Abstract
Sinopodophyllum hexandrum is an important medicinal plant whose genetic diversity must be conserved because it is endangered. The Qinling Mts. are a S. hexandrum distribution area that has unique environmental features that highly affect the evolution of the species. To provide the reference data for evolutionary and conservation studies, the genetic diversity and population structure of S. hexandrum in its overall natural distribution areas in the Qinling Mts. were investigated through inter-simple sequence repeats analysis of 32 natural populations. The 11 selected primers generated a total of 135 polymorphic bands. S. hexandrum genetic diversity was low within populations (average He = 0.0621), but higher at the species level (He = 0.1434). Clear structure and high genetic differentiation among populations were detected by using the unweighted pair group method for arithmetic averages, principle coordinate analysis and Bayesian clustering. The clustering approaches supported a division of the 32 populations into three major groups, for which analysis of molecular variance confirmed significant variation (63.27%) among populations. The genetic differentiation may have been attributed to the limited gene flow (Nm = 0.3587) in the species. Isolation by distance among populations was determined by comparing genetic distance versus geographic distance by using the Mantel test. Result was insignificant (r = 0.212, P = 0.287) at 0.05, showing that their spatial pattern and geographic locations are not correlated. Given the low within-population genetic diversity, high differentiation among populations and the increasing anthropogenic pressure on the species, in situ conservation measures were recommended to preserve S. hexandrum in Qinling Mts., and other populations must be sampled to retain as much genetic diversity of the species to achieve ex situ preservation as a supplement to in situ conservation.
Introduction
Sinopodophyllum hexandrum (Royle) Ying, family Berberidaceae, the only species of this genus in China, commonly known as Himalayan mayapple, is an endangered and medicinal perennial herb native to the Himalayan regions at elevations ranging from 2 700 m to 4 500 m [1]–[4]. Plants provide us with many important medicaments, including anticancer and antiinfective agents [5], and traditional Chinese medicine has contributed to identifying these substances [6]. S. hexandrum is a traditional Chinese medicine that has been used in folk medicine [7]. The roots and rhizomes of S. hexandrum contain large amounts of lignans. The most important lignan for human health is arguably the most active cytotoxic aryltetralin lignan, podophyllotoxin, with three times the podophyllotoxin levels compared to the American species Podophyllum peltatum [8], [9], [10], as a precursor for the semi-synthesis of the anticancer pharmaceuticals, such as etoposide (VP-16), teniposide (VM-26), GP-7, NK-611, etopophos, GL-331 and TOP-53 [9]–[15]. The destructive harvest of these plants added S. hexandrum to the endangered species list of the Convention on International Trade in Endangered Species of Wild Fauna and Flora [16]. S. hexandrum was classified as an endangered species (grade 3) in 1987 by the Chinese Plant Red Book [4].
Currently, with the enhanced awareness of its medicinal value and superior efficacy in clinical applications, the wild S. hexandrum populations in China has been noted to be very small and to be rapidly declining. The availability of podophyllotoxin from plants has become increasingly limited due to intense collection, habitat fragmentation, low natural regeneration rate, and the lack of organized cultivation. Wild S. hexandrum populations could become extinct without timely and effective protective measures, costing humans an ideal drug against cancer. Knowledge of the genetic diversity at intraspecific levels is an important prerequisite for species conservation and a rational exploitation program planning. However, previous studies have mainly focused on the identification and separation of the chemical components [17]–[25], biological properties [26]–[32], and micropropagation of S. hexandrum [33]. Only studies on genetic diversity in S. hexandrum from the Northwestern Himalayan region are available [34], [35], [36]. In particular, two areas where S. hexandrum is grown, namely, western Sichuan Province and Himalaya–Hengduan Mt. region in China, were investigated to identify genetic diversity of this species through inter-simple sequence repeats (ISSR) and amplified fragment length polymorphism (AFLP) markers [37], [38], [39], respectively. These reports with same results showed that S. hexandrum populations had relatively high genetic diversity (He = 0.2944∼0.3377). However, there is a consensus that alpine plants are faced with pollinator restriction [40]. The unclear extent of the species dispersal mechanisms makes it interesting to study the relationships between populations. Furthermore, many reports about genetic diversity of other medicinal plants, such as Calamagrostis porteri ssp. insperata [41], Aegiceras corniculatum [42], Sonneratia alba [43], Coscinium fenestratum [44], and Lilium pumilum [45], have been published based on the ISSR approach.
Molecular markers are very useful tools for genetic diversity studies. ISSR markers are molecular markers especially suited to genetic polymorphisms analyses of species without available sequence information [46], [47]. Studies on the population relationships, genetic diversity and conservation of S. hexandrum in Qinling Mts. are requisite because climate change and local overexploitation may cause unknown endangering mechanisms. The present study aims to establish management strategies for the conservation genetics of S. hexandrum by (1) examining the levels of genetic variability within and among S. hexandrum populations sampled from Qinling Mts. where historical records showed S. hexandrum having grown naturally, (2) assessing the possible factors that affect the genetic variation observed, and (3) comparing these within and among S. hexandrum populations with data published for itself or other plant taxa with similar characteristics.
Materials and Methods
Ethics statement
The endangered species were collected, and research activities were scientifically conducted under the permits issued by the local forestry department. A detailed description of the experimental material collection and procedures is provided. The experimental procedures were approved by the Ethics Committee for Plant Experiments of Northwest A & F University and the State Forestry Administration, P. R. China. The names of the authorities that issued the permit for each location were listed in Table S1 in Text S1.
Study area
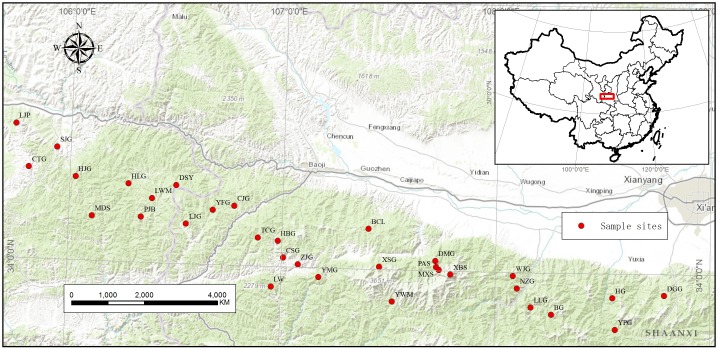
This study was performed in the Qinling Mts. (32°41′ to 34°59′ N, 103°54′ to 110°34′ E), which are located in central of China (Figure 1). The Qinling Mts., a 1 500 Km mountain chain, run east–west and act as an important watershed divider between two great Chinese rivers, the Yangtze River and the Yellow River, which constitute a transitional zone between the northern subtropical zone and warm-temperate zone. The Qinling Mts. were considered to be a biodiversity hotspot in China [48]. As one of the distribution areas of S. hexandrum, Qinling Mts. have unique environmental features which have high impact on the evolution of the species.
Figure 1. Locations of the 32 S. hexandrum populations in Qinling Mts. sampled for this study.
Plant materials
S. hexandrum distribution pattern and extent of in Qinling Mts. were investigated from 2010 to 2011. The S. hexandrum populations are distributed in small and scattered patches. According to the field survey information, a total of 32 wild S. hexandrum populations were sampled for DNA analysis between July 19, 2012, and September 17, 2012, to ensure collection period consistency (Table 1). S. hexandrum has a wide geographic distribution throughout the Qinling Mts. (Figure 1). The altitude of the sample sites ranged from 1 013 m to 2 883 m (Table1). Geographical distances between populations ranged from 5.5 km to 276.8 km. 20 plants were sampled from each population. The horizontal and vertical distances between sampled plants within each population were over 20 and 5 m, respectively, to increase the likelihood of sampling inter-individual variation within each population [37], [38]. About 2 g to 10 g of fresh young leaves per plant was immediately frozen in liquid nitrogen and then kept at −80 °C until DNA isolation. The key information on S. hexandrum populations in all sampling sites is summarized in Table 1.
Table 1. Sample information of the 32 sampling sites.
