Abstract
Addition of fresh medium to stationary cells of Arabidopsis suspension culture induces increased phosphorylation of the S6 ribosomal protein and activation of its cognate kinase, AtS6k. Analysis of the activation response revealed that medium constituents required for S6 kinase activation were the phytohormones 1-naphthylacetic acid (auxin) and kinetin. Pretreatment of cells with anti-auxin or PI3-kinase drugs inhibited this response. Consistent with these findings, LY294002, a PI3-kinase inhibitor, efficiently suppressed phytohormone-induced S6 phosphorylation and translational up-regulation of ribosomal protein S6 and S18A mRNAs without affecting global translation. These data indicate that (1) activation of AtS6k is regulated by phytohormones, at least in part, via a lipid kinase-dependent pathway, that (2) the translational regulation of ribosomal proteins appears to be conserved throughout the plant and animal kingdom, and that (3) these events are hallmarks of a growth-related signal transduction pathway novel in plants.
The Arabidopsis genome contains two nearly identical genes, termed AtS6k1/Atpk1/ATPK6 and AtS6k2/Atpk2/ATPK19 (Zhang et al., 1994b; Mizoguchi et al., 1995; Turck et al., 1998), that encode proteins that share high sequence homology with the mammalian 40S ribosomal protein kinases S6K1 and S6K2 (Volarevic and Thomas, 2001). The plant kinase genes are arranged as a direct repeat within the Arabidopsis genome. Previously, we demonstrated that AtS6k2, ectopically expressed and isolated from mammalian cells, specifically phosphorylated ribosomal protein S6 derived from either plants or mammals (Turck et al., 1998). Moreover, exploiting the natural resistance of AtS6k2 to rapamycin, we established that transient expression of AtS6k2 substitutes for the loss of S6 kinase function in rapamycin-treated human 293 cells by rescuing S6 phosphorylation in vivo (Turck et al., 1998).
The above data indicated that activation of plant and mammalian S6 kinases could be regulated, at least in part, by similar cellular processes. However, our knowledge about the signaling pathway leading to the activation of plant S6 kinases, in contrast to the mammalian homologs, is very limited. The activity of mammalian S6K1 increases in response to mitogenic stimuli (Dufner and Thomas, 1999). In addition, it has been shown that the enzyme is tightly controlled by the nutritional status of cells, notably amino acid (Fox et al., 1998) and ATP levels (Dennis et al., 2001). Although the regulation of S6K2 has been studied to a lesser extent, it shares high sequence homology to S6K1 and appears to be coregulated (Shima et al., 1998). Activation of S6K1/S6K2 is brought about by multiple phosphorylations at two distinct sets of sites. The first set displays Ser/Thr-Pro motifs and is clustered in the carboxy terminus of mammalian S6K1/S6K2 (Ferrari et al., 1992). These sites, as well as the entire carboxy terminus of S6K1/K2, are lacking in the Arabidopsis homologs. The second set of key regulatory sites, which is perfectly conserved in the plant homologs, is flanked by large aromatic residues (Pearson et al., 1995) and is the target of S6K1 selective dephosphorylation and inactivation by the immunosuppressant rapamycin, as well as the fungal metabolite wortmannin (Pearson et al., 1995). The Ser/Thr-aromatic phosphorylation sites are positioned within different subdomains of S6K1/S6K2, Thr-229 in the activation loop and Thr-389 in the hydrophobic carboxy-terminal linker domain of S6K1 (Pearson et al., 1995). Despite their common characteristics, different upstream kinases are involved in the phosphorylation of Thr-229 and Thr-389. Phosphorylation occurs in a hierarchical order, with Thr-389 phosphorylation prerequisite to Thr-229 phosphorylation (Pearson et al., 1995). Several reports have shown that Thr-229 phosphorylation is mediated by the phosphatidyl-dependent kinase 1 (PDK1; Alessi et al., 1998; Pullen et al., 1998), and it is now generally accepted that the mammalian target of rapamycin (mTOR) is the relevant upstream kinase for Thr-389 (Thomas, 2002). The Arabidopsis genome encodes for AtPDK1 (Deak et al., 1999) and AtTOR (Menand et al., 2002), which appear to be orthologs of the mammalian protein kinases. However, their relationship to AtS6k has not yet been under investigation.
In mammals, S6 phosphorylation appears to be involved in the selective translational up-regulation of mRNAs, which encode components of the translational apparatus and are characterized by a tract of polypyrimidines at the transcriptional start site, termed a 5′TOP (Levy et al., 1991). The suppressive effects of rapamycin on 5′TOP translation can be rescued by coexpression of recombinant rapamycin-resistant S6K1 mutants, providing a causal link between S6K1 activation and translational regulation of 5′TOP mRNAs (Jefferies et al., 1997). However, other data suggest that mechanisms independent of S6 phosphorylation and S6 kinase activation also play a role in the regulation of 5′TOP mRNA translation (Stolovich et al., 2002).
The up-regulation of ribosomal biogenesis is considered to facilitate G1 progression during the cell cycle (Kozma and Thomas, 2002). Genetic studies in Drosophila melanogaster point to the fact that the regulation of cell division is a consequence of growth regulation mediated by the S6 kinase pathway. Deletion of the D. melanogaster S6 kinase homolog generated a dwarfed fly phenotype caused by a general reduction of cell size rather than cell number (Montagne et al., 1999).
Thus, S6Ks appear to play an important role in the growth and, consequently, mitogenic response, yet an analogous role in plants has not been described. To elucidate the role of AtS6k1 and AtS6k2, we have examined the activity of the kinases and the status of S6 phosphorylation in Arabidopsis suspension-cultured cells under conditions that induce cell growth. In particular, we studied the relation between transcript levels and total kinase activity. In addition, we characterized components of the growth medium that are required for kinase activation. The use of pharmacological inhibitors allowed us to gain insight into the signal transduction pathway mediating kinase activation in plant cells. Finally, we also determined whether the pathway has an impact on the biogenesis of the translational machinery.
