2001). TLKs have also recently been shown to be inhibited by the ATM (ataxia telangiectasia mutated)- and the Chk1 protein kinase-DNA damage checkpoint pathways in response to double-strand breaks; possibly, transient inhibition of TLK-dependent activation of Asf1 allows chromatin restructuring prior to nucleosome reassembly during repair (Groth et al., 2003). Histone H3 is another potential TLK phosphorylation target; TLK1 phosphorylates histone H3 in vitro, and it complements a yeast mutation lacking the major histone H3 kinase, Ipl1, even though Ipl1 is not a TLK (Li et al., 2001). However, in C. elegans, it appears that RNA polymerase II large subunit, and not histone H3, may be a key TLK substrate (Han et al., 2003).
These results from animal systems suggest that TLKs participate in chromatin assembly, DNA repair, and in the regulation of gene expression. Although no mutations have been described in Asf1-like genes in Arabidopsis, several mutations in other chromatin assembly genes such as FAS1 and FAS2 (subunits of CAF-1) and cosuppressed AtMSI1 (another CAF-1 subunit) plants show pleiotropic defects in shoot, flower, and in some cases root development (Leyser and Furner, 1992; Kaya et al., 2001; Hennig et al., 2003). tsl loss-of-function mutations cause both leaf and flower defects; only approximately half of the normal complement of floral organs is formed due to a decrease in the number of organ primordia that are initiated, and the young floral meristems are abnormal in shape (Roe et al., 1993). To further examine the possible function of TSL in nuclear function in Arabidopsis, we tested whether TSL protein levels and kinase activity are cell cycle regulated, and we examined the expression pattern of the mitosis specific CycB1;1::GUS transgene (Colon-Carmona et al., 1999) in a tsl mutant. We demonstrate that TSL expression is fairly constant across the cell cycle in Arabidopsis cells, with higher activity seen in the G2/M-phase and G1-phase than S-phase, and that tsl mutants show ectopic expression of the cyclin B gene, CycB1;1. We also demonstrate the interaction of TSL with Asf1b/SGA1, one of two Arabidopsis Asf1 homologs, and with a novel SANT/myb-domain protein TKI1, which, together with other results, suggests that TSL may be involved in gene repression and/or chromatin regulation in Arabidopsis.
RESULTS
TSL and the Cell Cycle
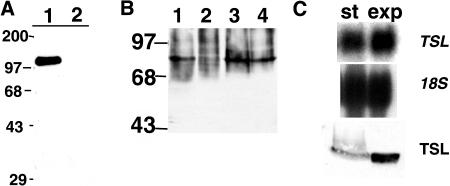
To follow expression, protein levels, and kinase activity, we first prepared a TSL-specific antisera. Anti-peptide antibody to the C-terminal TSL peptide (C)YNQEDRPDVLTMAQDPYLAYS was generated and affinity purified as described in “Materials and Methods”. The peptide sequence is not found in kinases other than TLK family members (BLAST analysis, data not shown). The antibody specifically recognizes a GST-TSL fusion protein purified from yeast (Fig. 1A), and detects a single 80-kD protein in Arabidopsis flower protein extracts by western-blot analysis (Fig. 1B). A band at the same Mr is not detected in extracts from tsl-1 flowers (Fig. 1B, lane 2), where no TSL protein is expected due to the T-DNA insertion in the gene, which inactivates gene expression. Equal amounts of protein extract were loaded in each lane, as determined by Coomassie Blue staining (data not shown). TSL protein is detected in tsl-2 mutants, as expected, where a single point mutation in the TSL gene affects only one amino acid in the predicted protein.
Figure 1.
TSL is more highly expressed in dividing cells. A, Western-blot analysis using α-TSL-peptide affinity-purified antisera of fusion proteins purified from yeast: lane 1, GST-TSL; lane 2, GST. B, Equal amounts of protein extracts from wild-type (lane 1, Wassilewskija ecotype, and lane 3, Columbia ecotype) and tsl (lane 2, tsl-1, and lane 4, tsl-2, in Wassilewskija and Columbia ecotype backgrounds, respectively) flowers were analyzed by western-blot analysis with α-TSL antibody. C, Northern- and western-blot analysis of TSL expression in stationary (st; 10 d after dilution) and exponentially (exp) growing (3 d) Arabidopsis suspension culture cells. Top, Northern-blot analysis of 20 μg total RNA hybridized with TSL cDNA probe (TSL). Hybridization of the same blot after stripping for the presence of 18S rRNA serves as a loading control (middle; 18S). Bottom, TSL protein was detected by western-blot analysis on equal amounts of protein extracts with α-TSL peptide antibody (TSL). Molecular weight markers (numbers) are in kD.
To determine if TSL is preferentially expressed in dividing cells, Arabidopsis suspension culture cells were harvested from cultures allowed to progress to stationary phase (10 d after subculture) and from cultures in the exponential phase of growth (3 d after subculture) and assessed for TSL expression. TSL mRNA levels are slightly higher when the cells are rapidly dividing, and TSL protein is present in higher steady-state levels in exponentially growing cells than in stationary cells arrested in G0/G1 (Fig. 1C).
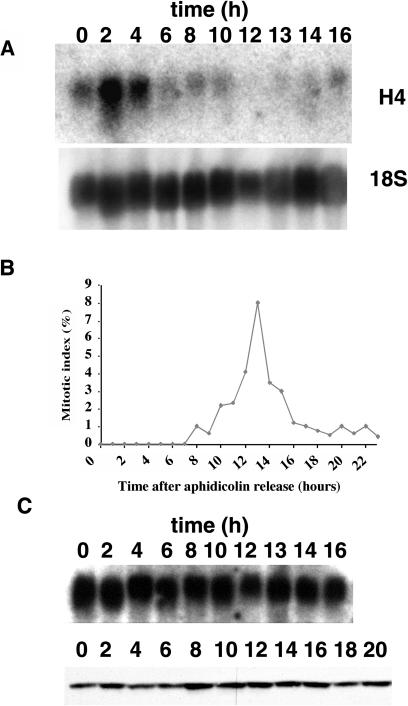
We next asked if the cell cycle expression of TSL is regulated in dividing cells. Arabidopsis suspension culture cells were synchronized with an aphidicolin block and release protocol which arrests cells in early S-phase, followed by synchronous progression through the cell cycle after release from the drug block (for review, see Planchais et al., 2000). Aliquots of cells were harvested at time points after the release. The culture contains cells in S-phase from time point 0 through hour 4, as shown by the northern blot detecting histone H4 mRNA levels, as expected (Fig. 2A). As cells progress out of S-phase during hours 6 to 8, the histone H4 mRNA levels had declined. Mitotic index was also determined by DNA staining and chromosome visualization, and mitotic cells were observed in samples from time points between 12 and 16 h, with a peak at 13 h (Fig. 2B). The percentage of cells in mitosis is underestimated with this method, as only the stages with visibly highly condensed chromosomes are counted; in fact, these numbers agree well with other published reports of synchronization in Arabidopsis cells that showed peak levels of 12% mitotic index (Menges et al., 2002; Menges and Murray, 2002). In contrast to histone H4 expression, TSL mRNA levels remain constant at all time points in the synchronized culture. Western-blot analysis on extracts across the time course with the TSL peptide antibody reveals that TSL protein is also present at fairly constant levels (Fig. 2C).
Figure 2.