| No. | Population | Prefecture | Coordinates | N | Altitude/m | SO/SD/° | Vegetation type |
| 1 | Caotangou | Maiji | E105°48′N34°21′ | 20 | 1 899 | SE70 | Shrub-grass |
| 2 | Tancaogou | Fengxian | E106°53′N34°6′ | 20 | 1 728 | SW45 | Shrub-grass |
| 3 | Chunshugou | Fengxian | E107°1′N34°2′ | 20 | 1 512 | SW65 | Shrub-grass |
| 4 | Hougou | Fengxian | E108°33′N33°55′ | 20 | 1 439 | N75 | Shrub-grass |
| 5 | Xiaoshagou | Taibai | E107°27′N34°1′ | 20 | 2 332 | NW60 | Forest-edge, Shrub-grass |
| 6 | Doumugong | Meixian | E107°43′N34°2′ | 20 | 2 748 | NW75 | Forest-edge, Shrub-grass |
| 7 | Pinganssi | Meixian | E107°43′N34°1′ | 20 | 2 815 | NW75 | Forest-edge, Shrub-grass |
| 8 | Mingxingsi | Meixian | E107°44′N34°0′ | 20 | 2 637 | E75 | Forest-edge, Shrub-grass |
| 9 | Xiabansi | Meixian | E107°47′N33°59′ | 20 | 2 883 | SE50 | Forest-edge, Shrub-grass |
| 10 | Laojungou | Liangdang | E106°33′N34°9′ | 20 | 1 902 | SE60 | Cliff, edge of forest |
| 11 | Youfanggou | Fengxian | E106°40′N34°12′ | 20 | 1 545 | SW70 | Shrub-grass |
| 12 | Baicaoling | Taibai | E107°24′N34°9′ | 20 | 1 373 | S60 | Forest-edge, Shrub-grass |
| 13 | Nianzigou | Zhouzhi | E108°6′N33°56′ | 20 | 1 821 | SW75 | Meadow, edge of forest |
| 14 | Wenjiagou | Zhouzhi | E108°5′N33°59′ | 20 | 1 765 | S80 | Meadow, edge of forest |
| 15 | Liulingou | Zhouzhi | E108°10′N33°52′ | 20 | 1 013 | NW75 | Meadow, edge of forest |
| 16 | Beigou | Zhouzhi | E108°16′N33°51′ | 20 | 1 579 | NW85 | Meadow, edge of forest |
| 17 | Dagangou | Huxian | E108°47′N33°55′ | 20 | 1 335 | NW65 | Shrub-grass |
| 18 | Yaowangmiao | Taibai | E107°31′N33°53′ | 20 | 1 779 | SE40 | Forest-edge, Shrub-grass |
| 19 | Yingpangoukou | Huxian | E108°34′N33°47′ | 20 | 1 487 | SE60 | Shrub-grass |
| 20 | Maiduoshigou | Liangdang | E106°7′N34°10′ | 20 | 1 520 | SE70 | Cliff, edge of forest |
| 21 | Panjiaba | Liangdang | E106°20′N34°10′ | 20 | 1 976 | SW45 | Cliff, edge of forest |
| 22 | Zhangjiagou | Fengxian | E107°5′N34°1′ | 20 | 1 469 | SW75 | Shrub-grass |
| 23 | Longwangmiao | Fengxian | E106°57′N33°55′ | 20 | 1 475 | SW75 | Shrub-grass |
| 24 | Huangbaigou | Fengxian | E106°59′N34°6′ | 20 | 1 745 | SW80 | Shrub-grass |
| 25 | Dashuiyugou | Maiji | E106°30′N34°18′ | 20 | 1 878 | W75 | Shrub-grass |
| 26 | Chenjiagou | Fengxian | E106°46′N34°13′ | 20 | 1 515 | SW45 | Shrub-grass |
| 27 | Longwangmiao | Maiji | E106°23′N34°15′ | 20 | 1 736 | NW60 | Shrub-grass |
| 28 | Yinmagou | Taibai | E107°11′N33°58′ | 20 | 2 252 | S60 | Forest-edge, Shrub-grass |
| 29 | Hualingou | Maiji | E106°17′N34°18′ | 20 | 1 793 | SW55 | Shrub-grass |
| 30 | Huojigou | Maiji | E106°2′N34°19′ | 20 | 1 718 | SE65 | Shrub-grass |
| 31 | Shijiagoucun | Maiji | E105°56′N34°25′ | 20 | 1 366 | SE60 | Shrub-grass |
| 32 | Liujiaping | Maiji | E105°44′N34°30′ | 20 | 1 690 | SW30 | Shrub-grass |
Note: N means sample size; SO/SD means slope orientation/degree.
DNA extraction
Total genomic DNA was extracted from frozen leaves by using a plant genomic DNA rapid extraction kit (Spin-column) (BioTek Corporation, Beijing, China; http://bioteke.biogo.net/). The extracted DNA was quantified by comparing with known DNA of standard quantity (Lambda DNA) through electrophoresis in ethidium bromide-stained 1.0% agarose gels (Gene Genius Bio Imaging System; Synegene), and the extracted DNA was diluted in TE buffer to a final concentration of 50 ng/µL and stored at−20°C before PCR amplification.
Primer screening and ISSR-PCR amplification
A total of 100 ISSR primers (synthesized by Shanghai Sheng Gong Biotechnology CO. LTD, China) were screened based on the primer set published by the Biotechnology Laboratory, University of British Columbia, Canada (UBC set No. 9) and the studies on Himalayan mayapple [34]–[39]. An optimum reaction system was obtained by screening DNA, Mg2+, dNTP, primer (UBC900 was used for preliminary test), and Taq DNA polymerase concentrations and annealing temperature, and reaction conditions. The optimization showed that 20 µL of reaction system is ideal. Each 20 µL amplification reaction consisted of 1×PCR buffer (10 mM Tris-HCl at pH 8.3, 50 mM L–1 KCl, 0.001% gelatin, and l.5 mmol L–1 MgCl2), 1.6 mmol L–1 dNTP mix, 0.6 µmol L–1 primer (UBC900 was used for preliminary test), 15 ng of template DNA, and 1.0 U Taq DNA polymerase (TaKaRa Biotechnology, Dalian, China), using a cycling profile of initial 5 min at 94°C, followed by 45 cycles of 30 s at 94°C, 45 s annealing at 50°C, and 90 s extension at 72°C, ending with a final extension of 7 min at 72°C.
The optimized PCR experiment conditions were applied for primer screening in a PTC100TM Programmable Thermal Controller (MJ Research, Waltham, MA, USA). Six populations (TCG, LJG, BG, MDS, YMG and CTG) with observable variations (morphology, habitat, etc) were selected to initially screen 100 primers by using 10 samples for each population. Primers that generated scorable bands and high levels of polymorphisms were selected by genotyping all populations. The amplification products were electrophoresed on 1.0% agarose gels buffered with 1.0×TBE for 2.5 h at 100 V and were detected through ethidium bromide staining, and the gels were imaged in the Gene Genius Bioimaging System. Band size was estimated from a 0.1 kbp DNA ladder (TaKaRa Biotechnology, Dalian, China). Each primer was amplified in triplicate to confirm reliability and reproducibility. A reaction without DNA was used as negative control.
Data analyses
Amplification results were scored according to the positions of the DNA bands from electrophoresis, being labeled “1” for presence of the bands and “0” for absence of the bands in the data matrix. Only stable bands with repeatable differences were considered valid for polymorphism loci.
The resulting ISSR phenotype data matrix (binary matrix from 0 to 1) was analyzed using the POPGENE software (version 1.31) [49] to compute genetic diversity parameters, such as allele frequencies, percentage of polymorphic bands (PPB), number of alleles per locus (Ao), effective number of alleles per locus (Ae), total gene diversity (Ht), the level of gene flow (Nm), Nei’s gene diversity (He), gene distance (GD), Shannon’s information index (Ho), within-population diversity (Hs), and mean coefficient of gene differentiation (Gst). An unweighted pair groups mean arithmetic (UPGMA) mean dendrogram was constructed using PowerMarker 3.23 to examine the genetic relationship at the species level [50]. A bootstrap (resampling) test was performed 1 000 times to determine distances between the populations using PHYLIP version 3.69 (PHYLogeny Inference Package) programs [51]. Bayesian analysis of population structure was performed as implemented in STRUCTURE (version 2.2) to infer the most likely number of population genetic clusters (K) in the ISSR dataset [52]. K ranged from 1 to 10, with 10 replicate runs for each K, and a burn-in period of 2×105 and 5×104 iterations. The “no admixture model” and independent allele frequencies were chosen for this analysis. The most likely number of clusters was estimated according to the model values (ΔK) based on the second order rate of change, with respect to K, of the likelihood function [53]. To detect within-group structure, subsequent runs were performed for each obtained clusters using the same settings as previous. Population similarity was also explored and visualized through principle coordinate analysis (PCoA) using NTSYSpc 2.10e [54]. Analysis of molecular variance (AMOVA), which partitions total phenotypic variance within and among populations, was performed using WIN AMOVA (version 1.55), which was provided by the Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland [55]. The AMOVA input files with the Euclidean distance matrix were created using AMOVA-PREP 1.01 [56]. Significance level was tested by comparing the frequency distributions from the original data and the data generated by a set of 1 000 computer simulations. A Mantel test was performed using Tools for Population Genetic Analysis (TFPGA) [56] for computing 5 000 permutations to test the isolation by distance (IBD) among populations by comparing genetic distance between all pairwise combinations of populations versus geographic distance. Geographical distance, Lab, was computed using the following formula: Lab = Arccos [cos(LATa)COS(LONGa)cos(LATb)cos(LONGb) + cos(LATb)sin(LONGb)cos(LATb)sin(LONGb) + sin(LATa)sin(LATb)]×R [57]. LONGa, LATa and LONGb, LATb are the longitudes and latitudes (in radians) of sampling sites a and b, respectively; R indicates the radius of the earth, which is 6 378 km; and Lab represents the geographic distance between sampling sites a and b.
Results
Genomic DNA amplification results
For polymorphism testing of the 640 S. hexandrum individuals from 32 populations, forty-eight ISSR primers amplified visible bands and were chosen from the initial set of 100 primers to screen for reproducible markers. PCR amplification results (Table 2) show that 11 primers produced 241 clear and replicated bands (250 bp to 2 000 bp), of which 135 were polymorphic (56.02%) with 100% reproducibility. Individual primers detected between 19 (UBC825) and 29 (UBC900) loci can amplify clear bands, with an average of 21.91. The percentage of polymorphism ranged from 42.86% (UBC834) to 68.97% (UBC900), indicating that the selected primers are highly polymorphic across S. hexandrum populations. The Ht was 0.1434, whereas Hs was found to be 0.0599. The Gst value of 0.5823 indicated that 41.77% of the genetic diversity resided within the populations. The Nm among the sampled populations was calculated as 0.3587 using Gst through the formula (0.5(1−Gst)/Gst).
