Abstract
SCF complexes are the largest and best studied family of E3 ubiquitin protein ligases that facilitate the ubiquitylation of proteins targeted for degradation. The SCF core components Skp1, Cul1, and Rbx1 serve in multiple SCF complexes involving different substrate-specific F-box proteins that are involved in diverse processes including cell cycle and development. In Arabidopsis, mutations in the F-box gene UNUSUAL FLORAL ORGANS (UFO) result in a number of defects in flower development. However, functions of the core components Cul1 and Rbx1 in flower development are poorly understood. In this study we analyzed floral phenotypes caused by altering function of Cul1 or Rbx1, as well as the effects of mutations in ASK1 and ASK2. Plants homozygous for a point mutation in the AtCUL1 gene showed reduced floral organ number and several defects in each of the four whorls. Similarly, plants with reduced AtRbx1 expression due to RNA interference also exhibited floral morphological defects. In addition, compared to the ask1 mutant, plants homozygous for ask1 and heterozygous for ask2 displayed enhanced reduction of B function, as well as other novel defects of flower development, including carpelloid sepals and an inhibition of petal development. Genetic analyses demonstrate that AGAMOUS (AG) is required for the novel phenotypes observed in the first and second whorls. Furthermore, the genetic interaction between UFO and AtCUL1 supports the idea that UFO regulates multiple aspects of flower development as a part of SCF complexes. These results suggest that SCF complexes regulate several aspects of floral development in Arabidopsis.
An Arabidopsis flower has four concentric whorls that contain four sepals, four petals, six stamens, and two carpels. After the transition from vegetative to reproductive development, the Arabidopsis apical meristem (inflorescence meristem) produces the floral meristem, which in turn undergoes a series of developmental stages to form a flower (Smyth et al., 1990). Genetic and molecular studies have uncovered a large number of genes that control different steps in flower development including flowering time, flower meristem identity, and flower organ identity (Zhao et al., 2001a). In particular, the ABC model has been proposed for the specification of floral organ identity (Coen and Meyerowitz, 1991; Ma, 1994; Weigel and Meyerowitz, 1994; Ma and dePamphilis, 2000). The combinatorial expression of ABC genes defines the organ type that differentiates in each whorl: A function alone specifies the sepal identity; A and B function together controls petal identity; B and C function together specifies stamen identity; and C function alone directs carpel identity.
The UNUSUAL FLORAL ORGANS (UFO) gene is involved in multiple aspects of floral development, including regulating floral meristem identity and floral organ development (Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995). One known function of UFO in floral organ development is a positive regulation of the expression of B function gene APETALA3 (AP3) in cooperation with the floral meristem identity gene LEAFY (Lee et al., 1997; Zhao et al., 2001b). Recently, a novel role of UFO in early petal formation was uncovered through the analysis of newly isolated ufo alleles (Durfee et al., 2003) and the transient restoration of UFO function in the strong ufo-2 mutant (Laufs et al., 2003).
The UFO gene encodes one of the approximately 700 F-box proteins that are believed to be components of the SCF-type E3 ubiquitin ligase (Gagne et al., 2002; Kuroda et al., 2002). The ubiquitin ligase (E3) functions in a pathway with a ubiquitin-activating enzyme (E1) and a ubiquitin-conjugating enzyme (E2) to catalyze the ubiquitylation of proteins targeted for degradation (Koepp et al., 1999; Pickart, 2001). The SCF complexes are members of the largest and best-studied family of E3 ubiquitin ligases. In addition to the substrate-recognition factor F-box protein, an SCF complex consists of Skp1, Cul1/Cdc53, and a RING finger protein Rbx1/Hrt/Roc1. Cul1 functions as a scaffold protein linking Skp1 with Rbx1, which acts to recruit the E2 enzyme. Skp1 serves as an adaptor that bridges Cul1/Cdc53 and the F-box protein (Deshaies, 1999; Schulman et al., 2000; Jackson and Eldridge, 2002; Zheng et al., 2002).
Homologs of the core components of the SCF ubiquitin ligase have been found in Arabidopsis. At least five Cul homologs are expressed in the Arabidopsis genome. Among these, AtCUL1 and AtCUL2 are able to interact with ASK1 and F-box proteins in a yeast (Saccharomyces cerevisiae) two-hybrid assay (Risseeuw et al., 2003). However, AtCUL1 is the only Cul1 homolog in Arabidopsis that has been verified to be part of SCF complexes in vivo (Gray et al., 1999; Xu et al., 2002). In addition, a null mutant allele of AtCUL1 exhibited embryo arrest at the single cell stage (Shen et al., 2002). Consistent with its broad expression pattern (Farras et al., 2001; del Pozo et al., 2002b; Shen et al., 2002), reduced AtCUL1 functions cause severe auxin related defects throughout plant development (Hellmann et al., 2003). Thus, it is likely that AtCUL1 is a component of multiple SCF complexes that play critical roles in development.
Two Rbx1 homologs were uncovered in the Arabidopsis genome. The two Rbx1 proteins are highly similar to each other as well as to that of the human Rbx1 proteins. However, based on expression levels, AtRbx1a seems to be the dominant participant in SCF complexes (Gray et al., 2002; Lechner et al., 2002). Altered expression of AtRbx1 causes severe defects in plant growth and development (Gray et al., 2002; Lechner et al., 2002; Schwechheimer et al., 2002; Xu et al., 2002), indicating that its function is essential.
Among the 21 Arabidopsis Skp1 homologs (called ASK) in the Arabidopsis genome, at least 18 were found to be expressed under normal growth conditions, with a large subset of them detected in the inflorescence (Zhao et al., 2003b). The ask1-1 mutant is male sterile and defective in both vegetative and reproductive development (Yang et al., 1999; Zhao et al., 1999). Recently, we have shown through mutations in ASK1 and ASK2 that these two genes are essential for normal embryo and seedling development (Liu et al., 2004). The relatively weak floral phenotypes of the ask1-1 null mutant compared to that of strong ufo alleles suggest that other ASK genes might also interact with UFO to regulate flower development. ASK1 and ASK2 are very similar in sequence and expression patterns (Zhao et al., 2003b), and both can interact with a similar set of F-box proteins (including UFO) in yeast two-hybrid assays (Gagne et al., 2002; Risseeuw et al., 2003). Thus, ASK1 and ASK2 are also likely to share redundant functions in flower development.
ASK1 interacts genetically and in yeast two-hybrid assays with UFO (Samach et al., 1999; Zhao et al., 1999, 2001b). In addition, ASK1 and UFO interact with LEAFY genetically to positively regulate B function gene expression (Zhao et al., 2001b). Furthermore, UFO interacts with ASK1 and AtCUL1 in an immunoprecipitation assay (Wang et al., 2003), supporting a role of the SCFUFO complex in flower development. However, the functions of the SCF core components, Cul1, Rbx1, and ASK2, in flower development have not previously been demonstrated. We report here the role of AtCUL1 and AtRbx1 in flower development. We also provide the first genetic evidence that UFO interacts with AtCUL1 to regulate several aspects of flower development. Moreover, genetic studies indicate that ASK1 and ASK2 share redundant functions in flower development, including the regulation of B function gene expression. Finally, we describe genetic evidence that supports a novel role of SCFUFO in regulating C function.
