Abstract
The molecular mechanism by which GA regulates plant growth and development has been a subject of active research. Analyses of the rice (Oryza sativa) genomic sequences identified 77 WRKY genes, among which OsWRKY71 is highly expressed in aleurone cells. Transient expression of OsWRKY71 by particle bombardment specifically represses GA-induced Amy32b α-amylase promoter but not abscisic acid-induced HVA22 or HVA1 promoter activity in aleurone cells. Moreover, OsWRKY71 blocks the activation of the Amy32b promoter by the GA-inducible transcriptional activator OsGAMYB. Consistent with its role as a transcriptional repressor, OsWRKY71 is localized to nuclei of aleurone cells and binds specifically to functionally defined TGAC-containing W boxes of the Amy32b promoter in vitro. Mutation of the two W boxes prevents the binding of OsWRKY71 to the mutated promoter, and releases the suppression of the OsGAMYB-activated Amy32b expression by OsWRKY71, suggesting that OsWRKY71 blocks GA signaling by functionally interfering with OsGAMYB. Exogenous GA treatment decreases the steady-state mRNA level of OsWRKY71 and destabilizes the GFP:OsWRKY71 fusion protein. These findings suggest that OsWRKY71 encodes a transcriptional repressor of GA signaling in aleurone cells.
GA regulates several aspects of plant development, such as germination, growth, and flowering. Studies using cell biology techniques suggest that perception of GA in barley (Hordeum vulgare) and oat (Avena sativa) aleurone cells occurs on the plasma membrane (Ritchie and Gilroy, 1998; Lovegrove and Hooley, 2000). Heterotrimeric G-proteins have been shown to be involved in the transduction of GA signals in oat (Jones et al., 1998) and rice (Oryza sativa; Ueguchi-Tanaka et al., 2000). Secondary messengers governing GA responses include Ca2+, pH, and cGMP (for review, see Olszewski et al., 2002). Studies with constitutive GA-responsive mutants reveal that Arabidopsis Spy and its barley ortholog HvSpy encode a Ser/Thr O-linked GlcNAc transferase (OGT) and RGA, a repressor of GA signaling (Jacobsen et al., 1996; Robertson et al., 1998; Silverstone et al., 2001). Wheat (Triticum sativum) Rht-B1 and Rht-D1, maize (Zea mays) D8, and barley and rice Slender genes are orthologs of RGA and GAI (Peng et al., 1999; Silverstone and Sun, 2000; Gubler et al., 2002; Itoh et al., 2002). Studies of recessive GA-unresponsive dwarf mutants have identified positive regulators in GA signaling, such as semidwarf rice dwarf1, barley gse (for GA sensitivity), and Arabidopsis sleepy1 (Steber et al., 1998) and pickle (Mitsunaga et al., 1994; Chandler and Robertson, 1999; Ogas et al., 1999). The dwarf1 and gse mutations prevent the GA induction of α-amylase gene expression in aleurone layers (Mitsunaga et al., 1994; Fujisawa et al., 1999).
cis-Acting elements necessary and sufficient for GA response of high-pI and low-pI α-amylase genes and protease genes in cereal crops have been defined (Skriver et al., 1991; Gubler and Jacobsen, 1992; Lanahan et al., 1992; Rogers and Rogers, 1992; Rogers et al., 1994; Tanida et al., 1994; Cercós et al., 1999). In the low-pI α-amylase promoter, Amy32b, five elements, namely O2S (also called Box 2), pyrimidine (Pyr) box, GA response element (GARE), amylase box (also called Amy Box and Box I), and down-stream amylase element, are essential for a high level of GA-induced expression (Lanahan et al., 1992; Rogers and Rogers, 1992; Rogers et al., 1994; Gómez-Cadenas et al., 2001a), although the O2S-Pyr-GARE and GARE-Pyr-GARE are sufficient for a significant level of GA induction (Rogers and Rogers, 1992). GARE appears to be bound by GA-inducible nuclear proteins (Sutliff et al., 1993), such as GAMYB (Gubler et al., 1995) and HRT (a zinc finger protein; Raventós et al., 1998). The O2S element of the barley Amy32b promoter (Lanahan et al., 1992) is similar to the Box 2 of the oat α-Amy2/A and α-Amy2/D genes (Willmott et al., 1998), both containing the sequence GATTGACTTGACC. Interestingly, this element includes two consensus sequences of the W box, TGAC(C/T), which is recognized by WRKY proteins (Eulgem et al., 2000). WRKY genes have been shown to be involved in biotic (bacterial and fungal diseases) and abiotic (wounding and freezing) stresses, as well as in anthocyanin biosynthesis, senescence, and trichome development (for review, see Eulgem et al., 2000). In addition, two wild oat (A. sativa subsp. fatua) WRKY proteins (ABF1 and ABF2) bind to the Box 2 (W box) of the GA-regulated α-Amy2 promoter (Rushton et al., 1995). Herein, we report that rice OsWRKY71 gene encodes a transcription factor that represses GA induction of the Amy32b promoter in aleurone cells.
RESULTS
OsWRKY71 mRNA Levels Decline in Response to Exogenous GA Treatment
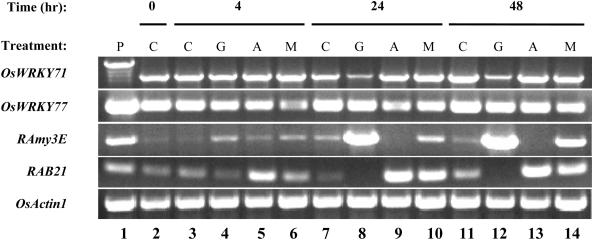
Analyses of available rice genome sequences (Goff et al., 2002; Yu et al., 2002) reveals 77 OsWRKY genes (Supplemental Figs. 1 and 2). To investigate which of these genes are expressed in aleurone cells, RNA was isolated from rice aleurone layers treated with no hormone, 1 μm GA3, 20 μm abscisic acid (ABA), or 1 μm GA3 plus 20 μm ABA for 4 h, 24 h and 48 h. RT-PCR was carried out with gene-specific primers (Supplemental Table I) for several OsWRKY genes, including OsWRKY71, OsWRKY77, and other control genes such as RAmy3E, RAB21, and OsActin1, using the same set of RNA samples. Primers for OsWRKY71 were designed to amplify the entire coding region of this gene including all three introns. Hence, should a RNA preparation be contaminated with genomic DNA, a larger PCR fragment would be produced, as in the genomic DNA control (Fig. 1, lane 1). Clearly, none of the RNA samples was contaminated with a detectable amount of genomic DNA. The mRNA level of OsWRKY71 is abundant in aleurone cells at the onset of hormone treatments (Fig. 1, lane 2). Treatments without hormone (Fig. 1, lane 3) or with GA3, ABA, and GA3 plus ABA for 4 h (Fig. 1, lanes 4–6) had little effect on the abundance of OsWRKY71 mRNA level. Prolonged GA3 treatments, for 24 h (Fig. 1, lane 8) and 48 h (Fig. 1, lane 12), decreased the steady-state mRNA level of OsWRKY71 by 55% and 67%, respectively. As controls, the steady mRNA level of OsWRKY77 didn't appear to be regulated by GA and that of RAmy3E, which encodes a GA-inducible α-amylase (Karrer et al., 1991), was drastically enhanced in the same samples treated with GA3 for 24 and 48 h (Fig. 1, lanes 8 and 12). ABA or GA3 plus ABA treatments had little effect on the expression of the OsWRKY71 gene. By contrast, the mRNA level of RAB21, a reported ABA-inducible gene (Mundy and Chua, 1988), increased greatly under the same ABA treatments (Fig. 1, lanes 5, 9, and 13). As expected, the mRNA levels of OsActin1, a constitutively expressed rice gene, were similar in all samples, suggesting that an equal amount of RNA was used for all RT-PCR reactions. These data show that OsWRKY71 is down-regulated by GA at either transcriptional and/or posttranscriptional levels. Alternatively, this mRNA could be under constant turnover, and reduction in its synthesis led to reduction of the mRNA level.
Figure 1.
RT-PCR analysis of OsWRKY gene expression in aleurone cells. Total RNA was isolated from rice aleurone layers treated with no hormone, 1 μm GA3, 20 μm ABA, or 1 μm GA3 plus 20 μm ABA for 4, 24, and 48 h, respectively. Genomic DNA was isolated from rice 10-d-old seedlings using the cetyl-trimethyl-ammonium bromide method. P, Positive control, with rice genomic DNA as the template; C, No hormone control; G, GA treatment; A, ABA treatment; M, GA + ABA treatment. Expected RT-PCR fragment of the intron-containing genomic sequence for OsWRKY71 is 1393 bp; the corresponding cDNA is 1047 bp in length. RAmy3E, a GA-inducible gene; RAB21, an ABA-inducible gene; OsActin1, a constitutively expressed actin gene.