Table 2. Description of the11 selected primers used for ISSR amplification.
| No. | Primer | Sequences 5′→3′ | Nt | Np | Pr % | Ht | Hs | Gst |
| 1 | UBC825 | (AC)8T | 19 | 12 | 63.16 | 0.1715 | 0.0835 | 0.5132 |
| 2 | UBC834 | (AG)8YT | 21 | 9 | 42.86 | 0.1035 | 0.0440 | 0.5748 |
| 3 | UBC844 | (CT)8AGC | 22 | 11 | 50.00 | 0.1160 | 0.0517 | 0.5543 |
| 4 | UBC845 | (CT)8AGG | 23 | 12 | 52.17 | 0.1192 | 0.0494 | 0.5855 |
| 5 | UBC853 | (CT)8AGT | 20 | 9 | 45.00 | 0.1121 | 0.0352 | 0.6858 |
| 6 | UBC855 | (AC)8YT | 21 | 11 | 52.38 | 0.1347 | 0.0645 | 0.5212 |
| 7 | UBC857 | (AC)8CTG | 20 | 13 | 65.00 | 0.1763 | 0.0763 | 0.5672 |
| 8 | UBC867 | (GGC)6 | 19 | 10 | 52.36 | 0.1251 | 0.0564 | 0.5492 |
| 9 | UBC873 | (GACA)4 | 22 | 11 | 50.00 | 0.1165 | 0.0441 | 0.6214 |
| 10 | UBC895 | AGAGTTGGTAGCTCTTGATC | 25 | 17 | 68.00 | 0.1875 | 0.0765 | 0.5920 |
| 11 | UBC900 | ACTTCCCCACAGGTTAACACA | 29 | 20 | 68.97 | 0.2150 | 0.0773 | 0.6404 |
| Sum | 241 | 135 | ||||||
| Mean | 21.91 | 12.27 | 56.02 | 0.1434 | 0.0599 | 0.5823 |
Note: Nt-No. of total amplified bands; Np-No. of polymorphic bands; Pr-Polymorphism rate; Ht- Total gene diversity among population; Hs-within population diversity; Gst-Mean coefficient of gene differentiation.
Population genetic diversity
Detailed statistical analyses were performed on the ISSR amplification results (Table 3). The numbers of polymorphic bands are different among populations. The highest PPB (39.17%) was observed in the DMG population. However, only 20.73% of the bands were polymorphic in the SJG population. The PPB was 56.02% at the species level, whereas those of the single populations ranged from 20.73% (SJG) to 39.17% (DMG), with an average of 27.33%. Ae ranged from 1.0182 (HJG) to 1.2643 (BG), with 1.1630 at the population level and 1.3732 at the species level. He within populations was lower (0.0621) than that of the species level (0.1434). Within each population, the He of most populations ranged from 0.0226 to 0.0971. Only in the population HG, a high He (0.1229, Table 3) was observed. Ho ranged from 0.0244 to 0.1038, with an average of 0.0637 at the population level and 0.2362 at the species level. Allele frequencies calculated using Popgene software were shown in Table S2 in Text S2.
Table 3. Descriptive statistics summary of the S. hexandrum populations.
| Population | Code | PPB (%) | Ao | Ae | He | Ho |
| Tancaogou | TCG | 23.63 | 1.3546 | 1.1684 | 0.0559 | 0.0737 |
| Chunshugou | CSG | 22.62 | 1.2687 | 1.1556 | 0.0337 | 0.0689 |
| Hougou | HG | 25.66 | 1.5642 | 1.2357 | 0.1229 | 0.1038 |
| Xiaoshagou | XSG | 25.32 | 1.5721 | 1.1957 | 0.0777 | 0.0733 |
| Doumugong | DMG | 39.17 | 1.3905 | 1.1404 | 0.0296 | 0.0255 |
| Pinganssi | PAS | 33.95 | 1.4397 | 1.1833 | 0.0567 | 0.0442 |
| Mingxingsi | MXS | 26.94 | 1.4720 | 1.1667 | 0.0381 | 0.0667 |
| Xiabansi | XBS | 23.51 | 1.4986 | 1.231 | 0.0542 | 0.0614 |
| Laojungou | LJG | 28.52 | 1.2199 | 1.1937 | 0.0426 | 0.0566 |
| Youfanggou | YFG | 21.95 | 1.3688 | 1.1846 | 0.0533 | 0.0755 |
| Baicaoling | BCL | 28.84 | 1.5579 | 1.1795 | 0.0603 | 0.0715 |
| Nianzigou | NZG | 25.18 | 1.473 | 1.2166 | 0.0868 | 0.0508 |
| Wenjiagou | WJG | 31.99 | 1.4238 | 1.1737 | 0.0797 | 0.0321 |
| Liulingou | LLG | 26.85 | 1.5053 | 1.2000 | 0.0882 | 0.0733 |
| Beigou | BG | 29.01 | 1.5319 | 1.2643 | 0.0670 | 0.0680 |
| Dagangou | DGG | 27.07 | 1.5912 | 1.2128 | 0.0448 | 0.0781 |
| Yaowangmiao | YWM | 32.03 | 1.5309 | 1.2024 | 0.0728 | 0.0972 |
| Yingpangoukou | YPG | 24.93 | 1.6054 | 1.2290 | 0.0778 | 0.0799 |
| Maiduoshigou | MDS | 24.63 | 1.1576 | 1.0445 | 0.0226 | 0.0656 |
| Panjiaba | PJB | 32.95 | 1.1872 | 1.1293 | 0.0252 | 0.0277 |
| Zhangjiagou | ZJG | 26.17 | 1.2364 | 1.1722 | 0.0523 | 0.0464 |
| Longwangmiao | LW | 24.46 | 1.2953 | 1.2199 | 0.0498 | 0.0636 |
| Huangbaigou | HBG | 24.07 | 1.3276 | 1.1913 | 0.0684 | 0.0994 |
| Dashuiyugou | DSY | 31.74 | 1.2435 | 1.0573 | 0.0804 | 0.0704 |
| Chenjiagou | CJG | 28.75 | 1.4232 | 1.2048 | 0.0643 | 0.0544 |
| Longwangmiao | LWM | 33.62 | 1.2577 | 1.0735 | 0.0422 | 0.0722 |
| Yinmagou | YMG | 23.52 | 1.4565 | 1.2381 | 0.0971 | 0.061 |
| Hualingou | HLG | 23.34 | 1.1842 | 1.1088 | 0.0387 | 0.0603 |
| Huojigou | HJG | 30.96 | 1.0761 | 1.0182 | 0.0541 | 0.0244 |
| Shijiagoucun | SJG | 20.73 | 1.1253 | 1.0611 | 0.0412 | 0.0431 |
| Caotangou | CTG | 26.42 | 1.1088 | 1.0826 | 0.0315 | 0.0533 |
| Liujiaping | LJP | 25.93 | 1.2165 | 1.0802 | 0.0573 | 0.0961 |
| Average | 27.33 | 1.3645 | 1.1630 | 0.0621 | 0.0637 | |
| Total | 56.02 | 1.7801 | 1.3732 | 0.1434 | 0.2362 |
Note: Ao, observed number of alleles per locus; Ae, effective number of alleles per locus; He, Nei’s gene diversity; Ho, Shannon’s information index; PPB, percentage of polymorphic bands.
Genetic structure and differentiation of the populations
UPGMA cluster analysis
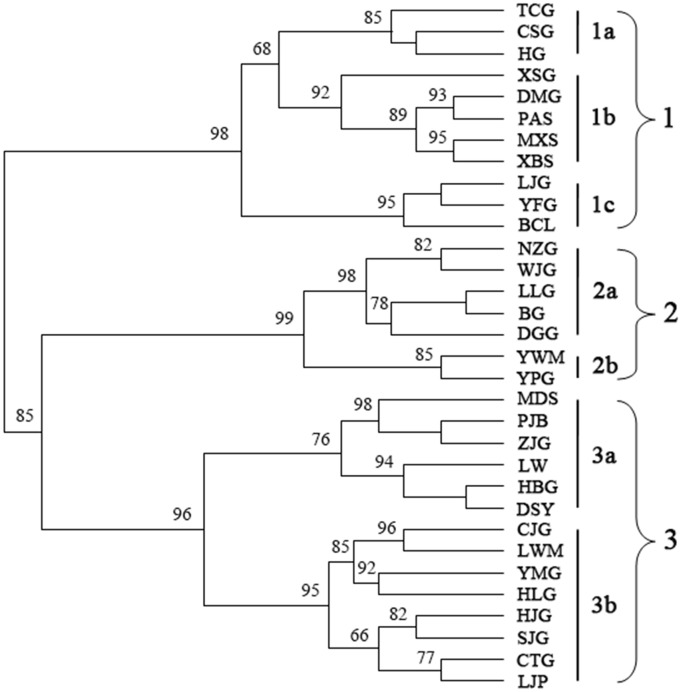
UPGMA clustering analysis defined three major groups among 32 populations (Figure 2). Group 1 included 11 populations, which were further divided into three subgroups. Populations TCG, CSG and HG were in subgroup 1a; populations XSG, DMG, PAS, MXS and XBS were in subgroup 1b; and populations LJG, YFG, and BCL were in subgroup 1c. Populations NZG, WJG, LLG and BG (Zhouzhi County), population DGG (Huxian County), population YWM (Taibai County) and population YPG (Huxian County) were grouped in cluster 2. Group 3 contained 14 populations, which were sampled from two adjacent cities, Baoji and Tianshui, and were subdivided into two clusters. Populations MDS, PJB, ZJG, LW, and HBG (Baoji) and DSY (Tianshui) were in subgroup 3a. Populations LWM, HLG, HJG, SJG, CTG, and LJP (Tianshui) and CJG, YMG (Baoji) were in subgroup 3b. Seven populations including DSY, LWM, HLG, HJG, SJG, CTG, and LJP in Group 3 were from Gansu Province. The other populations clustered in Groups 1, 2, and 3 were from Shaanxi Province. The S. hexandrum population was not clustered on the UPGMA tree according to geographic distance, which may indicate no obvious correlation between geographic distribution and genetic distance.