RESULTS
Floral Phenotypes of axr6-2, a Point Mutant in AtCUL1
A previous study showed that a null mutation in the AtCUL1 gene causes embryo lethality (Shen et al., 2002); therefore, the null mutant cannot be used to investigate a possible AtCUL1 function in flower development. Recently, a previously identified auxin resistant mutant, axr6-2, was found to carry a point mutation (replacement of Phe-111 by isoleucine) in AtCUL1 that results in a reduced physical interaction between ASK1 and AtCUL1 (Hobbie et al., 2000; Hellmann et al., 2003). Furthermore, viable plants homozygous for the weak axr6-2 allele have been obtained (G. Badrajan and L. Hobbie, unpublished), enabling analysis of the flowers. We have examined flower development in the homozygous axr6-2 mutant and found that axr6-2 flowers exhibited reduced organ numbers and/or various defects in all four whorls (with an average total organ number of 11.34 per flower; Table I).
Table I.
Comparison of floral organs
| Ler | axr6-2 | ufo-6 | axr6-2, ufo-6a | ask1-1 | ask2-1 | ask1-1, ask2-1/+ | |
|---|---|---|---|---|---|---|---|
| Whorl 1 | |||||||
| Sepal | 4.00 ± 0.00 | 3.72 ± 0.35 | 4.00 ± 0.00 | 3.35 ± 0.81 | 4.00 ± 0.00 | 4.00 ± 0.00 | 3.75 ± 0.61 |
| Carpel-like | 0 | 0 | 0 | 0.05 ± 0.21 | 0 | 0 | 0.07 ± 0.30 |
| Whorl 2 | |||||||
| Petal | 4.00 ± 0.00 | 1.51 ± 0.88 | 1.70 ± 0.42 | 0.07 ± 0.26 | 2.94 ± 0.61 | 4.00 ± 0.00 | 0.14 ± 0.41 |
| Sepal-like petal | 0 | 0 | 0.10 ± 0.21 | 0.02 ± 0.15 | 0 | 0 | 0 |
| Whorls 2 and 3 | |||||||
| Petal-stamen chimera | 0 | 0 | 2.06 ± 0.48 | 0.19 ± 0.45 | 0.76 ± 0.54 | 0 | 0.08 ± 0.29 |
| Filament | 0 | 0.13 ± 0.30 | 0.38 ± 0.62 | 0.86 ± 0.97 | 0 | 0 | 0.43 ± 0.70 |
| Stamen | 6.00 ± 0.00 | 4.02 ± 0.72 | 3.80 ± 0.88 | 3.16 ± 1.15 | 4.88 ± 0.61 | 6.00 ± 0.00 | 4.77 ± 1.39 |
| Carpel-likeb | 0 | 0 | 0.05 ± 0.18 | 0.30 ± 0.56 | 0 | 0 | 0.23 ± 0.55 |
| Whorl 4 | |||||||
| Carpel | 2.00 ± 0.00 | 1.96 ± 0.10 | 2.03 ± 0.14 | 2.16 ± 0.37 | 2.00 ± 0.00 | 2.00 ± 0.00 | 2.08 ± 0.27 |
| Sum of all organs | 16.00 ± 0.00 | 11.34 ± 1.46 | 14.17 ± 0.73 | 10.16 ± 1.59 | 14.59 ± 0.69 | 16.00 ± 0.00 | 11.53 ± 1.52 |
All plants were grown under the same conditions and the average number of organs per flower is given ± se. Unless otherwise indicated, the first 10 flowers on each given plant were analyzed, and a total of 100 flowers from 10 plants were examined.
A total of 43 flowers were analyzed.
Mosaic organs include carpel/sepal, carpel/stamen, carpel/filament, and carpel/sepal/stamen.
Compared with the wild-type flower (Fig. 1A), a typical axr6-2 mutant flower (Fig. 1B) has a slight reduction in the number of sepals, which are occasionally fused together (Table I). In the second whorl, the number of petals is reduced, and occasionally the size of petals is also reduced (Fig. 1B; Table I). In the third whorl, there are fewer than the normal number of stamens, and the stamen filaments are shorter than those of the wild type (data not shown). In addition, filamentous structures were also observed in the third whorl (Fig. 1B; Table I). Unlike ufo and ask1 flowers, no petal/stamen chimeric organs were observed in the axr6-2 flower (Table I), and whorls are clearly defined in the axr6-2 flower. In the fourth whorl, the number of carpels seems to be normal. However, about 25% of the gynoecia are curled (Fig. 1B), and occasionally carpels are not fused.
Figure 1.
The phenotypes of Arabidopsis wild-type and mutant flowers. One sepal was removed to show the interior organs in B, E, F, K, L, M, O, R, S, and V. A, A wild-type flower of the Columbia ecotype. B, An axr6-2 flower with reduced number of petals and stamens, a filament (arrow), and curled gynoecium. C, An aborted axr6-2 flower that did not open. The right flower is the same flower as the left one, but three sepals were removed to show the interior organs. D, A Dex-treated pTA2 control flower of WS ecotype. E, A Dex-treated AtRbx1 RNAi flower with three petals and ovule-like organs which fuse to the fourth whorl carpel (arrow). F, A Dex-treated AtRbx1 RNAi flower showing a second flower (arrow). G, A wild-type flower of Ler ecotype. H, An ask1 (Ler ecotype) flower with small staminoid petals (arrow). I, An ask2 flower (WS ecotype) with normal floral organs. J to N, ask1/ask1 ASK2/ask2 flowers with no petals, short stamens, and a carpel-like anther (arrow in J), a filament, a carpel-like sepal (arrow in K), curled carpels (K), a filament fused to a carpel (arrowhead in L) and fused stamens (arrow in L), papillae-like structures in the first whorl (arrow in M), or fused sepals and unfused carpels (N). O, An ASK1 RNAi flower (Ler ecotype) with papillae-like structures in the first whorl (arrow), one petal, and curled carpel. P, A normal ask1/ask1 ASK2/ask2 tASK1 flower. Q, An ask1/ask1 ASK2/ask2 35S:ASK2 flower with ask1-like phenotypes. R, an axr6-2 ask1 flower with carpelloid first whorl sepals (arrow) and curled, unfused fourth whorl carpels (arrowhead). S, A ufo-2 flower with sepal-like petals, filaments, and carpel-like structure in the middle whorls. T, A ufo-6 flower showing reduced petal number and staminoid petals U, A ufo-6 axr6-2 flower with ovule-like organ (arrow) in the first whorl, greatly reduced organ number in middle whorls and aborted carpel. V, A ufo-6 axr6-2 flower showing papillae-like structures (arrow). W, A ufo-6 axr6-2 inflorescence with a filament-like structure instead of a normal flower. X, A terminated ufo-6 axr6-2 inflorescence with pistil-like structure occupying the inflorescence meristem. Scale bars = 0.5 mm. A–V have the same magnification; W and X have the same magnification.
In addition, we also observed some small flower buds in the axr6-2 mutant which never opened (Fig. 1C). These flowers were found to contain aborted petals, stamens, and carpels inside relatively normal sepals.
Floral Phenotypes of Inducible AtRbx1 RNAi Plants
Continuous silencing of AtRbx1 expression by double-strand RNA interference caused severe defects in plant growth and development (Lechner et al., 2002; Xu et al., 2002; data not shown). Therefore, we examined the effects of reducing AtRbx1 expression during flower development by using inducible AtRbx1 RNAi plants previously generated by Genschik and colleagues (Lechner et al., 2002). In agreement with previous results, we found that the transgenic plants treated for a period of 5 to 7 d with dexamethasone (Dex) can recover and develop further after being transferred to a Dex-free environment (see “Materials and Methods”). Therefore, this system provides another opportunity to study the role of SCF complexes in flower development.