TSL expression levels are constant throughout the cell cycle in synchronized cells. A, Top, Suspension culture cells were incubated with aphidicolin for 24 h, and then washed extensively and fresh culture medium added. RNA was isolated from cells at time points (h) following release from aphidicolin block and subjected to northern-blot analysis. Total RNA (20 μg) was loaded per lane, and the blot was hybridized with a histone H4 cDNA probe. Bottom, The same blot was stripped and hybridized with 18S rRNA sequences as a control for RNA loading (after hybridizing with TSL cDNA in C). B, Samples from aphidicolin synchronized cell populations, as in A, were analyzed for mitotic index. C, Top, Northern-blot analysis of Arabidopsis TSL across the cell cycle. The same blot after stripping as in A was hybridized with a TSL cDNA probe. Bottom, TSL protein accumulation in synchronized Arabidopsis cell suspension culture. Total protein extract (300 μg) from samples of aphidicolin released cells collected at 2-h time intervals was run on a 10% SDS-polyacrylamide gel and then analyzed by western-blot analysis with α-TSL peptide antibody.
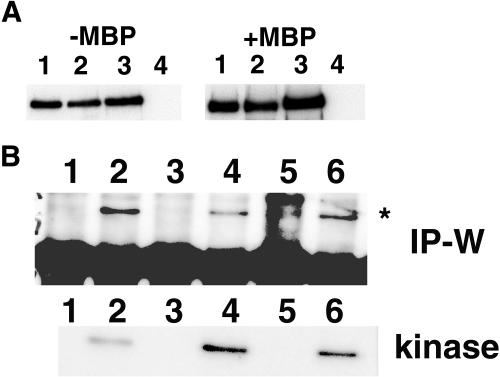
To assess when TSL is active across the cell cycle, we analyzed the protein kinase activity of TSL in immunoprecipitates from aphidicolin-treated and released cultures against the substrate myelin basic protein (MBP). Unfortunately, a highly active contaminating protein kinase showed high background activity in the preimmune controls. To avoid this contaminating kinase, in-gel kinase assays on immunoprecipitates were performed with gels containing MBP incorporated into the gel followed by addition of [32P-γ]ATP. We first tested purified TSL fusion proteins in the assay (Fig. 3A) and found that GST-TSL showed activity, whereas GST-K438E (contains a single point mutation that converts Lys-438 to Glu and inactivates kinase activity; Fig. 3A, lane 4) and GST alone (data not shown) did not. Also, we could detect TSL activity immunoprecipitated from exponentially growing cells (not shown). The assay was then performed on immunoprecipitates of extracts from aphidicolin-synchronized Arabidopsis suspension culture cells enriched in S-phase, G2/M-phase, and G1-phase (Fig. 3B). TSL could be identified specifically in this assay by its presence at 80 kD and absence in the nonrelated antibody control lanes. TSL is active in all the fractions but shows the highest level of activity in the enriched G2/M-phase sample, and more activity in the G1-enriched sample compared to the S-phase sample (Fig. 3B).
Figure 3.
TSL protein kinase activity is higher during G2/M- and G1-phase than S-phase. A, In-gel kinase assay on TSL fusion proteins purified from induced yeast extracts. GST-TSL was purified on glutathione-agarose and eluted by addition of sample buffer (lane 1), sample buffer followed by boiling (lane 2), or 50 mm glutathione (lane 3). Purified GST-K438E (lane 4) is the negative control. In-gel kinase assays were performed in SDS-polyacrylamide gel (10%) containing MBP as a substrate (1 mg/mL; right) with [32P-γ]ATP. For autophosphorylation studies, no MBP was added to the gel (left). Autoradiography of gel is shown. B, Comparison of TSL activity in different phases of the cell cycle in synchronized Arabidopsis cells. Samples were withdrawn from aphidicolin-treated and released cultures at different time points (S = 2 h, lanes 1 and 2; G2/M = 12 h, lanes 3 and 4; G1 = 20 h, lanes 5 and 6). Equivalent amounts of protein from crude extracts of cells (600 mg) were used for immunoprecipitation with α-TSL antibody (lanes 2, 4, and 6). An unrelated anti-T-span peptide antibody was used in parallel as a control (lanes 1, 3, and 5). Immunoprecipitated samples were split into two equal parts; one for western analysis (IP-W) with α-TSL antibody, and the other for TSL in-gel kinase activity assay (kinase) followed by autoradiography. The asterisk in the IP-W indicates the position of the TSL protein at 80 kD, and the large band below in all the lanes is heavy chain from the immunoprecipitation detected by the secondary antibody.
Mitotic Marker Gene Is Misexpressed in tsl Mutants
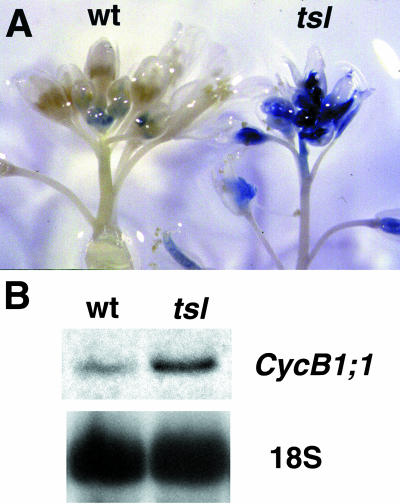
tsl mutants show abnormalities in meristem development (Roe et al., 1993). To assess potential cell cycle effects, the mitotic marker gene, CDG (Colon-Carmona et al., 1999), was introduced into tsl-2 mutants. This marker gene consists of the promoter and 5′ transcribed sequences from the Arabidopsis cyclin B gene CycB1;1 (Hemerly et al., 1992) cloned in frame with a uidA gene, which results in the production of a cyclin destruction box (CDB)-β-glucuronidase (GUS) fusion protein, CDG. The CDG marker gene allows for the visualization of cells in late G2 and mitosis, as the CycB1;1 promoter is activated in G2 just prior to mitosis, and the CDG enzyme that is synthesized is specifically degraded at the end of M via the CDB (Colon-Carmona et al., 1999; Menges and Murray, 2002). Thus, when plants carrying the marker gene are stained for GUS activity, only the cells in late G2 and M should stain. Inflorescences from tsl-2 plants carrying CDG were stained for GUS activity and compared to wild-type siblings carrying CDG (Fig. 4A). In wild type, the GUS activity was confined to a small population of cells in the youngest floral meristems where cell divisions are occurring, as expected (Fig. 4A). As the floral meristems develop and cells stop dividing, expression of the marker gene is lost, with the last expression seen in approximately stage 10 gynoecium on the carpel walls and ovule primordia. In contrast, tsl-2 meristems show greatly increased levels of GUS activity and the staining persists in older flowers, even after organ growth is near completion (Fig. 4A). Eventually, staining is lost in the outer organs. Cauline leaves showed only slightly increased GUS staining in mutant inflorescences compared to wild-type (not shown). This suggests that either more cells in G2 or M are present in the developing tsl flowers, or that deregulation of CycB1;1 expression or turnover is occurring.
Figure 4.
Ectopic CycB1;1 expression in tsl mutants. A, GUS activity staining was performed on wild-type (left) and tsl-2 (right) inflorescences carrying the mitotic marker gene CDG, which contains the CycB1;1 promoter and N-terminal coding sequence fused to the uidA gene (Colon-Carmona, 1999; 455). The branch contains flower buds of increasing developmental stage progressing down the stem. Mature flowers are evident at the lowest positions. B, Northern-blot analysis of CycB1;1 gene expression in wild-type and tsl-1 mutant flowers. Total RNA (20 μg) is loaded per lane, and blot was hybridized with CycB1;1 coding sequence cDNA probe. 18S rRNA was hybridized to the same blot after stripping as a loading control.