RESULTS
Characterization of AtS6k1 and AtS6k2 Activities, Transcript Levels, and Translational Efficiencies in Stationary and Refed Suspension Culture Cells
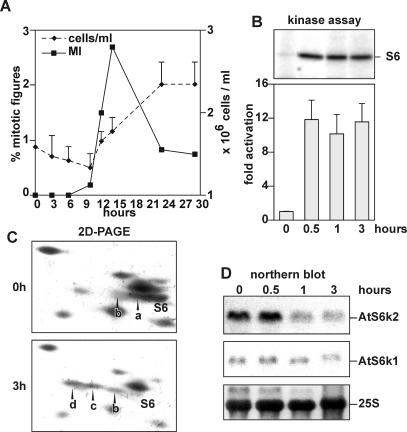
Mammalian S6K1 is rapidly activated following mitogenic stimulation of quiescent mammalian cells to proliferate (Dufner and Thomas, 1999). A large increase in S6 phosphorylation and a dramatic increase in the rate of protein synthesis parallel this increase (Dufner and Thomas, 1999). Similarly, addition of fresh medium to Arabidopsis stationary-phase cell cultures led to a sharp rise in the mitotic index after 12 h, resulting in an almost doubling of cell number after 22 h (Fig. 1A). Under such conditions, S6 kinase activity cannot be detected in a direct phosphorylation assay in plant extracts using ribosomal protein S6 as substrate (data not shown). However, after immunoprecipitation with antibodies against a common portion of AtS6k1 and AtS6k2, S6k activity was detected and increased approximately 10-fold as early as 30 min after addition of fresh medium (Fig. 1B). The antibodies used for immunoprecipitation do not cross-react with mammalian S6K1 (Turck et al., 1998), which is more related to AtS6k1 and AtS6k2 than any Arabidopsis kinase. In the following, we will refer to the activity in the precipitate as AtS6k activity. AtS6k activation was paralleled by a sharp increase in S6 phosphorylation, as measured by the appearance of the more highly phosphorylated S6 derivatives, S6c and d (Turck et al., 1998), resolved by two-dimensional PAGE (Fig. 1C). Endogenous kinase levels present in Arabidopsis cell extracts are below the detection limit by western-blot analysis before (Turck et al., 1998) and after stimulation (data not shown). However, previous reports highlighted transcriptional regulation of AtS6k1 and 2 (Mizoguchi et al., 1995), therefore we assessed whether transcript accumulation of these genes can be correlated with the detected changes in phosphorylation of S6 protein and kinase activity. Interestingly, despite the increase in kinase activity and S6 phosphorylation, the level of AtS6k2 transcripts rapidly decreased under growth-stimulating conditions, whereas AtS6k1 transcript levels remained constant (Fig. 1D).
Figure 1.
A, Effects of fresh medium supply on cell-cycle progression. An equal volume of fresh MSMO medium was added to stationary-phase Arabidopsis suspension cell cultures. Over a period of 24 h, aliquots were sampled and the mitotic index (left axis, black squares) and the average cell number (right axis, black diamonds) were determined. B, Effects of fresh medium supply on S6 kinase activity. AtS6k was immunoprecipitated from stationary-phase Arabidopsis suspension culture cells (0 h) or after refeeding of the cells with an equal volume of fresh MSMO medium for the indicated time. Kinase activity was measured using rat 40S ribosomes as substrate. The top shows the autoradiograph of a representative assay gel. The bar diagram shows average values of a duplicated assay quantified by PhosphorImager. Error bars represent average derivation of duplicates. C, S6 phosphorylation is increased after addition of fresh medium. Ribosomal proteins were extracted from stationary-phase Arabidopsis suspension cell cultures (0 h) and after the addition of fresh culture medium (3 h). The phosphorylation state of ribosomal protein S6 was analyzed by two-dimensional gel electrophoresis of total ribosomal proteins. Proteins were visualized by Coomassie Blue staining. The label S6 is found underneath the nonphosphorylated S6 signal, whereas a, b, c, and d represent higher phosphorylated forms of S6. The experiment was performed twice with similar results. D, Steady-state mRNA levels of AtS6k1 and AtS6k2 drop rapidly after induction of kinase activity. Total mRNA from parallel samples as in B were analyzed by northern blot for expression for AtS6k1 (middle) and AtS6k2 (top). Equal loading of samples was verified by methylene blue staining of the blot prior to hybridization (bottom). The experiment was performed three times with similar results.
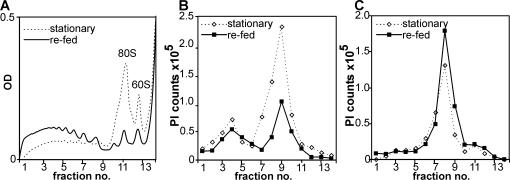
To test the translational efficiencies of AtS6k1 and AtS6k2 mRNAs, we analyzed polysome profiles for their distribution. In stationary-phase cultures, ribosomes accumulate as monosomes, indicating a down-regulation of overall cellular translation (Fig. 2A, dashed line). After stimulation with fresh medium, monosomes nearly disappear from the profiles since most ribosomes are now engaged in the translation of cellular mRNAs (Fig. 2A, solid line). Translation efficiency of AtS6k2 mRNA is increased during refeeding, but significant translation occurs already in arrested cells. Indeed, since AtS6k2 mRNA levels are substantially higher in arrested cells, more transcript accumulates in polysomes of such cells as compared to refed cells (Fig. 2B). Moreover, polysomal distribution of AtS6k1 indicates that AtS6k1 contributes little to the cellular pool of AtS6ks in the Arabidopsis suspension cell line used in our experiment since its mRNA is poorly translated in all treatments (Fig. 2C).
Figure 2.
Polysomal distribution of AtS6k1 and AtS6k2 mRNA. Polysomes were fractionated on 17% to 58% hyperbolic Suc gradients in extracts prepared from stationary-phase cells or 2 h after addition of fresh growth medium. The global distribution of polysomes was assessed by spectrophotometric detection of nucleic acids during fractionation (A) for stationary-phase (dashed line) and refed (solid line) cells. RNA was extracted from 14 recovered fractions and analyzed on northern blots for the expression of AtS6k2 (B) and AtS6k1 (C) transcripts using gene-specific antisense RNA probes. Plotted are arbitrary units of northern-blot signals as quantified by PhosphorImager scanning. The experiment was performed twice with similar results.
Thus, although the Arabidopsis genome encodes for two highly related kinases, AtS6k1 may be translationally repressed in the suspension-cultured cells used in our experiment. By contrast, AtS6k2 is expressed both in arrested and refed cells at a comparable level. Similar to mammalian cells, it seems that increased S6 phosphorylation is brought about by AtS6k activation rather than higher amount of these kinases when Arabidopsis stationary-phase cells are stimulated to proliferate. However, only in plants is the activation associated with decreasing levels of AtS6k2 transcripts.