All plants produce normal flowers without Dex treatment. Control plants transformed with an empty vector produced normal flowers after Dex treatment (Fig. 1D), except for occasional reductions in stamen number. In contrast, the flower of Dex-treated Rbx1 RNAi plants exhibited slightly reduced petal number and reduced stamen filament length (Fig. 1E). In addition, petal/stamen chimeras, filaments, and carpelloid organs were also observed in the middle whorls of the Dex-treated Rbx1 RNAi flowers (Fig. 1E; and data not shown), indicating a reduction of B function. Sometimes a curled gynoecium (Fig. 1E), or an axillary flower (Fig. 1F) was also observed in the AtRbx1 RNAi flowers.
Genetic Interaction of ASK1 and ASK2
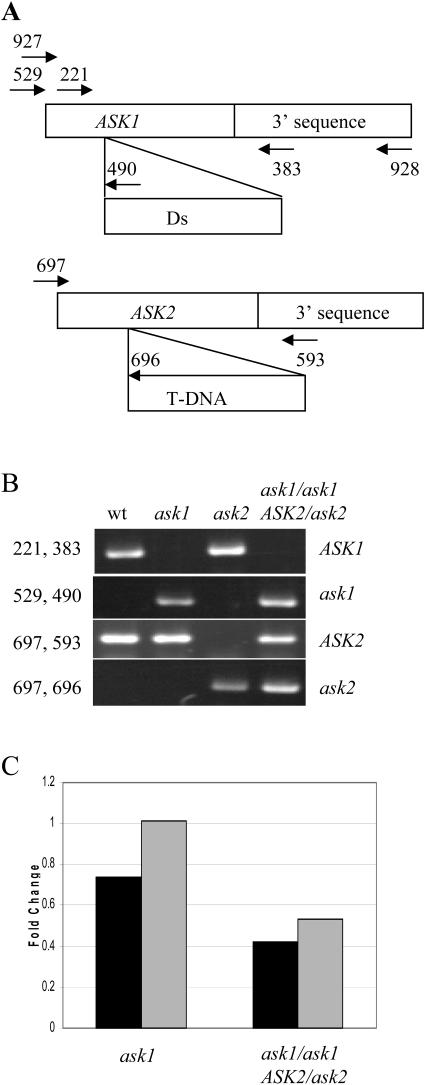
The ask1-1 mutant contains a Ds insertion in the middle of the coding region (Fig. 2A; Yang et al., 1999). Recently a mutation in the coding region of the ASK2 gene (ask2-1) was isolated from a T-DNA insertional population (Fig. 2A; Liu et al., 2004). ASK1 and ASK2 expression was not detected by RNA blot in the respective single mutant. To test for possible genetic interactions between ASK1 and ASK2, we compared flower development in different genotypes with wild-type and mutant alleles of these two genes (Fig. 2, A and B, see “Materials and Methods”). Consistent with previous results in our laboratory, we observed a slightly reduced number of petals and stamens, staminoid petals, and short stamen filaments in the flowers of the ask1-1 mutant (Fig. 1H; Table I; Zhao et al., 1999).
Figure 2.
Characterization of ask2 and ask1 ASK2/ask2. A, Schematic map of the ask1-1 and ask2-1 mutations and primers for identifying genotypes in F2 plants. B, Specificity of the primer combinations (left side of the panel) used in PCR reaction to amplify either ASK1, ask1-1, ASK2, or ask2-1 alleles (right side). C, Relative mRNA expression levels of ASK2 gene in mutants determined by real-time PCR. Black and gray bars represent results from two independent experiments. ASK2 expression in the wild type was considered as 1.
The flower of ask2-1 (Fig. 1I; Table I) was indistinguishable from that of the wild type. In the F2 progeny of a cross between ask1 and ask2, we found that the ask1 ask2 double mutant is defective in embryo development and is seedling lethal (Liu et al., 2004). Further examination revealed that some F2 plants displayed a novel floral phenotype characterized by the near absence of petals. From 400 F2 plants, we observed 22 (close to 1/15) plants with the ask1-1-like flower and 46 (close to 2/15) plants with this novel phenotype. All other plants were similar to the wild type. PCR analysis demonstrated that all F2 plants with the ask1-like flower were ask1 single mutants (homozygous for the wild-type allele at the ASK2 locus), whereas all F2 plants with the novel petalless phenotype were of the ask1/ask1 ASK2/ask2 genotype. The ASK1/ask1 ask2/ask2 plants produced normal flowers. In addition, we performed PCR analysis on 192 F2 plants with no apparent phenotype, and none of them were ask1/ask1 ASK2/ask2. The RNA expression levels of the ASK2 gene were further tested using real-time PCR. Compared to the wild type, ASK2 gene expression is close to normal in the ask1 mutant, whereas its expression is reduced to about 48% in the ask1/ask1 ASK2/ask2 plant (Fig. 2C). Residual ASK2 expression was also detected in the ask2-1 mutant using primers C-terminal relative to the T-DNA insertion (data not shown).
As mentioned above, the most dramatic phenotype in the ask1/ask1 ASK2/ask2 plants is the absence of second whorl organs in most flowers (Fig. 1, J–N; Table I), with an average number of 0.14 petals per flower. In addition, carpelloid stamens and filaments were observed in the third whorl of the ask1/ask1 ASK2/ask2 flowers (Fig. 1, J and K; Table I). Other defects that were also observed in whorls 2 and 3 include a variable number of stamens, both fewer and more than six, and chimeras of petal/stamen and sepal/carpel (Fig. 1K). Sometimes organs in the same whorl, or between different whorls, were fused together (Fig. 1L). The first and fourth whorls of the ask1/ask1 ASK2/ask2 flower were mostly normal, with a slightly reduced number and variable size of sepals, and a slightly increased number of carpels (Table I; and data not shown). Occasionally, carpelloid sepals (Fig. 1M; Table I), fused sepals (Fig. 1N), curled carpels or unfused carpels (Fig. 1, K and N) were also observed; these defects were more severe in late flowers (data not shown). The carpelloid sepals and absence of organs in whorl 2 in the ask1/ask1 ASK2/ask2 flower are similar to the phenotype of ap2 mutants, in which the C function gene AGAMOUS (AG) expands to the first and second whorls (Kunst et al., 1989; Bowman et al., 1991; Drews et al., 1991).
We have also generated ASK1 RNAi transgenic plants (Zhao et al., 2003b). Similar floral phenotypes to those mentioned above in the ask1/ask1 ASK2/ask2 plants were also observed in strong ASK1 RNAi plants, including a further reduction in petal number compared to the ask1 single mutant; carpelloid organs in the first and middle whorls; and enlarged, curled, or unfused carpels (Fig. 1O; and data not shown; Zhao et al., 2003b). In the inflorescences of strong ASK1 RNAi plants, ASK1 expression was not detected, while ASK2 expression was reduced (Zhao et al., 2003b).
To further verify that the enhanced and novel phenotypes in ask1/ask1 ASK2/ask2 flowers were caused by the combination of ask1 and ask2 mutations, we carried out a functional complementation of the ask1/ask1 ASK2/ask2 mutant with either a genomic ASK1 or a fusion of the ASK2 cDNA with the 35S promoter of the cauliflower mosaic virus (CaMV). The flowers of ask1/ask1 ASK2/ask2 mutant with a transgenic genomic ASK1 were restored to normal (Fig. 1P; and data not shown). In addition, the flowers of ask1/ask1 ASK2/ask2 mutant with the 35S-ASK2 construct exhibited phenotypes similar to those of the ask1 single mutant, including less reduction in petal number compared to the ask1/ask1 ASK2/ask2 flowers and correct organ identities for sepal, stamen, and carpel (Fig. 1Q). However, filaments were still occasionally observed in these plants, and the petal number was less than that in the ask1 mutant. This may be due to a possible difference in the expression pattern of the 35S promoter from that of the endogenous ASK2 regulatory elements. In conclusion, our results demonstrated that reduced expression of ASK1 and ASK2 is responsible for the ap2-like phenotype in the outer two whorls, the enhanced reduction of B function in the third whorl, and the defects in carpel identity in the fourth whorl of the ask1/ask1 ASK2/ask2 flower.