Figure 2.
Protein sequence alignment of OsWRKY71, AfABF2, and AtWRKY40 and domain structure of OsWRKY71. A, Alignment of the rice, wild oat, and Arabidopsis WRKY proteins. The deduced amino acid sequences were aligned by using ClustalW. Identical residues are shaded in black and residues chemically similar in gray. The putative nuclear localization signal (KRIR) and WRKY amino acid residues are labeled with rectangles, and amino acid residues potentially interacting with zinc ligands are pointed to with arrows. B, The OsWRKY71 protein sequence was analyzed for protein domain structures. Only three identified domains, nuclear localization signal (NLS), WRKY, and the zinc finger motif (Zn-F), are shown.
OsWRKY71 Shares the Highest Sequence Similarity with Wild Oat ABF2 and Arabidopsis WRKY40
The deduced amino acid sequences of the 77 OsWRKY proteins were aligned by using the ClustalW program (http://www.ebi.ac.uk/clustalw/) with WRKY proteins from other species (Supplemental Fig. 2). Among all OsWRKY proteins, it appears that OsWRKY71 shares the highest sequence similarity with the wild oat ABF2 and Arabidopsis WRKY40 proteins. Detailed comparisons were carried out by using the water (Smith-Waterman local alignment) program from the EMBOSS package (www.emboss.org), with the default parameters (gap opening penalty score 10.0 and gap extension penalty score 0.5). This analysis indicated that the identity between OsWRKY71 and ABF2 is 68.2%, and the similarity is 72.8%. The multiple sequence alignment of OsWRKY71, ABF2, and AtWRKY40 is shown in Figure 2A. OsWRKY71 contains one WRKY domain, a zinc finger-like motif (C-X4-5-C-X23-H-X-H) and a nuclear localization signal (Fig. 2B, NLS). The homology with AtWRKY40 is lower, with only 36.9% identity and 54.7% similarity.
OsWRKY71 Proteins Are Targeted to Nuclei of Aleurone Cells
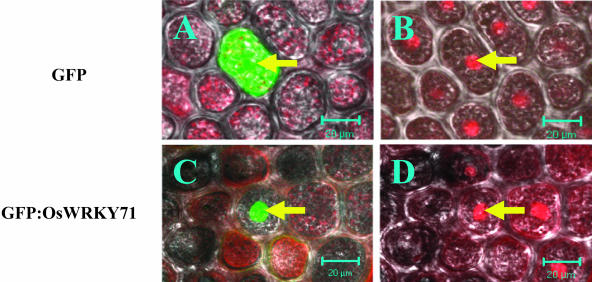
To investigate the subcellular localization of OsWRKY71 protein, the cDNA sequence of the OsWRKY71 gene was fused in frame to the 3′ end of the green fluorescent protein (GFP) gene that is driven by the constitutive maize Ubiquitin (UBI) promoter (Bruce et al., 1989). The fusion construct, UBI-GFP:OsWRKY71 and the control construct UBI-GFP were introduced, respectively, into barley aleurone cells by bombardment. Confocal microscopic observations indicated that GFP fluorescence was detected in the whole cell bombarded with the control construct UBI-GFP (Fig. 3A). By contrast, the GFP:OsWRKY71 fusion protein was targeted to the nucleus (Fig. 3C). Aleurone cell nuclei were stained with SYTO17, which released red fluorescence (Fig. 3, B and D).
Figure 3.
OsWRKY71 proteins are targeted to nuclei of aleurone cells. Aleurone cells were transformed with UBI-GFP (A and B) or UBI-GFP:OsWRKY71 (C and D). After incubation for 24 h, the aleurone cells were stained with SYTO17 for nuclear localization (B and D), followed by examination of GFP expression (A and C). Arrows in A and B point to the same cell; so do those in C and D. The bars represent 20 μm.
OsWRKY71 Specifically Represses GA Induction of the Amy32b α-Amylase Promoter in Aleurone Cells
Data presented above suggest that the OsWRKY71 gene might encode a transcription factor modulating GA response in aleurone cells. To test this hypothesis, we investigated the effect of this gene on the expression of an α-amylase promoter. Conventionally, cDNA clones of effector genes are used in this type of transient expression study. In this study, we decided to use genomic clones instead of cDNA clones. Technically, it is easier to clone a genomic fragment by PCR. The lack of a genome sequence limited the practice of this approach in the past. Now that the rice genome sequence is available, we decided to employ rice genomic effector genes. Introns from one cereal species appear to be effectively spliced out in another cereal, although those from a dicotyledonous plant might not be spliced out precisely in a monocotyledonous plant cell (Hanley and Schuler, 1988). The maize ubiquitin intron appears to be excised out correctly in barley (Bruce et al., 1989; Rogers and Rogers, 1992; Shen et al., 1993; Gómez-Cadenas et al., 1999). Based on these published data, it is conceivable that rice introns could be spliced correctly and efficiently in barley. Before we tested the intron-containing OsWRKY gene, experiments were carried out to show that the intron-containing OsPKABA1 and OsGAMyb indeed were as effective as the corresponding cDNA clones of TaPKABA1 (Gómez-Cadenas et al., 1999) and HvGAMyb (Gubler et al., 1995) in suppressing and activating the GA induction pathway respectively (Supplemental Fig. 3). Therefore, a genomic clone of OsWRKY71 was bombarded into barley aleurone cells in this study. We used barley aleurone cells because the expression level of the reporter construct in the rice aleurone cells is typically 10 times lower than that obtained in barley aleurone cells (Z.-L. Zhang and Q.J. Shen, unpublished data), hence lowering the sensitivity of detecting the difference of expression levels.
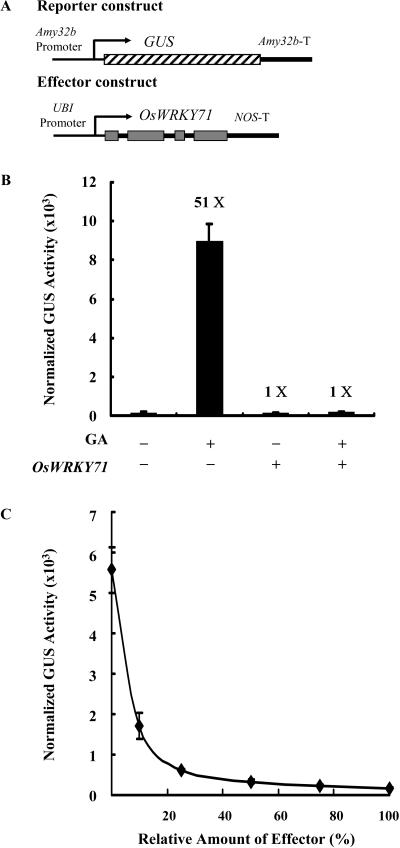
The exogenous GA treatment resulted in 51-fold enhancement of the β-glucuronidase (GUS) activity over that found with the control (Fig. 4B). Coexpression of the effector construct UBI-OsWRKY71 (at 1:1 molar ratio of effector:reporter) completely blocked GA induction of Amy32b-GUS. The repressing effect of OsWRKY71 was further confirmed by a dosage experiment, in which the amount of reporter plasmid was always constant, whereas that of the effector was included in different ratios (Fig. 4C). When the Amy32b-GUS construct was transformed alone, the treatment with 1 μm GA3 strongly induced the expression of this gene construct (62-fold). Expression of Amy32b-GUS in response to the GA treatment reduced gradually with increasing amount of the effector construct. When the relative amounts of the effector were higher than 25%, the GA induction of the Amy32b promoter was almost completely blocked, with GUS activities close to the basal level.
Figure 4.
