Abstract
Although numerous physiological studies have addressed the interactions between brassinosteroids and auxins, little is known about the underlying molecular mechanisms. Using an Affymetrix GeneChip representing approximately 8,300 Arabidopsis genes, we studied comprehensive transcript profiles over 24 h in response to indole-3-acetic acid (IAA) and brassinolide (BL). We identified 409 genes as BL inducible, 276 genes as IAA inducible, and 637 genes in total. These two hormones regulated only 48 genes in common, suggesting that most of the actions of each hormone are mediated by gene expression that is unique to each. IAA-up-regulated genes were enriched in genes regulated in common. They were induced quickly by IAA and more slowly by BL, suggesting divergent physiological roles. Many were early auxin-inducible genes and their homologs, namely SAUR, GH3, and IAA. The comprehensive comparison also identified IAA- and BL-specific genes, which should help to elucidate the specific actions of each hormone. The identified genes were classified using hierarchical clustering based on the similarity of their responses to the two hormones. Gene classification also allowed us to analyze the frequency of cis-elements. The TGTCTC element, a core element of the previously reported auxin response element, was not enriched in genes specifically regulated by IAA but was enriched in the 5′-flanking region of genes up-regulated by both IAA and BL. Such gene classification should be useful for predicting the functions of unknown genes, to understand the roles of these two hormones, and the promoter analysis should provide insight into the interaction of transcriptional regulation by the two hormones.
Auxins play critical roles in the major growth responses during plant development. At the cellular level, auxin acts as a signal for division, expansion, and differentiation throughout the plant life cycle. At the level of the whole plant, auxin plays an important role in root formation, apical dominance, the tropic response, and senescence. By contrast, less attention has been directed to brassinosteroids (BRs) since Grove et al. (1979) isolated the first BR, brassinolide (BL), from oilseed rape (canola [Brassica napus]) in 1979. BRs promote stem elongation and inhibit root elongation in various plant species. Nevertheless, only a small number of researchers accepted the hormonal status of BRs before BR mutants were discovered (Clouse, 1996, 1997, 2002) because BRs have activity similar to that of other plant hormones, especially auxins. BRs also interact synergistically with auxin in hypocotyl elongation in several plant species (for review, see Sasse, 1999) and in ethylene production (Arteca et al., 1988). Several authors have proposed that BR-induced effects are mediated via auxin, with BR treatment altering the levels of endogenous auxin or enhancing the sensitivity to auxin (Mandava, 1988; Sasse, 1999). Although numerous physiological studies have addressed the interactions between BRs and auxins, little is known about the underlying molecular mechanisms. Clouse and colleagues made extensive comparisons of the physiological effects of BRs and auxins and conducted molecular analyses of auxin-inducible genes and auxin-insensitive mutants in soybean (Glycine max), tomato (Lycopersicon esculentum), and Arabidopsis. In soybean and tomato, members of the small auxin up RNAs (SAUR) and GH3 gene families are not induced rapidly during BR-promoted cell expansion but are induced by BR subsequently, even after the beginning of cell elongation, with different kinetics than those induced by auxin treatment (Clouse et al., 1992; Zurek et al., 1994). Mass spectrometry analysis of the free indole-3-acetic acid (IAA) levels in BR-treated tissues showed that the free IAA levels decreased in BR-treated soybean epicotyls (Zurek et al., 1994). It was concluded that BR does not stimulate SAUR gene transcription via increased IAA levels. The auxin-insensitive tomato mutant dgt (Zurek et al., 1994) and the Arabidopsis mutant axr1-3 (Clouse et al., 1993) are sensitive to BR. Several studies have concluded that auxin signaling pathways are unlikely to mediate the promotion of cell elongation in soybean and tomato by BR or the inhibition of root elongation in Arabidopsis by BR (Clouse et al., 1992, 1993; Zurek et al., 1994). Conversely, McKay et al. (1994) reported that IAA levels are reduced in the youngest internodes of the pea (Pisum sativum) BR-insensitive mutant lka and the BR-deficient mutant lkb (Nomura et al., 1997) as compared with the wild type (WT) using mass spectrometry. This suggested that the endogenous BRs might increase the endogenous IAA content. Therefore, the mechanism of the interaction of these two hormones is still controversial.
The microarray technique is a powerful tool for obtaining an overview of hormone actions using inducible genes as molecular markers. Recently, several microarray studies have examined auxin- and BR-regulated genes. Tian et al. (2002) showed how SHY2/IAA3 affects the expression of auxin-related genes using IAA or mock-treated WT and shy2 mutants and identified 100 auxin-regulated genes 2 h after IAA treatment. We identified IAA-responsive genes at 15 min (Sawa et al., 2002) and suggested that auxin signals are mediated by a set of diverse transcription factors. BR-responsive genes have also been examined in comprehensive studies of BR-regulated genes in CPD antisense, dwf1-6 (Müssig et al., 2002), det2, and bri1 (Goda et al., 2002) mutants shortly after BR treatment or in comparison with WT plants. These genes have also been studied in the characterization of bes1-D, which shows constitutive BR response phenotypes (Yin et al., 2002a). Interestingly, all three reports revealed quick up-regulation of the early auxin-inducible genes in response to BR and indicated overlap of auxin- and BR-regulated genes. By contrast, BRs did not induce all of the early auxin-inducible genes within 3 h (Goda et al., 2002), and it was not clear whether these BR-insensitive auxin-responsive genes (referred to here as auxin-specific genes) are induced subsequently with BR treatment or vice versa (i.e. auxin-insensitive BR-response genes [referred to here as BR-specific genes] are induced subsequently with auxin treatment). To our knowledge, no study has attempted to compare the genes responding to these two hormones comprehensively or the comprehensive time-course response to either of these hormones. To reveal the relationship between auxin and BR actions, we studied the time course of auxin- and BR-regulated genes using an Affymetrix GeneChip representing 8,300 Arabidopsis genes. The results allowed the most comprehensive comparison of auxin- and BR-responsive gene expression to date under the same experimental conditions. This paper presents the kinetics of the auxin and BR responses using responsive genes as molecular markers, revealing the common and distinct actions of these two hormones.
RESULTS AND DISCUSSION
Identification of IAA-Regulated or BR-Regulated Genes
Previously, we showed that BR-regulated genes generally respond to BL in a similar fashion in the WT and a BR-deficient mutant, det2, but that the det2 response to BL is stronger than that of the WT (Goda et al., 2002). Consequently, we used det2 here and exposed plants to 10 nm BL for up to 24 h to identify BL-regulated genes. Conversely, we exposed WT (Colombia) Arabidopsis to 1 μm IAA for up to 24 h to identify IAA-regulated genes. Transcript abundance was then compared with the mock-treated samples at each time point using the Affymetrix Arabidopsis Genome Array, which represents about 8,300 genes. Hybridization was performed with biotin-labeled cRNA samples prepared from different plant samples in independent hormone-treatment experiments. The signal log ratio (SLR), which is the ratio of the hybridization signals of mock- and hormone-treated plants on a log scale (base 2), was calculated using Affymetrix Microarray Suite (MAS) version 5.0 software. An SLR of 1, for example, indicates a 2-fold increase in the transcript level, and −1 indicates a 2-fold decrease. We extracted genes with expression ratios greater than 2 (i.e. SLR < −1 or SLR > 1) as compared with the mock treatment at each time point. We also used Detection and Change values calculated with MAS to exclude false-positive signals resulting from cross-hybridization or noise. Considering SLR and these two parameters, genes that were reproducibly regulated by BL or IAA in independent experiments were identified as BL- or IAA-regulated genes, respectively. These genes were divided into three groups: genes specifically regulated by IAA (Table I), genes specifically regulated by BL (Table II), and genes regulated by both BL and IAA (Table III). They are listed with the mean and se of SLR both before and after signal amplification with phycoerythrin-streptavidin (Supplemental Tables I–III; available at www.plantphysiol.org). We classified genes that responded to hormone treatment within 3 h as early inducible genes (including moderate genes) and those responding after 3 h as late inducible genes. The genes were further classified by the direction (up and down) and time (early, late, or both) of their response. Consequently, we extracted 409 BL-inducible genes, 276 IAA-inducible genes, and 637 genes in total.
Table I.
Genes specifically regulated by IAA
| Affymetrix No. | Arabidopsis Genome Initiative Identification (AGI ID) | Gene Name or Comment | Group |
|---|---|---|---|
| Genes Down-Regulated at Early Stage | |||
| 19084_AT | At2g07400 | Putative retroelement pol polyprotein | A |
| 20675_AT | At2g20750 | β-expansin At-EXPB1 | H |
| 13463_AT | At4g33790 | Male sterility 2-like protein | H |
| 20007_AT | At4g18610 | Putative protein | H |
| 20420_AT | At4g19810 | Putative chitinase | J |
| 19008_S_AT | At2g28470 | Putative β-galactosidase | J |
| 19942_AT | At1g08190 | Vacuolar assembly protein vps41, putative | J |
| 20555_S_AT | At4g12280 | ACC synthase (AtACS-6) | J |
| 19847_S_AT | At4g19030 | Putative water channel | J |
| 14111_S_AT | At4g13900 | Putative disease resistance protein | L |
| 14626_AT | At5g35840 | Phytochrome C (sp P14714) | L |
| 17331_AT | At4g02420 | Ser/Thr kinase-like protein | L |
| Genes Down-Regulated at Early and Late Stages | |||
| 14076_AT | At2g20520 | Putative pollen surface protein | G |
| 19672_AT | At1g43160 | AP2 domain containing protein, putative | I |
| Genes Down-Regulated at Late Stage | |||
| 16832_AT | At1g05660 | Putative polygalacturonase | G |
| 20045_AT | At2g33790 | Putative extensin | G |
| 20269_AT | At2g45220 | Putative pectinesterase | G |
| 16630_S_AT | At4g25820 | XTR9 | G |
| 19294_AT | At4g28850 | XTR18 | G |
| 12365_AT | At4g37160 | Pectinesterase-like protein | G |
| 15208_AT | At4g40090 | Arabinogalactan-protein (AGP3) | G |
| 16489_AT | At5g46900 | Extensin-like protein | G |
| 18533_AT | At5g65730 | XTR10 | G |
| 15954_AT | At1g66270 | β-glucosidase | G |
| 17100_S_AT | At2g32300 | Putative uclacyanin I | G |
| 12746_I_AT | At4g11320 | Cys proteinase-like protein | G |
| 19348_AT | At4g26220 | Caffeoyl CoA O-methyltransferase-like protein | G |
| 13686_AT | At2g44110 | Similar to Mlo proteins from Hordeum vulgare | G |
| 14013_AT | At4g11210 | Putative disease resistance protein | G |