To test if the increased expression of the CDG marker gene in tsl floral meristems reflects ectopic expression of the endogenous CycB1;1 promoter, we performed northern-blot analysis on mRNA from wild-type and tsl-1 mutant flowers using a CycB1;1 cDNA as a probe (Fig. 5B). 18S rRNA levels were monitored to control for loading consistency (Fig. 4B). In comparing the level of transcripts in RNA isolated from wild-type and tsl flowers, a higher steady-state level of CycB1;1 mRNA is present in tsl flowers than in flowers of wild-type siblings. CycB1;1 mRNA levels were also slightly higher in tsl leaves than in wild-type (data not shown). Expression of a second cyclin B gene, CycB1;2, was also tested, as it is much more highly expressed in flowers than CycB1;1 (Day et al., 1996), and in contrast, the levels of CycB1;2 mRNA are comparable in the two samples (data not shown). Thus, CycB1;1 expression appears to be upregulated in tsl mutants. CycB1;1 protein levels could not be assessed, as an antibody that gave consistent results could not be obtained.
Figure 5.
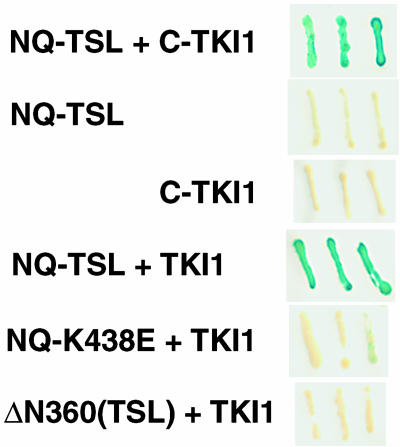
TKI1 interacts with active forms of TSL. Yeast transformants carrying two-hybrid plasmids expressing fusion proteins (left, TSL-Gal4 DBD fusions; right, TKI1-Gal4 ACT fusions) were lifted and assayed for β-galactosidase activity. The top two panels were stained overnight and the bottom four for 5 h; blue color indicates activity. NQ, TSL N-terminal deletion containing amino acids 73-688; K438E, point mutation changing amino acid 438 from Lys to Glu; ΔN360, TSL N-terminal deletion containing amino acids 360-688; C-TKI1, C terminus of TKI1, amino acids 546-741; TKI1, full-length TKI1.
Isolation of TKI1 as a TSL-Interacting Protein
To further investigate TSL function, a two-hybrid screen in yeast was performed to identify proteins that interact with TSL (Fields and Sternglanz, 1994). A TSL bait clone was constructed that encodes a near full-length TSL protein (amino acids 73-688) as a fusion protein with the DNA binding domain (DBD) of Gal4. The clone contains a centromere-based (CEN) origin of replication (Durfee et al., 1999) that was found to overcome the toxic effects of TSL expression from a 2-μ based plasmid vector in yeast (data not shown). As described previously, full-length TSL fused to Gal4-DBD weakly activates the reporter gene alone, and thus the N-terminal deletion was used instead.
The yeast strain YD116 was transformed with the TSL bait vector pCD-NQ-TSL, followed by a cDNA library in a Gal4-activation domain (ACT) plasmid, and clones encoding proteins that interact with TSL were selected for by growth on medium lacking Trp, Leu, and Ura. A total of 1 × 106 transformants were screened and putative positives were retested with the bait vector in a different strain, Y153, for activation of a UASGal4::lacZ reporter gene. A number of clones were eliminated as false positives, and several were able to activate transcription alone (data not shown) and also were eliminated. One clone, however, showed strong reporter gene activation that was completely dependent on the presence of the TSL bait vector (Fig. 5). Furthermore, it was shown that the clone, pC-TKI1 (TSL-kinase interacting protein), did not show activation when cotransformed with other DBD-fusion protein vectors such as lamin (data not shown). So, it appears that the TKI1 gene encodes a polypeptide that specifically interacts with TSL.
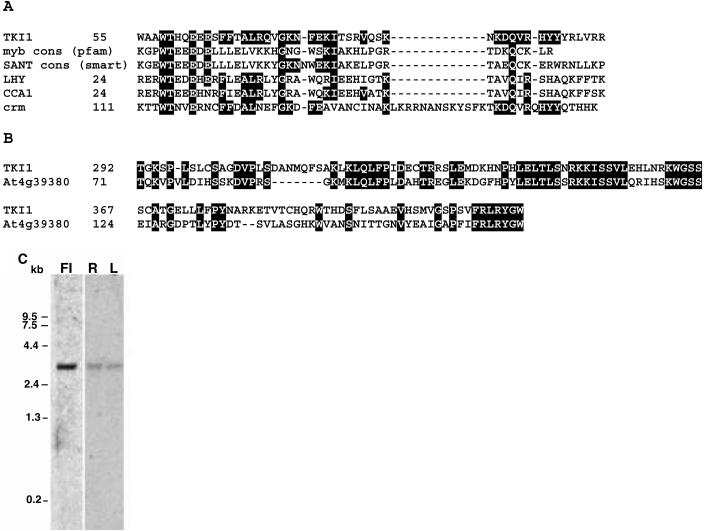
DNA sequence analysis of the TKI1 cDNA insert revealed that the clone isolated in the two-hybrid screen is not a full-length cDNA and encodes the C-terminal region of the TKI1 polypeptide. PCR on a cDNA library was used to complete the full-length coding region (GenBank accession no. AF530160) corresponding to locus At2g36960, which differed only slightly from the predicted open reading frame (GenBank accession no. AC006922) and a complete cDNA sequence already present in the database (GenBank accession no. AY064068). The TKI1 protein sequence encoded by the cDNA is 741 amino acids (predicted Mr = 82 kD) in length. TKI1 contains several regions that share homology with other proteins; these were identified by BLAST analysis (Altschul et al., 1997) on the GenBank database and by Reverse Position Specific BLAST analysis on the Conserved Domain Database (Marchler-Bauer et al., 2002). The first region is a single SANT/myb domain shared with the human oncogene Myb and other related proteins (Fig. 6A; Aasland et al., 1996). The proteins most homologous to TKI1 include the putative Arabidopsis transcription factors LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), but only the SANT domain is shared. These two factors play a role in circadian rhythm control (Schaffer et al., 1998; Wang and Tobin, 1998). LHY, CCA1, and TKI1 have only one myb repeat (the SANT/myb domain) and thus are not members of the large class of plant R2R3 myb domain proteins, which each have two myb domains (Romero et al., 1998). TKI1 also shares homology with the Polycomb-group D. melanogaster protein cramped (crm; GenBank accession no. Y13674; Yamamoto et al., 1997) in its SANT domain. The SANT domain is found in a number of proteins that are part of complexes that regulate transcription (Boyer et al., 2002, and see “Discussion and Conclusion”). We found that when swapped into the Gal4-DBD plasmid, the C terminus of TKI1 (amino acids 546-741), lacking the SANT/myb domain, acts as a strong transcriptional activator in yeast (Fig. 7).
Figure 6.
TKI1 is a SANT/myb domain protein expressed in flowers, roots, and leaves. A, Alignment of SANT/myb domain of TKI1 with a consensus myb (pfam00249) and SANT (smart00395) domain, as well as with the Arabidopsis proteins LHY (GenBank accession no. AJ006404), CCA1 (GenBank accession no. U28422), and the D. melanogaster protein cramped (crm; GenBank accession no. Y13674), which show the most homology to TKI1. Numbers represent amino acid number. B, Alignment of TKI1 domain that shares homology with a predicted protein from Arabidopsis chromosome 4. C, Total RNA (20 μg) isolated from flowers (Fl), roots (R), and leaves (L) was analyzed by northern-blot analysis using a TKI1 partial cDNA probe.