OsWRKY71 represses GA induction of the Amy32b promoter activity in a dosage-dependent manner. A, Schematic diagrams of the reporter and effector constructs used in the cobombardment experiment. Rectangles in the effector construct represent the exons of OsWRKY71 gene; bars between the rectangles denote the introns of this gene; thick lines represent terminators. B, The reporter construct, Amy32b-GUS, and the internal construct, UBI-Luciferase, were cobombarded into barley aleurone cells either with (+) or without (−) effector construct UBI-OsWRKY71 by using the same molar ratio of effector and reporter constructs. GUS activity was normalized in every independent transformation relative to the luciferase activity. Bars indicate GUS activity ±se after 24 h of incubation of the bombarded aleurone cells with (+) or without (−) 1 μm GA3. Data are means ± se of four replicates. C, The effector construct, UBI-OsWRKY71, was cobombarded into barley aleurone cells along with the reporter construct Amy32b-GUS and the internal control construct UBI-Luciferase. The amount of reporter and internal control plasmid DNA was always constant, whereas that of effector varied with respect to the reporter as shown in the x axis. 100% means the same molar ratio of effector and reporter DNA was used. GUS activity was normalized in every independent transformation relative to the luciferase activity. The line indicates GUS activity ±se after 24 h of incubation of the bombarded aleurone cells with 1 μm GA3. Data are means ± se of four replicates.
The rice RAmy1A promoter is extremely similar to that of the barley Amy32b in both the number of the cis-acting elements and their order. In RAmy1A, the structure is W Box-N4-Pyr-N60-GARE-N10-Amy-N91-TATA Box, and that in Amy32b is W Box-N17-Pyr-N4-GARE-N6-Amy-N69-TATA Box. Note that the sequences of the cis-acting elements are: W Box/O2S, TGACCT; Pyr Box, CTTTT; GARE, TAAC(G/A)(G/A); Amy Box, TATCCA; and TATA Box, TATAAATAC. The distances between the elements are shown with a letter N followed by the number of base pairs. Therefore, OsWRKY71 should repress GA-induced expression of the RAmy1A promoter, too. To test this hypothesis, RAmy1A-GUS reporter construct was prepared and cobombarded into barley half-seeds with (+) or without (−) the effector constructs (UBI-OsWRKY71). As shown in Supplemental Figure 4, GA3 treatment (1 μm) resulted in about 7-fold induction of the RAmy1A promoter. The induction was almost completely blocked by OsWRKY71.
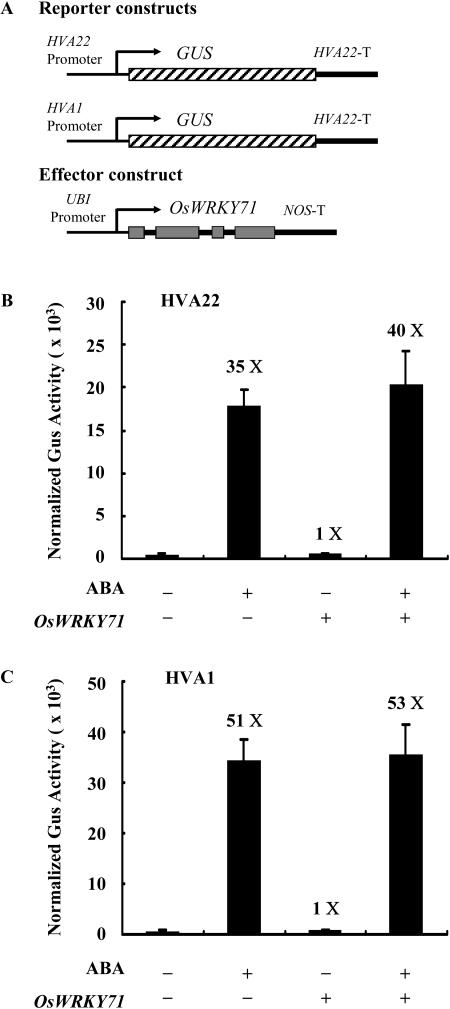
To investigate whether the suppressing effect of OsWRKY71 is specific on the GA signal transduction pathway, its function was also tested with two ABA-inducible promoters, HVA22 and HVA1 (Shen and Ho, 1995; Shen et al., 1996). ABA treatment led to a 35-fold induction of HVA22-GUS (Fig. 5B). The GUS activity remained more or less the same when the effector construct UBI-OsWRKY71 was cobombarded. Similar results were obtained with the ABA-inducible HVA1 promoter (Fig. 5C). All of these data indicated that OsWRKY71 doesn't interfere with ABA responses in the aleurone cells.
Figure 5.
OsWRKY71 has little effect on the ABA signaling in aleurone cells. A, Schematic diagrams of the reporter and effector constructs used in the transient experiment. HVA22 and HVA1 are two ABA inducible genes in barley aleurone cells. B, The reporter construct HVA22-GUS and the internal construct UBI-Luciferase were cobombarded into barley aleurone cells either with (+) or without (−) effector construct UBI-OsWRKY71 by using the same molar ratio of effector and reporter constructs. GUS activity was normalized in every independent transformation relative to the luciferase activity. Bars indicate GUS activity ±se after 24 h of incubation of the bombarded aleurone cells with (+) or without (−) 20 μm ABA. Data are means ± se of four replicates. C, The reporter construct HVA1-GUS and the internal construct UBI-Luciferase were cobombarded into barley aleurone cells either with (+) or without (−) effector construct UBI-OsWRKY71 by using the same molar ratio of effector and reporter constructs. GUS activity was normalized in every independent transformation relative to the luciferase activity. Bars indicate GUS activity ±se after 24 h of incubation of the bombarded aleurone cells with (+) or without (−) 20 μm ABA. Data are means ± se of four replicates.
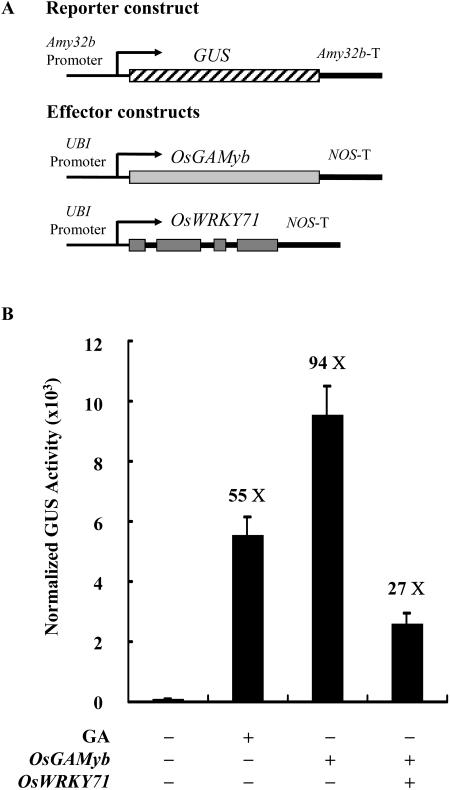
OsWRKY71 Blocks GA Signaling by Functionally Interfering with OsGAMYB
Several signaling molecules mediating the pathway between GA and α-amylase, such as GAMYB and PKABA1, have been reported (Gubler et al., 1995; Gómez-Cadenas et al., 1999). We studied the relationship between OsWRKY71 and OsGAMYB. The effector constructs UBI-OsGAMyb and UBI-OsWRKY71 were cobombarded along with the Amy32b-GUS reporter construct (Fig. 6A). Cotransformation of UBI-OsGAMyb resulted in 94-fold induction of the Amy32b promoter in the absence of GA (Fig. 6B). The OsGAMYB-activated expression of Amy32b-GUS decreased by 70% when OsWRKY71 was coexpressed (Fig. 6B), indicating that OsWRKY71 blocks GA signaling by interfering with the OsGAMYB function.
Figure 6.
OsWRKY71 blocks the transactivation of Amy32b by OsGAMYB. A, Schematic diagrams of the reporter and effector constructs used in the cobombardment experiment. OsGAMyb is from a rice cDNA clone. B, OsWRKY71 suppresses OsGAMYB-activated expression of Amy32b. The reporter construct (Amy32b-GUS), effector constructs (UBI-OsGAMyb and UBI-OsWRKY71), and the internal construct (UBI-Luciferase), at an equal molar ratio, were cobombarded into barley aleurone cells. Transformed aleurone cells were incubated for 24 h without GA3. GUS activity was normalized in every independent transformation relative to the luciferase activity. Data are means ± se of four replicates.
Interestingly, upon OsWRKY71 coexpression, the extent of the decrease was much less for the GAMYB-induced Amy32b promoter activity (Fig. 6B) than that for the GA3-induced activity (Fig. 4). It is possible that the level of GA3-induced GAMYB is less than that obtained in the overexpression study, hence leading to a higher level of the Amy32b activity (Fig. 6) that requires more OsWRKY71 for complete repression. To test this hypothesis, we studied the dosage response of OsWRKY71 repression. The GAMYB-activated expression of Amy32b-GUS dropped gradually with the increasing amount of OsWRKY71 (Supplemental Fig. 5). When the amount of OsWRKY71 was twice (molar ratio) as much as that of OsGAMyb, the activation effect of GAMYB was almost completely abolished.