| 14550_AT | At4g23690 | Putative disease resistance response protein | G |
| 12463_AT | At4g29690 | Nucleotide pyrophosphatase-like protein | G |
| 12933_R_AT | At4g33720 | Pathogenesis-related protein 1 precursor | G |
| 20442_I_AT | At1g16410 | CYP79F1 | G |
| 14366_AT | At1g67110 | CYP709A2 | G |
| 18852_AT | At2g25160 | CYP82F1 | G |
| 17932_AT | At1g05250 | Class III peroxidase PER2 | G |
| 17102_S_AT | At1g05260 | Class III peroxidase PER3 | G |
| 19621_AT | At5g42180 | Class III peroxidase PER64 | G |
| 19622_G_AT | At5g42180 | Class III peroxidase PER64 | G |
| 15101_S_AT | At3g14940 | Phosphoenolpyruvate carboxylase (PEPC) | G |
| 18743_F_AT | At5g07690 | Putative transcription factor | G |
| 17572_S_AT | At1g64780 | Ammonium transport protein (AMT1) | G |
| 17571_AT | At3g24300 | Ammonium transporter | G |
| 16229_AT | At4g12030 | Putative transport protein | G |
| 15666_AT | At5g59520 | Putative zinc transporter ZIP2 | G |
| 19737_AT | At1g01580 | Putative protein | G |
| 14901_AT | At1g62280 | Putative protein | G |
| 12758_AT | At2g01530 | Putative protein | G |
| 12021_AT | At2g25260 | Putative protein | G |
| 15137_S_AT | At2g44790 | Putative protein | G |
| 18814_AT | At2g45750 | Putative protein | G |
| 19911_AT | At2g48080 | Putative protein | G |
| 17748_AT | At4g20460 | UDP-glucose 4-epimerase-like protein | H |
| 20524_AT | At1g62560 | Similar to flavin-containing monooxygenase (sp P36) | H |
| 13623_R_AT | At4g20820 | Reticuline oxidase-like protein | H |
| 15775_AT | At4g29740 | Cytokinin oxidase (CKX4) | H |
| 14932_AT | At2g01880 | Putative purple acid phosphatase | H |
| 19068_I_AT | At1g14185 | Putative protein | H |
| 20446_S_AT | At1g05570 | Putative glucan synthase | I |
| 20448_AT | At4g00680 | Putative actin-depolymerizing factor | I |
| 15049_AT | At4g02270 | Extensin-like protein | I |
| 17485_S_AT | At4g16260 | β-1,3-glucanase class I precursor | I |
| 20431_AT | At4g22460 | Extensin-like protein | I |
| 20597_AT | At1g53940 | Fatty acids and isoprenoids lipase-like protein | I |
| 20341_AT | At2g29750 | Putative flavonol 3-O-glucosyltransferase | I |
| 13685_AT | At1g61560 | Mlo protein, putative | I |
| 16048_AT | At1g73330 | Dr4 (protease inhibitor) | I |
| 18983_S_AT | At4g12520 | pEARLI 1-like protein | I |
| 16045_AT | At4g15390 | HSR201-like protein | I |
| 20367_S_AT | At1g30870 | Class III peroxidase PER7 | I |
| 18150_AT | At2g39040 | Class III peroxidase PER24 | I |
| 16971_S_AT | At3g01190 | Class III peroxidase PER27 | I |
| 15562_AT | At4g26010 | Class III peroxidase PER44 | I |
| 12400_AT | At5g19890 | Class III peroxidase PER59 | I |
| 20296_S_AT | At5g67400 | Class III peroxidase PER73 | I |
| 18459_AT | At4g40010 | Putative protein kinase | I |
| 16005_AT | At4g17340 | Membrane channel-like protein | I |
| 12004_AT | At4g35060 | Farnesylated protein (ATFP6) | I |
| 18888_AT | At1g15380 | Putative protein | I |
| 16016_AT | At2g01520 | Putative protein | I |
| 20514_I_AT | At2g15370 | Putative protein | I |
| 20176_AT | At2g36100 | Putative protein | I |
| 20698_AT | At2g40330 | Putative protein | I |
| 19195_AT | At2g44380 | Putative protein | I |
| 15021_AT | At4g25220 | Putative protein | I |
| 16873_I_AT | At2g32530 | Putative cellulose synthase | K |
| 19374_AT | At2g28670 | Putative disease resistance response protein | K |
| 12139_AT | At4g13580 | Putative protein | K |
| 13538_AT | At4g20780 | Calcium-binding protein-like | K |
| 16483_AT | At5g65210 | Putative transcription factor | K |
| 12341_S_AT | At4g20110 | Vacuolar sorting receptor-like protein | K |
| 20180_AT | At4g26320 | Putative protein | K |
| 19592_AT | At3g49960 | Class III peroxidase PER35 | N |
| 20366_AT | At5g22410 | Class III peroxidase PER60 | N |
| 15851_I_AT | At2g27370 | Putative protein | N |
| Genes Up-Regulated at Early Stage | |||
| 20488_AT | At4g34770 | SAUR-1 | B |
| 14032_AT | At4g37370 | CYP81D8 | C |
| 12891_AT | At4g11280 | ACC synthase (AtACS-6) | C |
| 19409_AT | \NULL | IAA5 | C |
| 18216_AT | At1g27730 | Putative zinc finger protein | C |
| 17303_S_AT | At2g38470 | Putative WRKY-type DNA binding protein | C |
| 14711_AT | At2g40140 | Putative zinc finger protein | C |
| 16539_S_AT | At4g17490 | Ethylene-responsive element binding factor (AtERF6) | C |
| 15288_AT | At2g42430 | Putative protein | C |
| 17573_AT | At1g70940 | Auxin transport protein REH1 | D |
| 16610_S_AT | At1g19050 | Putative protein | D |
| 15665_AT | At5g04340 | Putative C2H2 zinc finger transcription factor | F |
| 19695_AT | At4g38840 | SAUR-14 | K |
| 16712_AT | At2g35710 | Putative glycogenin | M |
| 20144_AT | At4g25390 | Putative protein kinase | M |
| 18258_AT | At2g18210 | Putative protein | M |
| Genes Up-Regulated at Early and Late Stages | |||
| 20297_AT | At1g05680 | Putative indole-3-acetate β-glucosyltransferase (UGT74E2) | A |
| 16278_AT | At2g37030 | SAUR-46 | A |
| 17107_AT | At2g22810 | 1-aminocyclopropane-1-carboxylate synthase (ACS4) | B |
| 13661_AT | At1g52830 | IAA6 | B |
| 16807_AT | At2g34650 | Putative protein kinase | B |
| 14112_AT | At2g41820 | Putative protein kinase | B |
| 13439_AT | At4g22780 | Translation factor EF-1α-like protein | B |
| 12553_AT | At2g14960 | AtGH3-1 | C |
| 13293_S_AT | At2g33310 | IAA13 | C |
| 13297_AT | At3g23030 | IAA2 | C |
| 13289_S_AT | At4g14560 | IAA1 | C |
| 16989_AT | At4g27260 | AtGH3-5 | C |
| 13291_AT | At4g28640 | IAA11 | C |
| 16878_AT | At1g51170 | Putative protein kinase | C |
| 18950_AT | At5g47370 | HAT2 | C |
| 12090_AT | At2g39370 | Putative protein | C |
| 16381_AT | At2g42800 | Putative protein | C |
| Genes Up-Regulated at Late Stage | |||
| 20265_AT | At1g22880 | Putative endo-1,4-β-glucanase | A |
| 17268_AT | At2g43860 | Putative polygalacturonase | A |
| 18912_AT | At4g13210 | Pectate lyase-like protein | A |
| 16974_AT | At4g15160 | Cell wall protein-like | A |
| 14346_AT | At4g25240 | Pollen-specific protein precursor-like | A |
| 14356_AT | At5g59370 | Actin-4 (ACT4) | A |
| 18930_AT | At1g23730 | Putative carbonic anhydrase | A |
| 19840_S_AT | At1g30720 | Putative reticuline oxidase-like protein | A |
| 13406_AT | At2g23540 | Putative GDSL-motif lipase/hydrolase | A |
| 19640_AT | At2g29460 | Putative glutathione S-transferase | A |
| 16843_AT | At2g44460 | Putative β-glucosidase | A |
| 17326_AT | At2g44570 | Putative glucanase | A |
| 17428_AT | At4g37870 | Phosphoenolpyruvate carboxykinase (ATP)-like protein | A |
| 18201_AT | At2g19990 | Pathogenesis-related protein (PR-1) | A |
| 18151_AT | At2g35770 | Putative Ser carboxypeptidase II | A |
| 19355_S_AT | At2g41280 | Late embryogenesis abundant M10 protein | A |
| 14893_AT | At5g12330 | Lateral root primordia (LRP1) | A |
| 16993_AT | At5g58860 | CYP86A1 | A |
| 18960_AT | At1g68850 | Class III peroxidase PER11 | A |
| 16481_S_AT | At2g18980 | Class III peroxidase PER16 | A |
| 12355_AT | At2g35380 | Class III peroxidase PER20 | A |
| 13662_AT | At3g23050 | IAA7 | A |
| 12444_S_AT | At1g04310 | Putative ethylene receptor (ERS2) | A |
| 18908_I_AT | At2g04160 | Subtilisin-like Ser protease AIR3 | A |
| 20462_AT | At3g13380 | Putative protein kinase | A |
| 20343_S_AT | At1g34670 | Myb-related protein, putative | A |
| 20424_AT | At2g47260 | Putative WRKY-type DNA binding protein | A |
| 20143_AT | At4g30080 | Putative transcription factor | A |
| 20720_AT | At1g22990 | Putative metal-binding protein | A |
| 17041_S_AT | At3g51895 | Sulfate transporter ATST1 | A |
| 12083_AT | At1g23060 | Putative protein | A |
| 20646_AT | At1g77000 | Putative protein | A |
| 19025_AT | At1g77280 | Putative protein | A |
| 15347_AT | At2g03830 | Putative protein | A |
| 19386_AT | At2g22510 | Putative protein | A |
| 18405_S_AT | At2g38480 | Putative protein | A |
| 19162_AT | At4g16670 | Putative protein | A |
| 19415_AT | At4g20390 | Putative protein | A |
| 16514_AT | At4g38080 | Putative protein | A |
| 18911_AT | At1g04680 | Putative pectate lyase A11 | B |
| 16867_AT | At2g32610 | Putative cellulose synthase | B |
| 12515_AT | At2g39700 | Putative expansin At-EXP4 | B |
| 12415_AT | At1g49430 | Acyl-CoA synthetase, putative | B |
| 15653_S_AT | At1g78970 | Lupeol synthase | B |
| 18198_AT | At2g45400 | Putative flavonol reductase | B |
| 20238_AT | At3g13790 | β-fructofuranosidase | B |
| 20239_G_AT | At3g13790 | β-fructofuranosidase | B |
| 13210_AT | At1g11000 | AtMlo-h1-like protein | B |
| 15720_AT | At2g03200 | Putative chloroplast nucleoid DNA binding protein | B |
| 16440_AT | At2g40000 | Putative nematode-resistance protein | B |
| 16963_AT | At2g38390 | Class III peroxidase PER23 | B |
| 17299_S_AT | At4g25420 | Gibberellin 20-oxidase (AtGA20ox1) | B |
| 16099_AT | At4g09460 | Putative transcription factor | B |
| 19835_AT | At1g59740 | Oligopeptide transporter, putative | B |
| 16816_AT | At1g19230 | Putative protein | B |
| 19145_AT | At2g28350 | Putative protein | B |
| 15046_S_AT | At2g39710 | Putative protein | B |
| 20550_AT | At2g47860 | Putative protein | B |
| 19564_AT | At3g46810 | Putative protein | B |
| 13016_AT | At4g17350 | Putative protein | B |
| 15438_AT | At4g22610 | Putative protein | B |
| 14410_AT | At4g24140 | Putative protein | B |
| 12821_AT | At4g32460 | Putative protein | B |
| 20302_AT | At4g13710 | Putative pectate lyase A11 | C |
| 14267_AT | At1g30760 | Putative reticuline oxidase-like protein | C |
| 13793_AT | At4g26790 | Putative APG protein | C |
| 17179_AT | At1g49450 | En Spm-like transposon protein, putative | C |
| 17249_AT | At2g19970 | Putative pathogenesis-related protein | C |
| 20122_AT | At2g23060 | Similar to hookless1 (HLS1) | C |
| 20322_AT | At5g14130 | Class III peroxidase PER55 | C |
| 16247_AT | At2g45420 | Putative protein | C |
| 17697_AT | At2g46740 | Putative protein | C |
| 19565_AT | At3g02885 | Putative protein | C |
| 14828_AT | At4g30850 | Putative protein | C |
| 14606_AT | At2g32990 | Putative glucanase | D |
| 12115_AT | At4g22470 | Extensin-like protein | D |
| 14733_S_AT | At2g39800 | δ-1-pyrroline 5-carboxylase synthetase (P5C1) | D |
| 14025_S_AT | At2g04160 | Subtilisin-like Ser protease AIR3 | D |
| 19743_AT | At1g65680 | Pollen allergen | F |
| 16810_AT | At2g41850 | Putative polygalacturonase | F |
| 14446_AT | At2g43670 | β-1,3-glucanase-like protein | F |
| 15621_F_AT | At2g22240 | Putative myoinositol 1-phosphate synthase | F |
| 18447_AT | At2g40370 | Putative laccase | F |
| 17028_S_AT | At1g10460 | Germin-like protein (GLP7) | F |
| 13603_F_AT | At4g21650 | Subtilisin proteinase-like | F |
| 19045_AT | At2g46950 | CYP709B2 | F |
| 17514_S_AT | At3g23240 | Ethylene response factor 1 (ERF1) | F |
| 16234_AT | At1g49960 | Permease homolog (AtPER-X) | F |
| 12506_AT | At2g37360 | Putative ABC transporter | F |
| 14790_AT | At1g23560 | Putative protein | F |
| 13956_AT | At2g38110 | Putative protein | F |
| 18428_AT | At4g35420 | Putative protein | F |
| 17885_AT | At4g37900 | Putative protein | F |
| 12323_AT | At2g43870 | Putative polygalacturonase | J |
| 20328_AT | At2g22420 | Class III peroxidase PER17 | J |
| 12130_AT | At2g44310 | Putative protein | J |
| 19602_AT | At1g49570 | Class III peroxidase PER10 | K |
| 18596_AT | At1g62570 | Flavin-containing monooxygenase, putative | L |
| 13048_S_AT | At2g02850 | Putative basic blue protein | L |
| 18786_AT | At4g03140 | Putative alcohol dehydrogenase | N |
Table II.