Figure 7.
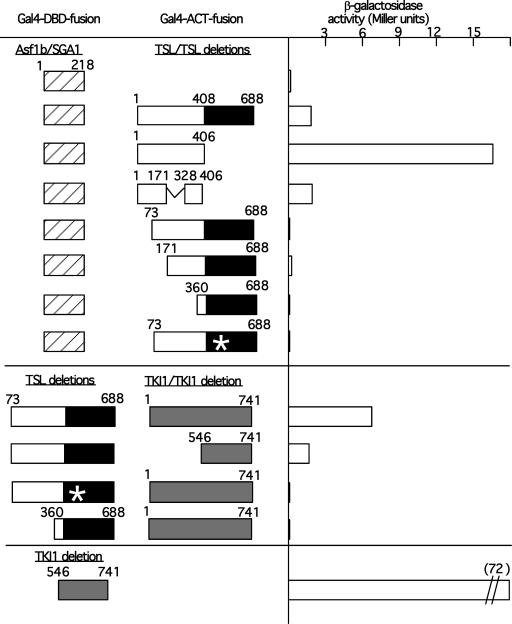
TSL interacts with Asf1b/SGA1 and TKI1 in different domains. Yeast transformants carrying two hybrid plasmids expressing Gal4-fusion proteins were grown and assayed for β-galactosidase activity using ONPG as a substrate after cell solubilization. Results shown are the average of two experiments, each performed in duplicate with each of two different transformants. Numbers represent amino acid number.
Several proteins with weaker homology to other regions in TKI1 were found, including a predicted protein on chromosome 4 (At4g39380; GenBank accession no. NM_120098; Fig. 6B), the human protein mitofilin (not shown; GenBank accession no. NM_006839; Odgren et al., 1996), and a predicted protein on chromosome 1 (At1g78650; GenBank accession no. NM_106512; not shown). The importance of these similarities is unknown. TKI1 is expressed in wild-type Arabidopsis flowers, roots, and leaves (Fig. 6C), and a transcript of 3.2 kb is detected.
The interaction of TKI1 and TSL was investigated further using deletion and mutant versions of TSL (Fig. 5). TSL interacts strongly with both the original C-terminal portion of TKI (amino acids 546-741) and the full-length TKI protein in filter lift assays (Fig. 5). In quantitative assays (Fig. 7), TSL interacts approximately fourfold more strongly with the full-length protein. Furthermore, TKI1 interacts only with active kinase forms of TSL and does not bind inactive mutants such as an N-terminal deletion that removes the coiled-coil domain (ΔN360) or a point mutation (K438E) that changes a single amino acid in TSL and inactivates the ATP-binding site in the catalytic domain (Figs. 5 and 7). The K438E mutant is expressed in normal levels in the yeast cell, as has been shown previously by a strong interaction with both itself and wild-type TSL in two-hybrid assays (Roe et al., 1997a).
TSL Interacts with Asf1b/SGA1, an Arabidopsis Asf1 Silencing Factor Homolog
Sillje and Nigg (2001) described the interaction of human TLK1 and TLK2 with two human homologs of the yeast silencing factor Asf1 and their ability to phosphorylate the Asf1-like proteins. Also, TLK activity paralleled Asf1 phosphorylation status in synchronized human cells. Two highly similar homologs of Asf1 are present in the Arabidopsis genome: Asf1a/SGA2 and Asf1b/SGA1 (locus numbers At1g66740 and At5g38110, respectively, and see http://chromdb.biosci.arizona.edu). We tested the latter for interaction with TSL in the two-hybrid system, because a cDNA database entry confirmed its validity as an expressed locus. The coding region of Asf1b/SGA1 was cloned into a two-hybrid vector, creating a fusion protein of Asf1b/SGA1 with the Gal4-DBD (pCD1-Asf1b); this fusion did not show autoactivation (Fig. 7). pCD1-Asf1b was used for two hybrid interaction tests with different forms of TSL cloned as fusion proteins with the Gal4 ACT. o-Nitrophenyl β-d-galactopyranoside (ONPG) assays on cell extracts were used to quantify the interactions observed in colonies stained for β-galactosidase activity (Fig. 7, and data not shown). Asf1b/SGA1 interacted strongly with full-length TSL, and even more strongly with the N terminus of TSL alone (amino acids 1-406), including a clone that was missing the coiled-coil domain (Fig. 7). N-terminal deletions of TSL lacking the first 73, 171, and 360 showed little to no binding activity, as did the active site mutant K438E (Fig. 7). These results suggest that Asf1b/SGA1 binds to the most N-terminal portion of the TSL protein, which may explain why it was not found in the library screen. The screen used an N-terminal deletion of TSL, lacking the putative Asf1b/SGA1 binding site. Thus, TSL binding to TKI1 and Asf1b/SGA1 appears to be dependent on different domains; Asf1b/SGA1 binds in the most N-terminal domain of TSL whereas TKI1 may bind to the active catalytic C-terminal domain.
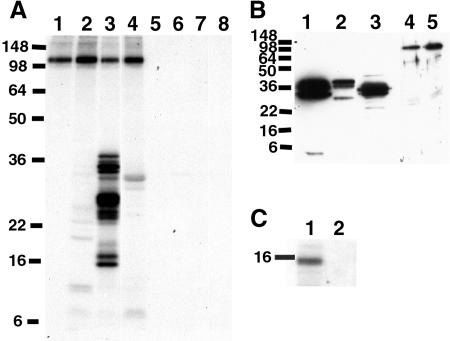
To test if TKI1 and Asf1b/SGA1 are potential TSL substrates, in vitro kinase assays were performed using purified GST-TSL and purified His-tagged TKI1 and Asf1b fusion proteins (Fig. 8). Both Asf1b and the C terminus of TKI1 acted as efficient substrates of TSL in vitro, in contrast to purified His-GFP as a control (Fig. 8A, lanes 1–4). The C-TKI1 lane shows not only phosphorylation of the full C-terminal TKI1 protein but also a number of degradation products as well (Fig. 8A, lane 3). A kinase dead mutant of TSL showed no activity (Fig. 8A, lanes 5–8). All of the fusion proteins could be detected by western-blot analysis (Fig. 8B), except full-length TKI1 which did not give a high yield of fusion protein and thus is not shown. GST-TSL also phosphorylated histone H3 in vitro (Fig. 8C).
Figure 8.
TKI1, Asf1b, and histone H3 are substrates of TSL in vitro. A, GST-TSL (lanes 1–4) and GST-K438E (lanes 5–8) fusion proteins were purified from yeast and incubated without (lanes 1 and 5) or with purified His-tagged fusion proteins His-GFP (lanes 2 and 6), His-C-TKI (lanes 3 and 7), and His-Asf1b (lanes 4 and 8) in a kinase reaction. Autoradiography is shown. Autophosphorylated GST-TSL appears at approximately 106 kD. B, Western-blot analysis of purified fusion proteins detected by anti-His (lanes 1–3) or anti-GST (lanes 4–5) primary antibody. Lane 1, His-GFP; lane 2, His-C-TKI; lane 3, His-Asf1b; lane 4, GST-TSL; and lane 5, GST-K438E. C, Purified GST-TSL and GST-438E were incubated with 1.25 μg histone H3 in a kinase reaction. The lower portion of the autoradiography is shown. Numbers on the left indicate Mr of markers.