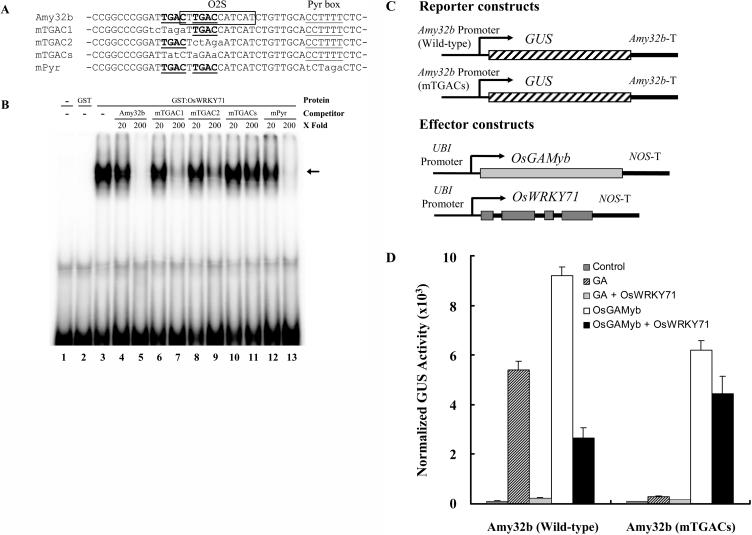
OsWRKY71 Binds to TGAC Motifs in the Amy32b Promoter to Suppress Its Activation by OsGAMYB
To determine whether OsWRKY71 can bind to the Amy32b promoter region, we performed electrophoretic mobility-shift assays with recombinant glutathione S-transferase (GST)-OsWRKY71 proteins. A DNA fragment containing the O2S (W box) and Pyr box of the Amy32b promoter was used as a probe, and competition assays were carried out using wild-type and various mutant DNA fragments. As indicated in Figure 7A, there are two TGAC cores in this region. No binding signal was detected for two controls: with no protein and GST only, respectively (Fig. 7B, lanes 1 and 2). OsWRKY71 bound to the wild-type probe Amy32b (Fig. 7, lane 3) and excess amount of nonlabeled Amy32b competed with the in vitro binding (Fig. 7, lanes 4 and 5). Similar competition was observed with fragments that had only one TGAC element mutated, e.g. mTGAC1 (Fig. 7, lanes 6 and 7) and mTGAC2, respectively (Fig. 7, lanes 8 and 9). However, a competitor with both elements mutated (mTGACs) could not abolish the binding of the protein to the probe (Fig. 7, lanes 10 and 11). A DNA fragment with the adjacent Pyr box mutated (mPyr) was also included in the experiment to further demonstrate that OsWRKY71 binds specifically to TGAC elements (Fig. 7, lanes 12 and 13). Taken together, these data indicate that OsWRKY71 can indeed recognize the Amy32b promoter in vitro, binding specifically to TGAC-containing cis-elements.
Figure 7.
OsWRKY71 interferes with OsGAMYB transactivation of Amy32b via binding to the TGAC cores within the promoter region. A, Partial nucleotide sequence of the Amy32b promoter and its mutant versions used as competitors in electrophoretic mobility-shift assays. Amy32b, wild-type Amy32b promoter; mTGAC1, the Amy32b promoter with the first W box mutated; mTGAC2, the Amy32b promoter with the second W box mutated; mTGACs, the Amy32b promoter with both W boxes mutated; mPyr, the Amy32b promoter with the Pyr box mutated. The W boxes (TGAC cores) and Pyr box are underlined. The O2S element is indicated with a rectangle. The mutated nucleotides are in lowercase. B, Electrophoretic mobility-shift assay with recombinant OsWRKY71. A 61-bp fragment probe containing the W box and Pyr box of the Amy32b promoter and 0.5 μg of GST:OsWRKY71 protein were used in each binding reaction. GST indicates the control binding reaction with GST only. The molar excess of each competitor used (20- and 200-fold) is indicated above each lane. The arrow indicates the DNA-protein complex. C, Schematic diagrams of the reporter and effector constructs used in the cobombardment experiment. D, The reporter constructs Amy32b-GUS and Amy32b(mTGACs)-GUS and the internal construct UBI-Luciferase were cobombarded into barley aleurone cells either with or without effector constructs UBI-OsGAMyb and UBI-OsWRKY71 by using the same molar ratio of effector and reporter constructs. GUS activity was normalized in every independent transformation relative to the luciferase activity. Bars indicate GUS activity ±se after 24 h of incubation of the bombarded aleurone cells with or without 1 μm GA3. Data are means ± se of four replicates. Control means no GA and effector genes were included.
Because the in vitro binding experiment suggested that OsWRKY71 could bind to either the TGAC core within the O2S element or the other TGAC core located immediately upstream from O2S but not to the double mutant (Fig. 7B), mutagenesis experiments were carried out to mutate both TGAC elements within the Amy32b promoter region, generating a new reporter construct named Amy32b(mTGACs)-GUS (Fig. 7C). Consistent with the data presented above, OsWRKY71 blocked the induction of wild-type Amy32b promoter by GA and the transactivation of this promoter by OsGAMYB (Fig. 7D, left). It has been reported that mutation within the O2S element of Amy32b reduced the induction level of this gene by GA (Lanahan et al., 1992; Gómez-Cadenas et al., 2001a). However, a significant level of GA induction was still observed with this mutant (JR335), in which only one TGAC core was mutated (Lanahan et al., 1992; Gómez-Cadenas et al., 2001a). In this study, GA induction of the double mutant, Amy32b(mTGACs)-GUS, was almost completely abolished (Fig. 7D, right). Also, OsGAMYB transactivated Amy32b(mTGACs)-GUS at a level about two-thirds of that obtained with Amy32b-GUS. Furthermore, OsWRKY71 was much less effective in suppressing OsGAMYB transactivation of Amy32b(mTGACs)-GUS; it blocked OsGAMYB transactivation of Amy32b-GUS by as much as 71%, but for Amy32b(mTGACs)-GUS the suppression level was only 28% (Fig. 7D).
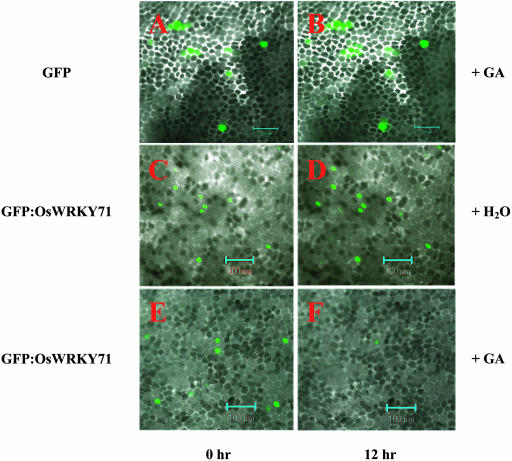
GA Treatment Destabilizes GFP:OsWRKY71 Fusion Proteins in Aleurone Cells
GA has been shown to promote the degradation of some DELLA proteins, such as RGA in Arabidopsis (Silverstone et al., 2001), SLN1 in barley (Fu et al., 2002; Gubler et al., 2002), and SLR1 in rice (Itoh et al., 2002). To investigate whether GA destabilizes OsWRKY71 protein, aleurone cells bombarded with UBI-GFP:OsWRKY71 fusion construct were treated with 100 μm GA3 for 12 h. As a control, the UBI-GFP construct was also included. For UBI-GFP, the GFP fluorescence at 12 h was as strong as that at 0 h (Fig. 8, A and B). Similarly, nucleus-localized GFP fluorescence from UBI-GFP:OsWRKY71 remains more or less the same in the water-treated control (Fig. 8, compare C with D). By contrast, nucleus-localized GFP fluorescence from UBI-GFP:OsWRKY71 disappeared within 12 h of GA treatment (Fig. 8, E and F). These observations suggest that GA application destabilized the GFP:OsWRKY71 fusion proteins in aleurone cells. This data helps explain why a detectable amount of RAmy3E mRNA was observed (Fig. 1, lane 3, row RAMY3E), while that of OsWRKY71 didn't appear to be affected by GA at the 4-h time point (Fig. 1, lane 3, row OsWRKY71)—it might due to the GA-promoted degradation of the OsWRKY71 repressor protein or result from the activity of a signaling pathway independent of OsWRKY71.
Figure 8.