Genes specifically regulated by BL
| Affymetrix No. | AGI ID | Gene Name or Comment | Group |
|---|---|---|---|
| Genes Down-Regulated at Early Stage | |||
| 20271_AT | At4g37310 | CYP81H1 | A |
| 14448_AT | At2g45210 | SAUR-36 | A |
| 17039_AT | At3g26220 | CYP71B3 | D |
| 18190_AT | At2g46660 | CYP78A6 | E |
| 16603_S_AT | At4g15550 | Indole-3-acetate glucosyltransferase-like protein (UGT75D1) | E |
| 20174_AT | At2g43060 | Transcription factor-like protein | E |
| 18780_AT | At2g43440 | Transcription factor-like protein | E |
| 17576_AT | At1g23080 | PIN7 | E |
| 12372_AT | At1g77380 | Amino acid carrier | E |
| 16163_S_AT | At4g22200 | AKT3 | E |
| 18290_AT | At1g49500 | Putative protein | E |
| 19977_AT | At3g48360 | Putative protein | E |
| 13656_AT | At4g01870 | Putative protein | E |
| 18295_S_AT | At1g03880 | Putative cruciferin 12S seed storage protein | F |
| 13198_I_AT | At4g28520 | 12S cruciferin seed storage protein | F |
| 20362_AT | At1g71030 | Putative transcription factor | J |
| Genes Down-Regulated at Early and Late Stages | |||
| 16119_S_AT | At2g30070 | AtKUP1 | D |
| 14240_S_AT | At1g77760 | Nitrate reductase 1 (NR1) | E |
| 12998_AT | At3g47800 | Aldose 1-epimerase-like protein | E |
| 14856_AT | At2g34490 | CYP710A | E |
| 13870_AT | At3g50660 | DWF4 | E |
| 16535_S_AT | At4g36380 | ROT3 | E |
| 16042_S_AT | At5g05690 | CPD | E |
| 14630_S_AT | At1g09530 | PIF3 | E |
| 19221_AT | At4g36780 | Putative protein | E |
| 19398_AT | At4g37540 | Putative protein | E |
| Genes Down-Regulated at Late Stage | |||
| 19684_AT | At4g34970 | Actin depolymerizing factor-like protein | A |
| 17795_AT | At2g14050 | Putative DNA replication licensing factor | A |
| 15669_S_AT | At1g06570 | 4-hydroxyphenylpyruvate dioxygenase (HPD) | D |
| 15142_AT | At1g22360 | UDP-glucose glucosyltransferase | D |
| 19759_AT | At1g23020 | Putative superoxide-generating NADPH oxidase flavo | D |
| 15190_S_AT | At2g26740 | Epoxide hydrolase (ATsEH) | D |
| 12798_AT | At2g38230 | Similar to SOR1 from the fungus Cercospora nicotia | D |
| 17002_AT | At3g51600 | Nonspecific lipid transfer protein | D |
| 14663_S_AT | At4g24040 | Trehalase-like protein | D |
| 12815_AT | At4g27450 | Amino acid biosynthesis Gln-dependent Asp synthetase | D |
| 13242_AT | At4g37980 | Cinnamyl-alcohol dehydrogenase ELI3-1 | D |
| 15144_S_AT | At5g14740 | CARBONIC ANHYDRASE 2 | D |
| 13824_AT | At5g23310 | Iron superoxide dismutase 3 | D |
| 19815_AT | At1g14210 | Ribonuclease | D |
| 13286_S_AT | At2g04030 | Putative heat shock protein | D |
| 20256_S_AT | At2g22990 | Proteolysis Ser-type carboxypeptidase-like protein | D |
| 18005_AT | At3g61620 | Exonuclease RRP41 | D |
| 18699_I_AT | At5g15970 | Cold-regulated protein COR6.6 (KIN2) | D |
| 17566_AT | At5g40160 | Ankyrin repeat protein EMB506 | D |
| 19730_AT | At4g39480 | CYP96A9 | D |
| 13385_AT | At1g14030 | Putative Rubisco oxy | D |
| 15153_AT | At3g27690 | Lhcb2 protein (Lhcb2:4) | D |
| 15793_AT | At4g23940 | FtsH protease, putative | D |
| 14039_AT | At2g19590 | 1-aminocyclopropane-1-carboxylate oxidase | D |
| 14557_AT | At1g02280 | Putative GTP-binding protein | D |
| 19749_AT | At1g31230 | Putative protein kinase | D |
| 16258_AT | At2g39510 | Nodulin-like protein | D |
| 16124_S_AT | At2g47590 | Photolyase/blue light photoreceptor PHR2 (PHR2) | D |
| 20120_AT | At1g03970 | G-box binding factor, GBF4 | D |
| 13168_I_AT | At2g45050 | Putative GATA-type zinc finger transcription factor | D |
| 19059_AT | At2g47520 | Putative AP2 domain transcription factor | D |
| 13642_AT | At1g23180 | Nuclear transport AtKAP α | D |
| 18800_AT | At1g60160 | Potassium transporter AtKT5p | D |
| 16613_S_AT | At2g40540 | Putative potassium transporter | D |
| 17042_S_AT | At4g02700 | Sulfate transporter protein | D |
| 12772_AT | At1g03220 | Putative protein | D |
| 13868_AT | At1g15440 | Putative protein | D |
| 17672_AT | At1g24340 | Putative protein | D |
| 19266_AT | At1g47580 | Putative protein | D |
| 13680_AT | At1g55020 | Putative protein | D |
| 18716_AT | At1g75830 | Putative protein | D |
| 13085_I_AT | At1g78820 | Putative protein | D |
| 13181_AT | At2g02160 | Putative protein | D |
| 12768_AT | At2g15890 | Putative protein | D |
| 15702_S_AT | At2g17250 | Putative protein | D |
| 18396_AT | At2g34640 | Putative protein | D |
| 13382_AT | At2g42750 | Putative protein | D |
| 13700_AT | At3g04550 | Putative protein | D |
| 16637_S_AT | At4g14690 | Putative protein | D |
| 20117_AT | At4g16370 | Putative protein | D |
| 14476_AT | At4g17940 | Putative protein | D |
| 15357_AT | At4g33560 | Putative protein | D |
| 13654_AT | At4g39040 | Putative protein | D |
| 20615_AT | At2g29390 | Putative C-4 sterol methyl oxidase | E |
| 19215_AT | At2g43910 | Putative methyl chloride transferase | E |
| 13573_AT | At4g37550 | Formamidase-like protein | E |
| 12526_AT | At4g27710 | CYP709B3 | E |
| 20389_AT | At5g65310 | Homeobox-Leu zipper protein ATHB-5 (HD-zip pro) | E |
| 14068_S_AT | At2g36950 | Putative farnesylated protein | E |
| 17832_AT | At2g16060 | Class I nonsymbiotic hemoglobin (AHB1) | F |
| 16253_AT | At2g17845 | Putative protein | F |
| 19826_AT | At1g12040 | Putative extensin | G |
| 13449_AT | At4g36700 | Globulin-like protein | G |
| 13197_R_AT | At4g27170 | Putative protein | G |
| 13278_F_AT | At5g12030 | Heat shock protein 17.6A | H |
| 19060_AT | At1g70300 | Potassium transporter | H |
| 12340_AT | At1g10450 | Putative protein | L |
| Genes Up-Regulated at Early Stage | |||
| 19905_AT | At4g19420 | Putative pectinacetylesterase | K |
| 12335_AT | At2g47060 | Putative protein kinase | K |
| 19992_AT | At4g01950 | Putative protein | K |
| 19142_AT | At1g23030 | Putative protein | L |
| 19211_AT | At4g27720 | 12S cruciferin seed storage protein | N |
| 13812_AT | At4g03400 | AtGH3-10 | N |
| 15271_AT | At2g34510 | Putative protein | N |
| Genes Up-Regulated at Early and Late Stages | |||
| 20689_AT | At2g43290 | Putative calcium-binding protein | B |
| 18300_AT | At5g37770 | TCH2 | B |
| 17961_AT | At1g01120 | Fatty acid elongase 3-ketoacyl-CoA synthase 1 (KCS1) | K |
| 17960_AT | At1g65310 | XTR1 | K |
| 19660_AT | At2g40610 | AtExp8 | K |
| 14612_AT | At4g02330 | Putative pectin methylesterase | K |
| 20537_AT | At4g13340 | Extensin-like protein | K |
| 15892_AT | At2g19620 | Putative SF21 protein (Helianthus annuus) | K |
| 12251_AT | At2g34930 | Putative disease resistance protein | K |
| 17966_AT | At4g00360 | CYP86A2 | K |
| 12356_AT | At5g06720 | Class III peroxidase PER53 | K |
| 18253_S_AT | At1g76680 | 12-oxophytodienoate reductase (OPR1) | K |
| 13322_AT | At4g38860 | SAUR-16 | K |
| 17440_I_AT | At1g78860 | Protein kinase | K |
| 12584_AT | At2g44500 | Similar to axi 1 protein from tobacco (Nicotiana tabacum) | K |
| 19857_AT | At4g31000 | Putative calmodulin-binding protein | K |
| 13806_AT | At2g17040 | NAM (no apical meristem)-like protein | K |
| 16438_AT | At1g03870 | Putative protein | K |
| 12046_AT | At1g30690 | Putative protein | K |
| 13916_AT | At2g19800 | Putative protein | K |
| 15403_S_AT | At2g31730 | Putative protein | K |
| 19880_AT | At2g47440 | Putative protein | K |
| 12027_AT | At4g20170 | Putative protein | K |
| 14947_AT | At4g37450 | Putative protein | K |
| 19976_AT | At4g38400 | Putative pollen allergen | N |
| 15107_S_AT | At5g10430 | AtAGP4 | N |
| 14077_AT | At4g08950 | Putative phi-1-like phosphate-induced protein | N |
| 17196_AT | At4g28780 | Pro-rich APG-like protein | N |
| 19288_AT | At2g27690 | CYP94C1 | N |
| 14779_AT | At2g30010 | Putative protein | N |
| Genes Up-Regulated at Late Stage | |||
| 18180_AT | At2g15310 | Putative ADP-ribosylation factor | A |
| 17338_AT | At2g47550 | Putative pectinesterase | B |
| 13706_AT | At2g18700 | Putative trehalose-6-phosphate synthase | B |
| 18567_AT | At2g47130 | Putative alcohol dehydrogenase | B |
| 18818_AT | At2g12210 | Putative TNP2-like transposon protein | B |
| 17403_AT | At2g45550 | CYP76C4 | B |
| 13209_S_AT | At1g04250 | IAA17 | B |
| 13022_AT | At1g34750 | Protein phosphatase type 2C, putative | B |
| 15085_AT | At4g23010 | GOG5-GDP-mannose transporter | B |
| 15556_AT | At1g21820 | Putative protein | B |
| 20194_AT | At2g17500 | Putative protein | B |
| 20616_AT | At2g32560 | Putative protein | B |
| 17262_AT | At2g15510 | Putative non-LTR retroelement reverse transcriptase | F |
| 18670_G_AT | At4g17090 | Putative β-amylase | H |
| 20277_I_AT | At4g13310 | CYP71A20 | I |
| 18922_AT | At3g07850 | Exopolygalacturonase | J |
| 12514_AT | At4g19750 | Chitinase-like protein | J |
| 12508_I_AT | At4g19760 | Chitinase-like protein | J |
| 13926_AT | At2g27920 | Putative carboxypeptidase | J |
| 16666_AT | At4g32540 | Dimethylaniline monooxygenase-like protein | J |
| 18422_AT | At2g01790 | Similarity to human ubiquitin-specific protease | J |
| 16701_AT | At2g02310 | Putative phloem-specific lectin | J |
| 19753_AT | At2g14300 | Putative helicase | J |
| 18997_S_AT | At2g23500 | Mutator-like transposase | J |
| 12564_AT | At2g30810 | Gibberellin-regulated protein homolog | J |
| 20630_I_AT | At2g40290 | Putative eukaryotic translation initiation factor | J |
| 12264_I_AT | At4g13610 | DNA (cytosine-5-)-methyltransferase-like protein | J |
| 16781_AT | At2g19130 | Putative protein kinase | J |
| 16360_AT | At4g21380 | Putative protein kinase | J |
| 20262_AT | At1g61140 | Potassium transporter AtKT5p | J |
| 19270_AT | At5g23270 | Monosaccharide transporter | J |
| 13374_AT | At1g23570 | Putative protein | J |
| 19676_AT | At2g22620 | Putative protein | J |
| 15859_AT | At2g28570 | Putative protein | J |
| 20124_AT | At2g29860 | Putative protein | J |
| 15287_S_AT | At3g47280 | Putative protein | J |
| 12502_AT | At4g19720 | Putative protein | J |
| 17305_AT | At1g53830 | Putative pectin methylesterase | K |
| 17386_AT | At2g21140 | Extensin-like protein | K |
| 12577_AT | At2g28630 | Putative fatty acid elongase | K |
| 12364_AT | At3g57240 | β-1,3-glucanase (BG3) | K |
| 18265_AT | At4g12730 | Putative pollen surface protein | K |
| 17899_AT | At4g15610 | Cell wall protein-like | K |
| 12239_AT | At4g29020 | Biogenesis of cell wall (cell envelope) Gly-rich protein | K |
| 16052_AT | At5g23860 | β-8 tubulin (TUB8) | K |
