Abstract
Fluorescence microscopy of live cells has become an integral part of modern cell biology. Fluorescent protein tags, live cell dyes, and other methods to fluorescently label proteins of interest provide a range of tools to investigate virtually any cellular process under the microscope. The two main experimental challenges in collecting meaningful live cell microscopy data are to minimize photodamage while retaining a useful signal-to-noise ratio, and to provide a suitable environment for cells or tissues to replicate physiological cell dynamics. This chapter aims to give a general overview on microscope design choices critical for fluorescence live cell imaging that apply to most fluorescence microscopy modalities, and on environmental control with a focus on mammalian tissue culture cells. In addition, we provide guidance on how to design and evaluate fluorescent protein constructs by spinning disk confocal microscopy.
1. Fluorescence Microscopy Basics
Fluorescence imaging relies on illumination of fluorescently labeled proteins or other intracellular molecules with a defined wavelength of light ideally near the peak of the fluorophor excitation spectrum, and detection of light emitted at a longer wavelength. An important question is how much excitation light is actually needed to obtain a useful image. At the objective front lens, the light power of our spinning disk confocal microscope with a 100 mW 488 nm solid state laser at 100% illumination is approximately 6 mW (measured with an X-Cite XR2100 light power meter, EXFO Photonic Solutions; Chapter 4). Divided by the area of the spinning disk aperture of ~6000 µm2 at 100× magnification this results in an irradiance of ~100 W cm−2. At lower magnification, the excitation light is spread over a larger area, thus the irradiance decreases proportional to the square of the magnification ratio (i.e. ~36 W cm−2 for 60×). For comparison, the direct solar irradiance at ground level on a bright sunny day at noon is ~1000 W m−2 (i.e. 0.1 W cm−2) across all wavelengths or ~1–1.5 W m−2 nm−1 for specific wavelengths within the visible part of the spectrum1. Although this should be considered a rough estimate, it shows that the maximum light intensity in a spinning disk confocal is ~1000 times higher compared with the total irradiance of direct sunlight, and one million times higher at a specific excitation wavelength. Similar calculations can be made for widefield epifluorescence illumination and result in similar values depending on the light source. Because laser scanning confocal microscopes utilize a focused beam to illuminate a very small area at a time, typical irradiance values can be several orders of magnitude higher.
This difference in specimen irradiance between spinning disk and laser scanning confocal microscopes explains partly why spinning disk confocal microscopes are the better choice for live cell imaging. Fluorescence emission is linearly related to the excitation light intensity as long as the majority of fluorescent molecules in a population are not in the excited state. At higher rates of photon flux, however, that are quite easily reached in laser scanning confocal microscopes, a large proportion of fluorophors populates the excited state and thus can no longer absorb additional photons (Wang, Babbey, & Dunn, 2005). This is referred to as ground-state depletion, and additional excitation light will only yield sub-proportional increases in fluorescence signal, but still contribute to photodamage. Because in spinning disk confocal microscopes, the excitation laser light is spread over thousands of pinholes that scan across the specimen rapidly (Chapter 9), ground-state depletion is not reached even with excitation lasers with hundreds of mW power output. It is interesting to note that ground state depletion can be used to achieve PALM/STORM-type superresolution (Lalkens, Testa, Willig, & Hell, 2012).
Although high intensity light itself is damaging to cells (especially in the near UV range that can induce DNA damage), the main phototoxic effects in live cell fluorescence microscopy result from fluorophor photobleaching. Each time a fluorescent sample is illuminated a fraction of the fluorophor population will be irreversibly destroyed. In addition to decreasing the available fluorescence signal with each exposure, photobleaching generates free radicals and other highly reactive breakdown products (a fact exploited in photoinactivation techniques such as CALI (Jacobson, Rajfur, Vitriol, & Hahn, 2008)). The degree of phototoxicity depends to a large extent on the fluorophor. For example, fluorescent proteins (FPs) tend to be less phototoxic because the photobleaching chemistry is contained within the β-barrel structure. The only certain way to reduce photobleaching and associated photodamage is to reduce excitation light exposure by limiting exposure time and light intensity as much as possible while retaining a useful signal-to-noise ratio required for the specific experimental question (Fig. 1).