Figure 2. UPGMA clustering of S. hexandrum populations in Qinling Mts.
Principle coordinate analysis
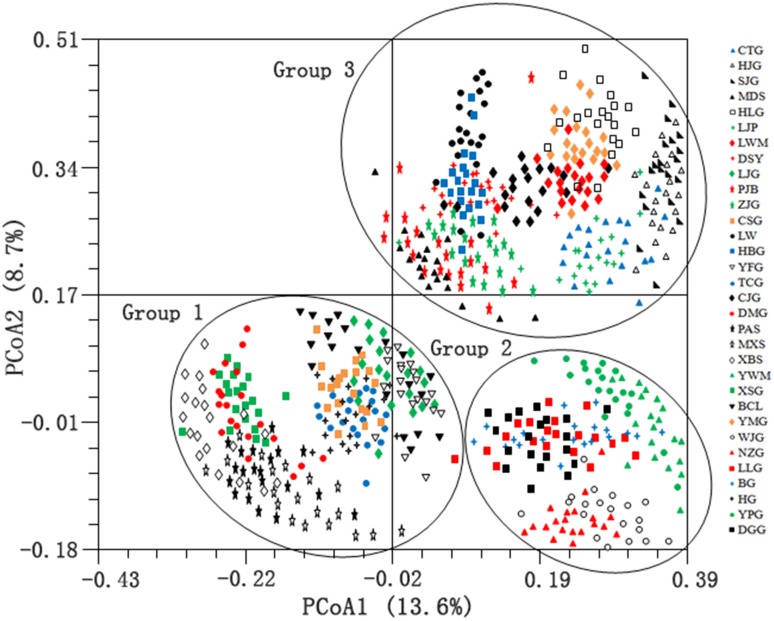
PCoA was used for ordination and exploration of the similarity between populations. All samples were clearly separated into three major groups on the first (PCo1) and second (PCo2) principal coordinates (Figure 3). PCo1 explained 13.6% of the total variance, and PCo2 explained 8.7% of the total variance. Groups 1 and 2 are clustered within each other’s vicinity even though clearly separated, indicating higher similarity between these two groups. The clustering of the populations was in agreement with the UPGMA dendrogram.
Figure 3. Distribution of individuals of the 32 S. hexandrum populations from Qinling Mts., according to the first (PCo1) and second (PCo2) principal coordinates.
PCo1 and PCo2 account for 13.6 and 8.7% of the total variation, respectively.
Mantel test for IBD
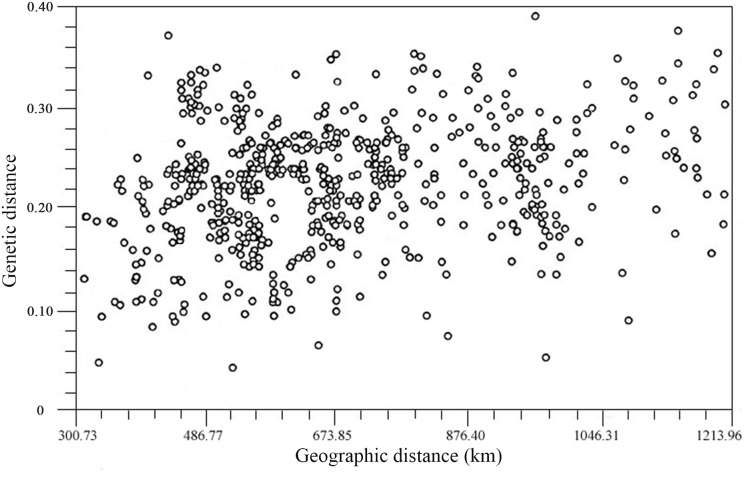
A Mantel test for IBD was performed to assess the correlation between the genetic distance matrix and the corresponding geographic distance matrix of the wild S. hexandrum populations. The correlation coefficient r of genetic distance and geographic distance was 0.212 (P = 0.287), and the correlation analysis diagram (Figure 4) was comprised of many disordered and scattered points, indicating that the IBD of the wild S. hexandrum populations in Qinling Mts. was not significant at the level of 0.05. The Mantel test did not indicate correlation between genetic distance and geographical provenance.
Figure 4. Mantel regression of the pairwise relationship between genetic and geographical distances for S. hexandrum populations.
Bayesian clustering
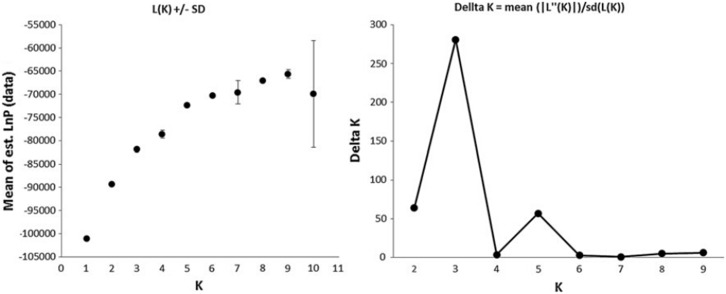
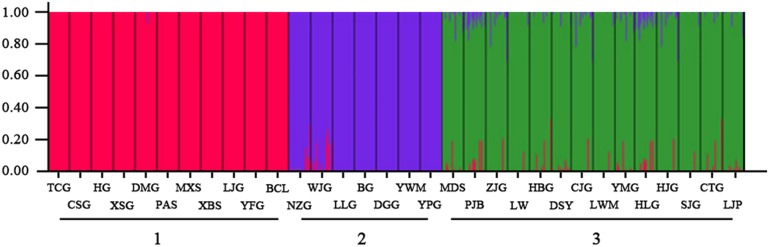
The genetic structure of the S. hexandrum samples were further analyzed using Bayesian clustering algorithm in the STRUCTURE software. The ΔK method indicated that the most likely K value was 3 (Figure 5). Sharp divisions were observed for the three clusters (Figure 6). The assignments of the populations to Groups 1, 2, and 3 were stable and consistent with UPGMA and PCoA clustering.
Figure 5. The probable K value estimated by likelihood of the probability of data L(K) and ad hoc quantity ΔK.
Figure 6. Bayesian clustering for infering population structure of S. hexandrum populations from Qinling Mts.
Analysis of molecular variance
The pairwise GD values (Table 4) were small and ranged from 0.0467 (XSG and DMG) to 0.3997 (BG and LW), indicating low differentiation within populations. Low population differentiation indicated that gene flow within each clustered group may be high or that isolation time was recent.
Table 4. Pairwise genetic distance estimates from 241 ISSR markers in S. hexandrum.