| 18968_AT | At5g57550 | XTR3 (EXGT-A5) | K |
| 19199_AT | At1g24170 | Putative glycosyl transferase | K |
| 16981_S_AT | At1g45145 | Thioredoxin, putative | K |
| 12277_AT | At1g47600 | Thioglucosidase, putative | K |
| 20391_AT | At2g23560 | Putative acetone-cyanohydrin lyase | K |
| 17008_AT | At2g24850 | Putative Tyr aminotransferase | K |
| 19129_AT | At2g30670 | Putative tropinone reductase | K |
| 16017_AT | At3g16370 | Putative APG protein | K |
| 12574_AT | At3g60140 | β-glucosidase-like protein | K |
| 20305_AT | At4g01070 | Putative flavonol glucosyltransferase | K |
| 16444_AT | At4g13890 | Gly hydroxymethyltransferase-like protein | K |
| 17449_AT | At4g14440 | Carnitine racemase-like protein | K |
| 13908_S_AT | At4g20860 | Berberine bridge enzyme-like protein | K |
| 12539_S_AT | At4g39640 | Putative γ-glutamyltransferase | K |
| 18250_AT | At5g16990 | Quinone oxidoreductase-like protein | K |
| 19178_AT | At5g20230 | Blue copper binding protein | K |
| 19339_I_AT | At2g10140 | Putative TNP2-like transposon protein | K |
| 14635_S_AT | At2g14610 | Pathogenesis-related PR-1-like protein | K |
| 19863_AT | At2g14900 | Gibberellin-regulated protein homolog | K |
| 16730_AT | At2g16040 | Ac-like transposase | K |
| 13004_AT | At2g17840 | Putative senescence-associated protein 12 | K |
| 13498_S_AT | At2g32450 | Putative O-GlcNAc transferase | K |
| 20268_S_AT | At3g46840 | Subtilisin-like proteinase | K |
| 16021_AT | At4g20260 | Endomembrane-associated protein | K |
| 16482_S_AT | At4g32940 | γ-VPE (vacuolar processing enzyme) | K |
| 16465_AT | At5g02490 | dnaK-type molecular chaperone hsc70.1-like | K |
| 20278_S_AT | At4g13290 | CYP71A19 | K |
| 12342_AT | At1g24650 | Putative protein kinase | K |
| 20227_S_AT | At1g52030 | Myrosinase-binding protein, putative | K |
| 16790_AT | At1g53700 | Putative protein kinase | K |
| 19434_AT | At2g04300 | Putative protein kinase | K |
| 16393_S_AT | At2g13790 | Putative protein kinase | K |
| 12497_AT | At2g31880 | Putative protein kinase | K |
| 17752_AT | At2g32800 | Putative protein kinase | K |
| 12958_AT | At2g33580 | Putative protein kinase | K |
| 12353_AT | At2g37710 | Putative protein kinase | K |
| 15475_S_AT | At2g40270 | Putative protein kinase | K |
| 17917_S_AT | At2g41090 | Calcium-binding protein (CaBP-22) | K |
| 13217_S_AT | At3g50770 | Calmodulin-like protein | K |
| 17291_AT | At4g13000 | Putative protein kinase | K |
| 17989_S_AT | At4g14640 | Calmodulin | K |
| 20232_S_AT | At4g23130 | Putative protein kinase | K |
| 20246_S_AT | At4g23250 | Putative protein kinase | K |
| 20373_AT | At4g39890 | GTP-binding protein GB2 | K |
| 17113_S_AT | At5g58670 | Phosphoinositide-specific phospholipase C | K |
| 19936_AT | At1g70000 | DNA binding protein MybSt1 | K |
| 13432_AT | At2g25000 | Putative WRKY-type DNA binding protein | K |
| 20382_S_AT | At2g30250 | Putative WRKY-type DNA binding protein | K |
| 20619_AT | At2g37430 | Putative transcription factor | K |
| 14507_AT | At2g38610 | Putative RNA-binding protein | K |
| 12471_S_AT | At4g03110 | Putative ribonucleoprotein | K |
| 16298_AT | At4g21850 | Putative transcription factor | K |
| 13672_AT | At5g11060 | HOMEOBOX PROTEIN KNOTTED-1 LIKE 4 (KNAT4) | K |
| 16488_AT | At1g11260 | Glucose transporter | K |
| 17278_AT | At1g30900 | Vacuolar sorting receptor-like protein | K |
| 19450_AT | At1g71880 | Sucrose transport protein SUC1 | K |
| 19122_AT | At2g29330 | Putative tropinone reductase | K |
| 15987_AT | At2g39010 | Putative aquaporin | K |
| 15934_I_AT | At3g01930 | HXT6 high-affinity hexose transporter | K |
| 18328_AT | At3g19930 | HXT7 high-affinity hexose transporter | K |
| 20369_S_AT | At4g13510 | Ammonium transport protein (AMT1) | K |
| 20521_AT | At4g18910 | Major intrinsic protein (MIP)-like | K |
| 17451_AT | At4g24120 | Putative oligopeptide transporter | K |
| 12943_AT | At1g03370 | Putative protein | K |
| 18881_AT | At1g12080 | Putative protein | K |
| 12105_AT | At1g22890 | Putative protein | K |
| 15338_AT | At1g23840 | Putative protein | K |
| 16202_AT | At1g47730 | Putative protein | K |
| 20469_AT | At1g60030 | Putative protein | K |
| 14964_AT | At1g65500 | Putative protein | K |
| 20594_AT | At1g70230 | Putative protein | K |
| 15846_AT | At2g14560 | Putative protein | K |
| 12642_AT | At2g15390 | Putative protein | K |
| 14916_AT | At2g16630 | Putative protein | K |
| 19369_AT | At2g17120 | Putative protein | K |
| 12392_AT | At2g23290 | Putative protein | K |
| 15540_AT | At2g24860 | Putative protein | K |
| 19856_AT | At2g25300 | Putative protein | K |
| 14924_AT | At2g28400 | Putative protein | K |
| 13428_AT | At2g31120 | Putative protein | K |
| 16422_AT | At2g33830 | Putative protein | K |
| 18287_AT | At2g37940 | Putative protein | K |
| 12990_AT | At2g41170 | Putative protein | K |
| 19363_AT | At2g42610 | Putative protein | K |
| 12084_AT | At2g43340 | Putative protein | K |
| 18635_AT | At2g43920 | Putative protein | K |
| 12037_AT | At2g44130 | Putative protein | K |
| 20017_AT | At2g44290 | Putative protein | K |
| 13539_I_AT | At3g47380 | Putative protein | K |
| 12171_AT | At3g52500 | Putative protein | K |
| 20429_AT | At4g14400 | Putative protein | K |
| 14401_AT | At4g15630 | Putative protein | K |
| 15815_AT | At4g17070 | Putative protein | K |
| 12561_AT | At4g19120 | Putative protein | K |
| 14946_AT | At4g21620 | Putative protein | K |
| 14431_AT | At4g23810 | Putative protein | K |
| 14400_AT | At4g25260 | Putative protein | K |
| 12696_AT | At4g26250 | Putative protein | K |
| 12209_AT | At4g26950 | Putative protein | K |
| 19182_AT | At4g33050 | Putative protein | K |
| 12443_AT | At4g34480 | Putative protein | K |
| 15084_AT | At4g35320 | Putative protein | K |
| 15817_AT | At4g37240 | Putative protein | K |
| 13055_AT | At4g38030 | Putative protein | K |
| 14882_AT | At4g39670 | Putative protein | K |
| 12118_AT | At4g39840 | Putative protein | K |
| 16897_I_AT | At5g15350 | Putative protein | K |
| 16053_I_AT | At1g02920 | Glutathione S-transferase, putative | L |
| 17372_AT | At1g62040 | Symbiosis-related protein, putative | L |
| 14362_AT | At2g30310 | Putative GDSL-motif lipase/hydrolase | L |
| 13014_AT | At2g30550 | Putative lipase | L |
| 13977_AT | At2g41540 | Glycerol-3-phosphate dehydrogenase | L |
| 19636_AT | At3g25110 | Acyl-(acyl carrier protein) thioesterase | L |
| 13942_AT | At3g50760 | Glycosyltransferase-like protein | L |
| 19171_AT | At2g43510 | Putative trypsin inhibitor | L |
| 19993_AT | At1g78490 | CYP708A3 | L |
| 15982_S_AT | At2g37130 | Class III peroxidase PER21 | L |
| 12333_AT | At4g36430 | Class III peroxidase PER49 | L |
| 16350_AT | At1g61390 | Putative protein kinase | L |
| 12276_AT | At2g28960 | Putative protein kinase | L |
| 16990_AT | At2g37640 | Nodulin-like protein | L |
| 15972_AT | At4g16190 | Cys proteinase | L |
| 20346_AT | At4g35600 | Putative protein kinase | L |
| 20027_AT | At1g50420 | Scarecrow-like 3 | L |
| 18933_AT | At2g40300 | Putative ferritin | L |
| 16296_AT | At4g04770 | Putative ABC transporter | L |
| 16031_AT | At5g01600 | Ferritin 1 precursor | L |
| 15032_AT | At1g61250 | Putative protein | L |
| 19709_I_AT | At1g62430 | Putative protein | L |
| 14959_AT | At1g79450 | Putative protein | L |
| 12062_AT | At2g01650 | Putative protein | L |
| 19207_AT | At2g01670 | Putative protein | L |
| 14381_AT | At2g02810 | Putative protein | L |
| 17448_AT | At2g16530 | Putative protein | L |
| 14423_AT | At2g25190 | Putative protein | L |
| 18267_AT | At2g32210 | Putative protein | L |
| 15854_AT | At2g36820 | Putative protein | L |
| 13475_AT | At2g38200 | Putative protein | L |
| 14972_AT | At2g38740 | Putative protein | L |
| 13941_AT | At4g12850 | Putative protein | L |
| 12540_AT | At4g14390 | Putative protein | L |
| 14825_AT | At4g21240 | Putative protein | L |
| 12995_AT | At4g24970 | Putative protein | L |
| 12968_AT | At4g28270 | Putative protein | L |
| 15938_AT | At4g33100 | Putative protein | L |
| 14884_AT | At4g33910 | Putative protein | L |
| 16968_AT | At4g34131 | Putative protein | L |
| 15877_AT | At4g35330 | Putative protein | L |
| 13059_AT | At4g36550 | Putative protein | L |
| 13955_AT | At5g44810 | Putative protein | L |
| 16700_AT | At2g02250 | Lectin-like protein | M |
| 14355_AT | At1g80390 | IAA15 | M |
| 18758_AT | At4g17660 | Putative protein kinase | M |
| 13659_AT | At4g23150 | Putative protein kinase | M |
| 20489_AT | At2g44840 | Putative ethylene-response element binding protein (EREBP) | M |
| 19340_S_AT | At4g03900 | Putative transposon protein | M |
| 16439_AT | At1g31580 | Putative protein | M |
| 18828_AT | At1g55660 | Putative protein | M |
| 16199_AT | At1g65160 | Putative protein | M |
| 14348_AT | At4g13700 | Putative protein | M |
| 15111_S_AT | At2g06850 | EXGT-A1(EXT) | N |
| 19017_AT | At4g37800 | XTR15 | N |
| 16279_AT | At2g04570 | Putative GDSL-motif lipase hydrolase | N |
| 14089_AT | At2g32150 | Putative hydrolase | N |
| 15601_S_AT | At2g34770 | Fatty acid hydroxylase (FAH1) | N |
| 13577_S_AT | At4g24510 | CER2 | N |
| 16791_AT | At4g39830 | Putative l-ascorbate oxidase | N |
| 16365_AT | At2g32680 | Putative disease resistance protein | N |
| 13625_S_AT | At3g50950 | Putative disease resistance protein | N |
| 13177_AT | At4g12720 | Growth factor-like protein | N |
| 19502_AT | At4g39510 | CYP96A12 | N |
| 13857_AT | At2g21220 | SAUR-12 | N |
| 16348_AT | At1g65790 | Putative protein kinase | N |
| 19290_AT | At2g21540 | Putative phosphatidylinositol/phophatidylcholine transfer protein | N |
| 17990_AT | At3g51920 | Putative calmodulin | N |
| 12360_AT | At4g23210 | Putative protein kinase | N |
| 15779_G_AT | At3g46090 | Zinc finger protein ZAT7 | N |
| 19750_AT | At2g16960 | Putative importin | N |
| 18844_AT | At2g29120 | Putative ligand-gated ion channel protein | N |
| 20128_AT | At1g16420 | Putative protein | N |
| 19825_AT | At1g65550 | Putative protein | N |
| 19065_AT | At2g37440 | Putative protein | N |
| 19985_I_AT | At2g47080 | Putative protein | N |
| 14249_I_AT | At3g52430 | Putative protein | N |
| 20199_AT | At3g52480 | Putative protein | N |
| 17653_AT | At4g39030 | Putative protein | N |
Table III.