Exogenous GA3 treatment promotes OsWRKY71 degradation in aleurone cells. Embryoless half-seeds were transformed with UBI-GFP or UBI-GFP:OsWRKY71. After incubation for 24 h, the aleurone layers were treated with water or 100 μm GA3 for 12 h, followed by examination of GFP fluorescence under confocal microscope. A, C, and E, The green fluorescence from GFP control and GFP:OsWRKY71 fusion protein, respectively, before GA treatment. Other sections show the images of the same scopes after GA treatment (B and F) or water-treated control (D). The bars represent 100 μm.
DISCUSSION
This work demonstrated that one of 77 OsWRKY genes modulates GA signaling in aleurone cells. Several lines of evidence suggest that OsWRKY71 encodes a transcriptional repressor of the GA signaling in aleurone cells: (1) GA treatment decreased the steady-state mRNA level of OsWRKY71 in aleurone cells by more than 50% (Fig. 1); (2) the OsWRKY71 proteins were targeted to the nuclei of aleurone cells (Fig. 3); (3) overexpression of OsWRKY71 blocked GA induction of the Amy32b promoter (Fig. 4B) in a dosage-dependent manner (Fig. 4C); (4) OsWRKY71 bound to the TGAC-cores (O2S/W box) in the Amy32b promoter (Fig. 7B) and suppressed OsGAMYB transactivation of Amy32b (Figs. 6 and 7D); and (5) GA treatment destabilized the OsWRKY71 protein (Fig. 8). It is important to note that the repression activity of OsWRKY71 is specific for the GA pathway because it has no effect on ABA induction of two other promoters, HVA1 and HVA22 (Fig. 5).
There are two groups of negative regulators involved in GA responses based on whether they bind to a GA-responsive promoter. The non-DNA-binding group includes SPY in Arabidopsis and barley (Jacobsen et al., 1996; Robertson et al., 1998), PKABA1 (Gómez-Cadenas et al., 1999), the DELLA subfamily including GAI/RGA/RGL in Arabidopsis (Peng et al., 1997; Silverstone et al., 1997; Wen and Chang, 2002), SLR1 in rice (Ikeda et al., 2001), SLN1 in barley (Chandler et al., 2002; Gubler et al., 2002), D8 in maize (Peng et al., 1999), and RHT-B1 and RHT-D1 in wheat (Peng et al., 1999). Also, a GAMYB-binding protein, KGM, a member of Mak-subgroup of cdc2- and MAP-kinase-related protein kinases, has been recently shown to specifically repress α-amylase promoter activity (Woodger et al., 2003). The second group, which has been shown or implicated to bind to promoter sequences, includes: SHI that contains a putative zinc-binding RING finger motif (Fridborg et al., 2001); HRT, a zinc finger protein with a C-X8-9-C-X10-C-X2-H consensus sequence, which can bind to TAACAAA element (GARE) and represses GA induction of α-amylase (Raventós et al., 1998); and WRKY proteins (this work).
There are several transcriptional repression mechanisms. (1) A repressor can mask the activation domain of a transcriptional activator by interacting with this activation domain and blocking its contact with the basal transcription machinery (Lodish et al., 2000). (2) A repressor can compete with a transcriptional activator to interact with the components of the transcription machinery to attenuate the transcription strength (Lodish et al., 2000). (3) A repressor can compete with an activator to bind to the same cis-acting element in the promoter (Lodish et al., 2000). (4) A repressor can block assembly of the transcription machinery, as in the cases of Srb10 (Hengartner et al., 1998) and Dr1/DrAp1-mediated repression (Song et al., 2002). (5) A repressor can promote the degradation of a transcriptional activator, as exemplified in COP1-mediated degradation of HY5 factor (Osterlund et al., 2000). (6) A negative regulator (such as 14-3-3) can shuttle a transcriptional activator, such as RSG, out of nuclei (Igarashi et al., 2001). Finally, a repressor can modify chromatin structure by, in the case of retinoblastoma protein, recruiting the Sin3 histone deacetylase complex to alter local chromatin structure, hence repressing transcription (Yang et al., 2001), or recruiting a histone methylase and a methyl-Lys binding protein HP1 to repress the cyclin E promoter through the methylation of histone 3 (Nielsen et al., 2001). Some repressors, such as Ssn6-Tup1 (Smith and Johnson, 2000), use a combination of some mechanisms described above to block transcription.
Binding of repressors to cis-acting elements could be involved in the first three repression mechanisms described above. Because OsWRKY71 (Fig. 7B) and its homolog ABF2 (Rushton et al., 1995) have been shown to bind to TTGAC(C/T) elements in α-amylase gene promoters, OsWRKY71 might repress GA responses via one of these three mechanisms. Our data indicate that OsWRKY71 functionally interferes with OsGAMYB on regulating α-amylase gene expression (Fig. 6). GAMYB and OsWRKY71 interact with different cis-acting elements on the α-amylase promoters, with the former binding to GARE (Gubler et al., 1995) and the latter to the TGAC cores within and upstream from O2S (Fig. 7, A and B). Therefore, the third mechanism is unlikely involved. Instead, OsWRKY71 might block GA responses by interacting with the activation domain of GAMYB (the first mechanism) or by competing with GAMYB to interact with components of the basal transcription machinery (the second mechanism). Of course, there are other possible modes of action for OsWRKY71. GAMYB and OsWRKY71 might compete for interaction with the same cotranscription factors. Also, OsWRKY71 might block the expression of α-amylase genes by enforcing the repression of KGM (Woodger et al., 2003) and HRT (Raventós et al., 1998) or interfering with binding of transcriptional activators to other cis-acting elements in α-amylase promoters. In the last regard, it is interesting to note that OsDOF3, which binds to the Pyr box in the promoter of the rice α-amylase gene, RAmy1A, physically interacts with OsGAMYB to enhance the transactivation of GAMYB (Washio, 2003). In the Amy32b and RAmy1A promoters, one Pyr box is located between O2S (W box) and GARE, and this element is essential for the response of these promoters to GA (Lanahan et al., 1992; Gómez-Cadenas et al., 2001a; Washio, 2003). Therefore, it is possible that OsWRKY71 interferes with the interaction of DOF and GAMYB.
It is interesting to note when OsWRKY71 was overexpressed, the extent of the decrease was much less for the OsGAMYB-induced promoter activity (Fig. 6) than that for the GA3-induced activity (Fig. 4). How to reconcile this difference? It has been shown that among the cis-acting elements essential for a high level of GA induction of the Amy32b promoter, GARE is the only indispensable element for HvGAMYB transactivation of the α-amylase promoter (Gómez-Cadenas et al., 2001a). It is possible that under normal conditions with no exogenous proteins or foreign DNA present in the cell, the rest of transcription factors (OsDOF3, OsMYBS1, OsMYBS2, etc.) binding to other cis-acting elements (Pyr and Amy, etc.) could function by stabilizing the GAMYB-GARE interaction. Using cobombardment of the GAMyb effector construct driven by a constitutive promoter probably led to the production of GAMYB protein in the cell to such a high level that no further stabilization of the DNA-protein interaction will be needed. Maybe due to the same reason, OsWRKY71 was less effective on repressing OsGAMYB-induced promoter activity than on repressing the GA3-induced activity. In supporting this, we have observed that increasing amount of OsWRKY71 can indeed completely block OsGAMYB-induced promoter activity (Supplemental Fig. 5).
It appears to be contradictory to show that the two TGAC cores within and upstream from the O2S element are essential for GA induction of Amy32b (Fig. 7D; Lanahan et al., 1992; Gómez-Cadenas et al., 2001a), yet these elements were bound by the OsWRKY71 transcriptional repressor (Fig. 7B). Similar paradoxical observation has been made for the Pyr box. Like O2S, the Pyr box is essential for high level of GA-induced expression of barley and rice α-amylases (Lanahan et al., 1992; Gómez-Cadenas et al., 2001a; Washio, 2003) and a barley cathepsin B-like protease (Mena et al., 2002). However, a DOF protein, namely BPBF, binds to the Pyr box (with a CTTT(A/T) core) and suppresses the induction of the cathepsin B-like protease promoter by GA or overexpression of HvGAMYB (Mena et al., 2002). The puzzle for the Pyr box has been partially solved by a recent study, which shows that another DOF, SAD, transactivates this protease promoter (Isabel-LaMoneda et al., 2003). Like BPBF (Diaz et al., 2002), the C terminus of the SAD protein physically interacts with HvGAMYB protein (Isabel-LaMoneda et al., 2003). In another case, three novel rice Myb genes (unlike GAMYB, each containing only one R repeat) regulate the expression of amylase gene in opposite way: OsMYBS1 and OsMYBS2 bind to the TATCCA (Amy Box) element and transactivate this TATCCA-containing promoter, whereas OsMYBS3 represses transcription of the same promoter under sugar starvation (Lu et al., 2002). Therefore, it is possible that each of the five cis-acting elements on the Amy32b promoter could be bound by at least a transactivator and a transrepressor, and the ratio of the activators and repressors controls the expression level of Amy32b. If this is correct, we should be able to identify OsWRKY or other genes that encode transactivators binding to the O2S element.