We have also made a cross between axr6-2 and ask1-1. Similar to the ask1/ask1 ASK2/ask2 flower, carpelloid stamens (data not shown), carpelloid first whorl sepals, and unfused fourth whorl carpels (Fig. 1R) were also observed in the axr6-2 ask1 flower.
Genetic Interaction of UFO and AtCUL1
Previous studies in our laboratory have shown that UFO interacts with ASK1 genetically to positively regulate B function gene expression (Zhao et al., 2001b). To further investigate if UFO interacts with AtCUL1 to regulate flower development, we generated a double mutant between axr6-2 and ufo-6 (a weak ufo allele) and compared the floral phenotypes of the double mutant with those of the single mutants and ufo-2, a strong ufo allele. Our results from the ufo single mutants are in agreement with previous reports (Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995).
The strong ufo-2 mutant flower exhibited obvious defects in organ identity in the middle whorls, with sepal-like petals and filament-like structures occupying most of the organ positions in the second and third whorl, respectively (Fig. 1S). The first whorl sepals are largely normal. Occasionally two sepals are fused together. Enlarged or increased number of carpels was observed in the fourth whorls. In the weak ufo-6 mutant, sepals and carpels are generally normal, whereas the number and identity of petals and stamens are somewhat altered (Fig. 1T; Table I). Filaments and occasionally sepal-like petals were observed (Table I), suggesting a slightly reduced B function. In addition, petal/stamen chimeric organs were frequently observed. The inflorescence of the ufo-6 mutant is normal.
In ufo-6 axr6-2 double mutant flowers, fused sepals were more frequently observed compared to the axr6-2 single mutant. Occasionally carpel-like sepals were observed in the first whorl (Fig. 1U; Table I). Compared to the axr6-2 and two ufo single mutants, the number of second whorl organs in the ufo-6 axr6-2 double mutant was greatly reduced, similar to those found in ask1/ask1 ASK2/ask2 flowers (0.09 per flower; Table I). Most flowers in the double mutant had no petals (Fig. 1, U and V). Occasionally a petal, or sepal-like petal, was observed in the second whorl (Table I). In the third whorl, the number of stamens was further reduced compared to the ufo-6 and axr6-2 single mutants, whereas carpel-like structures and filaments were increased in the double mutant (Fig. 1V; Table I). In the fourth whorl, the phenotypes of double mutant carpels were similar to those of axr6-2 single mutant. The increased carpelloid organs and filaments in the third whorl of ufo-6 axr6-2 double mutant flower support the hypothesis that these two genes act together to promote B function; there also seems to be a unique aspect of the phenotypes that may not result from B function, suggesting additional roles for AtCUL1, particularly in organ formation and carpel development.
Moreover, we observed inflorescence defects in the ufo-6 axr6-2 double mutant, including a filament-like structure instead of a normal flower (Fig. 1W), and termination of an inflorescence by a pistil-like structure (Fig. 1X), after the production of only four to five flowers. These defects were not observed in ufo-6 or axr6-2 single mutant.
AP3 and AG Expression in Wild-Type and Mutant Flowers
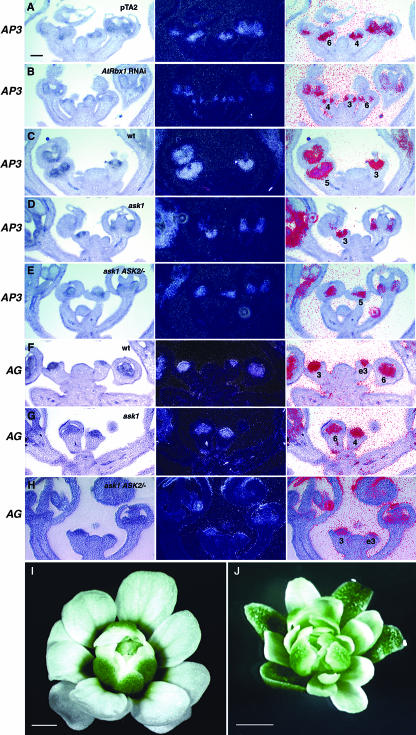
Our phenotypic analysis of Dex-treated AtRbx1 RNAi and ask1/ask1 ASK2/ask2 plants suggests that B function gene expression may be reduced in these plants. To evaluate this possibility, we performed RNA in situ hybridization with an AP3 probe. In the inflorescence of Dex-treated pTA2 control plants, AP3 RNA was detected at a high level in petal and stamen primordia of a young floral bud (Fig. 3A). In the young buds of Dex-treated AtRbx1 RNAi plants, the expression of AP3 was detected at a lower level (Fig. 3B). In addition, compared to the wild type (Fig. 3C), the ask1-1 flower showed slightly reduced AP3 signals (Fig. 3D; Zhao et al., 2001b). Similarly, the ask1/ask1 ASK2/ask2 flower also exhibited slightly reduced AP3 signals (Fig. 3E).
Figure 3.
Expression of AP3 and AG RNA in the inflorescence of wild type and mutants, and genetic interaction with AG. A to H, In situ RNA hybridization with an AP3 probe (A–E) and an AG probe (F–H). The left panel in each triplet is a bright field image showing the tissues, the central panel is a dark field image, and the right panel is a composite image of both. Numbers indicate floral stages, and e3 means early stage 3. The sections in the following panels have been hybridized and developed at the same time: A and B; C, D, and E; F, G, and H. A, pTA2 control transgenic plant. B, Rbx1 RNAi plant. C, Wild type. D, ask1-1. E, ask1/ask1 ASK2/ask2. F, Wild type. G, ask1-1. H, ask/ask1 ASK2/ask2. I, An ag-1 flower with the third whorl stamens converted to petals and a new flower initiated in the fourth whorl. J, An ask1 ASK2/ask2 ag-1 flower with normal organ identity in the first whorl and restored petal development in the second whorl. Scale bars = 50 μm (A–H), 0.5 mm (I and J).
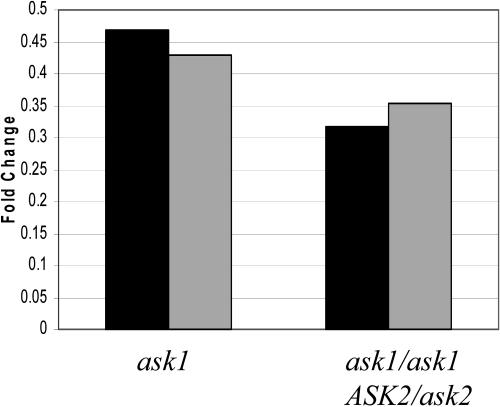
To further confirm a reduction of AP3 RNA expression in the ask1 mutant and an enhanced reduction of AP3 expression in the ask1ask1/ ASK2/ask2 plant, we performed real-time PCR using the RNA isolated from young floral buds up to stage 8 (Smyth et al., 1990) from the wild-type, ask1, and ask1/ask1 ASK2/ask2 plants. Compared to the wild type, AP3 expression was reduced to approximately 45% in the ask1 mutant, and further reduced to about 33% in the ask1/ask1 ASK2/ask2 plant (Fig. 4).
Figure 4.