DISCUSSION AND CONCLUSION
TSL Expression Is Increased in Dividing Cells and TSL Activity Oscillates during the Cell Cycle
TSL is expressed in Arabidopsis suspension culture cells, and both TSL mRNA and protein expression levels increase when stationary cells are diluted and cells resume division. To follow expression of TSL during the cell cycle, Arabidopsis suspension culture cells were synchronized with aphidicolin. The results we obtained are highly similar to the results described in the recently reported synchronization of Arabidopsis culture cells (Menges et al., 2002; Menges and Murray, 2002). TSL mRNA and protein expression levels are stable throughout the cell cycle. These data are similar to results obtained in human cell cycle studies where TLK mRNA and protein levels remain constant, although we did not observe clear differentially mobile forms of TSL during the cell cycle as is seen in humans (Sillje et al., 1999). Also, TSL expression was not found to be cell cycle regulated in the microarray analysis of synchronized Arabidopsis cells by Menges et al. (2002), confirming these results. TSL is a unique gene in Arabidopsis, and thus its expression can be clearly determined in these experiments. In contrast, however, TSL protein kinase activity oscillates during the cell cycle in Arabidopsis cells, showing higher activity during G2/M-phase and G1-phase compared to S-phase. This contrasts with the data in humans, where TLK activity was found to be the highest during S-phase (Sillje et al., 1999). The reason for this difference is unknown. It is possible that the in-gel kinase assay does not reflect the cellular TSL activity, as the proteins are first denatured during SDS-PAGE and coactivators/inhibitors might have been removed. Posttranslational modifications should be intact, however, and the assay should reflect changes in activity due to such modifications.
We also did not find direct evidence for TSL acting specifically during DNA replication, as was found in humans (Sillje et al., 1999), although TSL is active during S-phase in Arabidopsis. In humans, aphidicolin inhibited TLK activation (Sillje et al., 1999), whereas no such inhibition was seen in the plant cells (H. Ehsan, unpublished data). Perhaps TSL activity in Arabidopsis is influenced by other factors or signals.
Cell Cycle Marker Gene Is Aberrantly Expressed in tsl Meristems
The CDG mitotic marker gene is highly overexpressed in tsl floral meristems. This result may reflect that cells are progressing more slowly through late G2- and/or M-phase when transcription of CycB1;1 is highest (Colon-Carmona et al., 1999; Menges and Murray, 2002), and thus positive cells accumulate in the tsl floral meristems. Alternatively, the greater abundance of the CDB-GUS fusion protein in the tsl meristems could be due to either increased transcription from the CycB1;1 promoter or to decreased turnover of the protein in the cells. Endogenous CycB1;1 mRNA levels are increased in tsl inflorescences, suggesting that the CycB1;1 promoter has been ectopically activated/derepressed. The effect is most pronounced in flowers, consistent with the more dramatic effect of tsl mutations on flower rather than leaf development (Roe et al., 1993). Transgenic plants overexpressing CycB1;1 do not show a tsl phenotype (Doerner et al., 1996), and thus its ectopic expression is probably not the primary defect in tsl mutants. In addition, expression of a nondegradable mutant version of CycB1;1 in cultured tobacco cells does not perturb progression through the cell cycle (Criqui et al., 2000).
Ectopic expression of CycB1;1 in tsl loss of function mutations suggests that TSL may participate in repression of this gene. The increase in TSL activity that we observe in late G2/M-phase (12 h) coincides with the time of peak expression of CycB1;1 in synchronized Arabidopsis cells, as measured by northern-blot analysis (Menges and Murray, 2002). This gene did not show up as one of a group of several mitosis-specific, oscillating mitotic cyclin genes in the microarray analysis of synchronized cells (Menges et al., 2002), probably because it is also expressed at a detectable level in S-phase, and its expression does not decline greatly after the peak in G2/M (Menges and Murray, 2002). In the earlier study, at approximately 14 h following aphidicolin release (end of M), CycB1;1 mRNA levels begin to decline (Menges and Murray, 2002), perhaps due to gene repression. The mechanism of cell cycle-regulated expression of CycB1;1 is not fully understood, although a set of Myb-like transcription factors have been identified in tobacco that participate in both activation and repression of tobacco CycB1;1 (Ito et al., 1998, 2001). Interestingly, TKI is a Myb-like factor, although other than in the single SANT/myb domain, the rest of the TKI1 sequence is not related to these tobacco Myb-like factors, however, which are R1R2R3 myb domain proteins.
Asf1b/SGA1, TKI1, and Histone H3 Are Potential TSL Substrates
Recent reports have shown that Asf1 and histone H3 are potential targets of TLKs in humans (Li et al., 2001; Sillje and Nigg, 2001) and that TLKs may therefore be involved in chromatin regulation. Very little is known about the two Arabidopsis Asf1-like genes in Arabidopsis. Arabidopsis Asf1b/SGA1 does interact with TSL and is phosphorylated in vitro by TSL and could therefore be a substrate that leads to gene silencing via nucleosome assembly or chromatin regulation via interaction with specific protein complexes. It will be interesting to compare the tsl phenotype with a loss-of-function mutation in the two Asf1-like genes.
TSL also interacts with and can phosphorylate a novel SANT/myb domain protein, TKI1, that can act as a transcriptional activator as a fusion protein in yeast. While SANT/myb domains have been implicated in DNA binding as in Myb (Aasland et al., 1996), more evidence is accumulating that these domains play critical roles in the interaction of proteins in complexes involved in chromatin remodeling by interacting with both histone N-terminal tails and enzymes such as both histone acetyltransferase and histone deacetylase (Boyer et al., 2002; Sterner et al., 2002; Barbaric et al., 2003; Yu et al., 2003). Interestingly, Asf1 also interacts with a MYST acetyltransferase during chromatin silencing in yeast (Meijsing and Ehrenhofer-Murray, 2001; Osada et al., 2001), and these proteins are potentially in functionally similar complexes. The two interactors appear to bind differently to TSL. Whereas Asf1b/SGA1 binds strongly to the N-terminal regulatory domain, TKI1 binds only to active forms of the kinase and not to the regulatory domain alone. The SANT/myb domain of TKI1 is not required for TSL binding.
In addition to Asf1, histone H3 has been identified as a potential substrate of human TLK1 (Li et al., 2001). Phosphorylation of histone H3 modulates its interaction with chromatin, is likely involved during chromosome segregation, and a number of kinases have been identified that participate in this regulation (Jenuwein and Allis, 2001). TSL does phosphorylate histone H3 in vitro. However, histone H3 phosphorylation was not detectable in extracts of flower buds, either wild-type or mutant, by western-blot analysis using a Ser10-P specific antibody (J.L. Roe, unpublished data), and thus we could not confirm its importance in vivo. It will be interesting to follow the patterns of phosphorylation of all of these proteins in Arabidopsis, especially during nucleosome formation, transcription initiation, chromosome segregation, and DNA repair.
Chromatin Assembly/Modification Requirement during Plant Development
A number of genes that are likely involved in chromatin metabolism in Arabidopsis have been identified, and several mutations have been described which inactivate key regulatory genes. The effect is often a pleiotropic disruption of developmental events. Loss of either the FAS1 or FAS2 genes (CAF-1 subunits) results in a highly fasciated shoot meristem and abnormal flower and root development (Leyser and Furner, 1992; Kaya et al., 2001). Another mutation, splayed (syd), has a similar phenotype as tsl mutants, and the SYD gene encodes a SWI/SNF chromatin remodeling factor homolog (Wagner and Meyerowitz, 2002). The syd mutant has altered leaf shape, altered floral organ composition, and abnormal gynoecium and ovule development, and the mutation enhances weak leafy (lfy) and unusual floral organs (ufo) mutations, all features also caused by the tsl mutation (Roe et al., 1993; Wagner and Meyerowitz, 2002; J.L. Roe, unpublished data). tsl also shows a genetic interaction with leunig (Roe et al., 1997b); the LUG gene was identified as a putative transcriptional corepressor (Conner and Liu, 2000). These as well as other mutations in chromatin regulatory genes, such as those in Polycomb group genes, have shown the importance of transcriptional silencing and chromatin remodeling in plant development (for review, see Wagner, 2003), including the control of flowering time (Noh and Amasino, 2003; Pineiro et al., 2003; for review, see Sung et al., 2003). It is interesting that one of the proteins with the most homology to TKI1 is the Polycomb group protein, cramped, which is likely involved in chromatin regulation during D. melanogaster development (Yamamoto et al., 1997), and further investigation of its function will show if it plays a similar role in Arabidopsis.