| Population | TCG | CSG | HG | XSG | DMG | PAS | MXS | XBS | LJG | YFG | BCL | NZG | WJG | LLG | BG | DGG | YWM | YPG | MDS | PJB | ZJG | LW | HBG | DSY | CJG | LWM | YMG | HLG | HJG | SJG | CTG | LJP |
| TCG | *** | |||||||||||||||||||||||||||||||
| CSG | 0.1362 | *** | ||||||||||||||||||||||||||||||
| HG | 0.0712 | 0.1379 | *** | |||||||||||||||||||||||||||||
| XSG | 0.1756 | 0.1444 | 0.1064 | *** | ||||||||||||||||||||||||||||
| DMG | 0.1833 | 0.1433 | 0.1343 | 0.0467 | *** | |||||||||||||||||||||||||||
| PAS | 0.1654 | 0.1373 | 0.1312 | 0.0513 | 0.1514 | *** | ||||||||||||||||||||||||||
| MXS | 0.1532 | 0.1920 | 0.1255 | 0.0568 | 0.2222 | 0.0634 | *** | |||||||||||||||||||||||||
| XBS | 0.1743 | 0.1636 | 0.1168 | 0.0764 | 0.3867 | 0.0754 | 0.0822 | *** | ||||||||||||||||||||||||
| LJG | 0.1503 | 0.1520 | 0.1205 | 0.0608 | 0.1290 | 0.0879 | 0.1075 | 0.0732 | *** | |||||||||||||||||||||||
| YFG | 0.1853 | 0.1585 | 0.1484 | 0.0654 | 0.1640 | 0.0819 | 0.1015 | 0.0797 | 0.1755 | *** | ||||||||||||||||||||||
| BCL | 0.1897 | 0.1574 | 0.1453 | 0.0709 | 0.1684 | 0.1575 | 0.1262 | 0.0786 | 0.1799 | 0.2085 | *** | |||||||||||||||||||||
| NZG | 0.1974 | 0.1514 | 0.1396 | 0.0905 | 0.1461 | 0.1786 | 0.1278 | 0.3726 | 0.1876 | 0.1907 | 0.1586 | *** | ||||||||||||||||||||
| WJG | 0.2095 | 0.2061 | 0.1309 | 0.0680 | 0.3582 | 0.1546 | 0.1093 | 0.1273 | 0.1697 | 0.1790 | 0.1555 | 0.3824 | *** | |||||||||||||||||||
| LLG | 0.1673 | 0.1777 | 0.2213 | 0.1457 | 0.3891 | 0.1896 | 0.1158 | 0.0989 | 0.1830 | 0.1994 | 0.1498 | 0.2020 | 0.1616 | *** | ||||||||||||||||||
| BG | 0.2448 | 0.1592 | 0.2445 | 0.1568 | 0.3483 | 0.1940 | 0.1147 | 0.3804 | 0.1885 | 0.1907 | 0.1411 | 0.3772 | 0.2163 | 0.2304 | *** | |||||||||||||||||
| DGG | 0.2727 | 0.1657 | 0.2449 | 0.1368 | 0.1980 | 0.2017 | 0.1087 | 0.0869 | 0.2081 | 0.1829 | 0.1422 | 0.1818 | 0.1879 | 0.2020 | 0.2035 | *** | ||||||||||||||||
| YWM | 0.2696 | 0.1646 | 0.2185 | 0.1356 | 0.2179 | 0.2138 | 0.1334 | 0.0858 | 0.1815 | 0.1772 | 0.1384 | 0.1873 | 0.1763 | 0.1835 | 0.1948 | 0.2076 | *** | |||||||||||||||
| YPG | 0.2639 | 0.1586 | 0.3831 | 0.1431 | 0.1844 | 0.1716 | 0.1350 | 0.0798 | 0.1861 | 0.1685 | 0.2015 | 0.2069 | 0.1828 | 0.1900 | 0.3893 | 0.2272 | 0.2040 | *** | ||||||||||||||
| MDS | 0.1552 | 0.2133 | 0.3086 | 0.0830 | 0.2136 | 0.2014 | 0.1142 | 0.1345 | 0.1624 | 0.1612 | 0.2136 | 0.1784 | 0.1817 | 0.1889 | 0.2172 | 0.2047 | 0.2251 | 0.2114 | *** | |||||||||||||
| PJB | 0.2520 | 0.1849 | 0.3223 | 0.1026 | 0.2202 | 0.1462 | 0.1207 | 0.1061 | 0.1453 | 0.1891 | 0.1696 | 0.3726 | 0.2137 | 0.1829 | 0.2141 | 0.2093 | 0.1991 | 0.2318 | 0.2197 | *** | ||||||||||||
| ZJG | 0.1799 | 0.1641 | 0.3050 | 0.0741 | 0.1530 | 0.1127 | 0.0896 | 0.3760 | 0.1674 | 0.1860 | 0.1694 | 0.0781 | 0.1853 | 0.2376 | 0.2084 | 0.1729 | 0.2341 | 0.2231 | 0.3793 | 0.1879 | *** | |||||||||||
| LW | 0.2768 | 0.1706 | 0.3384 | 0.0787 | 0.1413 | 0.0843 | 0.1136 | 0.0971 | 0.1927 | 0.1803 | 0.1524 | 0.0977 | 0.1405 | 0.2092 | 0.3997 | 0.2079 | 0.2385 | 0.2474 | 0.2108 | 0.1822 | 0.2160 | *** | ||||||||||
| HBG | 0.1778 | 0.1695 | 0.1430 | 0.0842 | 0.1617 | 0.0753 | 0.1683 | 0.3744 | 0.1667 | 0.2150 | 0.1747 | 0.0729 | 0.2006 | 0.1884 | 0.1905 | 0.2123 | 0.2162 | 0.1946 | 0.2154 | 0.1735 | 0.2206 | 0.2416 | *** | |||||||||
| DSY | 0.1574 | 0.1635 | 0.3868 | 0.1394 | 0.1530 | 0.0818 | 0.2399 | 0.0540 | 0.2017 | 0.1510 | 0.1711 | 0.0775 | 0.1307 | 0.1949 | 0.2184 | 0.2200 | 0.2283 | 0.2011 | 0.3709 | 0.1772 | 0.2261 | 0.2208 | 0.2214 | *** | ||||||||
| CJG | 0.1925 | 0.2182 | 0.1617 | 0.1110 | 0.3773 | 0.0807 | 0.1154 | 0.3891 | 0.2061 | 0.1556 | 0.1624 | 0.1916 | 0.1769 | 0.1938 | 0.2153 | 0.2021 | 0.2161 | 0.2000 | 0.2405 | 0.2051 | 0.2457 | 0.2273 | 0.2461 | 0.0884 | *** | |||||||
| LWM | 0.1963 | 0.1898 | 0.1586 | 0.0865 | 0.1169 | 0.0747 | 0.1219 | 0.0929 | 0.1838 | 0.1611 | 0.1569 | 0.2112 | 0.1676 | 0.1878 | 0.2096 | 0.1899 | 0.2372 | 0.1940 | 0.2139 | 0.2020 | 0.2087 | 0.2262 | 0.2177 | 0.0624 | 0.0477 | *** | ||||||
| YMG | 0.2042 | 0.1653 | 0.1529 | 0.1514 | 0.3719 | 0.1294 | 0.0908 | 0.1008 | 0.1959 | 0.1807 | 0.1848 | 0.1836 | 0.1757 | 0.2425 | 0.2009 | 0.2110 | 0.2145 | 0.2487 | 0.3815 | 0.1963 | 0.2152 | 0.2202 | 0.1631 | 0.0974 | 0.0756 | 0.1038 | *** | |||||
| HLG | 0.1864 | 0.1718 | 0.1442 | 0.1410 | 0.1563 | 0.1010 | 0.1148 | 0.0930 | 0.1837 | 0.1651 | 0.1817 | 0.1882 | 0.1880 | 0.2141 | 0.1936 | 0.1870 | 0.1941 | 0.2203 | 0.2240 | 0.1876 | 0.2141 | 0.2749 | 0.1910 | 0.1018 | 0.0725 | 0.0772 | 0.1090 | *** | ||||
| HJG | 0.1747 | 0.1707 | 0.1369 | 0.1481 | 0.1640 | 0.0825 | 0.1395 | 0.0919 | 0.2048 | 0.1697 | 0.1760 | 0.3937 | 0.1935 | 0.1896 | 0.2215 | 0.2220 | 0.2292 | 0.2148 | 0.2436 | 0.1844 | 0.2081 | 0.2465 | 0.0774 | 0.0795 | 0.0668 | 0.0818 | 0.0874 | 0.1415 | *** | |||
| SJG | 0.0991 | 0.1647 | 0.3648 | 0.1470 | 0.1761 | 0.0890 | 0.1111 | 0.0859 | 0.1821 | 0.1752 | 0.1673 | 0.2133 | 0.2131 | 0.1961 | 0.2184 | 0.2264 | 0.2330 | 0.2344 | 0.2159 | 0.2123 | 0.2628 | 0.2220 | 0.0717 | 0.0916 | 0.0581 | 0.0873 | 0.0939 | 0.1131 | 0.0940 | *** | ||
| CTG | 0.1864 | 0.1894 | 0.1617 | 0.1722 | 0.1339 | 0.1366 | 0.1021 | 0.1106 | 0.1617 | 0.1948 | 0.1581 | 0.1622 | 0.1975 | 0.1950 | 0.2127 | 0.2341 | 0.2409 | 0.2096 | 0.3624 | 0.2092 | 0.2344 | 0.2285 | 0.0839 | 0.0794 | 0.0526 | 0.1069 | 0.0928 | 0.0886 | 0.0880 | 0.0853 | *** | |
| LJP | 0.2107 | 0.1610 | 0.1560 | 0.1686 | 0.3655 | 0.1082 | 0.1086 | 0.3822 | 0.1968 | 0.1723 | 0.1860 | 0.1687 | 0.2021 | 0.1890 | 0.3834 | 0.2462 | 0.2231 | 0.2142 | 0.2213 | 0.2153 | 0.2700 | 0.2274 | 0.0894 | 0.1005 | 0.0805 | 0.0793 | 0.0868 | 0.0951 | 0.0847 | 0.0918 | 0.0907 | *** |
AMOVA (Table 5) was also performed for population differentiation to further evaluate genetic structure. Highly significant (P<0.000 2) genetic variance was expectedly observed among the populations and explained 63.27% of the total variance, supporting the results from the hierarchical and Bayesian clustering. Only 36.73% of the total genetic variance occurred within populations, indicating higher genetic differentiation between the populations than within each population and the emergence of genetic differentiation among populations. The Gst value (0.5823) (Table 2) showed there were more variation among populations than that within populations, confirming the AMOVA results.