AP3 transcript levels in mutant inflorescences. Relative mRNA expression levels of AP3 gene in mutants were determined by real-time PCR. Black and gray bars represent results from two independent experiments. AP3 expression in the wild type was considered as 1.
In addition, the ap2-1 like phenotype of ask1/ask1 ASK2/ask2 flowers suggests an expansion of C function to the first and second whorl. To test whether C function is altered at the transcription level in the ask1/ask1 ASK2/ask2 flower, we performed RNA in situ hybridization in the wild-type, ask1, and ask1/ask1 ASK2/ask2 flowers with an AG probe. AG RNA was detected in the central dome of wild-type flowers starting at early stage 3, and thereafter was restricted to stamen and carpel primordia (Fig. 3F). Nonspecific signal was detected in the upper part of sepals in late stage flowers (starting from stage 7), as observed previously (Drews et al., 1991; data not shown). The ask1 flower showed a normal spatial and temporal pattern of AG expression, and the expression level was also close to normal (Fig. 3G). The ask1/ask1 ASK2/ask2 flower exhibits a slightly reduced AG expression level in the center (Fig. 3H), whereas the expression pattern was close to normal. No significant AG signal was detected in early sepal primordia (Fig. 3H). Compared to the wild-type flower, slightly earlier and stronger signals seemed to be detected at the upper part of sepals in the mutant flower (data not shown). However it is not certain whether these signals reflect altered AG expression pattern or are only nonspecific signals.
Genetic Interaction with AG
To further test if AG is epistatic to ASK1 and ASK2 in terms of the ap2-like phenotypes in the ask1/ask1 ASK2/ask2 flower, we crossed the ask1/ask1 ASK2/ask2 into ag-1, a strong ag allele in which stamens are converted to petals, and whorl 4 is replaced by another flower (Bowman et al., 1989; Fig. 3I). The ask1/ask1 ASK2/ask2 ag-1 flower is similar to the ag-1 flower with normal whorl 1 organ identity and whorl 2 petals restored (Fig. 3J), suggesting that AG function is essential for the ap2-like phenotypes in the ask1/ask1 ASK2/ask2 flower. Floral organs in the ask1/ask1 ASK2/ask2 ag-1 plant are smaller than those in the ag-1 mutant, suggesting that the defect in organ size of the ask1/ask1 ASK2/ask2 flower is not dependent on AG function.
DISCUSSION
Ubiquitin-mediated protein degradation has been recognized as a very important mechanism for regulating many cellular events. In particular, the SCF ubiquitin-protein ligases are known to control cell cycle regulation, signal transduction, transcription, and other biological events (Bai et al., 1996; Hershko and Ciechanover, 1998; Schulman et al., 2000; DeSalle and Pagano, 2001; Conaway et al., 2002; Zheng et al., 2002). In Arabidopsis, several SCF complexes that are involved in hormone signaling or cell division have been characterized (Gray et al., 1999; del Pozo et al., 2002a; Xu et al., 2002). The existence of a large number of F-box genes in the Arabidopsis genome suggests that plants make extensive use of SCF complexes to regulate multiple biological processes (Gagne et al., 2002; Risseeuw et al., 2003).
Expression analysis indicates that many ASK genes are expressed in the inflorescence, suggesting that SCF complexes may play multiple roles in flower development (Zhao et al., 2003b). However, among the F-box and ASK genes, only UFO and ASK1 have been shown to have roles in flower development. Functional redundancy among genes in these families may partially explain this limited characterization. ASK1 and ASK2 were found to be capable of interacting with many F-box proteins (Gagne et al., 2002; Risseeuw et al., 2003). In addition, ASK1 and ASK2 are both highly expressed in all major tissues (Zhao et al., 2003b). Therefore, it is likely that ASK1 or ASK2 is a component of many SCF complexes, and they may share redundant functions throughout plant life cycle, including flower development. Indeed, we found that the ask2 single mutant was indistinguishable from the wild type, whereas the ask1 ask2 double homozygous mutant was seedling lethal (Liu et al., 2004), and the ask1/ask1 ASK2/ask2 flower exhibited enhanced and additional novel phenotypes compared to the ask1 flower. Although residual expression of the C terminus of the ASK2 gene was detected in the ask2-1 mutant by real-time PCR, the phenotypes in the ask1/ask1 ASK2/ask2 flowers are not likely caused by altered ASK2 protein structure, as these phenotypes were also observed from strong ASK1 RNAi plants, and were restored to normal with an additional copy of genomic ASK1.
Previous studies have suggested that AtCUL1 and AtRbx1a encode core components of many SCF complexes in Arabidopsis and that they play critical roles throughout development (Gray et al., 2002; Lechner et al., 2002; Schwechheimer et al., 2002; Shen et al., 2002; Xu et al., 2002; Hellmann et al., 2003). Therefore, studies on the mutants or transgenic plants with a reduced function of these core components may also reveal the diverse roles of multiple SCF complexes in flower development. Indeed, we found diverse floral phenotypes in plants that are homozygous for a point mutation in the AtCUL1 gene or carrying a Dex-induced AtRbx1 RNAi construct.
Although several similar floral phenotypes were observed in all of the ask1/ask1 ASK2/ask2, axr6-2, and AtRbx1 RNAi plants, distinctive floral phenotypes were also observed in plants of each of these genotypes. One of the explanations for the distinctive phenotypes is the partial functional redundancy within different members of ASK genes and cullin homologs. It is also possible that the AtCUL1 point mutation might affect its interaction with some F-box proteins more so than that with others. In addition, although all of the ASK1, ASK2, AtCUL1, and AtRbx1 genes encode core components of SCF complexes, we cannot rule out the possibility that these proteins can also function as a subunit of non-SCF complexes so that each gene may have its distinctive function.
Regulation of B Function Gene Expression by the SCFUFO Complex
The floral phenotypes of axr6-2, axr6-2 ask1, and Dex-induced AtRbx1 RNAi plants suggest a reduction of B function in these flowers. In situ results further suggested a reduction of AP3 gene expression in the Dex-induced strong AtRbx1 RNAi flower. Furthermore, the ufo-6 axr6-2 double mutant showed an increased number of carpelloid organs and filaments in the third whorl, suggesting a further reduction in B function compared to either single mutant. Previous studies in our laboratory indicated that UFO interacts with ASK1 and LEAFY genetically to regulate B function gene expression (Zhao et al., 1999, 2001b). In addition, UFO interacts with ASK1 and AtCUL1 physically (Samach et al., 1999; Wang et al., 2003). We have recently confirmed the physical interaction between UFO and ASK1 by coimmunoprecipitation using an anti-myc antibody and inflorescence extracts from 35S:UFO-myc transgenic plants (data not shown). Taken together, these results support the idea that B function gene expression is positively regulated by SCFUFO complex consisting of UFO, AtCUL1, AtRbx1, and ASK1.
Consistent with previous results, we observed a slight reduction of B function in the ask1-1 mutant flower (Zhao et al., 1999, 2001b). A further reduction of ASK2 levels in the ask1/ask1 ASK2/ask2 plants and the strong ASK1 RNAi plants produced an additional reduction of B function in the flowers, which was further confirmed by complementation with either the ASK1 or ASK2 transgene. Both ASK1 and ASK2 can interact with UFO in a yeast two-hybrid assay (Samach et al., 1999). Therefore, it is likely that ASK1 and ASK2 share a redundant function in promoting B function through their interaction with UFO. Our results also suggest that ASK1 plays a much more important role than ASK2 in promoting B function, as the ask1 flower exhibits phenotypes of reduced B function, whereas the ask2 flower as well as the ASK1/ask1 ask2/ask2 flower is normal.