Genes commonly regulated by IAA and BL
| Affymetrix No. | AGI ID | Gene Name or Comment | Group |
|---|---|---|---|
| Genes Down-Regulated by IAA and BL | |||
| 20547_AT | At5g04950 | Nicotianamine synthase | E |
| 17045_AT | At1g78090 | Trehalose-6-phosphate phosphatase (AtTPPB) | G |
| 16070_S_AT | At3g60280 | Uclacyanin 3 (UCC3) | G |
| 17849_S_AT | At1g09090 | Putative respiratory burst oxidase protein B | G |
| 17255_AT | At2g25980 | Similar to jasmonate-inducible proteins from Brassica | G |
| 12748_F_AT | At4g11320 | Cys proteinase-like protein | G |
| 14117_AT | At4g37410 | CYP81F4 | G |
| Genes Up-Regulated by IAA and BL | |||
| 18955_AT | At1g04220 | Putative β-ketoacyl-CoA synthase | B |
| 13301_AT | At1g04240 | IAA3 | B |
| 13660_I_AT | At1g15580 | IAA5 | B |
| 13781_AT | At2g18010 | SAUR-10 | B |
| 17894_AT | At2g18690 | Putative protein | B |
| 16995_AT | At2g23170 | AtGH3-3 | B |
| 12543_AT | At2g26710 | BAS1 | B |
| 15005_S_AT | At2g30040 | Putative protein kinase | B |
| 12330_AT | At2g34080 | Cys proteinase | B |
| 18885_AT | At2g36220 | Putative protein | B |
| 13296_AT | At3g15540 | IAA19 | B |
| 13999_AT | At4g03420 | Putative protein | B |
| 14951_AT | At4g09890 | Putative protein | B |
| 13395_AT | At4g13790 | SAUR-25 | B |
| 12501_AT | At4g21200 | Gibberellin 2-oxidase (AtGA2ox8) | B |
| 15431_AT | At4g27280 | Stress response calcineurin B-like protein | B |
| 12947_AT | At4g36110 | SAUR-9 | B |
| 13565_AT | At4g37390 | AtGH3-2 | B |
| 12608_I_AT | At4g38850 | SAUR-AC1 | B |
| 18946_AT | At5g39580 | Class III peroxidase PER62 | B |
| 20035_AT | At5g44440 | Berberine bridge enzyme-like protein | B |
| 17292_AT | At5g49630 | Amino acid permease 6 | B |
| 15985_AT | At5g64100 | Class III peroxidase PER69 | B |
| 20334_S_AT | At1g74650 | Putative transcription factor | J |
| 19490_AT | At1g10550 | Putative endoxyloglucan transferase | K |
| 16434_AT | At4g18970 | Putative protein | K |
| 13495_S_AT | At2g02850 | Basic blue protein | L |
| 15933_AT | At1g21830 | Putative protein | M |
| 20502_AT | At2g21200 | SAUR-7 | M |
| 18284_AT | At4g34150 | Putative protein | M |
| 17533_S_AT | At4g25810 | XTR6 | N |
| 16620_S_AT | At5g57560 | TCH4 | N |
| Genes Down-Regulated by IAA and Up-Regulated by BL | |||
| 16028_AT | At4g30170 | Class III peroxidase PER45 | I |
| 20608_S_AT | At2g44390 | Putative protein | I |
| 12438_AT | At4g18430 | Membrane-bound small GTP-binding-like protein | K |
| 18224_S_AT | At4g21830 | Putative transcription factor | K |
| 12953_AT | At4g01080 | Putative protein | K |
| Genes Up-Regulated by IAA and Down-Regulated by BL | |||
| 19177_AT | At5g22500 | Male sterility 2-like protein | A |
| 19281_I_AT | At2g23180 | CYP96A1 | A |
| 15434_AT | At4g35720 | Putative protein | A |
| 19346_AT | At4g01630 | Putative expansin At-EXP17 | D |
| 14643_S_AT | At2g03760 | Putative steroid sulfotransferase | D |
| 14517_AT | At2g41800 | Putative protein | D |
| 15178_S_AT | At4g14130 | XTR7 | E |
| 15098_S_AT | At4g35770 | Senescence-associated protein sen1 | E |
| 16078_AT | At3g16500 | Phytochrome-associated protein 1 (PAP1) | E |
| 17977_AT | At4g01680 | MYB55 | E |
| 14062_AT | At2g47780 | Putative protein | F |
Overview and Comparison of IAA and BL Induction
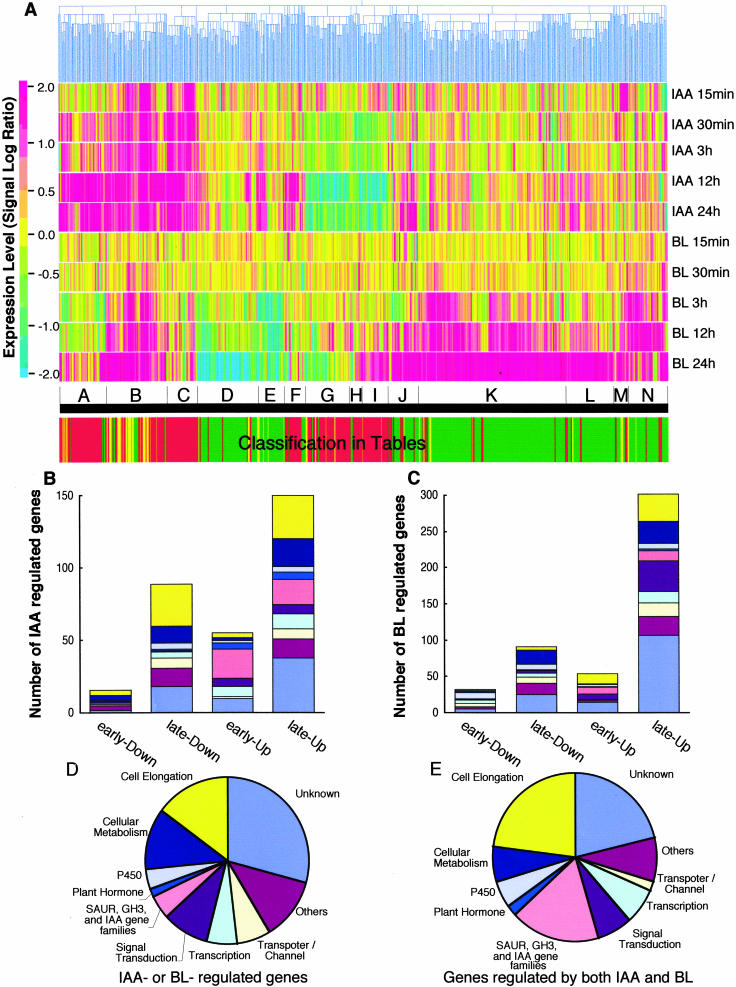
The expression profiles of the time-course experiments with the IAA or BL treatments were analyzed by hierarchical clustering (Eisen et al., 1998) using 637 BL- or IAA-regulated genes (listed in Tables I–III). The expression levels are indicated using color (Fig. 1A). The dendrogram represents the relationships between genes based on the similarity of their responses to the two hormones. In the dendrogram, the genes are roughly clustered into four groups (from left to right in Fig. 1A): those up-regulated by IAA (groups A–C), down-regulated by BL (groups D–G), down-regulated by IAA (groups G–I), and up-regulated by BL (groups I–N). The classification in Tables I to III is shown at the bottom of Figure 1A (red, green, and yellow). This classification is not necessarily consistent with the classification using the hierarchical clustering since the genes were classified in the tables using the criteria described above, whereas in the clustergram they were classified using similarity in their gene expression patterns (described in “Materials and Methods”). For example, group H included IAA-down- and BL-up-regulated genes. Most of them are listed as genes specifically regulated by IAA in Table I because their response to BL was less than 2 based on the SLR or was not significant based on the results of the MAS version 5 analysis. The most remarkable finding was that the majority of the genes regulated by both BL and IAA (listed in Table III; shown in yellow in the bottom line of Fig. 1A) were included in the cluster of IAA-up-regulated genes, and they were especially enriched in group B. Interestingly, these genes were up-regulated by BL.
Figure 1.
Gene expression patterns in response to BL and IAA treatment. Seven-day-old WT seedlings were treated with IAA, or det2 seedlings were treated with BL. Then transcript abundance was analyzed using an Affymetrix GeneChip representing about 8,300 Arabidopsis genes. A, The expression of 637 BL- or IAA-inducible genes (listed in Tables I–III). Colors (red to blue, defined to the left of the column) represent the magnitude of induction in SLR values relative to mock-treated samples. Genes were clustered hierarchically using GeneSpring and grouped into groups A to N (at the bottom of the column). The trees at the top (in blue lines) indicate similarity in the gene expression patterns. The horizontal color bars (at the bottom) represent the classification used in the tables, namely IAA-regulated genes (red), BL-regulated genes (green), and genes regulated by both BL and IAA (yellow). B to E, The frequencies of IAA- and BL-regulated genes. The IAA- and BL-regulated genes are classified into 10 functional categories (indicated in D and E) based on their established or putative functions. The genes induced more than 2-fold within 3 h of hormone treatment are defined as early inducible genes, and those induced between 12 h and 24 h are defined as late inducible genes. The numbers of IAA- (B) or BL- regulated genes (C) are shown. The frequencies of genes regulated by IAA or BL (D) or both (E) are shown.