MATERIALS AND METHODS
Arabidopsis Suspension Culture Cells
Arabidopsis suspension culture cells were grown at 22°C to 27°C, shaken at 120 rpm in the dark in Murashige and Skoog medium supplemented with Gamborg's B5 vitamins (Sigma, St. Louis), kinetin (1 μg/mL), and α-naphthaleneacetic acid (1 μg/mL), and subcultured weekly.
Cell Synchronization
For cell cycle synchronization, 5 μg/mL of aphidicolin (Sigma) was added to a freshly diluted (1:5) stationary culture of Arabidopsis suspension culture cells, and cells were incubated with shaking for 24 h to block the cells in S-phase. Cells were then washed by pouring through a mesh screen (150 μm), followed by three washing steps, all with sterile solutions. First, cells were washed with 3% sucrose (1 L); second with 1 L Murashige and Skoog medium; and finally with 500 mL Murashige and Skoog medium containing auxin and cytokinin as above. Cells were then suspended in 100 mL medium, still on the screen, and shaken gently but continuously 3 to 5 times for 2 min each. After resuspension in fresh medium, a t = 0 sample was removed, and cells were grown at 24°C with shaking in the dark. Aliquots of 5 to 10 mL were removed at time points following release, and cells were harvested by settling on ice. The medium was removed, and cells were quick-frozen in liquid nitrogen. Separate aliquots of cells were fixed with ethanol:acetic acid (3:1, v/v) and stained with 5 μm 4,6-diamidino-2-phenylindole (Sigma). Mitotic index was determined by counting the proportion of cells in late prophase to telophase compared to cells in interphase observed by UV fluorescence on a Zeiss Axiophot microscope (Zeiss, Jena, Germany). For each sample, >300 cells were counted. Some samples were stained with propidium iodide (50 μg/mL) or Sytox (0.5 μm, Molecular Probes, Eugene, OR) and observed by confocal fluorescence microscopy.
RNA and Protein Extraction
RNA was purified from pelleted cells or quick-frozen (liquid nitrogen) ground tissue with Trizol, according to the manufacturer's protocol (Life Technologies, Rockville, MD). Total protein extracts were obtained by pelleting cells and resuspending in Lysis250 plus protease inhibitors (50 mm Tris, pH 7.4, 250 mm sodium chloride, 5 mm EDTA, 0.1% IGEPAL CA 630 [Nonidet P-40 substitute; Sigma], 50 mm sodium fluoride) supplemented with phenylmethylsulfonyl fluoride (100 μg/mL), aprotinin (2 μg/mL), leupeptin (2 μg/mL), pepstatin A (1 μg/mL), dithiothreitol (1 mm), and benzamidine (1 mm) and after freeze-thaw, vortexing with glass beads to solubilize the cells. After centrifugation, soluble protein was quantitated by A280 measurement and by running aliquots on SDS-PAGE gels and staining with Coomassie Blue R-250. Flower extracts were obtained by grinding in liquid nitrogen and solubilizing in Laemmli sample buffer plus 6 m urea.
Antibodies
Anti-TSL antibody was obtained as follows. The peptide (C)YNQEDRPDVLTMAQDPYLAYS was synthesized, anti-peptide antisera was generated in rabbits, and peptide-specific antibody was affinity purified on peptide-conjugated support by Zymed Laboratories (South San Francisco, CA).
Western-Blot Analysis
Western-blot analysis was performed by standard techniques after transfer of SDS-PAGE gels to Immobilon-P membranes (Millipore, Bedford, MA). Blots were incubated overnight with anti-TSL affinity-purified anti-peptide antisera (12 μg IgG) or anti-His-horseradish peroxidase mouse monoclonal IgG (sc-8036, Santa Cruz Biotechnology, Santa Cruz, CA; 1:1000) or anti-GST (sc-459, Santa Cruz Biotechnology; 1:1000) rabbit polyclonal antisera in washing buffer (50 mm Tris, pH 7.5, 150 mm sodium chloride, nonfat dry milk, and 0.05% Tween 20), washed three times for 30 min with washing buffer, incubated for 2 h at room temperature with 1:1000 secondary antibody (goat anti-rabbit IgG conjugated to horseradish peroxidase; Zymed or peroxidase-labeled anti-mouse IgG; Vector Laboratories, Burlingame, CA), washed three times for 30 min each with washing buffer, and rinsed with 50 mm Tris, pH 7.5, 150 mm sodium chloride. Presence of the secondary antibody was detected by chemiluminescence by incubation of blots with SuperSignal (Pierce, Rockford, IL) followed by autoradiography.
Immunoprecipitation
Suspension culture cells were lysed by freezing in liquid nitrogen, grinding on dry ice, followed by vortexing in Lysis250 plus protease inhibitors and glass beads. After centrifugation, total soluble protein extract was incubated with affinity-purified antibody (12 μg immune or preimmune) for 2 h at 4°C with shaking. Affi-Prep Protein A Matrix (Bio-Rad Laboratories, Hercules, CA) was added (30-μL beads), incubated for 30 min at 4°C, and beads were washed three times in extraction buffer. For SDS-PAGE, samples were boiled in Laemmli sample buffer.
In-Gel Kinase Assay
In-gel kinase assays were performed as described in Zhang and Liu (2001). Briefly, SDS-PAGE gels were formed containing MBP (Life Technologies) at 0.25 mg/mL in the resolving gel. After electrophoresis, SDS was removed, proteins were renatured, and the gel was incubated in [32P-γ]ATP (NEN, Boston; 3,000 Ci/mmol). Following five washes in 5% (w/v) trichloroacetic acid/1% (w/v) sodium pyrophosphate, the gel was dried and autoradiography was performed.
Induction and Purification of Fusion Proteins and Kinase Assays
GST-fusion proteins were purified as described from yeast (Saccharomyces cerevisiae) after induction with galactose (Roe et al., 1997a). His-tagged fusion proteins were purified from IPTG-induced Escherichia coli strain BL21DE3plysS using Talon resin (Clontech, Palo Alto, CA). Cells were sonicated in extraction buffer (50 mm HEPES, pH 7.0, 250 mm NaCl plus protease inhibitors), centrifuged 20 min at 12,000g to remove insoluble protein, incubated with resin, washed in extraction buffer, and His-tagged proteins were eluted in 150 mm imidazole. Kinase assays are as described with [32P-γ]ATP (NEN, 3,000 Ci/mmol; Roe et al., 1997a). Purified histone H3 was obtained from Roche Diagnostics (Indianapolis).