Stationary Phase and AtS6k Activation
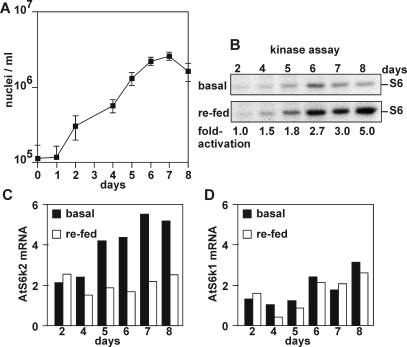
Cold, salt, and drought stress were reported to lead to increased AtS6k2 mRNA levels (Mizoguchi et al., 1995). The entry into stationary phase could also be viewed as a stress condition for cultured plant cells, therefore we monitored AtS6k activity and mRNA accumulation during an 8-d growth cycle of Arabidopsis cultures. Divisional growth was monitored by analysis of nuclei per volume (Fig. 3A). Exponential growth can be detected between day 1 and day 5 after subculture, whereas from day 5 the growth rate declines. Kinase activity was assessed every day, in the basal state as well as 1 h following stimulation with fresh medium (Fig. 3B). Under these conditions, basal kinase activity began to increase at day 5 as cells entered the stationary phase (Fig. 3B, top). Consistent with this finding, the levels of both AtS6k1 and AtS6k2 transcripts also increased (Fig. 3, C and D, black bars). Thus, as cells reach stationary phase, there is an increase in AtS6k1 and AtS6k2 mRNA correlated with an increase in basal kinase activity. Interestingly, the accumulation of AtS6k2 mRNA preceded accumulation of AtS6k1 mRNA by 1 d (Fig. 3, C and D, black bars), which indicates a lower threshold for response to growth phase changes. Significant kinase activation induced by fresh medium could not be observed before the cultures had reached the stationary phase (Fig. 3B, compare top and bottom). Consistent with the decrease in steady-state AtS6k2 mRNA levels, which follows stimulation of AtS6k activity by fresh medium (Fig. 1D), addition of fresh medium at any point during the approach to stationary phase led to an immediate drop in AtS6k2 transcript levels (Fig. 3C, compare black and white bars). In contrast to AtS6k2 transcripts, AtS6k1 mRNA expression was not affected by the addition of fresh medium within the chosen time window (Fig. 3D, compare black and white bars). This is consistent with the observation that the expression of this gene is regulated with kinetics different from that of AtS6k2 (Fig. 3, C and D, compare black bars).
Figure 3.
Influence of growth phase on AtS6k activation and expression. A, Growth curve of Arabidopsis suspension culture. Aliquots of suspension-cultured cells were sampled every day over a growth period of 8 d and analyzed for nuclei number. Data represent averages of two parallel samples. Error bars represent se as calculated from four independent counts per sample. B, AtS6k activity during the culture cycle. Immunoprecipitated AtS6k was assayed as described in samples either directly taken at the indicated day (basal, top) or after addition of an equal volume of MSMO medium for 1 h (refed, bottom). Autoradiograms show lanes from a representative assay. The intensity of the signals was quantified by PhosphorImager and fold stimulation per day after re-feeding is indicated in numbers below the autoradiographs. C, AtS6k2 expression during the culture cycle. Total RNA was prepared from parallel samples as in B and analyzed by northern blotting. The signal intensity was quantified by PhosphorImager and plotted in a bar diagram. The signals derived from unstimulated cells are shown with black bars, and signals from samples taken 1 h after addition of fresh medium are shown in white bars. D, AtS6k1 expression during the culture cycle. Parallel blots as for C were hybridized with an AtS6k1-specific antisense RNA probe. The experiment was performed twice with similar results.
Phytohormones Are Required for AtS6k Activation
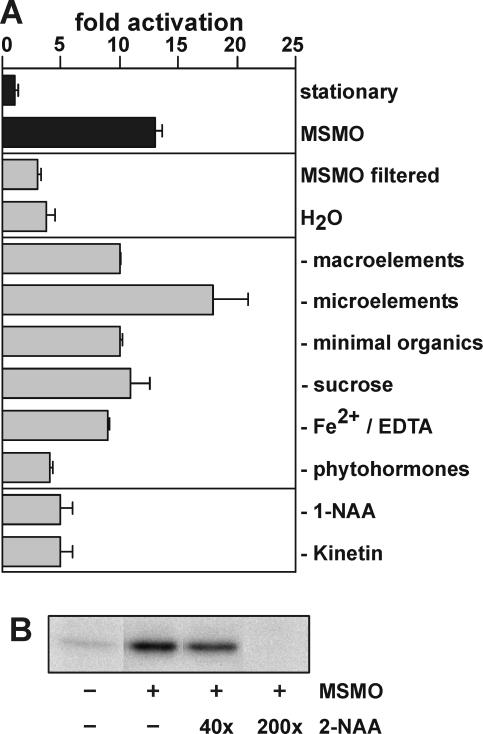
The results above suggest that, as cells approach the stationary phase, they increase their capacity to respond to fresh medium in terms of AtS6k activation and S6 phosphorylation. However, this response may be distinct from a growth response and may instead reflect the dilution of an inhibitory factor that accumulates in the culture medium. To address this issue, stationary cell cultures were either diluted in an equal volume of filtered medium from parallel cultures grown under identical conditions or water adjusted to the pH of stationary cell cultures. In both instances, there was little effect on AtS6k kinase activity as compared to that observed with complete medium (Fig. 4A). These findings indicated that one or more components in the medium, required for inducing AtS6k activation and S6 phosphorylation, had been depleted in stationary-phase cultures. To identify these components, one of each of the six defined constituents, (1) macroelements, (2) microelements, (3) minimal organics, (4) Suc as the carbon source, (5) Fe2+/EDTA, and (6) phytohormones (1-naphthylacetic acid [1-NAA] and kinetin), was omitted from the growth medium. The absence of each constituent from the medium was then tested for its effect on AtS6k activation. The data demonstrate that full activation of the kinase strongly depends on the presence of phytohormones, whereas no significant decrease in kinase activity could be detected between samples stimulated with medium lacking one of the other constituents of complete medium (Fig. 4A). To assess the role of 1-NAA and kinetin, stationary cultures were stimulated with fresh medium lacking one of the two hormones. The results show that the absence of either hormone prevented AtS6k activation (Fig. 4A). Consistent with this finding, 2-NAA, which has been reported to be a competitive antagonist of auxin signaling (Ray, 1977), inhibits AtS6k activation in a dose-dependent manner (Fig. 4B). However, addition of either phytohormone alone or in combination did not lead to comparable activation of the kinase (data not shown). Taken together, the results suggest the extent of AtS6k activation to be closely but not solely regulated by phytohormones.