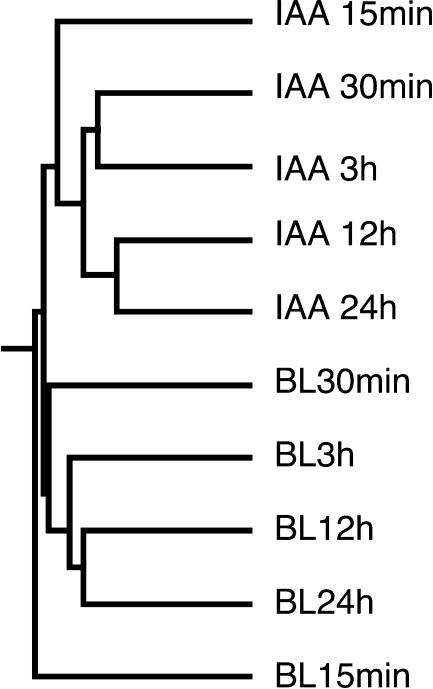
This clustergram represented the general trend of BL- and IAA-regulated genes well, i.e. IAA induction was quicker than BL induction for both up- and down-regulated genes. IAA-regulated genes were detected within 15 min, as we reported previously (Sawa et al., 2002), and the number of IAA-regulated genes peaked at 12 h (data not shown). By contrast, no genes responded in a reproducible manner to BL in 15 min, as we reported previously (Goda et al., 2002), and the number of BL-regulated genes increased continuously over time (Fig. 1A). The difference in induction speed with BL and IAA treatment is also conserved in the genes regulated by both BL and IAA. The lag period for BL-induced gene expression may be due to the time needed to induce auxin biosynthesis or to activate auxin sensitivity. If this is the case, the gene expression pattern in response to BL, especially at early time points, may be similar to the IAA response. To test this hypothesis, the relationship between the gene expression patterns at each time point of the BL and IAA treatments was hierarchically calculated using data on the expression of the 637 genes listed in Tables I to III. The dendrogram indicated that the BL and IAA treatments clustered independently (Fig. 2). In each cluster of the BL and IAA treatments, the continuous experiments were related vicinally. These results suggested that BL and IAA treatments induce gene expression independently. Consistent with this finding, only 48 genes (8%) were regulated by both BL and IAA (Table III); the majority of BL- and IAA-inducible genes are regulated by BL or IAA independently. These results suggest that BL regulates plant growth using a set of genes that is independent from IAA for most of its response.
Figure 2.
Relationships between the BL and IAA treatments. The dendrogram was calculated by hierarchical clustering using data on the expression of 637 BL- or IAA-regulated genes (listed in Tables I–III). The dendrogram represents the similarity of the gene expression profiles with the BL and IAA treatments at each time point.
To overview the functional overlap and divergence of BL- and IAA-inducible genes, the genes were classified into 10 categories based on their established or putative functions. The frequencies of BL- and IAA-inducible genes are shown in Figure 1, B to E (the categories are indicated in Fig. 1, D and E). The largest group of early down-regulated BL genes were P450 genes (Fig. 1C), while relatively few P450 genes were in IAA-regulated genes (Fig. 1B). BL induced more signal transduction-related genes (49 genes, 13.9%), especially at the late stage, than did IAA (17 genes, 4.7%; Fig. 1, C compared with B). IAA induced 17 transcription factor genes (12.7%) at 30 min, consistent with our previous report at 15 min (Sawa et al., 2002), whereas BL induced only one gene (1.9%) at the same time. These results may also reflect the different modes of action in the BL and IAA signal transduction systems. There were fewer down-regulated genes than up-regulated genes in both hormone treatments.
Regulation of the SAUR, GH3, and IAA Gene Families
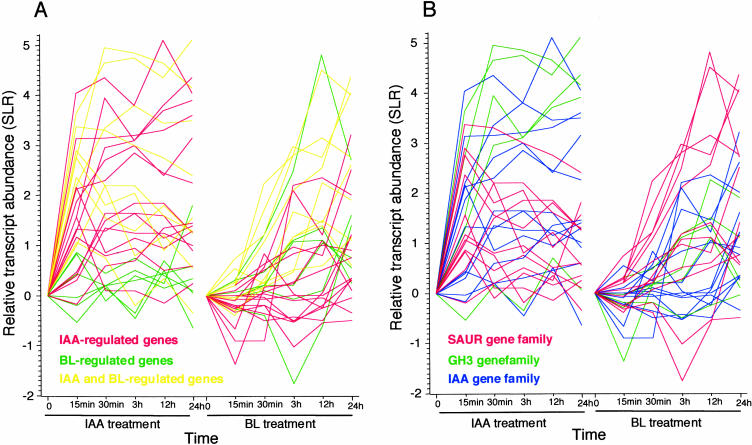
Genes that are induced by auxins within minutes of treatment are referred to as early auxin-inducible genes, and they form three major gene families, namely SAUR, GH3, and IAA (Hagen and Guilfoyle, 2002; Liscum and Reed, 2002). The SAUR, GH3, and IAA genes predominated in IAA-up-regulated genes, both early and late (Fig. 1B). This is consistent with previous DNA-microarray studies of IAA-inducible genes studied at early times (Sawa et al., 2002; Tian et al., 2002). These genes also predominated in BL-up-regulated genes (Fig. 1C). Interestingly, these families are relatively more enriched in genes regulated by both BL and IAA (Fig. 1, E compared with D). Previously, we showed that BL treatment induced a member from each gene family (SAUR-AC1, GH3-homolog BRU6, and IAA3/SHY2) after a lag period of 30 to 60 min (Goda et al., 2002). This comprehensive study also demonstrated that most genes in this category regulated by both IAA and BL are regulated quickly by IAA but more slowly by BL (Fig. 3A). A possible mechanism for the difference in induction speed is discussed below. The difference in induction speed between the two hormones suggests that auxin regulates rapid physiological responses, such as tropic responses, whereas BR regulates slower physiological responses, such as developmental regulation and more gradual responses to the environment.
Figure 3.
Induction kinetics of SAUR, GH3, and IAA genes. Transcript abundance of IAA- or BL-regulated SAUR, GH3, and IAA genes relative to mock-treated samples is given in SLR values. The data are given as the means of three or two independent hormone-treated plant samples. A, Colored according to hormone inducibility: regulated by IAA (red), BL (green), or both (yellow). Some genes that were induced more than 2-fold in both treatments were classified as being regulated by one hormone if the induction with the other hormone was not significant based on the results of the MAS (version 5) analysis. B, SAUR, GH3, and AUX/IAA family genes are shown in red, yellow, and blue, respectively. The value at 0 h is a theoretical value (0) and is not based on experimental results.
Of the three gene families, the SAUR genes had the strongest BL responses (SLR > 3) in the BL treatment (Fig. 3B, red lines). By contrast, the GH3 and IAA genes had the strongest IAA responses (Fig. 3B, green and blue lines, respectively). This complementary inducibility may be related to the synergism between BR and auxin. The expression of SAUR genes correlates well with auxin-induced elongation (McClure and Guilfoyle, 1987, 1989; Gee et al., 1991), although their functions are still unclear. Yang and Poovaiah (2000) demonstrated that the amino-terminal domain of SAUR proteins binds to calmodulin in maize (Zea mays), soybean, and Arabidopsis. Very recently, we demonstrated that the expression of SAUR-AC1 correlates well with BR-mediated elongation and that it is regulated by BRs independently of the endogenous auxin levels (Nakamura et al., 2003b). These findings, together with our finding that a number of genes encoding calcium-binding protein are regulated by BL or IAA (Tables I–III), suggest that the calcium and calmodulin system is an important target for studying BR and auxin signal interaction.
In this study, we also identified GH3 and IAA genes: IAA specifically regulated eight genes (AtGH3-1 and 5, and IAA1, 2, 6, 7, 11, and 13); BL specifically regulated three genes (AtGH3-10, IAA15, and IAA17/AXR3); and both BL and IAA regulated six genes (AtGH3-2 and 3, and IAA3, 5, 19, and 26). The IAA17/AXR3 gene, an auxin-inducible gene (Ouellet et al., 2001), was not identified as an IAA-responsive gene since its response to IAA was below the threshold (SLR = 0.7). Mutants in members of these gene families exhibit phenotypes with insensitivity to auxin and other hormones, as well as defects in light signaling and photomorphogenesis (Hagen and Guilfoyle, 2002; Liscum and Reed, 2002; Swarup et al., 2002). By contrast, we found that BL-induced IAA genes in a manner independent of the endogenous IAA level (Nakamura et al., 2003a). These findings suggest that IAA and GH3 genes are important cross talk points in BR, auxin, light, and other signaling pathways.
Regulation of Genes Involved in Cell Expansion or Cell Wall Organization
The regulation of tissue elongation is an important function of both BR and auxin. Synergistic interactions between BR and auxin occur in elongating tissues and cells in dicots (Yopp et al., 1981; Cohen and Meudt, 1983; Katsumi 1985; Sala and Sala, 1985) and monocots (Yopp et al., 1981; Takeno and Pharis, 1982), including bending responses. Tissue elongation or cell expansion is considered an important response for understanding interactions between auxins and BR, but the molecular mechanisms by which they interact and regulate plant tissue elongation are poorly understood. Xu et al. (1995) reported that the Arabidopsis TCH4 gene, which encodes a xyloglucan endotransglycosylase, was induced quickly by IAA but rather slowly by BL. We have also reported that potential cell wall-related genes (TCH4, AtExp8, and KCS1) are induced quickly by IAA but slowly by BL, and that BL regulates a number of cell wall-related genes (Goda et al., 2002).
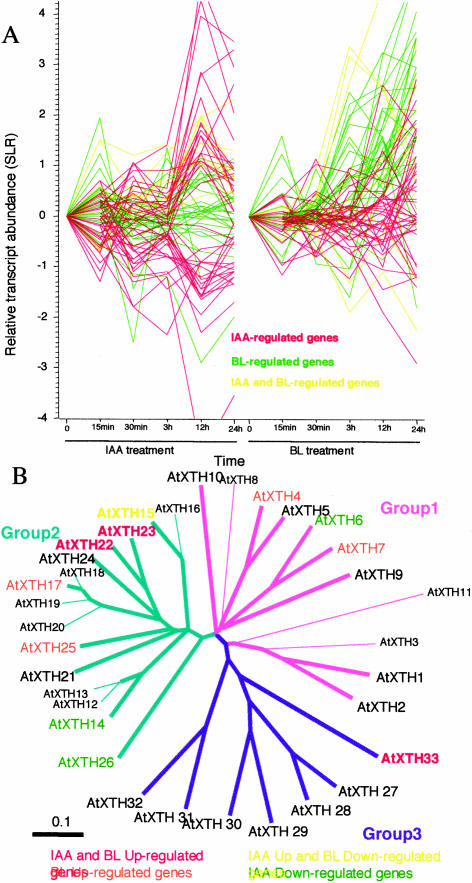
Here, we identified at least 100 genes potentially involved in cell wall organization as IAA or BL regulated. These genes include those encoding cell wall synthesis enzymes, cell wall modifying agents, cell wall component proteins, and wall rigidification and wax-related proteins and included all the functional subcategories necessary for the completion of cell wall organization. This revealed the global manner by which these hormones regulate cell wall-related genes. We observed overlap and divergence of IAA and BL in regulating the genes involved in cell wall organization and cell elongation. Genes in this category are mainly early BL-up-regulated genes (Figs. 4A, green lines, and 1C) and not early IAA-up-regulated genes (Figs. 4A, red lines, and 1B). The majority of BL-regulated genes were up-regulated, and only five were down-regulated, whereas the numbers of IAA genes up- and down-regulated were comparable (Fig. 4A). Some members (e.g. β-1,3-glucanase, chitinase, peroxidase, and Leu-rich repeat proteins with or without extensin region) in this category are annotated as pathogen-related or disease resistance-related genes in a database based on their research history. However, we classified them as cell wall-related genes since recent studies have revealed that these genes are involved in multiple biological processes (Baumberger et al., 2001; Hrmova and Fincher, 2001; Passarinho and deVries, 2002; Yoshida et al., 2003). Since many genes await characterization to understand cell wall biogenesis and cell expansion, the genes listed and classified in Tables I to III should prove useful for identifying novel cell wall-related genes and further understanding cell wall biogenesis.
Figure 4.
Regulation of genes involved in cell expansion or cell wall organization. A, The induction kinetics of genes involved in cell expansion or cell wall organization. Transcript abundance of IAA- or BL-regulated genes in this category relative to mock-treated samples is shown as SLR values. Genes regulated by IAA, BL, or both are shown in red, green, or yellow, respectively. The data are shown as the means of three or two independent hormone-treatment experiments. Some genes induced more than 2-fold by both hormone treatments were classified as specifically regulated by one hormone if induction with the other hormone was not significant based on the results of the MAS (version 5) analysis. The value at 0 h is a theoretical value (0) and is not based on experimental results. B, Regulation of XTH genes by IAA or BL. A phylogenetic tree of the Arabidopsis XTH gene family was generated using ClustalW and TreeViewPPC software based on the deduced amino acid sequences of all 33 Arabidopsis XTH genes. Genes from groups 1, 2, and 3 are shown with pink, green, and violet lines, respectively. The color coding of the letters is as follows: genes up-regulated by both IAA and BL (red letters); IAA-up- and BL-down-regulated genes (yellow); IAA-down-regulated genes (green); BL-up-regulated genes (orange); and genes not regulated by either IAA or BL (black). Genes not represented on the Affymetrix Arabidopsis Genome Array are shown in small letters.