Northern-Blot Analysis
Northern-blot analysis was performed using standard techniques. RNA samples were run on formaldehyde-agarose gels, blotted on Hybond N membranes (Amersham, Buckinghamshire, UK), and hybridized with probe labeled with [32P-α]dCTP (ICN Biomedicals, Costa Mesa, CA; 3,000 Ci/mmol) by random priming using Prime-it II kit (Stratagene, La Jolla, CA) and purified on NucTrap columns (Stratagene). After high stringency washing, blots were subjected to autoradiography. 18S rRNA and histone H4 were detected as described by Roe et al. (1993) and Ehsan et al. (1999), respectively. TSL was detected with a full-length cDNA probe, and TKI1 was detected with the insert from the two-hybrid clone, pC-TKI1 (see below). CycB1;1 coding sequence was synthesized by PCR using the oligonucleotides 5′-GCGGATCCATGGTGACTTCTCGTTCGATTG-3′ and 5′-GTCCATGGCAGATTCAGTTCCGGTCAAC-3′ on the two-hybrid ACT library (see below), digested with BamHI and partially with NcoI, and subcloned into a pBluescript-based vector which was sequenced for verification. An NcoI fragment was used as a probe which contained the full coding sequence.
Plant Material and GUS Activity Staining
tsl-1 and tsl-2 mutants are as described (Roe et al., 1993) and are in the Wassilewskija and Columbia ecotype, respectively. The CDG marker gene in Columbia ecotype was crossed into the tsl-2 background. Inflorescences from F2 individuals were stained for GUS activity using X-glucuronic acid as described by Sessions et al. (1999) overnight at 37°C, and destained in 70% ethanol.
Cloning of Asf1b/SGA1, TKI1, Fusion Protein, and Two-Hybrid Clones
Oligonucleotides 5′-CGGCATATGAGCTCTATCAATATCACT-3′ and 5′-CGCGGATCCTCATGTCTCCTGGAGATT-3′ were used to clone the coding sequence of Asf1b/SGA1 (locus At5g38110) and add an NdeI site and BamHI site by PCR on a cDNA two-hybrid library template (see below), followed by subcloning into pCD.1 (Durfee et al., 1999) as a 0.65-kb NdeI-BamHI fragment to form pCD1-Asf1b, which encodes a fusion protein of the DBD of Gal4 with Asf1b/SGA1. The sequence was identical to that of the full-length cDNA sequence (GenBank accession no. AY072420). Some TSL two-hybrid and GST-fusion protein clones and the K438E mutation are as described (Roe et al., 1997). To generate the TSL bait vector, the fragment in pAS-ΔN73-TSL (Roe et al., 1997a) was subcloned into the CEN plasmid pCD.1 (Durfee et al., 1999) to obtain the plasmid pCD-NQ-TSL which encodes amino acids 73-688 of TSL as a fusion protein with Gal4-DBD. Details of this cloning and other two-hybrid clones are available on request. pACTII (Bai and Elledge, 1996) was used for subcloning to obtain ACT plasmids. pC-TKI1 was isolated as a two-hybrid library plasmid in the vector pGAD424 (Ballas and Citovsky, 1997), which encoded amino acids 546-741 of TKI1. Full-length TKI1 cDNA was assembled by PCR amplification of the N-terminal coding sequences using the two-hybrid cDNA library followed by subcloning into pC-TKI1 (details available on request). The full-length TKI1 cDNA was then sequenced on both strands (GenBank accession no. AF530160). The TKI C-terminal coding region of pC-TKI and the coding regions of Asf1b and GFP were subcloned into pRSET (Invitrogen, Carlsbad, CA) to generate His-tagged fusion protein clones; details are available on request.
Two-Hybrid Library Screening and Two-Hybrid Assays
The TSL bait plasmid pCD-NQ-TSL was transformed into YD116 cells (Durfee et al., 1999; selected on Trp− plates). A cDNA library in the pGAD424 vector constructed using mRNA from aerial parts of Arabidopsis including flowers (gift of Vitaly Citovsky; Ballas and Citovsky, 1997) was introduced into cells from cultures of a bait-containing transformant and plated on selective Trp− Leu− Ura− plates. Colonies that grew were retested by isolation of the library plasmid, retransformation of Y153 cells (Durfee et al., 1993) with and without the TSL bait vector, followed by staining of colony lifts for β-galactosidase activity as described (Durfee et al., 1993). To quantitate two-hybrid interactions, ONPG assays were performed on transformants grown in liquid culture as described (Miller, 1972; Clontech, Yeast Protocols Handbook), and the data are reported in Miller units.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AF530160.
Acknowledgments
We gratefully acknowledge Leila Nyberg and Mitchell Bowen for excellent technical assistance, Peter Doerner for the CDG marker line, Vitaly Citovsky for the two-hybrid cDNA library, Dan Boyle for help with confocal microscopy, Scott Todd for the control anti-T-span peptide antibody, and Carol Rivin for valuable comments on the manuscript.
This work was supported by the U.S. Department of Agriculture (grant nos. USDA 98–35304–6676 and AES KS2453).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.038117.
References
- Aasland R, Stewart AF, Gibson T (1996) The SANT domain: a putative DNA binding domain in the SWI-SNF and ADA complexes, the transcriptional corepressor N-CoR and TFIIIB. Trends Biochem Sci 21: 87–88 [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Elledge SJ (1996) Gene identification using the yeast two-hybrid system. Methods Enzymol 273: 331–347 [DOI] [PubMed] [Google Scholar]
- Ballas N, Citovsky V (1997) Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc Natl Acad Sci USA 94: 10723–10728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric S, Reinke H, Horz W (2003) Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol Cell Biol 23: 3468–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL (2002) Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell 10: 935–942 [DOI] [PubMed] [Google Scholar]
- Carrera P, Moshkin YM, Gronke S, Sillje HHW, Nigg EA, Jackle H, Karch F (2003) Tousled-like kinase functions with the chromatin assembly pathway regulating nuclear divisions. Genes Dev 17: 2578–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimura T, Kuzuhara T, Horikoshi M (2002) Identification and characterization of CIA/ASF1 as an interactor of bromodomains associated with TFIID. Proc Natl Acad Sci USA 99: 9334–9339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Conner J, Liu Z (2000) LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc Natl Acad Sci USA 97: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criqui MC, Parmentier Y, Derevier A, Shen W-H, Dong A, Genschik P (2000) Cell cycle-dependent proteolysis and ectopic overexpression of cyclin B1 in tobacco BY2 cells. Plant J 24: 763–773 [DOI] [PubMed] [Google Scholar]
- Day IS, Reddy ASN, Golovkin M (1996) Isolation of a new mitotic-like cyclin from Arabidopsis: complementation of a yeast cyclin mutant with a plant cyclin. Plant Mol Biol 30: 565–575 [DOI] [PubMed] [Google Scholar]
- Doerner P, Jorgenesen J, You R, Steppuhn J, Lamb C (1996) Control of root growth and development by cyclin expression. Nature 380: 520–523 [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn AE, Lee W-H, Elledge SJ (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev 7: 555–569 [DOI] [PubMed] [Google Scholar]
- Durfee T, Draper O, Zupan J, Conklin DS, Zambryski PC (1999) New tools for protein linkage mapping and general two-hybrid screening. Yeast 15: 1761–1768 [DOI] [PubMed] [Google Scholar]
- Ehsan H, Roef L, Witters E, Reichheld J-P, Van Bockstaele D, Inze D, Van Onckelen H (1999) Indomethacin-induced G1/S phase arrest of the plant cell cycle. FEBS Lett 458: 349–353 [DOI] [PubMed] [Google Scholar]
- Emili A, Schieltz DM, Yates JR III, Hartwell LH (2001) Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell 7: 13–20 [DOI] [PubMed] [Google Scholar]
- Fields S, Sternglanz R (1994) The two-hybrid system: an assay for protein-protein interactions. Trends Genet 10: 286–292 [DOI] [PubMed] [Google Scholar]
- Groth A, Lukas J, Nigg EA, Sillje HHW, Wernstedt C, Bartek J, Hansen K (2003) Human Tousled like kinases are targeted by an ATM- and Chk1-dependent DNA damage checkpoint. EMBO J 22: 1676–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Saam JR, Adams HP, Mango SE, Schumacher JM (2003) The C. elegans Tousled-like Kinase (TLK-1) has an essential role in transcription. Curr Biol 13: 1921–1929 [DOI] [PubMed] [Google Scholar]