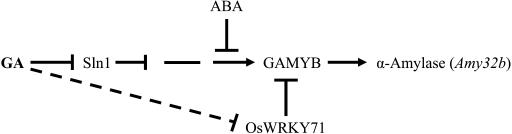
One of the most intriguing questions is how GA signal is perceived, resulting in induction or activation of positive signals and repression or degradation of negative regulators. A partial GA response pathway leading to expression of the α-amylase gene in aleurone cells has been proposed (Gómez-Cadenas et al., 2001b; Olszewski et al., 2002). In this work, we have shown that OsWRKY71 represses induction of the Amy32b α-amylase gene expression by OsGAMYB (Figs. 6B and 7D). Based on the data, the GA response pathway that mediates the expression of α-amylase genes, such as Amy32b in aleurone cells, is updated and shown in Figure 9. Perceiving of GA signal results in the degradation of the negative regulator SLN1 and subsequently activation of GAMYB and expression of the α-amylase gene. OsWRKY71 inhibits GAMYB transactivation of the Amy32b low-pI α-amylase gene, and this repressor is destabilized by GA (Fig. 8).
Figure 9.
Signaling components on the GA response pathway leading to expression of the Amy32b α-amylase gene in aleurone cells. SLN1 is a repressor, which is destabilized by GA treatment; GAMYB is a GA-inducible transcriptional activator. OsWRKY71 functions as a transcriptional repressor interacting with GAMYB to repress the GA response; GA down-regulates the expression of the OsWRKY71 gene at transcriptional or posttranscriptional level and destabilizes OsWRKY71 protein.
Two models have been proposed for the removal of repressor proteins by GA. One suggests that Arabidopsis SPY, which encodes an OGT, control the activities of RGA and GAI (Thornton et al., 1999; Sun, 2000). In the other model, it is proposed that a GA-controlled kinase phosphorylates GID2/SLY1, which then somehow leads to the phosphorylation of the DELLA protein (RGA/SLR1) and promotes the degradation of these proteins via ubiquitin-mediated proteolysis (Silverstone et al., 2001; Fu et al., 2002; Gubler et al., 2002; Itoh et al., 2002; McGinnis et al., 2003; Sasaki et al., 2003). We have shown that GA promotes the degradation of OsWRKY71 (Fig. 8). It will be very exciting to study whether the ubiquitin-mediated pathway is involved in the control of OsWRKY71 degradation.
It is generally believed that similar (if not identical) GA pathways are operating in different plant species. Indeed, a partial GA response pathway integrating GA signaling molecules from different plant species has been outlined in two elegant reviews published recently (Olszewski et al., 2002; Gomi and Matsuoka, 2003). We have shown that in barley aleurone cells, OsPKABA1 functions as effectively as TaPKABA1 on suppressing GA induction of Amy32b (Supplemental Fig. 3B). Likewise, OsGAMYB transactivates, as effectively as HvGAMYB, the expression of the Amy32b gene in barley aleurone layers (Gubler et al., 1997). GARE and Amy Box are highly conserved in the α-amylase promoters of barley, wheat, and rice (Huang et al., 1990a, 1990b; O'Neill et al., 1990; Itoh et al., 1995). Indeed, the wheat protease promoter has been shown to be responsible to GA in barley aleurone cells (Mena et al., 2002; Isabel-LaMoneda et al., 2003), and rice effecter genes are functional in barley aleurone cells (Gubler et al., 1997; Lu et al., 2002). Finally, just as in barley, oat, and wheat, W box (with a TGAC core) is present in the promoters of several rice α-amylase genes, such as OsAmy1A, OsAmy1B, OsAmy3A, OsAmy3D, and OsAmy3E (Z.-L. Zhang, Z. Xie, and Q.J. Shen, unpublished data). Therefore, it is likely that OsWRKY71 is also involved in regulation of these α-amylase genes in rice. Future experiments will be carried out with rice aleurone cells using the approach described recently (Sutoh and Yamauchi, 2003; Washio, 2003) to test this hypothesis. This work demonstrates an alternative approach to study rice gene functions in barley aleurone cells.
MATERIALS AND METHODS
Chemicals and Enzymes
T4 DNA ligase, DNA polymerase I large fragment (Klenow), Taq polymerase, and ImProm-II reverse transcriptase were obtained from Promega (Madison, WI). T7 DNA polymerase was purchased from New England Biolabs (Beverly, MA). Restriction enzymes were acquired from both Promega and New England Biolabs. Shrimp alkaline phosphatase was purchased from Roche Diagnostics (Mannheim, Germany). Luciferin was acquired from BD Pharmingen (San Diego). SYTO17 was purchased from Molecular Probes (Eugene, OR). KlenTaq LA DNA polymerase, AccuTaq LA DNA polymerase, and all other reagents including poly(dIdC), 4-methylumbelliferyl-β-d-glucuronide trihydrate, and 4-methylumbelliferone were obtained from Sigma (St. Louis).
Plant Materials
Rice (Oryza sativa L. subsp. japonica and indica) seeds were kindly provided by Dr. Kent McKenzie at the California Rice Experiment Station and Mr. Jack de Wit from DeWit Firm (Davis, CA). Barley (Hordeum vulgare cv Himalaya) seeds (1998 harvest) were purchased from Washington State University (Pullman, WA). The preparation and imbibition of the embryoless half-seeds were done as described (Shen et al., 1993).
Reverse Transcription and RT-PCR Analyses
Total RNA was isolated from rice aleurone cells with the LiCl precipitation method. After being imbibed for 2 d, rice half-seeds were continuously incubated in petri dishes containing 20 mm CaCl2, 20 mm sodium succinate without hormone (control) or with 1 μm GA3, 20 μm ABA, or 1 μm GA3 plus 20 μm ABA at 24°C for 4, 24, and 48 h, respectively. After incubation, 50 half-seeds were frozen in liquid nitrogen and homogenized to a fine powder in a mortar and pestle. The powder was added to 5 mL of RNA grinding buffer (0.1 m Tris, pH 9.0, 1% SDS, 10 mm EDTA, pH 8.0, and 50 mm β-mercaptoethanol). After centrifuging, the supernatant was extracted with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, v/v). Nucleic acids were precipitated with 1/9 volume of 3 m sodium acetate (pH 5.2) and 2.5 volume of absolute ethanol. The pellets were resuspended in diethyl pyrocarbonate-treated water and then precipitated with an equal volume of 4 m LiCl twice. After washing the RNA pellet with 70% (v/v) ethanol, the pellet was dissolved in water and stored at –80°C.
The first-strand cDNAs were synthesized using ImProm-II reverse transcriptase in a 50-μL reaction containing 2.5 μm oligo(dT) primers, 2.5 μm random heximer, and 2.5 μg of total RNA, according to the manufacturer's instructions. Five microliters of each reaction mixture was used as a template for PCR amplification in a 25-μL mixture containing 1.5 μm MgCl2, 200 μm dNTPs, 5% dimethyl sulfoxide, 2.5 units of Taq DNA polymerase, and 0.4 μm gene-specific primers (Supplemental Table I). The band intensities were quantified using the Multi-Analyst package from Bio-Rad (Hercules, CA).
Genomic DNA Isolation
Rice seeds were germinated on wet Whatman paper saturated with imbibing solution (20 mm CaCl2 and 20 mm sodium succinate) in dark at 26°C. Genomic DNA was isolated from 10-d-old seedlings. Briefly, sterile rice shoots were frozen in liquid nitrogen and ground into a powder. The frozen powder tissue was suspended in CTAB extraction buffer (55 mm cetyl-trimethylammonium bromide, 1.4 m NaCl, 100 mm Tris-HCl, pH 8.0, and 20 mm EDTA) plus 2% β-mercaptoethanol. The homogenate was incubated at 55°C for 30 to 45 min, cooled to room temperature, and extracted twice with an equal volume of chloroform. The DNA was precipitated with isopropanol and then redissolved in Tris-EDTA buffer plus RNase A (20 μg/mL). After incubation at 37°C for 1 h, the DNA was precipitated with ammonium acetate and ethanol and then dissolved in Tris-EDTA buffer.