Figure 4.
Influence of medium composition on the activation of AtS6k. A, Immunoprecipitated AtS6k activity was assayed as described in stationary-phase Arabidopsis suspension culture cells (basal), after addition of an equal volume of fresh MSMO medium (MSMO), after addition of medium filtered from a parallel culture flask (MSMO filtered), or after dilution of the cells with an equal volume of distilled water (H2O). Next, MSMO medium that lacked either macroelements, microelements, vitamins, Fe2+/EDTA, Suc, phytohormones (1-NAA and kinetin), or either 1-NAA or kinetin was added. The bar diagram represents average activity of representative duplicate assays as determined by PhosphorImager quantification. The experiment was performed three times with similar results. B, Antagonistic effect of 2-NAA on AtS6k activation. Stationary-phase suspension culture cells (−) were preincubated for 15 min with indicated ratios of the compound 2-NAA to 1-NAA before addition of an equal volume fresh MSMO medium (+). Immunoprecipitated AtS6k activity was assayed and quantified as described above. The experiment was performed two times with similar results.
Effects of Putative Upstream Inhibitors on AtS6k Activity
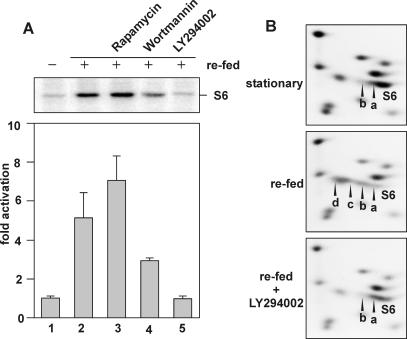
The elucidation of the mitogen-induced mammalian S6K1 signaling pathway has largely relied on the use of specific inhibitors to define individual upstream components linking it to the activated receptor (Dufner and Thomas, 1999). We have shown previously that AtS6k2 ectopically expressed in human 293 cells is resistant to rapamycin but is partially sensitive to wortmannin (Turck et al., 1998). As observed in transient transfection studies with AtS6k2 in mammalian cells, no decrease of AtS6k activation was observed when rapamycin was added together with fresh medium to Arabidopsis cells (Fig. 5A). Indeed, a small activation was detected in the presence of rapamycin, similar to that found when rapamycin-resistant forms of mammalian S6K1 were expressed in the presence of the macrolide (Dennis et al., 1996). In contrast to rapamycin, wortmannin attenuated AtS6k activation (Fig. 5A). Wortmannin is thought to block mitogen-induced mammalian S6K1 activation through inhibition of the class I PI3-kinase family members, which are hypothesized to be upstream elements of the S6K1 activation pathway (Dufner and Thomas, 1999). To date, no homologs of this class of PI3-kinase have been identified in plants (Mueller-Roeber and Pical, 2002), suggesting that wortmannin could exert its effect through another inhibitory target. To further examine this possibility, we tested a chemically unrelated mammalian class I PI3-kinase inhibitor, LY294002 (Vlahost et al., 1994). Addition of LY294002 to fresh medium completely abolished the activation of AtS6k at a concentration of 50 μm (Fig. 5A), consistent with results in mammalian cells (Vlahost et al., 1994). Addition of LY294002 also inhibited the increased phosphorylation of S6 induced by placing stationary cultures in fresh medium (Fig. 5B). Taken together, the results argue that AtS6k activation and S6 phosphorylation are mediated through a signal transduction cascade that is triggered by phytohormones and that an as yet unidentified plant lipid kinase may be a critical component of this cascade.
Figure 5.
A, Effects of PI3-kinase inhibitors on AtS6k activation. Stationary-phase suspension culture cells (day 8 of culture) were supplied with an equal volume of fresh MSMO medium, and AtS6k activity was measured prior to stimulation (lane 1) or after treating cells simultaneously with DMSO (lane 2), 20 nm rapamycin (lane3), 200 nm wortmannin (lane4), and 50 μm LY294002 (lane 5). Data were quantified and plotted as described. The experiment was performed three times with similar results. B, Effect of LY294002 treatment on S6 phosphorylation. Ribosomes were analyzed by two-dimensional gel electrophoresis and Coomassie Blue staining of ribosomal proteins from stationary-phase cultures (stationary) or after 2 h of addition of an equal volume of fresh growth medium in the absence (refed) or presence of 50 μm LY294002 (refed + LY294002). The experiment was performed twice with similar results.
Polysome Distribution of Individual mRNA Transcripts
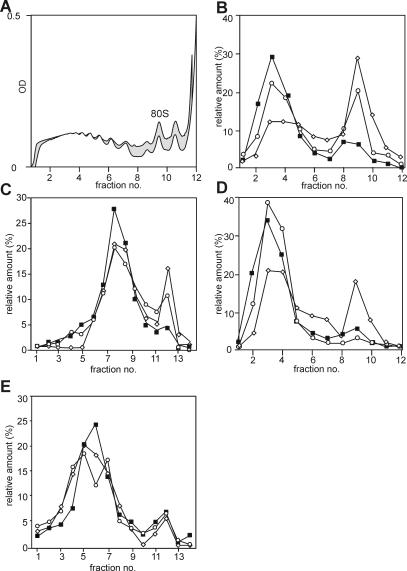
In mammalian cells, activation of S6K1 has been shown to correlate to the translational up-regulation of a family of mRNAs encoding for components of the protein synthetic apparatus, most notably ribosomal proteins (Jefferies et al., 1997). In general, a polypyrimidine tract at their transcriptional start site, which invariably begins with a cytosine, characterizes these transcripts. It has been reported that at least one plant ribosomal protein transcript contains a polypyrimidine tract (Gao et al., 1994), but a comparative analysis of ribosomal protein encoding sequences argues against their general presence in plants (Barakat et al., 2001). To test whether the translation of ribosomal protein transcripts in plants is up-regulated in parallel with AtS6k activation and S6 phosphorylation and whether the same signaling pathway regulates this, cells were stimulated with fresh medium in the absence and presence of LY294002. The distribution of several transcripts was followed on polysome profiles. As mentioned above, stimulation of stationary cells with fresh medium induces a large shift of monosomes into polysomes (Fig. 2A). This general shift of monosomes into polysomes is only slightly affected by the presence of the compound LY294002 (Fig. 6A), indicating that the compound is not a general inhibitor of translation and not toxic to the cells. Typical of mRNAs encoding ribosomal proteins (Meyuhas et al., 1996), a significant proportion of S6 transcripts reside within the disome peak of the Suc gradient in stationary cells (Fig. 6B, white diamonds). After stimulation of cells with fresh medium, ribosomal protein encoding transcripts participate in the general shift of monosomes into polysomes (Fig. 6B, black squares). However, the translational up-regulation of S6 transcripts is partially inhibited in the presence of LY294002 (Fig. 6B, white circles). This polysomal partitioning is not restricted to S6 mRNAs, as the mRNA encoding ribosomal protein S18A follows the same pattern (Fig. 6C). By contrast, the regulation of other transcripts is not affected by the compound: Arabidopsis small GTPase Ran1 mRNA shifts to larger polysomes after addition of fresh medium either in the presence or absence of LY294002 (Fig. 6D). By contrast, the translation of the Arabidopsis ubiquitin gene is not affected by either treatment (Fig. 6E). Similarly, no significant differences in the relative polysomal distribution of mRNAs encoding AtS6k2 and AtS6k1 can be observed after LY24002 treatment, indicating that no autoregulatory feedback on S6 kinase expression acts at the level of translational regulation (data not shown).