The majority (81%) of this gene category was composed of cell wall modifying agents, including xyloglucan endotransglucosylase/hydrolases (XTH), glucanase, polygalacturonase, pectin esterase, expansin, extensin, and chitinase. The most abundant cross-linking glycan in the primary cell wall of dicots is xyloglucan, which is thought to play an essential role in cell wall loosening and cell expansion. There are 33 XTH genes in the Arabidopsis genome (for review, see Rose et al., 2002), and they are classified into three major phylogenetic groups (Yokoyama and Nishitani, 2001). We found that 11 of them were regulated by IAA or BL. Interestingly, most of them belonged to group 1 or group 2. By contrast, only one exception (AtXHT33) belonged to group 3, although all the members of group 3 were represented on the array. This trend was reproduced in our whole-genome array experiments (H. Goda and Y. Shimada, unpublished data). Interestingly, members of groups 1 and 2 mediate transglucosylation between xyloglucans (Nishitani and Tominaga, 1992; Xu et al. 1996), while members of group 3 catalyze xyloglucan endohydrolysis (Fanutti et al., 1993). The responses of XTH genes in our data are consistent with previous studies of IAA regulation (Xu et al., 1995, 1996; Sawa et al., 2002) and BR regulation (Xu et al., 1995, 1996; Goda et al., 2002), except that some minor responses differed from those in a report by Yokoyama and Nishitani (2001). Although the reason is unclear at present, one possible explanation is that minor responses may depend on the experimental conditions, such as growth or hormone-treatment conditions.
Analysis of the Prompter Regions of IAA-Responsive and BL-Responsive Genes
Auxin response elements (AuxREs), which consist of a TGTCTC sequence and an adjacent or overlapping coupling element, were defined based on the auxin-responsive promoter of the soybean GH3 gene (Liu et al., 1994; Ulmasov et al., 1995). Gain-of-function experiments with minimal promoter-GUS (β-glucuronidase) reporter genes have shown that a single copy of an AuxRE is sufficient to confer auxin responsiveness to reporter genes (Ulmasov et al., 1995). DR5, an artificial AuxRE containing the TGTCTC element, has increased auxin responsiveness (Ulmasov et al., 1997). The GUS reporter gene fused to a minimal cauliflower mosaic virus 35S promoter and the DR5 AuxRE has been used widely as a marker to monitor the distribution of endogenous IAA, as it has been suggested that the resulting GUS activity coincides with this distribution (Sabatini et al., 1999; Casimiro et al., 2001).
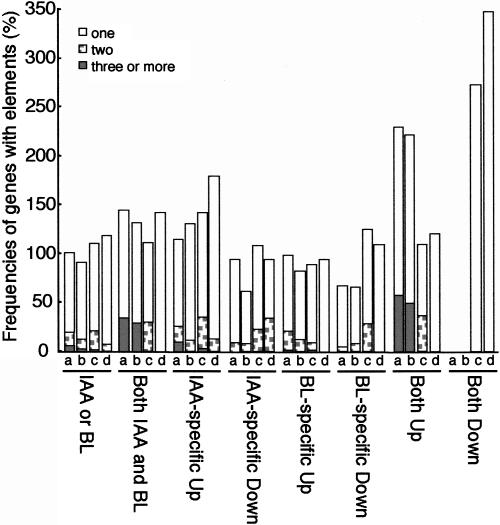
As we found that a number of the early auxin-inducible genes are induced in response to BL treatment, we tested the frequency of BL-inducible genes possessing the TGTCTC element in the 5′-flanking region. The 8,300 genes represented on the Arabidopsis Genome Array corresponded to 7,388 independent loci in the Arabidopsis genome. The numbers of IAA- and BL-regulated genes containing the TGTCTC element or its inverse (GAGACA) were counted and are given as a proportion of the 7,388 genes (Fig. 5). At least one TGTCTC element exists 5′ upstream from the start codon within 1,000 bp of 1,817 genes (25%) or within 500 bp of 1,071 genes (14%). Similarly, the inverse element, GAGACA, exists within 1,000 bp of 1,640 genes (22%) or within 500 bp of 863 genes (12%). Surprisingly, the TGTCTC element was most frequent for genes regulated by both IAA and BL, rather than in genes up-regulated specifically by IAA (Fig. 5). The frequency of genes with multiple TGTCTC elements was also highest in these genes. This is consistent with our recent finding that BL treatment induces the DR5-GUS gene in Arabidopsis (Nakamura et al., 2003a). We also demonstrated that the early auxin-inducible genes IAA3, GH3-2/BRU6, SAUR-AC1 (Goda et al., 2002; Nakamura et al., 2003b), IAA5, and IAA19 (Nakamura et al., 2003a) are induced with similar kinetics to the DR5-GUS gene in Arabidopsis, namely they are quickly and transiently induced by IAA and gradually and continuously induced by BL. Furthermore, BL induces SAUR-AC1 (Nakamura et al., 2003b), IAA5, IAA19, and DR5-GUS (Nakamura et al., 2003a) in a manner independent of the endogenous auxin levels. Consequently, we speculate that genes up-regulated by both BL and IAA are regulated by a common cis-regulatory element, which includes TGTCTC. Interestingly, the frequency of genes having the TGTCTC element was lower in genes down-regulated by both BL and IAA, although as there were only seven such genes, this result could be due to an artifact. However, this trend was also observed in early BL-down-regulated genes, late BL-down-regulated genes, and late IAA-down-regulated genes (data not shown). Furthermore, the inverse element (CGCACA) was not enriched in genes up-regulated by both IAA and BL but was enriched in genes down-regulated by both (Fig. 5), even though it is generally believed that the inverse element has the same function as the orthodromic element. This trend was also observed in early BL-down-regulated genes (data not shown). These findings will be useful for future studies to understand the roles of TGTCTC and the inverse element in BR- and auxin-regulated gene expression, as well as to identify novel cis-regulatory elements that are specific to BL or IAA regulation and to elements involved in down-regulation.
Figure 5.
Frequencies of genes with TGTCTC or GAGACA elements in the 5′-flanking region of IAA- or BL-regulated genes. The numbers of genes containing TGTCTC or its inverse element (GAGACA) in the 5′-flanking region (up to −500 or −1,000 bp) were calculated using GeneSpring. The frequencies of genes with these elements are given as a proportion of the 7,388 independent loci represented in the Arabidopsis Genome Array (about 8,300 genes corresponding to 7,388 independent loci). a, TGTCTC (−1,000 bp); b, TGTCTC (−500 bp); c, GAGACA (−1,000 bp); d, GAGACA (−500 bp). The shading in each bar indicates the ratios of the genes containing one, two, or three and more elements.
Other Interactions between BR and Auxin
We found the following responses, which may be important to further understanding auxin/BR interactions. Three genes potentially involved in signal transduction pathway were newly identified here as being induced by both BL and IAA: a homolog (At2g30040) of the brassinosteroid-insensitive 2 kinase gene (BIN2; Choe et al., 2002; Li and Nam, 2002; Perez-Perez et al., 2002), At4g27280 encoding calcineurin B-like protein, and At1g74650 encoding a putative transcription factor (Myb-like). We previously reported that PIN7, a homolog of the PIN1 and PIN2 genes for putative auxin-efflux carrier proteins (Galweiler et al., 1998; Muller et al., 1998), was repressed by BL treatment (Goda et al., 2002). This response was confirmed here. BRI1 is a critical component of the BR receptor (Wang et al., 2001). Three genes encode BRI1-like proteins in Arabidopsis: BRL1, BRL2, and BRL3. BRL1 and BRL3 are reported to bind BL (Yin et al., 2002b). In this study, BRL3 (At3g13380) was up-regulated in response to IAA treatment later on. Conversely, we observed that BAS1/CYP72B1, which encodes an enzyme that inactivates BRs (Neff et al., 1999), was increased by IAA treatment later on. These results suggest that auxin regulates BR signaling and catabolism.
As described above, P450 genes constituted the largest group of early BL-down-regulated genes (Fig. 1C), while relatively few P450 genes were IAA-regulated genes, perhaps because a number of P450 genes are involved in BR biosynthesis and catabolism (Fujioka and Yokota, 2003). Conversely, none of the genes involved in auxin metabolism were identified here as IAA regulated. BAK1 encodes the Leu-rich repeat receptor-like kinase belonging to the Leu-rich repeat receptor kinase II and X family (http://plantsp.sdsc.edu/plantsp/family/class). Overexpression of the BAK1 gene leads to a phenotype reminiscent of the BRI1-overexpression transgenic plant, and BAK1 protein interacts with BRI1 in vivo and in vitro (Li et al., 2002; Nam and Li, 2002). We found that a BAK1 homolog (At2g13790), the gene most closely related to BAK1 in the Leu-rich repeat receptor-like kinase gene family of Arabidopsis, was induced by BL treatment at a later time point. It will be interesting to test whether the At2g13790 gene functions in the BR signaling.
Cross Talk with Other Plant Hormone Signaling
Earlier studies reported that IAA and BR exhibit cross talk with other plant hormones. In Arabidopsis, BL induced the OPR1 (Goda et al., 2002) or OPR3 (Müssig et al., 2000) genes encoding 12-oxophytodienoic acid 10,11-reductase involved in jasmonate biosynthesis (Biesgen and Weiler, 1999). BL also induced the GA 20-oxidase gene (AtGA20ox1; Bouquin et al., 2001). IAA treatment induced the 1-aminocyclopropane-1-carboxylate (ACC) synthase gene (ACS; Abel et al., 1995). These hormone cross talk responses observed previously in Arabidopsis were confirmed here. In addition, BL induction of ACS has been reported in mung bean (Vigna radiata; Yi et al., 1999). Auxin regulation of the GA20ox gene has been well studied in the pea (Van Huizen et al., 1997; Ngo et al., 2002; O'Neill and Ross, 2002). These responses found in other species were confirmed here in Arabidopsis for the first time, to our knowledge. Namely, BL induced AtACS4, and IAA induced AtGA20ox1. In addition, we found that BL induced AtGA2ox8, which encodes GA-inactivating enzyme (Schomburg et al., 2003). IAA repressed the cytokinin oxidase gene (CKX4), which encodes an enzyme that inactivates cytokinin (Bilyeu et al., 2001). These novel findings could be clues to unravel complex phytohormone cross talk and plant signaling networks.
CONCLUSIONS
To our knowledge, this is the first comprehensive expression profiling study of either auxin or BR over time. In addition, this is, to our knowledge, the first report to investigate the relationship between the actions of auxin and BR using a comprehensive expression profiling approach. The time course experiment revealed overlap and divergence between the actions of these two hormones. We identified 637 genes regulated by IAA or BL. Of these, 48 were regulated by both IAA and BL. Most BR actions are mediated by the induction of genes that are independent of the auxin response. The SAUR, GH3, and IAA gene families were the largest group of genes regulated by both IAA and BL. A number of the early auxin-inducible genes are not specifically regulated by auxin, but are regulated by these two hormones in common. Conversely, this study revealed true auxin-specific and BR-specific genes. This classification of genes is important for understanding the functional divergence and interaction of auxin and BR. A previously reported TGTCTC element in AuxRE was not enriched in genes specifically regulated by IAA but was enriched in genes up-regulated by both BL and IAA. This observation is consistent with our previous findings that a synthetic AuxRE, DR5, responded to both IAA and BL with kinetics similar to those of IAA or SAUR genes, independent of the endogenous auxin level (Nakamura et al., 2003a, 2003b). Therefore, the DR5-GUS reporter system is not specific to auxin action, but is an important marker for studying the BR/auxin interaction. About 30% of IAA- or BL-regulated genes were classified in an unknown category. A classification based on expression analysis will be useful for elucidating the functions of these genes and should provide insight into the activities of auxin and BR. For example, since all known BR-biosynthetic genes were classified in group E, this group may include genes that are important for BR biosynthesis and action.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis ecotype Colombia (Col-0) was used as the WT in this study. The Arabidopsis mutant det2-1 (Chory et al., 1991) was used as a BR-deficient mutant. Seedlings were grown for 7 d at 22°C under continuous light in half-strength Murashige and Skoog (1962) liquid medium (Gibco BRL, Cleveland) supplemented with 1.5% (w/v) sucrose. The seedlings were then treated with 1 μm IAA or 10 nm BL or mock treated with dimethyl sulfoxide (final concentration 0.1%). Then, they were immediately frozen in liquid nitrogen and stored at −80°C until RNA isolation.