Regulation of C Function by the SCFUFO Complex and Other Possible SCF Complexes
The carpelloid sepals in the first whorl and near absence of petal in both the ufo-6 axr6-2 and ask1/ask1 ASK2/ask2 flowers were similar to those observed in ap2 mutants, in which expression of the C function gene AG expands to the first and second whorls (Bowman et al., 1991; Drews et al., 1991). The similar phenotypes in strong ASK1 RNAi flowers and genetic complementation of ask1/ask1 ASK2/ask2 plants further verified that ASK1 and ASK2 are responsible for the ap2-like phenotype. In addition, carpelloid sepals were also observed in the axr6-2 ask1 flowers. Genetic analysis suggests that AG function is essential for the ap2-like phenotypes in the ask1/ask1 ASK2/ask2 flowers. Recently, new ufo alleles characterized by the absence of petals were isolated, which uncovered an additional role for UFO in promoting organ formation in the second whorl (Durfee et al., 2003). Genetic data suggested that UFO functions to inhibit an AG-dependent activity to promote early petal formation (Durfee et al., 2003). Intriguingly, the ufo-6 mutation (P299 to L) maps immediately adjacent to the ufo-14 mutation (S298 to A), which is required for petal formation (Lee et al., 1997; Durfee et al., 2003). In conjunction with this, petal/stamen mosaic organs were frequently observed in the second whorl of the ufo-6 flower. One alternative explanation is that C function expands into the second whorl of the ufo-6 flower and that this phenotype was enhanced by the axr6-2 mutant. Taken together, these results support a role for the SCFUFO complex in promoting early petal formation through a negative regulation of AG function.
Unlike the ufo-14 mutant, flowers in the ufo-6 axr6-2 double mutant exhibited carpelloid sepals in the first whorl. Carpelloid sepals were also observed in strong ufo alleles in ecotype Landsberg erecta of Arabidopsis (Ler) background (Wilkinson and Haughn, 1995). Similar carpelloid sepals were also observed in the ask1/ask1 ASK2/ask2 flower. These results suggest that SCFUFO complex(es) may also contribute to the inhibition of C function in the first whorl. The similar weak phenotypes found for the strong ufo alleles, the ufo-6 axr6-2 double mutant, and the ask1/ask1 ASK2/ask2 plant all indicate that other SCF complexes are also likely to be involved in inhibition of C function in the first whorl. Alternatively, UFO may retain some residual function even in these strong ufo alleles.
Our in situ results suggest that AG RNA did not expand to the sepal primordia in the ask1/ask1 ASK2/ask2 flower. Similarly, AG RNA was not detected in the second whorl of the newly isolated petalless ufo alleles (Durfee et al., 2003), suggesting that AG might act non-cell autonomously to play a role in the first and second whorls in those mutants. Alternatively, AG RNA may have been expressed at a very low level, or at late stages in the sepal of the ask1/ask1 ASK2/ask2 flower. It is possible that the high level of nonspecific signals in the upper part of the late stage sepals makes it difficult to recognize the real AG signals.
SCFUFO and other SCF complexes can repress AG protein function in the first and second whorls either through a direct repression of AG function, or through an indirect repression by activating a repressor of AG function. Because ectopic expression of UFO throughout the flower does not lead to an ag phenotype (Lee et al., 1997), it is not likely that UFO can act directly on AG, at least in the third and fourth whorl. Further support for the idea that UFO does not repress AG function directly in the first and second whorls can be obtained from an analysis on the double transgenic plant of 35S:UFO and 35S:AG. In addition to AP2, many other genes also contribute to the inhibition of AG function in the first and second whorls, including AINTEGUMENTA (Krizek et al., 2000), LEUNIG (Liu and Meyerowitz, 1995), STERILE APETALA (Byzova et al., 1999), CURLY LEAF (Goodrich et al., 1997), INCURVATA2 (Serrano-Cartagena et al., 2000), and SEUSS (Franks et al., 2002). It is possible that the SCFUFO complex and other SCF complexes contribute to the inhibition of AG function in the first and second whorls through the degradation of transcription repressors or other proteins to activate one or more of the negative regulators of AG function, although the SCFUFO complex could also work separately to promote petal formation (Durfee et al., 2003; Laufs et al., 2003).
The curled or unfused carpel in the fourth whorl of the ask1/ask1 ASK2/ask2 flowers suggests a reduced C function, as supported by the in situ hybridization results. Unfused carpels were also found in the ufo-1 mutant when grown under short day (SD) condition (Wilkinson and Haughn, 1995), indicating that UFO might also participate in promoting C function in the fourth whorl. A role of UFO in the fourth whorl is also consistent with the expression of UFO in the center of a stage 2 flower primordia (Lee et al., 1997). Reduced C function was also observed in null mutants of FIM gene, the UFO homolog in Antirrhinum majus (Ingram et al., 1997). Thus, it is likely that the SCFUFO complex and other SCF complexes might also function in promoting C function in the center of the flower.
Regulation of Other Aspects of Flower Development by the SCFUFO Complex and Other SCF Complexes
The inflorescence of the ufo-6 axr6-2 double mutant exhibited a filament-like structure instead of a normal flower and terminal inflorescence meristem, similar to those found in strong ufo mutants (Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995). Thus, it is likely that UFO also regulates floral and inflorescence meristem through an SCFUFO complex containing AtCUL1.
An axillary flower similar to those found in ap1 mutants (Bowman et al., 1993) was sometimes found in the AtRbx1 RNAi flower. Similar secondary flowers were also found in A. majus fim null alleles (Ingram et al., 1997), indicating that the SCFUFO complex or other SCF complexes are also involved in regulating floral meristem identity.
The axr6-2 flower exhibited a reduction of floral organ number in all four whorls, indicating that SCF complexes also regulate floral organ number, probably through regulating cell division in the floral meristem. In addition, fused sepals, petals, and stamens were observed in ask1/ask1 ASK2/ask2 and strong ASK1 RNAi plants, which indicate that SCF complexes are also required for organ separation, probably through the regulation of cell division in each individual whorl. Furthermore, increased number or size of carpel was found in the ask1/ask1 ASK2/ask2 flower. Similar phenotypes were also observed in strong ufo mutants. UFO seemed to be required for the restriction of cell division in the center of a stage 2 flower (Samach et al., 1999). Our results support that UFO interacts with ASK1 or ASK2 to restrict cell division in the central region.
In conclusion, our results indicate that SCF complexes regulate several aspects of floral development in Arabidopsis. Further functional studies on additional F-box proteins in flower development, as well as target proteins regulated by these SCF complexes, will provide insights into the network of flower development regulation by SCF complexes in Arabidopsis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The axr6-2 mutant is in the Columbia background (Hobbie et al., 2000). The following mutants and transgenic plants were in the Landsberg erecta (Ler) background: the ufo-2, ufo-6 mutant (Levin and Meyerowitz, 1995), the ask1-1 mutant (Yang et al., 1999), the ASK1 RNAi plants (Zhao et al., 2003b), and the ag-1 mutant (Bowman et al., 1989). The Dex-inducible AtRbx1 RNAi lines (Lechner et al., 2002) and ask2-1 mutant were of the Wassilewskija (Ws) ecotype.
The axr6-2 mutant was backcrossed to the wild type eight times before phenotypic analysis. Among the rootless homozygous axr6-2 seedlings cultured on Murashige and Skoog medium for 10 d, the few seedlings that developed roots were transferred to soil for further characterization.