Figure 6.
Polysome distribution of S6 mRNA transcripts. Polysome profiles were analyzed on 17% to 58% hyperbolic Suc gradients in extracts prepared from stationary-phase cells or 2 h after addition of fresh growth medium in the absence or presence of 50 μm LY294002. A, The global distribution of polysomes was assessed by spectrophotometric detection of nucleic acids during fractionation and is shown for stimulated cells in absence (white shading) and presence of (gray shading) LY249002. B–E, RNA was extracted from 12 or 14 recovered fractions as indicated and analyzed on northern blots for the expression of AtS6 (B), AtS18A (C), AtRan (D), and AtUbiquitin4 (E). Polysomes from stationary-phase cells are labeled with white diamonds, from refed cells with black squares, and from cells that were refed in the presence of LY294002 with white circles. The intensities of detected northern-blot signals were quantified by PhosphorImager analysis. The total amount of detected signal per transcript was set to 100% for each treatment. Plotted are relative distributions of mRNAs on polysome profiles. The experiment was performed twice with similar results.
Thus, the results are consistent with the hypothesis that (1) translational up-regulation of ribosomal proteins resides on a common pathway with AtS6k activation and S6 phosphorylation and that (2) a LY294002 sensitive lipid kinase may be involved as upstream component of the translational pathway in higher plants.
DISCUSSION
Earlier studies have indicated that the S6 phosphorylation pathway is regulated by increased temperature in plants, as such treatment abolishes S6 phosphorylation in tomato (Lycopersicon esculentum; Scharf and Nover, 1982) and Arabidopsis cell (Turck et al., 1998) cultures. In addition, S6 dephosphorylation parallels AtS6k inactivation at elevated temperatures in Arabidopsis cells (Turck et al., 1998). As AtS6k1 and AtS6k2 transcripts are strongly up-regulated under cold stress conditions in Arabidopsis seedlings (Mizoguchi et al., 1995), it was proposed that the enzymes would play a role in cold stress responses, presumably by increasing the level of S6 phosphorylation. However, at least in the Arabidopsis suspension-cultured cells used in our studies, cold stress has no detectable effect on S6 phosphorylation and kinase activity (data not shown). By contrast, we detected an increased S6 phosphorylation and higher AtS6k activity after refeeding of Arabidopsis stationary cells with fresh medium. Thus, our data are more consistent with the model in which S6 phosphorylation functions in metazoan growth control (Kozma and Thomas, 2002).
Activation of mammalian S6K1 and S6K2 is highest when cells are stimulated to pass a G0-to-G1 transition of the cell cycle (Kozma and Thomas, 2002). Accordingly, we found that full AtS6k activation had two requirements: first, the accumulation of cells in a division arrested stage and second, the addition of growth-promoting conditions, including fresh nutrients and phytohormones (Fig. 3). Unlike the situation in animals, AtS6 K transcript and, presumably, protein levels are up-regulated as cells approach the stationary phase (Figs. 2 and 3), but the kinase is maintained in a quiescent state and S6 phosphorylation is low (Figs. 1 and 3). When the appropriate growth signals are applied, the kinase is activated but transcript levels return to those in growing cells (Figs. 1 and 3). Structurally, AtS6ks appear as simplified versions of the highly complex mammalian S6Ks. In fact, the plant kinases are structural equivalents of S6K1 double truncations (S6K1ΔNΔC; Pearson et al., 1995). S6K1ΔNΔC lacks the acidic amino-terminal as well as the autoinhibitory carboxy-terminal domain. Both S6K1ΔNΔC and AtS6k2 showed higher basal levels as S6K1 if expressed in mammalian cells (Turck et al., 1998). In our experiments in suspension cells, AtS6k activity varied within a window of a 10-fold stimulation of activated versus basal levels, which is by far less than observed for S6K1/S6K2 (Shima et al., 1998) but corresponds to the stimulation shown previously in mammalian cells (Turck et al., 1998).
The identity between AtS6k1 and AtS6k2 is approximately 87%, strongly implying that AtS6k1 is a second S6 kinase and that both genes arose through gene duplication. By contrast, baculovirus-expressed AtS6k1 has previously been reported not to phosphorylate S6 protein but other ribosomal substrates in vitro (Zhang et al., 1994a). AtS6k1 transcript levels show a delayed up-regulation compared to AtS6k2 levels during the approach of the stationary phase (Fig. 3) and a slower down-regulation during refeeding of stationary cells with fresh medium (Figs. 1 and 3). However, AtS6k1 transcripts are poorly translated in the cultured cells in all treatments, therefore these cells may not be the suitable material to study the role of AtS6k1.