Table 5. Analysis of molecular variance (AMOVA) for 640 individuals in 32 populations of S. hexandrum using 11 selected ISSR primers.
| Source of variation | df | Sum of squares | Mean squares | Variance component | Total variance (%) | P-value |
| Among population | 31 | 4,028.326 | 129.946 | 3.9128 | 63.27 | <0.000 2 |
| Within population | 608 | 950.912 | 1.564 | 1.564 | 36.73 | <0.000 2 |
Discussion
Genetic diversity of S. hexandrum in Qinling Mts
The ISSR markers developed in this study effectively revealed low genetic diversity within the S. hexandrum populations sampled in Qinling Mts. Populations are isolated given the population differentiation and clear clustering. The comparison of the average genetic diversity to that of S. hexandrum from Northwestern Himalayan region [34], [36], [38] and other Berberidaceae species [58], [59] based on the ISSR approach showed that S. hexandrum populations of Qinling Mts. have low genetic diversity (average He = 0.0621). The S. hexandrum in the Northwestern Himalayan region [34], [36], [38] showed high genetic variation (He = 0.2944, 0.092, respectively). S. hexandrum is native to the Himalayan region, growing in valleys with secondary vegetation, or under shrubs or around trees [2]. Their habitat is significantly different from that of Qinling Mts. S. hexandrum reproduces through vegetative reproduction and seeds. Insects and birds are limited in the high altitude regions, implying that S. hexandrum pollination is easier than in the Qingling Mts. The sizes of wild populations of S. hexandrum are very small and declines each year in Qinling Mts. because of habitat fragmentation and deterioration caused by human disturbance (overcollection due to economic interests). The rapid decrease in individuals in wild populations may also cause loss of the genetic diversity of this endangered species. Another Berberidaceae species, Dysosma versipellis, is an endangered species endemic to China and has been listed as a key protected wild plant in China due to habitat fragmentation or destruction. Qiu et al. found this species had high level of genetic diversity in China (He = 0.378) [58]. Dysosma pleiantha, a threatened medicinal plant species distributed in southeastern China, sexually and asexually reproduces. High He (0.364) was observed in this species [59]. The relatively high level of genetic variation observed within two species suggested that the balance between vegetative reproduction and sexual reproduction was more in favor of sexual reproduction in the populations D. versipellis and D. pleiantha than in the S. hexandrum populations. The He found in this study was 0.1434 at the species level, lower than those of some strictly self-pollinating soybean species (He = 0.1714) [60] and the self-pollinating Oryza granulata (He = 0.210) [61], which also indicated that potential selfing system in these populations reduced genetic diversity of S. hexandrum populations. Historical events are also responsible for the variation in genetic diversity [62].
Genetic diversity is affected by multiple factors, such as geographical distribution, mating system, life form, pollen and seed dispersal [63], [64]. Low genetic variation within populations could be attributed to seed dispersal and the predominant clonal reproduction in S. hexandrum in this high mountainous area. However, the genetic diversity of S. hexandrum is not much lower than other endangered species analyzed using ISSR markers, such as Leontice microrhyncha (Berberidaceae) (He = 0.021), which is a polycarpic perennial herb found in deciduous or coniferous forests in Korea and Northeast China [65]. For species L. microrhyncha, each pollinated flower produces an 8 mm berry and seed dispersal is restricted due to its heavy berry [66]. The genetic diversity of S. hexandrum was also higher than those of two other species, the endangered Pinus squamata (He = 0.020) [67] and the first-degree endangered species Manglietia decidua (He = 0.0637) [68]. P. squamata and M. decidua are extremely rare and endangered tree species in China. The extremely low genetic diversity of this two species could have resulted from the severe bottleneck effect during their evolutionary process. The gene drift and inbreeding may further decrease their genetic diversity in the shrinking populations. The weak competitive ability against broad - leaved trees and human activities may also accelerated the decrease of genetic variation.
S. hexandrum is reasonably long lived because its rhizomes easily reproduce, which could slow down the loss of genetic diversity. Pollen dispersal is generally restricted to a small region due to the large pollen size, which limits gene flow to increase or maintain genetic diversity. S. hexandrum is native to the Himalayan region, including China, India, Nepal, and Myanmar. No other genetic diversity studies of S. hexandrum exist in other locations. Further research should include more populations in other regions of China and countries.
High genetic differentiation and distinct genetic structure
High level of genetic differentiation and clear population structure was detected in this study. K = 3 in the Bayesian clustering as a meaningful value because group 1, 2 and 3 could be divided by detecting the within-group substructure. This result is also supported by distance-based clustering and PCoA. Groups 1 and 2 are genetically close but significantly different. Estimation of the number of clusters K should be treated with care because it is computationally difficult to obtain accurate estimates and the method merely provides an ad hoc approximation [52].
All sampled individuals were strongly assigned to their original populations, and all data strongly support the conclusion that the 32 S. hexandrum populations distributed in the Qinling Mts. are clustered into three major groups. These methods consistently showed that high genetic differentiation existed among S. hexandrum populations, which is consistent with genetic variation studies in certain selfing species [69]. This would mean that S. hexandrum should be a selfing species or a selfing predominant species, which is consistent with previous studies on S. hexandrum by Ma et al. [70]. Aside from the breeding system, the high genetic differentiation across populations may also be caused by genetic drift [71]. Wright [72] noted that genetic drift would lead a small population to emerge with a distinct genetic differentiation when the Nm value is lower than 1.0. The Nm of S. hexandrum (0.3587) determined using the POPGENE software was lower than 1.0 in the present study, which suggested that some genetic drift may have emerged among the populations of this species. The distribution of S. hexandrum populations obviously tend to fragment based on the field investigation, which is consistent with the possibility of genetic drift.
Migration of plant populations can occur through dispersal of pollen and seed [73]. But a number of factors such as fragmented geographical distribution, lack of pollinators or seed dispersers can be a barrier to gene flow between populations [74], [75]. Limited gene flow among S. hexandrum populations may be related to inbreeding of the species and limited seed propagation distance. Some studies have found that seed dispersal is the primary factor influencing variation of gene flow and population structure [76]. Heavy mature berries of S. hexandrum usually drop to the ground because of rain or wind, settling some seeds in the soil, whereas others are dispersed by cattle, birds, or humans. Therefore, the short distance of seed dispersal of S. hexandrum probably resulted in limited gene flow among populations. Mountain ranges and rivers are possible barriers to either dispersal of pollen or rhizomes of S. hexandrum, reproductively isolating the populations. The restriction of gene flow associated with geographical distance is consistent with the results of previous studies on this species [34], [35], [36].
Implications for conservation
S. hexandrum is a rare and threatened species [4] in China. Assessment of genetic diversity is important for designing conservation strategies for threatened and endangered species [77], [78]. The results of this study showed that there was low genetic diversity among S. hexandrum populations and genetic differentiation among populations was higher than within populations. Genetic diversity loss has deleterious effects on species fitness and threatens the population survival and could be the key reason that explains the endangerment of S. hexandrum in Qinling Mts. [79], [80]. The estimation of the genetic diversity and population genetic structure could provide bases for S. hexandrum conservation and its reasonable utilization. The results will help determine what to conserve and where and how to conserve this species.
The field survey showed that the habitats of some populations have been destroyed by human disturbance for great medical value. Damage to natural habitats would led to a decrease in population sizes and probably a subsequent increase in inbreeding, decreasing its genetic diversity. In situ conservation effectively and sustainably prevents this problem. The establishment of S. hexandrum reserves should be the primary method because the Qinling Mts. are situated in state forest conservation areas, where cutting and hunting are restricted. The management for the conservation of genetic variability in this species should aim to preserve not only large populations but also as many of the small populations outside nature reserves as possible. Reduced levels of genetic variation, especially in the smaller populations, will affect the species’ ability to adapt to changes in its habitat [81]. Positive correlations between population size, expected heterozygosity, and plant fitness were found in Gentiana pneumonanthe [82] and Arnica Montana [81]. Thus, policy plans should also be developed to stimulate seedling recruitment in the small populations (e.g., PAS and MXS). It may be dangerous to mix highly divergent populations because it could cause loss of adaptive diversity [45]. Therefore, it is necessary to improve gene flow among populations within each group through some artificial means, such as transplanting individuals (by seed, rhizomes from one population to another). Furthermore, to avoid human overcollection, greater awareness for S. hexandrum protections must be emphasized, and related forest departments should be encouraged to undertake conservation through an integrated conservation strategy based on demographic, ecological, and genetic aspects.
As a supplement to in situ conservation, ex situ conservation would also be feasible as underlined by other studies on endangered species [83], [84], [85]. Populations may be partially preserved through seed banks or in vitro germplasm collections. S. hexandrum has favorable sexual reproduction. Each plant produces approximately 60 seeds, with a maximum of approximately 180 seeds [70]. Seed collection is easier for S. hexandrum than other endangered species. Thus, a strategy involving extensive collection to ensure full sampling of genetic diversity, subsequent cultivation in a garden at least 1000 m above sea level [7], and reintroduction into their original wild habitats seems feasible, although S. hexandrum mainly grows wild on high altitude mountain ranges. For S. hexandrum populations in Qinling Mts., there are some preserved forest farms which could be used for relocation. However, ex situ conservation has many drawbacks because it is impossible to recreate the habitat as a whole. The new environment may have important ecological differences compared with the original habitat, and the approach is technically challenging and is often expensive. Therefore, ex situ conservation is recommended only to supplement in situ conservation or as a last resort. In vitro techniques are also proven to be an effective alternative means of propagation that facilitates the recovery of the rare and endangered S. hexandrum [33]. At present, an effective protocol of in vitro propagation, involving multiple shoot formation from zygotic embryos and subsequent rooting, could be available for S. hexandrum. In vitro propagation may well be used as a means to rescue zygotic embryos for this species. In vitro techniques induce variability, but plants raised from tissue cultures may be screened for useful somaclonal variants and exploited to obtain plants or cultures with high podophyllotoxin contents, which possibly reduces the pressure on natural S. hexandrum populations.
Most of the genetic diversity of the important medicinal and endangered species S. hexandrum in Qinling Mts. must be guaranteed with these combined and sustained efforts.
Supporting Information
Description of the sampling procedures.
(DOC)
Allele frequencies per locus.
(DOC)
Acknowledgments
The authors are also grateful to Genlu Bai, Guowei Xia and all the colleagues in the same laboratory for the assistance in the work.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the program from the Forestry Research Foundation for the Public Service Industry of China (200904004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Editorial committee, Chinese Academy of Sciences (2010) Vol. 29, Flora of China. Beijing: Science Press. 249–251 p. (In Chinese.).