For floral phenotypic analysis of AtRbx1 RNAi plants, a strong line (dsRNA-2), a weak line (dsRNA-64), and a control line transformed with an empty vector were treated with Dex for 5 to 7 d at 2 to 3 weeks of age, then transferred to Dex-free conditions for further culture and analysis. Dex was dissolved in ethanol and kept at a concentration of 30 mm, and was directly added at a concentration of 1 μm to the medium or dripped onto plants in a solution at 10 μm (with 0.01% Tween 20). For RNA in situ hybridization, 3-week-old dsRNAi-2 plants and control plants were treated with Dex for 5 d, then fixed in formaldehyde-acetic acid fixative.
Plants were cultured on Murashige and Skoog medium, or grown on Metro-Mix 360 (Scotts-Sierra Horticultural Products, Marysville, OH) at 22°C (16 h light, 8 h dark).
Construction and Identification of Double Mutants
The single mutants used for phenotypic analysis and comparison were from self-pollination of either homozygous (ufo-2, ufo-6, ask2-1) or heterozygous (axr6-2, ask1-1, ag-1) plants. To construct a ufo-6 axr6-2 double mutant, a heterozygous axr6-2 plant was used as the male parent in a cross with a homozygous ufo-6 plant. The genotype of the double mutant was determined by sequencing PCR products. For determining the homozygous ufo-6, genomic DNA was amplified with primers oMC 834 (5′-CTTTGCCACGGCTTTGTAGCTTG-3′) and oMC 835 (5′-GACCCACAGCCAGCTTTTTCTCA-3′). For determining the homozygous axr6-2, DNA was amplified with primers oMC 836 (5′-TGTGGTTAGGTTTTGCCTGCGTT-3′) and oMC 837 (5′-AGCAGGGCCCTATCAATCTGCTC-3′).
To construct an ask1-1 ask2-1 double mutant, pollens from a homozygous ask2-1 plant were used to pollinate homozygous ask1-1 pistils. The genotypes of F2 plants were determined by PCR. The ask1-1 mutant harbors a Ds transposon in ASK1 at the position 237 bp downstream of the ATG start codon (Yang et al., 1999). The ask2-1 mutant carries a T-DNA in ASK2 at the position 318 bp downstream of the ATG start codon. To determine the genotypes of F2 plants, gene-specific and allele-specific primers were designed to amplify ASK1 (wild-type allele), ASK2, ask1-1 (mutant allele), and ask2-1 (Fig. 2). The wild-type ASK1 allele was amplified with primers oMC221 (5′-AAGGTGATCGAGTATTGCAAGAG-3′) and oMC383 (5′-GAAGATAGTCATGATTCATGAAG-3′). The ask1-1 allele was amplified with primers oMC529 (5′-TCACTAGTGAGCTCATAACCATGTCTGCGAAGAA-3′) and oMC490 (5′-CGTTCCGTTTTCGTTTTTTACC-3′) on Ds element. The wild-type ASK2 allele was amplified with primers oMC697 (5′-TCCACGTCGTCTCTAAACTCAG-3′) and oMC593 (5′-AAATGGGTCGAGGACATGAC-3′). The ask2-1 allele was amplified with primers oMC696 (5′-CCATCATACTCATTGCTGATCC-3′) on T-DNA boarder region and oMC697 (see above).
For the complementation of ask1/ask1 ASK2/ask2 floral phenotypes with ASK1, we crossed the ask2-1 mutant with an ask1-1 mutant harboring an ASK1 transgene (tASK1), which contains a 5,578 bp HindIII/EcoRV genomic fragment including the ASK1 gene and 4,183 bp upstream of the ATG (Zhao et al., 2003a). The F2 plants were first screened with Liberty herbicide (AgrEvo, USA Company, Montvale, NJ), then genotyped for the ask1/ask1 ASK2/ask2 allele. The wild-type ASK1 allele was identified with primers oMC927 (5′-GAGTTCCGATGGTGAATCTTTC-3′) at 27 bp downstream of the ASK1 ATG start codon, and oMC928 (5′-TAGCTCTTTTCGAGTGACCACA-3′) at 1,466 bp downstream of the ASK1 ATG. All other primers for identifying ask1, ASK2 and ask2 alleles were the same as above.
For the complementation of ask1/ask1 ASK2/ask2 with ASK2, we performed a cross between the ask2-1 mutant and an ask1-1 mutant harboring an ASK2 transgene (tASK2), which contains an ASK2 cDNA fused with the 35S promoter (Zhao et al., 2003a). The F2 plants were first screened with Liberty herbicide as above, then screened for the ask1/ask1 ASK2/ask2 genotype harboring tASK2. The tASK2 was identified with primers oMC570 (5′-CCGACAGTGGTCCCAAAGATGGA-3′) specific to 35S promoter and oMC593 (see above). All the primers for identifying the ask1/ask1 ASK2/ask2 genotype were the same as above.
To construct an ask1-1 axr6-2 double mutant, pollens from a heterozygous axr6-2 plant were used to pollinate homozygous ask1-1 pistils. To construct an ask1/ask1 ASK2/ask2 ag-1 mutant, pollens from a heterozygous ag-1 plant were used to pollinate ASK1/ask1 ask2/ask2 pistils. The genotypes of F2 plants were determined by PCR as described before.
In Situ RNA Hybridization
Inflorescences from wild-type and mutant plants were harvested from 3- to 4-week-old plants and immediately fixed in a formaldehyde-acetic acid fixative. RNA in situ hybridizations with radioactive probes were performed as previously described (Drews et al., 1991; Flanagan and Ma, 1994). The AP3 and AG antisense probes were synthesized using pD793 (digested with BglII) and pCIT565 (digested with HindIII) as template, respectively (Yanofsky et al., 1990; Jack et al., 1992). Both probes were synthesized with T7 RNA polymerase (Promega, Madison, WI).
RNA Quantitation by Real-Time PCR
RNA was isolated from the young inflorescence including stage 0 to 8 young floral buds (Smyth et al., 1990). Total RNA was isolated using the RNeasy mini kit (QIAGEN, Valencia, CA) and was treated with DNase I (Life Technologies/Gibco-BRL, Carlsbad, CA). One microgram of RNA from different tissues was reverse transcribed into cDNA with oligo(dT), 16 mer, using Super Script II reverse transcriptase (Life Technologies/Gibco-BRL) in a total volume of 20 μL. The cDNA was then diluted 100 times, and 5 μL of the diluted cDNA was used as a template for real-time PCR analysis. The primers were designed with Primer Express version 1.0 (ABI, Foster City, CA). The primer sequences were as follows: ASK2FW (oMC1531), 5′-GGACTGTTGGACTTGACTTGCC-3′; ASK2RV (oMC1532), 5′-GAGACACAAATGGGTCGAGGA-3′; AP3FW (oMC1529), 5′-GATGTCGATGTTTGGGCCAC-3′; AP3RV (oMC1530), 5′-AGATTTGAACTTGCGCTCGC-3′. The primers are expected to produce 201-bp products. Primers for ACTIN genes were used as an internal control to normalize the expression data for each gene. The primers were designed so that the two genes ACTIN2 and ACTIN8 were amplified simultaneously (Charrier et al., 2002). The sequence for the control primers are as follows: ACTINFW (oMC1533) 5′-GGTAACATTGTGCTCAGTGGTGG-3′; ACTINRV (oMC1534) 5′-AACGACCTTAATCTTCATGCTGC-3′. They are expected to produce a product of 108 bp.