Several lines of evidence point to the fact that AtS6k activity in plants is modulated by changes in the phosphorylation state of specific residues mediated by upstream signaling molecules. Previously, we showed that AtS6k2 expressed in mammalian cells does respond to serum stimulation in a similar fashion as the mammalian S6 kinase and that the increase in activity is dependent on the phosphorylation state of the molecule (Turck et al., 1998). Two key phosphorylation sites of the mammalian kinase, corresponding to S6K1 T229 and T389, are conserved between AtS6k and metazoan S6Ks. In mammals, PDK1 (Alessi et al., 1998) and mTOR (Burnett et al., 1998) were identified as the upstream kinases responsible for the phosphorylation of T229 and T389, respectively. The Arabidopsis genome encodes a homolog of PDK1 with a pleckstrin homology domain that binds several different phospholipids besides PIP3 (Deak et al., 1999). The plant kinase has been shown to phosphorylate and activate human PKB in a phospholipid-dependent manner (Deak et al., 1999). However, plants do not contain a homolog of PKB. mTOR, which is responsible for T389 phosphorylation in S6K1, has a putative ortholog in the Arabidopsis genome. Menand et al. (2002) reported that homozygous attor mutant embryos are aborted in the dermatogen stage, although cells undergo normal divisions. The authors propose that a defect in the transition of the endosperm from a merely endoreplicative to a growth stage is responsible for embryo arrest. This phenotype resembles that of growth-arrested D. melanogaster larvae with defects in protein synthesis, thus support a hypothesis in which AtTOR controls protein synthesis, possibly via regulation of AtS6ks.
Wortmannin is a reasonably specific class I PI3-kinase inhibitor in mammalian cells (Vanhaesebroeck et al., 1997), whereas LY294002 represents a second specific but structurally unrelated inhibitor of these kinases (Vlahost et al., 1994). However, both compounds have been shown to directly inhibit the protein kinase activity of mTOR at similar concentrations, as they have been shown to be specific for PI3-kinases (Brunn et al., 1996). Based on sequence similarity, plants do not contain class I PI3-kinase homologs (Mueller-Roeber and Pical, 2002). Although it cannot be excluded that other plant lipid kinases are sensitive to both wortmannin and LY294002, our data suggest that AtTOR is inhibited by both compounds and is involved in a signaling pathway upstream of AtS6k (Fig. 5).
The data presented here demonstrate that the phytohormones 1-NAA and kinetin play a determining role in the downstream signaling events, which mediate S6 kinase activation in plants (Fig. 4A). The importance of 1-NAA is further strengthened by the finding that the competitive antagonist 2-NAA (Ray, 1977) reduces kinase activity even below basal levels (Fig. 4B). At these concentrations, 2-NAA probably displaces residual auxin in the stationary-phase culture from active receptor sites, which could be responsible for the maintenance of basal kinase activity level.
Hormonal regulation does not appear to be confined to Arabidopsis suspension culture cells, as treatment of detached pumpkin (Cucurbita pepo) cotyledons with 6-benzylaminopurine, which induces rapid polysome formation, also leads to increased phosphorylation of a ribosomal protein of equivalent Mr to S6 (Yakovleva and Kulaeva, 1987). Indeed, abscisic acid, which induces polysome disassembly in the same protocol, inhibits the cytokinin-induced phosphorylation of the same protein (Yakovleva and Kulaeva, 1987). More recently, regulation of S6 phosphorylation has been described for intact plant tissues, such as the increase in ribosomal S6 protein phosphorylation in maize (Zea mays) seedlings by ZmIGF, a protein purified from the same source that cross-reacts with antibodies against bovine insulin (Garcia Flores et al., 2001). In addition, oxygen deprivation causes the loss of highly phosphorylated forms of ribosomal protein S6 in maize root tips, and the regeneration of these forms at stress recovery is greatly inhibited by the parallel addition of LY249002 in the recovery media (Williams et al., 2003). Obviously, these findings open the prospect of a new hormone-dependent signaling pathway in plants, which is related to the mammalian pathway.
In addition to the LY249002 sensitivity of S6 phosphorylation, plants and animals seem to share the effect that this pathway has on translation (Jefferies and Thomas, 1996; Meyuhas et al., 1996). This observation is supported by the finding that translation of S6 and S18 transcripts is up-regulated following supplementation of fresh growth media and that this up-regulation is specifically inhibited in the presence of LY294002 (Fig. 6, B and C). In contrast to mammalian cells, ribosomal protein transcripts in Arabidopsis do not generally begin with a polypyrimidine tract, which is also the case for protein S18A and both copies of ribosomal protein S6 detected in our study (Barakat et al., 2001). This is similar to D. melanogaster, in which ribosomal protein transcripts are also under translational control, yet many do not contain a 5′TOP (Meyuhas et al., 1996). It will be interesting to identify the structural requirements of LY249002-dependent translational regulation in plants.
MATERIALS AND METHODS
Maintenance of Suspension Cultures, Refeeding, and Cold and Chemical Treatments
Arabidopsis suspension cells were maintained as described (Turck et al., 1998). For refeeding experiments, 30 mL of a total of 60 mL of cell suspension was removed from a culture flask and replaced by an equal volume of fresh growth medium. Incomplete Murashige and Skoog Minimum Organics (MSMO) medium was prepared on the basis of commonly used Murashige and Skoog stock solutions by omitting one of the following categories. (1) Macroelements: 1.9 g/L of KNO3, 1.65 g/L of NH4NO3, 0.37 g/L of MgSO4·7H2O, 0.44 g/L of CaCl2·2H2O, and 0.17 g/L of KH2PO4; (2) microelements: 62 mg/mL of H3BO3, 223 mg/L of MnSO4·4H2O, 86 mg/L of ZnS04·7H2O, 2.5 mg/mL of Na2MoO4, 0.25 mg/L of CuSO4·5H2O, and 0.25 mg/L of CoCl2·6H2O; (3) minimal organics: 100 mg/L of myoinositol and 0.4 mg/L of thiamine hydrochloride; (4) 3% Suc; (5) Fe2+/EDTA: 37.2 mg/L of EDTA and 27.8 mg/L of FeSO4·7H2O; and (6) phytohormones: 0.5 mg/L of 1-NAA and 0.05 mg/L of kinetin. Rapamycin stock solution (2 mm in ethanol) was diluted to 1:100 in MSMO medium and added at the moment of refeeding to a concentration of 20 nm. Wortmannin (1 mm stock in dimethyl sulfoxide [DMSO]) and LY294002 (75 mm in DMSO) or the corresponding amount of carrier was added at the moment of refeeding for the indicated time to a final concentration of 200 nm and 50 μm, respectively. After treatments for the indicated time, cells were harvested by filtration, flash-frozen in liquid nitrogen, and stored at −80°C until use.