- Hemerly A, Bergounioux C, Van Montagu M, Inze D, Ferreira P (1992) Genes regulating the plant cell cycle: isolation of a mitotic-like cyclin from Arabidopsis thaliana. Proc Natl Acad Sci USA 89: 3295–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Taranto P, Walser M, Schonrock N, Gruissem W (2003) Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130: 2555–2565 [DOI] [PubMed] [Google Scholar]
- Hu F, Alcasabas AA, Elledge SJ (2001) Asf1 links Rad53 to control of chromatin assembly. Genes Dev 15: 1061–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A (2001) G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13: 1891–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Iwase M, Kodama H, Lavisse P, Komamine A, Nishihama R, Machida Y, Watanabe A (1998) A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10: 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kaya H, Shibahara K, Taoka K, Iwabuchi M, Stillman B, Araki T (2001) FASCIATA genes for Chromatin Assembly Factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131–142 [DOI] [PubMed] [Google Scholar]
- Le S, Davis C, Konopka JB, Sternglanz R (1997) Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13: 1029–1042 [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Furner IJ (1992) Characterization of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116: 397–403 [Google Scholar]
- Li Y, DeFatta R, Anthony C, Sunavala G, De Benedetti A (2001) A translationally regulated Tousled kinase phosphorylates histone H3 and confers radioresistance when overexpressed. Oncogene 20: 726–738 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res 30: 281–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijsing SH, Ehrenhofer-Murray AE (2001) The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-1 and Asf1 in Saccharomyces cerevisiae. Genes Dev 15: 3169–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello JA, Sillje HHW, Roche DMJ, Kirschner DB, Nigg EA, Almouzni G (2002) Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep 3: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH (2002) Cell cycle-regulated gene expression in Arabidopsis. J Biol Chem 277: 41987–42002 [DOI] [PubMed] [Google Scholar]
- Menges M, Murray JAH (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Moshkin YM, Armstrong JA, Maeda RK, Tamkun JW, Verrijzer P, Kennison JA, Karch F (2002) Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev 16: 2621–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munakata T, Adachi N, Yokoyama N, Kuzuhara T, Horikoshi M (2000) A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells 5: 221–233 [DOI] [PubMed] [Google Scholar]
- Noh Y-S, Amasino RM (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odgren PR, Toukatly G, Bangs PL, Gilmore R, Fey EG (1996) Molecular characterization of mitofilin (HMP), a mitochondria-associated protein with predicted coiled coil and intermembrane space targeting domains. J Cell Sci 109: 2253–2264 [DOI] [PubMed] [Google Scholar]
- Osada S, Sutton A, Muster N, Brown CE, Yates JR, Sternglanz R, Workman JL (2001) The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev 15: 3155–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro M, Gomez-Mena C, Schaffer R, Martinez-Zapater JM, Coupland G (2003) EARLY BOLTING IN SHORT DAYS is Related to Chromatin Remodeling Factors and Regulates Flowering in Arabidopsis by Repressing FT. Plant Cell 15: 1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchais S, Glab N, Inze D, Bergounioux C (2000) Chemical inhibitors: a tool for plant cell cycle studies. FEBS Lett 476: 78–83 [DOI] [PubMed] [Google Scholar]
- Roe JL, Durfee T, Zupan JR, Repetti PP, McLean BG, Zambryski PC (1997. a) TOUSLED is a nuclear serine/threonine protein kinase that requires a coiled-coil region for oligomerization and catalytic activity. J Biol Chem 272: 5838–5845 [DOI] [PubMed] [Google Scholar]
- Roe JL, Nemhauser JL, Zambryski PC (1997. b) TOUSLED participates in apical tissue formation during gynoecium development in Arabidopsis. Plant Cell 9: 335–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JL, Rivin CJ, Sessions RA, Feldmann KA, Zambryski PC (1993) The Tousled gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell 75: 939–950 [DOI] [PubMed] [Google Scholar]
- Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J (1998) More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J 14: 273–284 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Sessions A, Weigel D, Yanofsky MF (1999) The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J 20: 259–263 [DOI] [PubMed] [Google Scholar]
- Shalom S, Don J (1999) Tlk, a novel evolutionarily conserved murine serine threonine kinase, encodes multiple testis transcripts. Mol Reprod Dev 52: 392–405 [DOI] [PubMed] [Google Scholar]
- Sharp JA, Fouts ET, Krawitz DC, Kaufman PD (2001) Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol 11: 463–473 [DOI] [PubMed] [Google Scholar]
- Sillje HHW, Nigg EA (2001) Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr Biol 11: 1068–1073 [DOI] [PubMed] [Google Scholar]
- Sillje HHW, Takahashi K, Tanaka K, Van Houwe G, Nigg EA (1999) Mammalian homologues of the plant Tousled gene code for cell-cycle-related kinases with maximal activities linked to ongoing DNA replication. EMBO J 18: 5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahawald M, Gottschling DE (1998) Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Wang X, Bloom MH, Simon GM, Berger SL (2002) The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J Biol Chem 277: 8178–8186 [DOI] [PubMed] [Google Scholar]
- Sunavala-Dossabhoy G, Li Y, Williams B, De Benedetti A (2003) A dominant negative mutant of TLK1 causes chromosome missegregation and aneuploidy in normal breast epithelial cells. BMC Cell Biol 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung ZR, Chen L, Moon Y-H, Lertpiriyapong K (2003) Mechanisms of floral repression in Arabidopsis. Curr Opin Plant Biol 6: 29–35 [DOI] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen S-R, Kobayashi R, Kamakaka RT, Kadonaga JT (1999) The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402: 555–560 [DOI] [PubMed] [Google Scholar]
- Tyler JK, Collins KA, Prasas-Sinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, Kadonaga JT (2001) Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol 21: 6574–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D (2003) Chromatin regulation of plant development. Curr Opin Plant Biol 6: 20–28 [DOI] [PubMed] [Google Scholar]
- Wagner D, Meyerowitz EM (2002) SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr Biol 12: 85–94 [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Yamakawa A, Kameoka Y, Hashimoto K, Yoshitake Y, Nishikawa K, Tanihara K, Date T (1997) cDNA cloning and chromosomal mapping of genes encoding novel protein kinases termed PKU-α and PKU-β, which have nuclear localization signal. Gene 202: 193–201 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Girard F, Bello B, Affolter M, Gehring WJ (1997) The cramped gene of Drosophila is a member of the Polycomb-group, and interacts with mus209, the gene encoding proliferating cell nuclear antigen. Development 124: 3385–3394 [DOI] [PubMed] [Google Scholar]
- Yu J, Li Y, Ishizuka T, Guenther MG, Lazar MA (2003) A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J 22: 3403–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HS, Postigo AA, Dean DC (1999) Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4α, TGFβ, and contact inhibition. Cell 97: 53–61 [DOI] [PubMed] [Google Scholar]
- Zhang S, Liu Y (2001) Activation of salicyclic acid-induced protein kinase, a mitogen-activated protein kinase, induces multiple defense responses in tobacco. Plant Cell 13: 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]