Effector Construct Preparation
Plasmid pMBL022 (Lanahan et al., 1992), designated Amy32b-GUS in this article, was used as one of the reporter constructs. To facilitate oligonucleotide-directed mutagenesis (Kunkel et al., 1987), the Amy32b promoter-GUS-Amy32b terminator cassette from pMBL022 was subcloned into the HindIII and EcoRI sites of pBluescript KS (−) (Stratagene, La Jolla, CA). Single-stranded DNA was prepared from this construct for the mutation of the two TGAC cores in the Amy32b promoter by using primer P1 (Supplemental Table II), generating Amy32b (mTGACs)-GUS. The other two reporter constructs, HVA22-GUS and HVA1-GUS, were made by ligating a 49-bp HVA22 promoter sequence containing ABRC1 and a 68-bp HVA1 promoter sequence containing ABRC3 with the SmaI-digested MP64 progenitor, respectively (Shen et al., 1996). Plasmid pAHC18 (UBI-Luciferase), which contains the luciferase reporter gene driven by the constitutive maize ubiquitin promoter (Bruce et al., 1989), was used as an internal control construct (Lanahan et al., 1992; Shen et al., 1993) to normalize GUS activities of the reporter construct. For UBI-OsWRKY71 effector construct, the intron-containing coding region of OsWRKY71 was amplified by PCR with primers P2 and P3 (Supplemental Table II), with a BamHI site included to facilitate cloning. A silent mutation was included to eliminate the BamHI site in the OsWRKY71 gene by mutating the codon for the second amino acid from GAT to GAC. The PCR product was confirmed by several different restriction enzyme digestions and then was cloned into BamHI site of plasmid pAHC18 to replace the Luciferase gene. Then the OsWRKY71 sequence was further confirmed by sequencing. To prepare single-stranded DNA for oligonucleotide-directed mutagenesis, the f1 filamentous phage origin of replication was amplified from pBluescript SK(+) (Stratagene) by PCR with primers P4 and P5 (Supplemental Table II) and inserted into the HindIII site upstream of the UBI promoter. The resulting construct is named UBI-OsWRKY71. To facilitate the cloning of other effector genes, a new expression vector was prepared as follows: a linker containing AscI, PacI, EcoRV, NotI, and SacII restriction enzyme sites (annealed oligonucleotides P6 and P7; Supplemental Table II) was inserted into the BamHI site. In this expression vector, designated UBI-linker-NOS, the orientation of the f1 origin is opposite to that in UBI-OsWRKY71. The coding sequence of the OsGAMyb gene was amplified by RT-PCR with primers P8 and P9 (Supplemental Table II) and inserted into the AscI site of UBI-linker-NOS. The final effector construct was designated UBI-OsGAMyb.
Particle Bombardment and Transient Expression Assays
The detailed descriptions of transient expression procedure with the barley (Hordeum vulgare) aleurone system and the particle bombardment technique have been published before (Shen et al., 1993). Briefly, de-embryonated half-seeds of Himalaya barley were imbibed for 2.5 to 3 d, and then the pericarp and testa were removed. The DNA mixture (in 1:1 molar ratio) of a reporter construct and the internal control construct (UBI-Luciferase), along with or without an effector construct(s), was bombarded into barley embryoless half-seeds (four replicates per test construct). For each bombardment, eight prepared half-seeds were arranged in a small circle (about 1.8 cm in diameter) to maximize the bombarded surface area. After bombardment treatments, GUS assay and luciferase assay were done as published before (Shen et al., 1996).
Preparation of GFP Fusion Constructs and Confocal Microscopy
To make the GFP fusion constructs, mutagenesis was carried out with primer P10 to introduce a KpnI site (lowercase in the P10 sequence listed in Supplemental Table II) next to the AscI site in UBI-linker-NOS. The GFP gene was amplified from 35S-sGFP by PCR using primers P11 and P12 (Supplemental Table II). Then, the sGFP gene was inserted into the KpnI site in this modified vector to produce UBI-GFP. The coding sequence of the OsWRKY71 gene was amplified by RT-PCR with primers P13 and P14 (Supplemental Table II) and cloned into the AscI site in UBI-GFP to generate UBI-GFP:OsWRKY71.
Barley aleurone cells were bombarded with UBI-GFP and UBI-GFP:OsWRKY71 fusion constructs, respectively. After incubation at 24°C for 24 h, the aleurone layers were peeled from barley half-seeds and soaked in 5 μm SYTO17 solution. The stained samples were observed, and images of GFP fluorescent and SYTO17 staining were obtained simultaneously through Laser Scanning Microscope LSM 510 (Carl Zeiss, Jena, Germany) with 488-nm excitation and 505- to 530-nm emission wavelength for the green fluorescence and 633-nm excitation and 650-nm emission wavelength for the red fluorescence in separate channels. The acquired images were processed using Paint Shop Pro 7 (Jasc Software, Eden Prairie, MN).
For the analyses of GA effect on the stability of GFP:OsWRKY71 fusion protein, the aleurone layers were peeled from the bombarded barley half-seeds and treated with 100 μm of GA3 solution or water only for 12 h prior to GFP fluorescence observation.
Electrophoretic Mobility Shift Assay
The plasmid pGEX-KG, harboring the GST gene, was modified by inserting a linker (annealed oligonucleotides P15 and P16; Supplemental Table II), which contains an AscI into the XbaI and SalI sites of pGEX-KG. The OsWRKY71 coding sequence was then cloned into the AscI site of the modified pGEX-KG vector, generating GST:OsWRKY71. The GST:OsWRKY71fusion construct was then transformed in BL-21 (DE3) pLysS Escherichia coli. The GST fusion proteins were purified using glutathione-agarose beads (Sigma) following manufacturer's recommendations. In brief, overexpression of the fusion protein was induced by 1 mm isopropylthio-β-galactoside at 30°C in 2× YT medium. E. coli cells were lysed by sonication in a column buffer (20 mm Tris, 200 mm NaCl, 1 mm EDTA, 10 mm β-mercaptoethanol, 200 μm phenylmethylsulfonyl fluoride, 1 μg/mL of Leupeptin, 1 μg/mL of Pepstatin A, and 0.1% (v/v) Triton X-100). Following centrifugation (15,000g, 15 min, 4°C), the supernatants were mixed with glutathione-agarose beads, which were washed three times with 20 mL of ice-cold column buffer. Fusion proteins were eluted with 15 mm reduced glutathione/50 mm Tris-HCl (pH 8.0) solution. Protein concentrations were measured by using Bio-Rad protein assay kit. Purified proteins were stored in 10% (v/v) glycerol at −80°C.
The purified recombinant protein was used in electrophoretic mobility shift assays using a radiolabeled fragment of the Amy32b promoter that spans from position −167 to −111 and contains the O2S (W box) and the Pyr box (Gómez-Cadenas et al., 2001a). Synthetic oligonucleotides (P17 through P26; Supplemental Table II) were used for the Amy32b probe and competitors. The probe was labeled with α-32P-dATP by the Klenow fill-in reaction. Binding reactions (30 μL) containing 1.5 ng of probe, 1 μg of poly(dIdC), 10 mm Tris-HCl, pH 7.6, 50 mm KCl, 10% glycerol, and 0.5 μg of recombinant protein were incubated at room temperature for 30 min. Competitors were added in a 20- or 200-fold molar excess. All reaction mixtures were resolved by electrophoresis on a 5% polyacrylamide gel in 0.5× TBE (45 mm Tris, 45 mm boric acid, and 1 mm EDTA) buffer.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers BK005004–BK005080.
Supplementary Material
Acknowledgments
We thank people in the Shen Laboratory for their contributions to the improvement of this article. We also thank Dr. John Cushman at the University of Nevada, Reno, for the sGFP construct and Dr. Su-May Yu at Institute of Molecular Biology, Academic Sinica, Taiwan, for the rice Amy1A promoter.
This work was supported by the U.S. Department of Agriculture (grant no.02-35301-12066) and University of Nevada, Las Vegas, start-up funds (to Q.J.S.) and the National Science Foundation (grant no. IBN-9983126 to T.-H.D.H.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.034967.