DNA Microarray Analysis
The DNA microarray analysis was performed essentially as described previously (Goda et al., 2002). Total RNA was isolated from seedlings using the acid-guanidinium-phenol-chloroform method (Sambrook et al., 1989) and converted into double-stranded cDNA using a Super Script Choice cDNA synthesis kit (GIBCO BRL) with an oligo(dT)24 primer containing a T7 polymerase promoter site at its 3′ end (Amersham Pharmacia Biotech, Uppsala). Biotin-labeled cRNA was generated from the double-stranded cDNA using a BioArray HighYield RNA transcript labeling kit (Enzo Biochem, Farmingdale, NY) and was then purified using an RNeasy RNA purification kit (Qiagen USA, Valencia, CA). Each cRNA sample (20 μg) was fragmented and hybridized with the Arabidopsis Genome Array (Affymetrix, Santa Clara, CA) for 16 h at 45°C with rotation at 60 rpm. Each array was then washed and detected by consecutive exposure to phycoerythrin-streptavidin (Molecular Probes, Eugene, OR), biotinylated antibodies to streptavidin (Vector Laboratories, Burlingame, CA), and phycoerythrin-streptavidin, after which each array was washed again with a nonstringent wash buffer. All washing and staining procedures were performed with a Fluidics Station 400 (Affymetrix). The array was scanned using a confocal microscope scanner (HP Genome Array Scanner; Affymetrix) at a wavelength of 570 nm. To achieve a higher signal dynamic range, we scanned each chip before and after signal amplification using an anti-streptavidin antibody. Each chip was normalized relative to the sum of the signal values, and then the control and IAA- or BL-treated samples were compared at each time point using the GeneChip software MAS version 5 (Affymetrix). Genes that were assigned as up- or down-regulated on the basis of more than a 2-fold difference in their signal values and that were assigned as Increase or Decrease on the basis of their Change values were extracted. Furthermore, genes with Absent for the Detection value in the baseline data and Decrease for the Change value were excluded from the list. Similarly, genes with Absent for the Detection value in the experimental data and Increase for the Change value were also excluded from the list. To ensure the reproducibility of the results, we performed three (15 min for IAA treatment; 15 min, 30 min, and 3 h for BL treatment) or two (for other time points) independent hormone-treatment experiments with different plant samples. Genes that showed reproducible responses in all experiments were classified as genes regulated by IAA or BL at each time point of the treatments (Tables I–III). This threshold seems to be more stringent for false-positive genes than the conventional threshold based solely on a statistical analysis (Welch's t test, at a significance level of P < 0.05) of signal values in independent experiments, as described previously (Goda et al., 2002).
The SLR was imported into GeneSpring software (version 4; Silicon Genetics, Redwood, CA), and the genes were clustered hierarchically based on the angular separation of the expression vectors for each gene. Elements of the 5′-flanking regions were also analyzed using GeneSpring software.
Annotations and Database Analysis
Since the locus assignments and annotations of genes provided by Affymetrix contain errors, we used information provided by The Arabidopsis Information Resource (TAIR; files available at ftp://tairpub:tairpub@ftp.arabidopsis.org/home/tair/Microarrays/Affymetrix/). This information was produced by BLASTing the array element sequences downloaded from the Affymetrix Web site (http://www.affymetrix.com) against the Arabidopsis bacterial artificial chromosomes, chloroplast, and mitochondria genomes from The Institute for Genomic Research using BLASTN with an E value cutoff of 1e−6. The TAIR annotations were further revised manually using BLAST searches of the GenBank/EMBL/DNA Data Bank of Japan (DDBJ) databases, as well as reference searches on the ISI Web of Science.
Supplementary Material
Acknowledgments
We thank Dr. Joanne Chory for providing the BR mutants, Dr. Margarita Garcia-Hernandez and TAIR for providing information on Arabidopsis loci and annotations, and Dr. Masaki Fumoto and the DDBJ for creating the Arabidopsis genome sequence files. We thank Mr. Narumasa Miyauchi for his technical assistance with the molecular biological analyses.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.034736
References
- Abel S, Nguyen MD, Theologis A (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Arteca RN, Bachman JM, Tsai DS, Mandava NB (1988) Fusicoccin, an inhibitor of brassinosteroid-induced ethylene production. Physiol Plant 74: 631–634 [Google Scholar]
- Baumberger N, Ringli C, Keller B (2001) The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev 15: 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesgen C, Weiler EW (1999) Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta 208: 155–165 [DOI] [PubMed] [Google Scholar]
- Bilyeu KD, Cole JL, Laskey JG, Riekhof WR, Esparza TJ, Kramer MD, Morris RO (2001) Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol 125: 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J (2001) Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol 127: 450–458 [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzè D, Sandberg G, Casero PJ, Bennett M (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee M-O, Yoshida S, Feldmann KA, Tax FE (2002) Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semi-dominant and defective in a Glycogen Synthase Kinase 3β-like kinase. Plant Physiol 130: 1506–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA (1991) Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD (1996) Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J 10: 1–8 [DOI] [PubMed] [Google Scholar]
- Clouse SD (1997) Molecular genetic analysis of brassinosteroid action. Physiol Plant 100: 702–709 [Google Scholar]
- Clouse SD (2002) Brassinosteroids. In The Arabidopsis Book. American Society of Plant Biologists, http://www.aspb.org/publications/arabidopsis/toc.cfm DOI/10.1199/tab.0009
- Clouse SD, Hall AF, Langford M, McMorris TC, Baker ME (1993) Physiological and molecular effects of brassinosteroids on Arabidopsis thaliana. J Plant Growth Regul 12: 61–66 [Google Scholar]
- Clouse SD, Zurek DM, McMorris TC, Baker ME (1992) Effect of brassinolide on gene expression in elongating soybean epicotyls. Plant Physiol 100: 1377–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Meudt WJ (1983) Investigation on the mechanism of the brassinosteroid response. Plant Physiol 72: 691–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanutti C, Gidley MJ, Reid JSG (1993) Action of a pure xyloglucan endo-transglycosylase (formerly called xyloglucan-specific endo-(1→4)-beta-d-glucanase) from the cotyledons of germinated nasturtium seeds. Plant J 3: 691–700 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Galweiler L, Guan CH, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Gee MA, Hagen G, Guilfoyle TJ (1991) Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs. Plant Cell 3: 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove MD, Spencer GF, Rohwedder WK, Mandava N, Worley JF, Warthen JD, Steffens GL, Flippenanderson JL, Cook JC (1979) Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281: 216–217 [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Hrmova M, Fincher GB (2001) Structure-function relationships of β-d-glucan endo- and exohydrolases from higher plants. Plant Mol Biol 47: 73–91 [PubMed] [Google Scholar]
- Katsumi M (1985) Interaction of a brassinosteroid with IAA and GA3 in the elongation of cucumber hypocotyl sections. Plant Cell Physiol 26: 615–625 [Google Scholar]
- Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J, Wen JQ, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400 [PubMed] [Google Scholar]
- Liu ZB, Ulmasov T, Shi XY, Hagen G, Guilfoyle TJ (1994) Soybean Gh3 promoter contains multiple auxin-inducible elements. Plant Cell 6: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandava NB (1988) Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol 39: 23–52 [Google Scholar]
- McClure BA, Guilfoyle T (1987) Characterization of a class of small auxin-inducible soybean polyadenylated Rnas. Plant Mol Biol 9: 611–623 [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T (1989) Rapid redistribution of auxin-regulated RNAs during gravitropism. Science 243: 91–93 [DOI] [PubMed] [Google Scholar]
- McKay MJ, Ross JJ, Lawrence NL, Cramp RE, Beveridge CA, Reid JB (1994) Control of internode length in Pisum sativum. Plant Physiol 106: 1521–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Guan CH, Galweiler L, Tanzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15: 473–498 [Google Scholar]
- Müssig C, Biesgen C, Lisso J, Uwer U, Weiler E, Altmann T (2000) A novel stress-inducible 12-oxophytodienoate reductase from Arabidopsis thaliana provides a potential link between brassinosteroid-action and jasmonic-acid synthesis. J Plant Physiol 157: 143–152 [Google Scholar]
- Müssig C, Fischer S, Altmann T (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S (2003. a) Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 133: 1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Shimada Y, Goda H, Fujiwara MT, Asami T, Yoshida S (2003. b) AXR1 is involved in BR-mediated elongation and SAUR-AC1 gene expression in Arabidopsis. FEBS Lett 553: 28–32 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li JM (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, Chory J (1999) BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo P, Ozga JA, Reinecke DM (2002) Specificity of auxin regulation of gibberellin 20-oxidase gene expression in pea pericarp. Plant Mol Biol 49: 439–448 [DOI] [PubMed] [Google Scholar]
- Nishitani K, Tominaga R (1992) Endoxyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267: 21058–21064 [PubMed] [Google Scholar]
- Nomura T, Nakayama M, Reid JB, Takeuchi Y, Yokota T (1997) Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol 113: 31–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill DP, Ross J J (2002) Auxin regulation of the gibberellin pathway in pea. Plant Physiol 130: 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A (2001) IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarinho PA, de Vries SC (2002) Arabidopsis chitinase: a genomic survey. In The Arabidopsis Book. American Society of Plant Biologists, http://www.aspb.org/publications/arabidopsis/toc.cfm doi/10.1199/tab.0023
- Perez-Perez JM, Ponce MR, Micol JL (2002) The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 242: 161–173 [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, Scheres B (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sala C, Sala F (1985) Effect of brassinosteroid on cell-division and enlargement in cultured carrot (Daucus carota L.) cells. Plant Cell Rep 4: 144–147 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sasse J (1999) Physiological actions of brassinosteroids. In A Sakurai, T Yokota, SD Clouseeds, eds, Brassinosteroids: Steroidal Plant Hormones. Springer-Verlag, Tokyo, pp 137–161
- Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T (2002) The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J 32: 1011–1022 [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM (2003) Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Parry G, Graham N, Allen T, Bennett M (2002) Auxin cross-talk: integration of signalling pathways to control plant development. Plant Mol Biol 49: 411–426 [DOI] [PubMed] [Google Scholar]
- Takeno K, Pharis RP (1982) Brassinosteroid-induced bending of the leaf lamina of dwarf rice seedlings: an auxin-mediated phenomenon. Plant Cell Physiol 23: 1275–1281 [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ (1995) Composite structure of auxin response elements. Plant Cell 7: 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle T J (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Huizen R, Ozga JA, Reinecke DM (1997) Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiol 115: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Xu W, Campbell P, Vargheese AK, Braam J (1996) The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J 9: 879–889 [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J (1995) Arabidopsis Tch4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TB, Poovaiah BW (2000) Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J Biol Chem 275: 3137–3143 [DOI] [PubMed] [Google Scholar]
- Yi HC, Joo S, Nam KH, Lee JS, Kang BG, Kim WT (1999) Auxin and brassinosteroid differentially regulate the expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in mung bean (Vigna radiata L.). Plant Mol Biol 41: 443–454 [DOI] [PubMed] [Google Scholar]
- Yin YH, Wang ZY, Mora-Garcia S, Li JM, Yoshida S, Asami T, Chory J (2002. a) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yin YH, Wu DY, Chory J (2002. b) Plant receptor kinases: systemin receptor identified. Proc Natl Acad Sci USA 99: 9090–9092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K (2001) A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol 42: 1025–1033 [DOI] [PubMed] [Google Scholar]
- Yopp JH, Mandava NB, Sasse JM (1981) Brassinolide, a growth-promoting steroidal lactone: 1. Activity in selected auxin bioassays. Physiol Plant 53: 445–452 [Google Scholar]
- Yoshida K, Kaothien P, Matsui T, Kawaoka A, Shinmyo A (2003) Molecular biology and application of plant peroxidase genes. Appl Microbiol Biotechnol 60: 665–670 [DOI] [PubMed] [Google Scholar]
- Zurek DM, Rayle DL, McMorris TC, Clouse SD (1994) Investigation of gene-expression, growth-kinetics, and wall extensibility during brassinosteroid-regulated stem elongation. Plant Physiol 104: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.