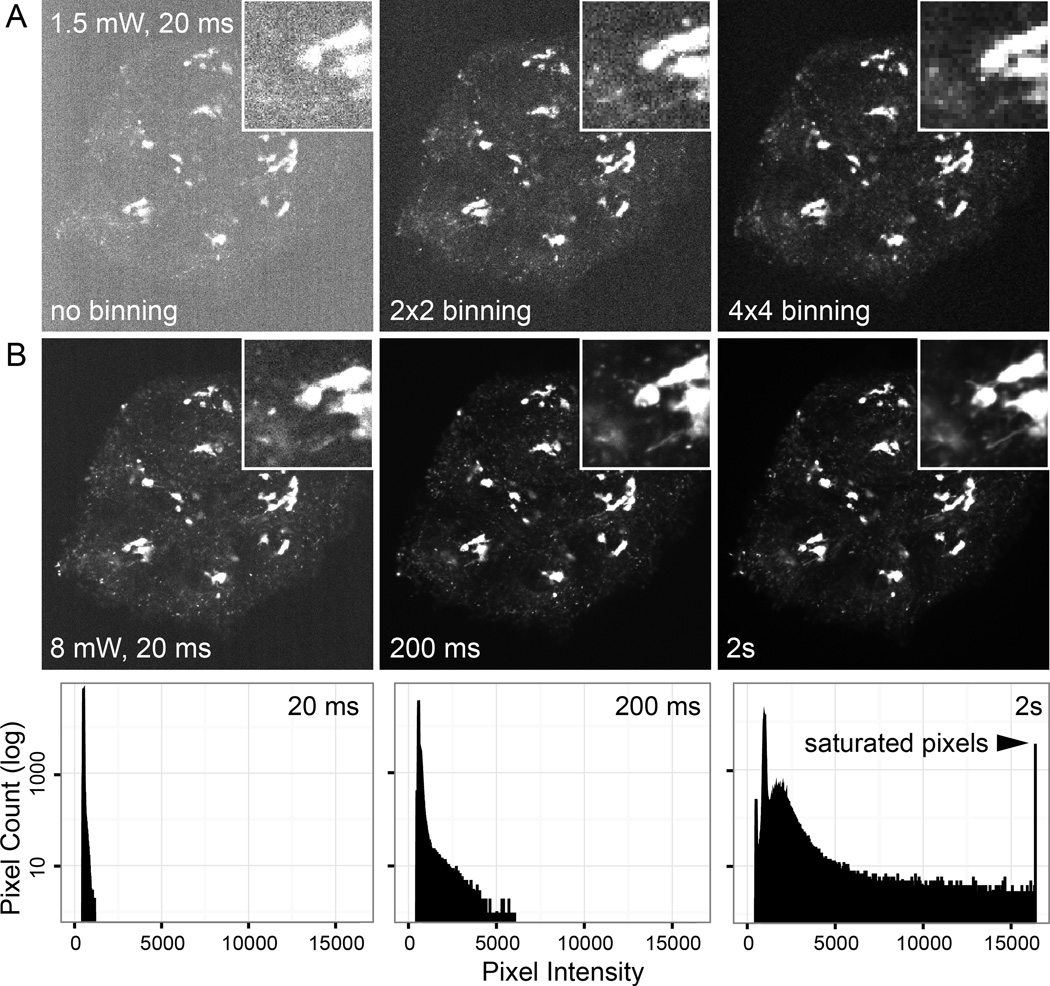
Figure 1. Dependence of signal-to-noise on effective pixel size and exposure.
Spinning disk confocal images of HaCaT cells expressing EGFP-Rab6A that localizes to the Golgi apparatus and intracellular vesicles acquired with 488 nm excitation and a 525/50 nm bandpass emission filter using a Nikon 60× CFI Apo TIRF N.A. 1.49 oil immersion objective. (A) Images acquired with different camera binning, but otherwise identical exposure settings (~1.5 mW light power at the objective; 20 ms exposure time). Although binning drastically increases signal-to-noise, it also decreases resolution. (B) Images with no binning at ~8 mW light power acquired at different exposure times. The graphs below show corresponding histograms of pixel intensities. At sufficient signal-to-noise ratio (somewhere between 20 and 200 ms; magnified insets) details such as EGFP-Rab6 tubules become visible representing optimal image settings for this sample. Longer exposure times (2 s) result in unnecessary photobleaching, blurring of fast moving vesicles without a significant increase in signal-to-noise, and camera saturation.