Determination of Cell Number and Mitotic Index
For cell density measurements, duplicate suspension aliquots (0.25 mL) were removed from the culture flasks at the times indicated. Cell walls were digested by addition of 0.25 mL of digestion buffer (2% cellulase, 10% pectinase, 0.6 m mannitol, 5 mm CaCl2, and 8 mm PIPES, pH 6.0) for 3 h at 37°C, and nuclei were counted in a hemocytometer. The mitotic index was determined after acid hydrolysis of cell aliquots (0.2 mL) in 2 mL of cold fixative (ethanol:acetic acid, 3:1) and refrigeration overnight. Cells were then washed once with ethanol and three times with 0.01 m citric acid-sodium citrate buffer including 10 mm EDTA. Nuclei were stained by adding 1 μg/mL of 4′,6-diamino-phenylindole, incubated for 15 min, and washed once with buffer. After squeezing and fixed mounting, mitotic figures of approximately 1,000 nuclei were counted per time point.
Extract Preparation and Kinase Assay
All steps were carried out at +4°C. Frozen cells (200 mg) were ground to a fine powder in liquid nitrogen and suspended in 1 mL of ice-cold plant cell extraction buffer (50 mm HEPES, pH 7.6, 50 mm pyrophosphate, 5 mm EDTA, 15 mm EGTA, 1 mm benzamidine, 25 mm NaF, 1 mm sodium molybdate, 1.5% polyvinylpolypyrrolidone [PVPP], 200 mm mannitol, 2 mm dithiothreitol, and 0.2 mm phenylmethylsulfonyl fluoride). After thawing, the extracts were cleared by two successive centrifugations for 10 min in an Eppendorf centrifuge at maximal speed. For immunoprecipitation, extracts (200 μg) were diluted to 1 mg/mL in plant cell extraction buffer without PVPP and incubated in the presence of 3 μL of AtS6k-specific antibody C (Turck et al., 1998) for 2 h. Incubation mixes were spun for 5 min in an Eppendorf centrifuge at maximal speed and the supernatants incubated with protein A Sepharose and processed as described previously (Pearson et al., 1995). S6 kinase activity was measured by using rat 40S subunits as substrate as described (Turck et al., 1998).
Preparation of Polysomes, Two-Dimensional Gel Electrophoresis, and Polysome Profiles
Analysis of plant ribosomes by two-dimensional electrophoresis was performed as described (Turck et al., 1998). For polysome analysis, 200 mg of a frozen cell pellet was ground into a fine powder in liquid nitrogen and then resuspended in 1 mL of ribosome lysis buffer (100 mm NaCl, 10 mm MgCl2, 5 mm EGTA, 20 mm Tris/HCl, pH 9.0, 1% DOC, 1% Triton X-100, 1 mm sodium molybdate, 1.5% PVPP, 1 mg/mL of heparin, 1 mm dithiothreitol, and 200 units/mL of RNAsin). Extracts were centrifuged twice for 10 min at 15,000 rpm in an Eppendorf centrifuge at +4°C. Equal amount (200 μg) of cell extracts was analyzed on exponential (17% to 41%) Suc gradients. Fractions of 440 μL were immediately collected into 160 μL of RNAse stop buffer (1.5 m NaCl, 150 mm Tris/HCl, pH 8.0, 15 mm EDTA, 10% SDS, 0.5% β-mercaptoethanol, and 5 mm aurin-tricarboxylic acid) and 600 μL of phenol/chloroform. RNA was precipitated overnight by the addition of isopropanol at −20°C and analyzed on northern blots.
RNA Extraction and Northern Blotting
Total mRNA was prepared from frozen plant tissue using a standard guanidium thiocyanate extraction buffer, fractionated (10 μg) on formaldehyde-agarose gels, blotted onto nylon filters (Hybond N, Amersham, Buckinghamshire, UK), and fixed by UV cross-linking. Antisense RNA probes were prepared by in vitro transcription with T7 RNA polymerase of linearized cDNAs in pBluescript vector (Stratagene, La Jolla, CA). The AtS6k1 antisense probe (linearization with BamHI) is complementary to 114 bases of AtS6k1 specific 3′ untranslated region and 78 bases of AtS6k1 coding region. The AtS6k2 derived probe (linearization with HindIII) is complementary to 123 bases of AtS6k2 specific 3′ untranslated region and 335 bases of AtS6k2 coding region. This probe hybridizes with AtS6k1 and AtS6k2 under the conditions employed (overnight hybridization in 50% formamide, 5× sodium chloride/sodium phosphate/EDTA, 0.5% SDS, 5× Denhardt's, and 100 μg/mL of tRNA at 65°C). AtS6k1 and AtS6k2 specific signals can be distinguished based on their different migration pattern on northern gels. AtS18A specific probe was generated as RNA antisense of the S18A 3′ untranslated region (van Lijsebettens et al., 1994). Hybridization probes for S6 (Turck et al., 1998), Ran1 (Haizel et al., 1997), and ubiquitin were prepared by random labeling of full-length coding sequences.
Acknowledgments
We thank M. van Lijsebettens and M. van Montagu for S18A-specific northern probes and B. Weisshaar for ubiquitin cDNA. We also thank J. Bailey-Serres, P.B. Dennis, J. Paszkowski, and H. Rothnie for their critical reading of the manuscript.
This work was supported the Deutscher Akademischer Austauschdienst (fellowship to F.T.) and partially by the Howard Hughes Medical Institute (International Research Scholar Award to F.N.), the Swiss National Foundation for Science (grant to S.C.K. and F.N.), and the Human Frontier Science Program and European Economic Community (grants to G.T.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.035873.