References
- Bruce WB, Christensen AH, Klein T, Fromm M, Quail PH (1989) Photoregulation of a phytochrome gene promoter from oat transferred into rice by particle bombardment. Proc Natl Acad Sci USA 86: 9692–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cercós M, Gómez-Cadenas A, Ho THD (1999) Hormonal regulation of a cysteine proteinase gene, EPB-1, in barley aleurone layers: cis- and trans-acting elements involved in the co-ordinated gene expression regulated by gibberellins and abscisic acid. Plant J 19: 107–118 [DOI] [PubMed] [Google Scholar]
- Chandler PM, Marion-Poll A, Ellis M, Gubler F (2002) Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol 129: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Robertson M (1999) Gibberellin dose-response curves and the characterization of dwarf mutants of barley. Plant Physiol 120: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martinez M, Isabel-La Moneda I, Carbonero P (2002) The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J 29: 453–464 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Fridborg I, Kuusk S, Robertson M, Sundberg E (2001) The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and barley. Plant Physiol 127: 937–948 [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Ait-Ali T, Hynes LW, Ougham H, Peng J, Harberd NP (2002) Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14: 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y (1999) Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA 96: 7575–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Gómez-Cadenas A, Verhey SD, Holappa LD, Shen Q, Ho THD, Walker-Simmons MK (1999) An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc Natl Acad Sci USA 96: 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas A, Zentella R, Sutliff TDH, Ho THD (2001. a) Involvement of multiple cis-elements in the regulation of GA responsive promoters: definition of a new cis-element in the Amy32b gene promoter of barley (Hordeum vulgare). Physiol Plant 112: 211–216 [DOI] [PubMed] [Google Scholar]
- Gómez-Cadenas A, Zentella R, Walker-Simmons MK, Ho THD (2001. b) Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13: 667–679 [PMC free article] [PubMed] [Google Scholar]
- Gomi K, Matsuoka M (2003) Gibberellin signalling pathway. Curr Opin Plant Biol 6: 489–493 [DOI] [PubMed] [Google Scholar]
- Gubler F, Chandler PM, White RG, Llewellyn DJ, Jacobsen JV (2002) Gibberellin signaling in barley aleurone cells: control of SLN1 and GAMYB expression. Plant Physiol 129: 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Jacobsen JV (1992) Gibberellin-responsive elements in the promoter of a barley high-pI alpha-amylase gene. Plant Cell 4: 1435–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV (1995) Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7: 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Watts RJ, Kalla R, Matthews P, Keys M, Jacobsen JV (1997) Cloning of a rice cDNA encoding a transcription factor homologous to barley GAMyb. Plant Cell Physiol 38: 362–365 [DOI] [PubMed] [Google Scholar]
- Hanley BA, Schuler MA (1988) Plant intron sequences: evidence for distinct groups of introns. Nucleic Acids Res 16: 7159–7176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA (1998) Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell 2: 43–53 [DOI] [PubMed] [Google Scholar]
- Huang N, Koizumi N, Reinl S, Rodriguez RL (1990. a) Structural organization and differential expression of rice alpha-amylase genes. Nucleic Acids Res 18: 7007–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Sutliff TD, Litts JC, Rodriguez RL (1990. b) Classification and characterization of the rice alpha-amylase multigene family. Plant Mol Biol 14: 655–668 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13: 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel-LaMoneda I, Diaz I, Martinez M, Mena M, Carbonero P (2003) SAD: a new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. Plant J 33: 329–340 [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Yamaguchi J, Huang N, Rodriguez RL, Akazawa T, Shimamoto K (1995) Developmental and hormonal regulation of rice α-amylase (RAmy1A)-gusA fusion genes in transgenic rice seeds. Plant Physiol 107: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE (1996) SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA 93: 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HD, Smith SJ, Desikan R, Plakidou-Dymock S, Lovegrove A, Hooley R (1998) Heterotrimeric G proteins are implicated in gibberellin induction of alpha-amylase gene expression in wild oat aleurone. Plant Cell 10: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer EE, Litts JC, Rodriguez RL (1991) Differential expression of alpha-amylase genes in germinating rice and barley seeds. Plant Mol Biol 16: 797–805 [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol 154: 367–382 [DOI] [PubMed] [Google Scholar]
- Lanahan MB, Ho THD, Rogers SW, Rogers JC (1992) A gibberellin response complex in cereal alpha-amylase gene promoters. Plant Cell 4: 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (2000) Repressors interfere directly with transcription initiation in several ways. In S Tenney, ed, Molecular Cell Biology. W.H. Freeman and Company, New York, p 392
- Lovegrove A, Hooley R (2000) Gibberellin and abscisic acid signalling in aleurone. Trends Plant Sci 5: 102–110 [DOI] [PubMed] [Google Scholar]
- Lu CA, Ho THD, Ho SL, Yu SM (2002) Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. Plant Cell 14: 1963–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-Box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P (2002) A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol 130: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsunaga S, Tashiro T, Yamaguchi J (1994) Identification and characterization of gibberellin-insensitive mutants selected from among dwarf mutants of rice. Theor Appl Genet 87: 705–712 [DOI] [PubMed] [Google Scholar]
- Mundy J, Chua N-H (1988) Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J 7: 2279–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561–565 [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill SD, Kumagai MH, Majumdar A, Huang N, Sutliff TD, Rodriguez RL (1990) The alpha-amylase genes in Oryza sativa: characterization of cDNA clones and mRNA expression during seed germination. Mol Gen Genet 221: 235–244 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al (1999) 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Raventós D, Skriver K, Schlein M, Karnahl K, Rogers SW, Rogers JC, Mundy J (1998) HRT, a novel zinc finger, transcriptional repressor from barley. J Biol Chem 273: 23313–23320 [DOI] [PubMed] [Google Scholar]
- Ritchie S, Gilroy S (1998) Gibberellins: regulating genes and germination. New Phytol 140: 363–383 [DOI] [PubMed] [Google Scholar]
- Robertson M, Swain SM, Chandler PM, Olszewski NE (1998) Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell 10: 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JC, Lanahan MB, Rogers SW (1994) The cis-acting gibberellin response complex in high pI alpha-amylase gene promoters. Requirement of a coupling element for high-level transcription. Plant Physiol 105: 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JC, Rogers SW (1992) Definition and functional implications of gibberellin and abscisic acid cis-acting hormone response complexes. Plant Cell 4: 1443–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol Biol 29: 691–702 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, Matsuoka M (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Shen Q, Ho THD (1995) Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Uknes SJ, Ho THD (1993) Hormone response complex of a novel abscisic acid and cycloheximide inducible barley gene. J Biol Chem 268: 23652–23660 [PubMed] [Google Scholar]
- Shen Q, Zhang P, Ho THD (1996) Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8: 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PY, Martinez EC, Sun TP (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Sun T (2000) Gibberellins and the green revolution. Trends Plant Sci 5: 1–2 [DOI] [PubMed] [Google Scholar]
- Skriver K, Olsen FL, Rogers JC, Mundy J (1991) Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc Natl Acad Sci USA 88: 7266–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RL, Johnson AD (2000) Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci 25: 325–330 [DOI] [PubMed] [Google Scholar]
- Song W, Solimeo H, Rupert RA, Yadav NS, Zhu Q (2002) Functional dissection of a Rice Dr1/DrAp1 transcriptional repression complex. Plant Cell 14: 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, Cooney SE, McCourt P (1998) Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T (2000) Gibberellin signal transduction. Curr Opin Plant Biol 3: 374–380 [DOI] [PubMed] [Google Scholar]
- Sutliff TD, Lanahan MB, Ho THD (1993) Gibberellin treatment stimulates nuclear factor binding to the gibberellin response complex in a barley alpha-amylase promoter. Plant Cell 5: 1681–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutoh K, Yamauchi D (2003) Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J 34: 635–645 [DOI] [PubMed] [Google Scholar]
- Tanida I, Kim JK, Wu R (1994) Functional dissection of a rice high-pI alpha-amylase gene promoter. Mol Gen Genet 244: 127–134 [DOI] [PubMed] [Google Scholar]
- Thornton TM, Swain SM, Olszewski NE (1999) Gibberellin signal transduction presents ellipsis the SPY who O-GlcNAc'd me. Trends Plant Sci 4: 424–428 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M (2000) Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA 97: 11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washio K (2003) Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol 133: 850–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen CK, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmott RL, Rushton PJ, Hooley R, Lazarus CM (1998) DNase1 footprints suggest the involvement of at least three types of transcription factors in the regulation of alpha-Amy2/A by gibberellin. Plant Mol Biol 38: 817–825 [DOI] [PubMed] [Google Scholar]
- Woodger FJ, Gubler F, Pogson BJ, Jacobsen JV (2003) A Mak-like kinase is a repressor of GAMYB in barley aleurone. Plant J 33: 707–717 [DOI] [PubMed] [Google Scholar]
- Yang SH, Vickers E, Brehm A, Kouzarides T, Sharrocks AD (2001) Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol Cell Biol 21: 2802–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.