- 2. Ying TS (1979) Study on Dysosma Woodson and Sinopodophyllum Ying (new genus) of the Berberidaceae. Acta Phytotax Sin 17: 17–23 (In Chinese with English abstract.) [Google Scholar]
- 3. Chatterjee R (1952) Indian podophyllum. Econ Bot 6: 342–354. [Google Scholar]
- 4.Fu LG (1992) Plant red book of China: Rare threatened plant. Beijing: Science Press. (In Chinese.).
- 5. Cragg GM, Newman DJ, Snader KM (1997) Natural products in drug discovery and development. J Nat Prod 60: 52–60. [DOI] [PubMed] [Google Scholar]
- 6. Dong JE, Ma XH, Wei Q, Peng SB, Zhang SC (2011) Effects of growing location on the contents of secondary metabolites in the leaves of four selected superior clones of Eucommia ulmoides . Ind Crop Prod 34: 1607–1614. [Google Scholar]
- 7. Li GM (1975) Introduction a medicine plant: Sinopodophyllum emodi Wall. var. Chinense Sprague . Journal of Botony 2: 28 (In Chinese with English abstract.) [Google Scholar]
- 8. Fay DA, Ziegler HW (1985) Botanical source differentiation of Podophyllum resin by high performance liqid chromatography. J Liq Chromatogr 8: 1501–1506. [Google Scholar]
- 9. Giri A, Narasu ML (2000) Production of podophyllotoxin from Podophyllum hexandrum: a potential natural product for clinically useful anticancer drugs. Cytotechnology 34: 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stahelin HF, Von Warhurg A (1991) The chemical and biological route from podophyllotoxin glucoside to etoposide: ninth cain memorial avard lecture. Cancer Res 51: 5–11. [PubMed] [Google Scholar]
- 11. Kamil WM, Dewick PM (1986) Biosynthetic relationship of aryltetralin lactone lignans to dibenzylbutyrolactone lignans. Phytochemistry 25: 2093–2102. [Google Scholar]
- 12. Holthuis JJM (1988) Etoposide and teniposide. Pharm Weekbl 10: 101–116. [DOI] [PubMed] [Google Scholar]
- 13. Imbert TF (1998) Discovery of podophyllotoxin. Biochimie 80: 207–222. [DOI] [PubMed] [Google Scholar]
- 14.Moraes RM, Lata H, Bedir E, Maqbool M, Cushman K (2002) The American mayapple and its potential for podophyllotoxin production. In: Janick J, Whipkey A, editors. Trends in new crops and new uses. Alexandria, VA: ASHS Press. pp. 527–532.
- 15. Yousefzadi M, Sharifi M, Behmanesh M, Moyano E, Bonfill M, et al. (2010) Podophyllotoxin: current approaches to its biotechnological production and future challenges. Eng Life Sci 4: 281–292. [Google Scholar]
- 16. Lata H, Moraes RM, Bertoni B, Pereira Ana MS (2010) In vitro germplasm conservation of Podophyllum peltatum L. under slow growth conditions. In Vitro Cell Dev Biol–Plant 46: 22–27. [Google Scholar]
- 17. Lin MC, Lin JH, Chen SK, Cheng YW, Cheng HW (2008) Simultaneous determination of podophyllotoxin, quercetin and kaempferol in podophyllin by liquid chromatography tandem mass spectrometry. J Food Drug Anal 16: 29–40. [Google Scholar]
- 18. Zhou Y, Jiang SY, Ding LS, Cheng SW, Xu HX, et al. (2008) Chemical fingerprinting of medicinal plants ‘‘Gui-jiu’’ by LC-ESI multiple-stage MS. Chromatographia 68: 781–789. [Google Scholar]
- 19. Purohit MC, Bahuguna R, Maithani UC, Rawat MSM (1999) Variation in podophylloresin and podophyllotoxin content in different populations of Podophyllum hexeandrum . Curr Sci India 77: 1078–1079. [Google Scholar]
- 20. Zhao CQ, Cao W, Nagatsu A, Ogihara Y (2001) Three new glycosides from Sinopodophyllum emodi (Wall.) Ying. Chem Pharm Bull 49: 1474–1476. [DOI] [PubMed] [Google Scholar]
- 21. Kong Y, Xiao JJ, Meng SC, Dong XM, Ge YW, et al. (2010) A new cytotoxic flavonoid from the fruit of Sinopodophyllum hexandrum . Fitoterapia 81: 367–370. [DOI] [PubMed] [Google Scholar]
- 22. Zhao CQ, Zhu YY, Chen SY, Ogihara Y (2011) Lignan glucoside from Sinopodophyllum emodi and its cytotoxic activity. Chinese Chem Lett 22: 181–184. [Google Scholar]
- 23. Sun YJ, Li ZL, Chen H, Liu XQ, Zhou W, et al. (2011) Three new cytotoxic aryltetralin lignans from Sinopodophyllum emodi. . Bioorg Med Chem Lett 21: 3794–3797. [DOI] [PubMed] [Google Scholar]
- 24. Qin Y, Gui MY, Yu LN, Ru H, Jin YR, et al. (2009) RP-HPLC determination of lignans in Sinopodophyllum emodi Wall. Chin J Pharm Anal 29: 1490–1493 (In Chinese with English abstract.) [Google Scholar]
- 25. Huang K, Jiang W, Zhao JF, Wang CH, Liu X, et al. (2012) Determination of podophyllotoxin and total lignans in Sinopodophyllum emodi . China journal of Chinese material medicia 37: 1360–1365 (In Chinese with English abstract.) [PubMed] [Google Scholar]
- 26. Inamori Y, Kubo M, Tsujibo H, Ogawa M, Baba K, et al. (1986) The biological activities of podophyllotoxin compounds. Chem pharm Bull 34: 3928–3932. [DOI] [PubMed] [Google Scholar]
- 27. Goel HC, Prasad J, Sharma A, Singh B (1998) Antitumour and radioprotective action of Podophyllum hexandrum . Indian J Exp Biol 36: 583–587. [PubMed] [Google Scholar]
- 28. Chattopadhyay S, Bisaria VS, Panda AK, Srivastava AK (2004) Cytotoxicity of invitro produced podophyllotoxin from Podophyllum hexandrum on human cancer cell line. Nat Prod Res 18: 51–57. [DOI] [PubMed] [Google Scholar]
- 29. Reddy PB, Paul DV, Agrawal SK, Saxena AK, Kumar HMS, et al. (2008) Design, synthesis, and biological testing of 4b-[(4-Substituted)-1,2,3-triazol-1-yl] podophyllotoxin analogues as antitumor agents. Arch Pharm Chem Life Sci 341: 126–131. [DOI] [PubMed] [Google Scholar]
- 30. Zhu CG, Yang J, Xiong Y (2004) Research progress in natural antineoplastic podophyllotoxin and its derivatives. Drug Eval 1: 306. [Google Scholar]
- 31. Wang DW, Guo FX, Ma XY (1997) The antitumor activity of Sinopodophyllum emodi . Chinese medicine matria 20: 571–574 (In Chinese with English abstract.) [PubMed] [Google Scholar]
- 32. Li GY (2005) Effects of ethanol extracts from Podophylum emodi var. on proliferation and apoptosis of breast careinoma cell line MCF–7. Chinese Journal of New Drugs 26: 2185–2188 (In Chinese with English abstract.) [Google Scholar]
- 33. Nadeem M, Palni LMS, Purohit AN, Pandey H, Nandi SK (2000) Propagation and conservation of Podophyllum hexandrum Royle: an important medicinal herb. Bio Conserv 92: 121–129. [Google Scholar]
- 34. Alam MA, Naik PK, Gulati P, Gulati AK, Mishra GP (2008) Characterization of genetic structure of Podophyllum hexandrum populations, an endangered medicinal herb of Northwestern Himalaya, using ISSR-PCR markers and its relatedness with podophyllotoxin content. Afr J Biotechnol 7: 1028–1040. [Google Scholar]
- 35. Alam MA, Gulati P, Aswini KG, Gyan PM, Pradeep KN (2009) Assessment of genetic diversity among Podophyllum hexandrum genotypes of Northwestern Himalayan region for podophyllotoxin production. Indian J Biotechnol 8: 391–399. [Google Scholar]
- 36. Naik PK, Alam MA, Singh H, Goyal V, Parida S, et al. (2010) Assessment of genetic diversity through RAPD, ISSR and AFLP markers in Podophyllum hexandrum: a medicinal herb from the Northwestern Himalayan region. Physiol Mol Biol Plants 16: 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiao M, Li Q, Guo L, Luo T, Duan WX, et al. (2006) AFLP analysis of genetic diversity of the endangered species Sinopodophyllum hexandrum in the tibetan region of Sichuan province, China. Biochem Genet 44: 47–59. [DOI] [PubMed] [Google Scholar]
- 38. Xiao M, Li Q, Wang L, Guo L, Li J, et al. (2006) ISSR analysis of the genetic diversity of the endangered species Sinopodophyllum hexandrum (Royle) Ying from Western Sichuan province, China. J Integr Plant Bio 48: 1140–1146. [Google Scholar]
- 39. Li Y, Zhai SN, Qiu YX, Guo YP, Ge XJ, et al. (2011) Glacial survival east and west of the ‘Mekong–Salween Divide’ in the Himalaya–Hengduan mountains region as revealed by AFLPs and cpDNA sequence variation in Sinopodophyllum hexandrum (Berberidaceae). Mole Phylogenet Evol 59: 412–424. [DOI] [PubMed] [Google Scholar]
- 40. Totland Ø (1999) Effects of temperature on performance and phenotypic selection on plant traits in alpine Ranunculus acris. . Oecologia 120: 242–251. [DOI] [PubMed] [Google Scholar]
- 41. Esselman EJ, Li J, Crawford DJ, Winduss JL, Wolfe AD (1999) Clonal diversity in the rare Calamagrostis porteri ssp. insperata (Poaceae): Comparative results for allozymes and random amplified polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR) marker. Mol Ecol 8: 443–451. [Google Scholar]
- 42. Ge XJ, Sun M (1999) Reproductive biology and genetic diversity of a cryptoviviparous mangrove Aegiceras corniculatum (Myrsinaceae) using allozyme and inter-simple sequence repeat (ISSR) analysis. Mol Ecol 8: 2061–2069. [DOI] [PubMed] [Google Scholar]
- 43. Li HS, Chen GZ (2004) Genetic diversity of Sonneratia alba in China detected by inter-simple sequence repeat (ISSR) analysis. Acta Bot Sin 46: 515–521. [Google Scholar]
- 44. Thriveni HN, Sumangala RC, Shivaprakash KN, Ravikanth G, Vasudeva R, et al. (2014) Genetic structure and diversity of Coscinium fenestratum: a critically endangered liana of Western Ghats, India. Plant Syst Evol 300: 403–413. [Google Scholar]
- 45. Tang N, Mo G, van Tuyl JM, Arens P, Liu JJ, et al. (2014) Genetic diversity and structure of Lilium pumilum DC. in southeast of Qinghai–Tibet plateau. Plant Syst Evol 300: 1453–1464. [Google Scholar]
- 46. Bornet B, Branchard M (2001) Nonanchored inter simple sequence repeat (ISSR) markers: reproducible and specific tools for genome fingerprinting. Plant Mol Biol Rep 19: 209–215. [Google Scholar]
- 47. Nagaoka T, Ogihara Y (1997) Applicability of inter-simple sequence repeat polymorphisms in wheat for use as DNA markers in comparison to RFLP and RAPD markers. Theor Appl Genet 94: 597–602. [Google Scholar]
- 48.Li JJ, Zhu ZC, Min ZL (1989) Comprehensive survey of the Taibai Mountain preserve (1st ed). Xi’an: Shaanxi Normal University Press. 1–9 p. (In Chinese.).