The cDNA was amplified using the SYBR Green PCR Master Mix (Stratagene, La Jolla, CA) on the ABI PRISM 7700 thermocycler (ABI). The PCR conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 60°C for 1 min, 72°C for 1 min. The cycle threshold values were used to calculate differences in fold changes. At the end of PCR cycles, the data were analyzed with the ABI Sequence Detection Systems (SDS) version 1.7 (ABI). To check the specificity of annealing of the PCR products, a dissociation kinetics was performed by the machine at the end of the experiment. In addition, PCR products were verified by sequencing directly. Negative control using the same amount of RNA did not produce any PCR product. In one experiment, at least three replications were performed for each sample. The experiments were repeated at least twice independently.
Acknowledgments
We thank P. Genschik for kindly providing the AtRbx1 RNAi lines, E. Risseeuw and W.L. Crosby for providing the tASK1 and 35S:ASK2 constructs, and N. Wei for the 35S:UFO-myc seeds. In addition, we thank Y. Hu for technical assistance, and A. Omeis and J. Wang for plant care. We are grateful for helpful comments from C.L. Hendrix, W. Hu, and L.M. Zahn. We thank two anonymous reviewers for their helpful comments.
This work was supported by the National Science Foundation (grant nos. MCB–9896340 and MCB–0092075 to H.M. and IBN–998926 to L.H.), and by funds from the Department of Biology and the Huck Institutes of Life Sciences at the Pennsylvania State University.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.031971.
References
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Byzova MV, Franken J, Aarts MG, de Almeida-Engler J, Engler G, Mariani C, Van Lookeren Campagne MM, Angenent GC (1999) Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev 13: 1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130: 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Conaway RC, Brower CS, Conaway JW (2002) Emerging roles of ubiquitin in transcription regulation. Science 296: 1254–1258 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002. a) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCFAtSKP2 pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M (2002. b) AXR1–ECR1–dependent conjugation of RUB1 to the Arabidopsis Cullin AtCUL1 is required for auxin response. Plant Cell 14: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSalle LM, Pagano M (2001) Regulation of the G1 to S transition by the ubiquitin pathway. FEBS Lett 490: 179–189 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol 15: 435–467 [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM (1991) Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002 [DOI] [PubMed] [Google Scholar]
- Durfee T, Roe JL, Sessions RA, Inouye C, Serikawa K, Feldmann KA, Weigel D, Zambryski PC (2003) The F-box-containing protein UFO and AGAMOUS participate in antagonistic pathways governing early petal development in Arabidopsis. Proc Natl Acad Sci USA 100: 8571–8576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farras R, Ferrando A, Jasik J, Kleinow T, Okresz L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C (2001) SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J 20: 2742–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan CA, Ma H (1994) Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant Arabidopsis flowers. Plant Mol Biol 26: 581–595 [DOI] [PubMed] [Google Scholar]
- Franks RG, Wang C, Levin JZ, Liu Z (2002) SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129: 253–263 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Hellmann H, Dharmasiri S, Estelle M (2002) Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14: 2137–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H, Hobbie L, Chapman A, Dharmasiri S, Dharmasiri N, Del Pozo C, Reinhardt D, Estelle M (2003) Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J 22: 3314–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hobbie L, McGovern M, Hurwitz LR, Pierro A, Liu NY, Bandyopadhyay A, Estelle M (2000) The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127: 23–32 [DOI] [PubMed] [Google Scholar]
- Ingram GC, Doyle S, Carpenter R, Schultz EA, Simon R, Coen ES (1997) Dual role for fimbriata in regulating floral homeotic genes and cell division in Antirrhinum. EMBO J 16: 6521–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697 [DOI] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG (2002) The SCF ubiquitin ligase: an extended look. Mol Cell 9: 923–925 [DOI] [PubMed] [Google Scholar]
- Koepp DM, Harper JW, Elledge SJ (1999) How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97: 431–434 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Prost V, Macias A (2000) AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell 12: 1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Klenz JE, Martinez-Zapater J, Haughn GW (1989) AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1: 1195–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Takahashi N, Shimada H, Seki M, Shinozaki K, Matsui M (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol 43: 1073–1085 [DOI] [PubMed] [Google Scholar]
- Laufs P, Coen E, Kronenberger J, Traas J, Doonan J (2003) Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development 130: 785–796 [DOI] [PubMed] [Google Scholar]
- Lechner E, Xie D, Grava S, Pigaglio E, Planchais S, Murray JA, Parmentier Y, Mutterer J, Dubreucq B, Shen WH, Genschik P (2002) The AtRbx1 protein is part of plant SCF complexes, and its down-regulation causes severe growth and developmental defects. J Biol Chem 277: 50069–50080 [DOI] [PubMed] [Google Scholar]
- Lee I, Wolfe DS, Nilsson O, Weigel D (1997) A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol 7: 95–104 [DOI] [PubMed] [Google Scholar]
- Levin JZ, Meyerowitz EM (1995) UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell 7: 529–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ni W, Griffith ME, Huang Z, Chang C, Peng W, Ma H, Xie D (2004) The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell 16: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Meyerowitz EM (1995) LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121: 975–991 [DOI] [PubMed] [Google Scholar]
- Ma H (1994) The unfolding drama of flower development: recent results from genetic and molecular analyses. Genes Dev 8: 745–756 [DOI] [PubMed] [Google Scholar]
- Ma H, dePamphilis C (2000) The ABCs of floral evolution. Cell 101: 5–8 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J 34: 753–767 [DOI] [PubMed] [Google Scholar]
- Samach A, Klenz JE, Kohalmi SE, Risseeuw E, Haughn GW, Crosby WL (1999) The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J 20: 433–445 [DOI] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408: 381–386 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Deng XW (2002) Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14: 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Cartagena J, Candela H, Robles P, Ponce MR, Perez-Perez JM, Piqueras P, Micol JL (2000) Genetic analysis of incurvata mutants reveals three independent genetic operations at work in Arabidopsis leaf morphogenesis. Genetics 156: 1363–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WH, Parmentier Y, Hellmann H, Lechner E, Dong A, Masson J, Granier F, Lepiniec L, Estelle M, Genschik P (2002) Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol Biol Cell 13: 1916–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Feng S, Nakayama N, Crosby WL, Irish V, Deng XW, Wei N (2003) The COP9 signalosome interacts with SCFUFO and participates in Arabidopsis flower development. Plant Cell 15: 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78: 203–209 [DOI] [PubMed] [Google Scholar]
- Wilkinson MD, Haughn GW (1995) UNUSUAL FLORAL ORGANS controls meristem identity and floral organ primordia fate in Arabidopsis. Plant Cell 7: 1485–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D (2002) The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Hu Y, Lodhi M, McCombie WR, Ma H (1999) The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc Natl Acad Sci USA 96: 11416–11421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]
- Zhao D, Han T, Risseeuw EP, Crosby WL, Ma H (2003. a) Conservation and divergence of ASK1 and ASK2 gene functions during male meiosis in Arabidopsis thaliana. Plant Mol Biol 53: 163–173 [DOI] [PubMed] [Google Scholar]
- Zhao D, Ni W, Feng B, Han T, Petrasek MG, Ma H (2003. b) Members of the ASK gene family exhibit a variety of expression patterns and may play diverse roles in Arabidopsis. Plant Physiol 133: 203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Yang M, Solava J, Ma H (1999) The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev Genet 25: 209–223 [DOI] [PubMed] [Google Scholar]
- Zhao D, Yu Q, Chen C, Ma H (2001. a) Genetic control of reproductive meristems. In MT McManus, B Veit, eds, Annual Plant Reviews: Meristematic Tissues in Plant Growth and Development. Sheffield Academic Press, Sheffield, UK, pp 89–142
- Zhao D, Yu Q, Chen M, Ma H (2001. b) The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development 128: 2735–2746 [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, et al. (2002) Structure of the Cul1-Rbx1-Skp1-F box Skp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]