References
- Alessi DR, Kozlowski MT, Weng QP, Avruch J (1998) 3-phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol 8: 69–81 [DOI] [PubMed] [Google Scholar]
- Barakat A, Szick-Miranda K, Chang I-F, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J (2001) The organization of cytoplasmatic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 127: 398–415 [PMC free article] [PubMed] [Google Scholar]
- Brunn GJ, Willams J, Sabers C, Wiederrecht G, Lawrence JC, Jr., Abraham RT (1996) Direct inhibition of the signalling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors wortmannin and LY294002. EMBO J 15: 5256–5267 [PMC free article] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM (1998) RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA 95: 1432–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak M, Casamayor A, Currie RA, Downes CP, Alessi DR (1999) Characterisation of a plant 3-phosphoinositide-dependent protein kinase-1 homologue which contains a pleckstrin homology domain. FEBS Lett 451: 220–226 [DOI] [PubMed] [Google Scholar]
- Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thoas G (2001) Mammalian TOR: a homeostatic ATP sensor. Science 294: 1102–1105 [DOI] [PubMed] [Google Scholar]
- Dennis PD, Pullen N, Kozma SC, Thomas G (1996) The principal rapamycin-sensitive p70s6k phosphorylation sites, T229 and T389, are differentially regulated by rapamycin-insensitive kinase-kinases. Mol Cell Biol 16: 6242–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufner A, Thomas G (1999) Ribosomal S6 kinase signaling and the control of translation. Exp Cell Res 253: 100–109 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Bannwarth W, Morley SJ, Totty NF, Thomas G (1992) Activation of p70s6k is associated with phosphorylation of four clustered sites displaying Ser/Thr-Pro motifs. Proc Natl Acad Sci USA 89: 7282–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox HL, Kimball SR, Jefferson LS, Lynch CJ (1998) Amino acids stimulate phosphorylation of p70S6k and organization of rat adipocytes into multicellular clusters. Am J Physiol 274: C206–C213 [DOI] [PubMed] [Google Scholar]
- Gao J, Kim SR, Lee JM, An G (1994) Developmental and environmental regulation of two ribosomal protein genes in tobacco. Plant Mol Biol 25: 761–770 [DOI] [PubMed] [Google Scholar]
- Garcia Flores C, Aguilar R, Reyes de la Cruz H, Albores M, Sanchez de Jimenes E (2001) A maize insulin-like growth factor signals to a transduction pathway that regulates protein synthesis in maize. Biochem J 358: 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haizel T, Merkle T, Pay A, Fejes E, Nagy F (1997) Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J 11: 93–103 [DOI] [PubMed] [Google Scholar]
- Jefferies HB, Thomas G (1996) Ribosomal protein S6 phosphorylation and signal transduction. In J.W.B. Hershey, M.B. Mathews, and N. Sonenberg, eds, Translational Control. Cold Spring Harbor Laboratory Press, Plainview, NY, pp 389–409
- Jefferies HBJ, Fumagalli S, Dennis PD, Reinhard C, Pearson RB, Thomas G (1997) Rapamycin suppresses 5′TOP mRNA translation through the inhibition of p70s6k. EMBO J 16: 3693–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma SC, Thomas G (2002) Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays 24: 65–71 [DOI] [PubMed] [Google Scholar]
- Levy S, Arni RP, Hariharan N, Perry RP, Meyuhas O (1991) Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc Natl Acad Sci USA 88: 3319–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O, Avni D, Shama S (1996) Translational control of ribosomal protein mRNAs in eukaryotes. In J.W.B. Hershey, M.B. Mathews, and N. Sonenberg, eds, Translational Control. Cold Spring Harbor Laboratory Press, Plainview, NY, pp 363–388
- Mizoguchi T, Hayashida N, Shinozaki YK, Kamada H, Shinozaki K (1995) Two genes that encode ribosomal-protein S6 kinase homologs are induced by cold or salinity stress in Arabidopsis thaliana. FEBS Lett 358: 199–204 [DOI] [PubMed] [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G (1999) Drosophila S6 kinase: a regulator of cell size. Science 285: 2126–2129 [DOI] [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C (2002) Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C1. Plant Physiol 130: 22–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson RC, Dennis PB, Han J-W, Williamson NA, Kozma SC, Wettenhall EH, Thomas G (1995) The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J 14: 5279–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen N, Dennis PD, Andjelkovic M, Dufner A, Kozma SC, Hemmings BA, Thomas G (1998) Phosphorylation and activation of p70s6k by PDK1. Science 279: 707–710 [DOI] [PubMed] [Google Scholar]
- Ray PM (1977) Specificity of auxin-binding sites on maize coleoptile membranes as possible receptor sites for auxin action. Plant Physiol 60: 585–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Nover L (1982) Heat-shock-induced alterations of ribosomal protein phosphorylation in plant cell cultures. Cell 30: 427–437 [DOI] [PubMed] [Google Scholar]
- Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC (1998) Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J 17: 6649–6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolovich M, Tang H, Hornstein E, Levy G, Cohen R, Baie SS, Birnbaum MJ, Meyuhas O (2002) Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol Cell Biol 22: 8101–8113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G (2002) The S6 kinase signalling pathway in the control of development and growth. Biol Res 35: 305–313 [DOI] [PubMed] [Google Scholar]
- Turck F, Kozma SC, Thomas G, Nagy F (1998) A heat sensitive Arabidopsis thaliana kinase substitutes for human p70s6k function in vivo. Mol Cell Biol 18: 2038–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Panayotou G, Waterfield MD (1997) Phosphoinositide 3-kinases: a conserved family of signal transducers. Trends Biochem Sci 22: 267–272 [DOI] [PubMed] [Google Scholar]
- van Lijsebettens M, Vanderhaegen R, de Block M, Bauw G, Villarroel R, van Montagu M (1994) An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J 13: 3378–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahost CJ, Matter WF, Hui KY, Brown RF (1994) A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem 269: 5241–5248 [PubMed] [Google Scholar]
- Volarevic S, Thomas G (2001) Role of S6 phosphorylation and S6 kinase in cell growth. Prog Nucleic Acid Res Mol Biol 65: 101–127 [DOI] [PubMed] [Google Scholar]
- Williams AJ, Werner-Fraczek J, Chang I-F, Bailey-Serres J (2003) Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol 132: 2086–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovleva LA, Kulaeva ON (1987) The effect of phytohormones on phosphorylation of ribosomal proteins in detached pumpkin cotyledons. Biochem Physiol Pflanz 182: 359–365 [Google Scholar]
- Zhang SH, Broome MA, Lawton MA, Hunter T, Lamb CJ (1994. a) Atpk1, a novel ribosomal protein kinase gene from Arabidopsis. Part 2. J Biol Chem 269: 17593–17599 [PubMed] [Google Scholar]
- Zhang SH, Lawton MA, Hunter T, Lamb CJ (1994. b) Atpk1, a novel ribosomal protein kinase gene from Arabidopsis. Part 1. J Biol Chem 269: 17586–17592 [PubMed] [Google Scholar]