The traditional fluorescence microscope design utilizing the same lens system both as condenser and objective is suboptimal in limiting light exposure of the specimen. Even in a confocal setup in which emitted out-of-focus light is rejected, the specimen above and below the focal plane is still illuminated, and thus subjected to photobleaching and toxicity. This greatly limits the number of images that can be acquired and in many cases makes it impossible to acquire high resolution, time-lapse series of 3D volumes. It will be exciting to see whether recent light sheet microscopy approaches (Chapter 11) in which excitation is limited to the focal plane that is currently imaged can revolutionize live imaging in cell biology as they have done in developmental biology. Most light sheet microscope designs are limited to low NA, low magnification objectives, which is sufficient to image cell movements in developmental processes, but does not routinely allow high resolution imaging of intracellular dynamics. Recent advances such as Bessel beam microscopy show great promise (Gao et al., 2012), but are currently only available to specialist labs, and this chapter will focus on more traditional fluorescence microscopy technology.
2. The Live Cell Imaging Microscope
Most modern widefield epifluorescence, spinning disk confocal or TIRF microscope setups rely on a similar set of optical and mechanical components, and all imaging modalities are often used for live cell imaging. In the following, we give a short overview of the most critical parameters when designing or optimizing a system for fluorescence live cell microscopy. While it may not be possible to optimize all components of a specific fluorescence microscope setup depending on the imaging modality and experiment, we outline important hardware factors that should be considered in the design of a live cell imaging microscope to limit light exposure as much as possible.
Excitation and Emission light path: The wavelengths of excitation and emission filters should be optimized to match the fluorophor used to limit unnecessary light exposure and optimize detection of fluorophor emission (see more below). Remove all unnecessary optical components from the light path. For example, a forgotten polarizer in the emission path will cut the signal reaching the camera in half. It is also important to reduce background light by turning off the room lights, and minimize scattered light in widefield imaging by closing the field diaphragm as much as possible.
Shutters: Fast, motorized shutters should be used to turn off the excitation light when not needed to take an image. It is particularly important to note that software-controlled shutters often have a significant overhead. Because of computer-induced delays in sending commands (and sloppy programing in almost all modern commercial software), shutters can open and close hundreds of milliseconds before and after an image is actually taken. Ideally, shutters are directly hardware-triggered by the camera. Most camera and shutter manufacturers support this option using a simple coaxial TTL trigger cable, but it is often not implemented by default. It is also important to note that switching between wavelengths will require some additional hardware to combine the camera trigger with a wavelengths selection signal from the imaging software (Stehbens, Pemble, Murrow, & Wittmann, 2012). In the absence of significant shutter overhead and at excitation light intensities well below fluorophor ground state depletion, the recorded signal and the degree of photobleaching should only depend on the total amount of light received by the specimen (i.e. 100 ms exposure at 10 mW should be the same as 1s exposure 1 mW excitation light power). Of note, LED-based light engines are becoming more and more common. An important advantage of LEDs is that they can be switched on and off very rapidly and may thus not require additional mechanical shutters.
Objective lens: As outlined above, specimen irradiance increases drastically with magnification. Thus, to limit photodamage to the specimen, the lowest magnification should be used as determined by the experimental question. However, it is important to note that sufficient sampling of the microscope optical resolution in many cases requires 100× magnification (Stehbens et al., 2012). Depending on how low of a fluorescent signal needs to be observed, one should also try to select the brightest possible objective. For example, phase contrast objectives transmit ~5% less light. We routinely use Nikon 60× and 100× 1.49 NA CFI Apochromat TIRF objectives for spinning disk microscopy to maximize light collection using standard immersion oil. For a more detailed discussion of objective lens characteristics see Chapter 2.
Camera: To detect dim fluorescent signals, it is essential to use cooled scientific grade cameras with the lowest readout noise available. Lower noise allows detection of dimmer signal. While Interline CCD cameras have historically shown the best performance for live cell imaging, the camera field has developed rapidly in recent years (see Chapter 3), and it is difficult to make a general recommendation. Ideally, different types of cameras should be tested with the specimen of interest before a purchasing decision is made to find the best compromise between price and performance. A note of caution regarding EM-CCD cameras that are often pushed for live cell imaging: The electron multiplication feature only discriminates against camera-inherent noise and thus only makes sense with specimens that have low signal and essentially no background fluorescence. For this reason, EM-CCDs are very powerful in single molecule imaging approaches. However, this is not the case for most live cell imaging experiments in which a substantial fluorescent background exists, for example from the pool of cytoplasmic protein. In this case, both the fluorescent background and the signal are amplified equally resulting in larger pixel intensity values, but essentially the same signal-to-noise ratio. Apparent gain in signal-to-noise compared with Interline CCD cameras largely results from the larger pixel size in EM-CCD cameras (the most commonly used EM-CCD chips have a 6× larger pixel area compared with Interline CCDs). Without having done a direct comparison, a very similar increase in signal-to-noise ratio (with an associated decrease in image resolution) can likely be achieved by binning of a regular CCD camera for a much lower cost (Fig. 1). More on cameras in Chapter 3.