- 49.Yeh FC, Yang RC, Boyle T, Ye ZH, Mao JX (1997) POPGENE: The user friendly shareware or population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Edmonton.
- 50. Liu KJ, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21: 2128–2129. [DOI] [PubMed] [Google Scholar]
- 51. Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 52. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 54. Jensen RJ (1989) Ntsys-Pc-numerical taxonomy and multivariate analysis system-version 1.40. Q Rev Biol 64: 250–252. [Google Scholar]
- 55. Exeoeffr L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: applications to human mitochondrial DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller MP (1997) Tools for Population Genetic Analysis (TFPGA), Vesion 1.3. USA: Department of Biological Sciences, Northern Arizona University, Ariozna.
- 57. Larson SR, Palazzo AJ, Jensen KB (2003) Identification of western wheatgrass cultivars and accessions by DNA fingerprinting and geographic provenance. Crop Sci. 43: 394–401. [Google Scholar]
- 58. Qiu YX, Li JH, Liu HL, Chen YY, Cheng XF (2006) Population structure and genetic diversity of Dysosma versipellis (Berberidaceae), a rare endemic from China. Biochem Syst Ecol 34: 745–752. [Google Scholar]
- 59. Zong M, Liu HL, Qiu YX, Yang SZ, Zhao MS, et al. (2008) Genetic diversity and geographic differentiation in the threatened species Dysosma pleiantha in China as revealed by ISSR analysis. Biochem genet 46: 180–196. [DOI] [PubMed] [Google Scholar]
- 60. Jin Y, Zhang WJ, Fu D X, Lu BR (2003) Sampling strategy within a wild soybean population based on its genetic variation detected by ISSR markers. Acta Bot Sin 45: 995–1002 (In Chinese with English abstract.) [Google Scholar]
- 61. Wu CJ, Chen ZQ, Huang XQ, Yin SH, Cao KM, et al. (2004) Genetic diversity among and within populations of Oryza granulata from Yunnan of China revealed by RAPD and ISSR markers: implications for conservation of the endangered species. Plant Sci 167: 35–42. [Google Scholar]
- 62.Karron JD (1991) Patterns of genetic variation and breeding systems in rare plant species. In: Falk DA, Holsinger, KE, editors. Genetics and conservation of rare plants. New York: Oxford University Press. pp. 87–98.
- 63. Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forest 6: 95–124. [Google Scholar]
- 64. Ohsawa T, Saito Y, Sawada H, Ide Y (2008) Impact of altitude and topography on the genetic diversity of Quercus serrata populations in the Chichibu Mountains, central Japan. Flora 203: 187–196. [Google Scholar]
- 65.Lee TB (1980) Illustrated flora of Korea. Seoul: Hyang-mun Pub. Co. (In Korean.).
- 66. Chang CS, Kim H, Park TY, Maunder M (2004) Low levels of genetic variation among southern peripheral populations of the threatened herb, Leontice microrhyncha (Berberidaceae) in Korea. Biol Conserv 119: 387–396. [Google Scholar]
- 67. Zhang ZY, Li DZ (2003) Conservation genetics of extremely endangered pine, Pinus squamata. . Acta Botanica Yunnanica 25: 544–55 (In Chinese with English abstract.) [Google Scholar]
- 68. Liao WF, Xia NH, Deng YF, Zheng QY (2004) Study on genetic diversity of Manglietia decidua . Acta Botanica Yunnanica 26: 58–64 (In Chinese with English abstract.) [Google Scholar]
- 69. Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Plant Ecol Evol Syst 3: 93–114. [Google Scholar]
- 70. Ma SB, Xu ZY, Hu ZH (1997) A contribution to the reproductive biology of Sinopodophyllum hexandrum (Role) Ying (Berberidaceae). Acta Bot Boreal – Occident Sin l7: 49–55 (In Chinese with English abstract.) [Google Scholar]
- 71.Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Khaler AL, Weir BS, editors. Plant population genetics, breeding and genetic resources. Sunderland: Sinauer Associates, Inc., Publisher. pp. 43–63. [Google Scholar]
- 72. Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19: 395–420. [Google Scholar]
- 73. Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plant-populations. Heredity 72: 250–259. [Google Scholar]
- 74. Slatkin M (1985) Gene flow in natural-populations. Annu Rev Ecol Syst 16: 393–430. [Google Scholar]
- 75. Zhou TH, Qian ZQ, Li S, Guo ZG, Huang ZH, et al. (2010) Genetic diversity of the endangered Chinese endemic herb Saruma henryi Oliv. (Aristolochiaceae) and its implications for conservation. Popul Ecol 52: 223–231. [Google Scholar]
- 76. Kalisz S, Hanzawa FM, Tonsor SJ, Thiede DA, Voigt S (1999) Ant-mediated seed dispersal alters pattern of relatedness in a population of Trillium grandifloru . Ecology 80: 2620–2634. [Google Scholar]
- 77.Hamrick JL (1983) The distribution of genetic variation within and among natural plant populations. In: Schonewald-Cox CM, Chambers SM, McBryde B, Thomas WL, editors. Genetics and conservation. Menlo Park: Benjamin Cummings Publishing Company. pp. 335–348.
- 78. Francisco-Ortega J, Santos-Guerra A, Kim SC, Crawford DJ (2000) Plant genetic diversity in the Canary Islands: a conservation perspective. Am J Bot 87: 909. [PubMed] [Google Scholar]
- 79. Malone CL, Knapp CR, Taylor JF, Davis SK (2003) Genetic consequences of pleistocene fragmentation: isolation, drift, and loss of diversity in rock iguanas (Cyclura). Conserv Genet 4: 1–15. [Google Scholar]
- 80. Reed DH (2003) Correlation between fitness and genetic diversity. Conserv Biol 17: 230–237. [Google Scholar]
- 81. Luijten SH, Dierick A, Oostermeijer JGB, Raijmann LEL, Den Nijs JCM (2000) Population size, genetic variation and reproductive success in the rapidly declining, self-incompatible perennial (Arnica montana) in the Netherlands. Conserv Biol 14: 1776–1787. [DOI] [PubMed] [Google Scholar]
- 82. Oostermeijer JGB, Van Eijck MW, Van Leeuwen NC, Den Nijs JCM (1995) Analysis of the relationship between allozyme heterozygosity and fitness in the rare Gentiana pneumonanthe L. J Evol Biol. 8: 739–757. [Google Scholar]
- 83. Bunn E (2005) Development of in vitro methods for ex situ conservation of Eucalyptus impensa, an endangered mallee from southwestern Western Australia. Plant Cell Tiss Org 83: 97–102. [Google Scholar]
- 84. Cochrane JA, Crawford AD, Monks LT (2007) The significance of ex situ seed conservation to reintroduction of threatened plants. Aust J Bot 55: 356–361. [Google Scholar]
- 85. Li QM, Xu ZF, He TH (2002) Ex situ genetic conservation of endangered Vatica guangxiensis (Dipterocarpaceae) in China. Biol Conserv 106: 151–156. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of the sampling procedures.
(DOC)
Allele frequencies per locus.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.