Although the above considerations generally also apply to non-fluorescent, transmitted light microscopy, light intensities in phase contrast or DIC are much lower and rarely problematic as long as heat-cutting filters and shutters to turn off illumination between exposures are used. In addition, less expensive objectives and cameras can be used for transmitted light. Environmental control of the specimen becomes much more important to ensure viability in time-lapse experiments monitoring long-term cell dynamics. Other automation that is equally useful for both fluorescent and transmitted live cell microscopy includes:
Hardware Autofocus: Focus drift is a notorious problem in high resolution time-lapse imaging. To some extent focus drift can be minimized by using good experimental practice. For example, by making sure that the specimen is securely seated on the stage and allowing a sufficient period of time for thermal equilibration before starting a time-lapse experiment. However, in experiments that last longer than a few minutes this will likely not be enough. Hardware autofocus systems mostly rely on detecting a reflection of a near IR light beam from the interface between coverslip glass and tissue culture medium, and use drift of this reflection as a feedback for the motorized focus drive. This principle, similar to TIRF, relies on a sufficient refractive index mismatch at the interface and can maintain satisfactory focus even at high magnification for days (Haynes, Srivastava, Madson, Wittmann, & Barber, 2011). Software autofocus may be used on a transmitted light image, but not in a fluorescence channel because the focusing algorithms will need to acquire multiple additional images per time point to determine focus thus resulting in rapid photobleaching.
Motorized Stage: Multi-point acquisition allows parallel data collection from many fields of view, which is especially important for longer time-lapse experiments or drug treatments that can only be done once per specimen. For high magnification experiments the accuracy with which a motorized stage returns to a previous position is key. In high-precision stages equipped with linear encoders to measure stage position independent of the stepper motor, this repeatability should be in the order of hundreds of nanometers. Thus, at high magnification, some inaccuracy in returning to previous position is inevitable and will be visible as slight jitter in time-lapse sequences. However, this often can be corrected by correlation-based image alignment algorithms.
3. Microscope Environmental Control
Cultured cells and tissues will only behave normally in a physiological environment, and control of factors such as temperature and tissue culture medium composition is thus critically important to obtaining meaningful data in live cell imaging experiments. The conditions required to successfully maintain cell health on the microscope stage obviously depend on the organism, and in this section we will provide a general overview of options available to maintain environmental control with a focus on live cell imaging of mammalian cell types.
3.1. Temperature
The most basic level of environmental control is maintaining correct temperature to ensure that observed cell dynamics are an accurate representation of in vivo cell behavior. For cells from warm-blooded animals, the specimen thus needs to be warmed. Although the most commonly used temperature is 37°C for mammalian cell lines, it should be noted that physiological body temperatures can vary considerably (i.e. 42 °C in chickens), and depending on the experiment one may want to take this into consideration. In contrast, Drosophila cells will not behave normally or even survive at 37°C. In general, biochemical reaction kinetics is temperature-dependent, and one would expect that most dynamic cell processes occur at slower rates at lower temperature. Thus, in addition to maintaining cell health, accurate temperature control will also limit variability between experiments. Each of the designs used to achieve temperature control has its advantages and disadvantages, and the choice of equipment will be determined by experimental question and type of specimen:
Air stream incubators: The simplest way to control temperature is by blowing warm air across the specimen. Such glorified hair dryers with highly accurate temperature control are available commercially (Nevtek), but can also be home built. While this design has the advantage of being compatible with virtually any microscope stage and specimen, it is difficult to precisely control temperature at the specimen. In addition, heat fluctuations and vibration negatively influence focus stability, and the stream of warm air will very rapidly evaporate tissue culture media from open cell culture dishes.
Stage-top incubators: Heated stage incubators are available from many manufacturers and are growing in popularity. These range from relatively simple heated inserts for existing microscope stages to more complex incubation chambers that combine temperature and gas control and also allow perfusion of different types of media (Okolab, Tokai Hit, Warner Instruments and others). A general problem with this design is that the central hole in the heated plate through which the specimen is observed is not heated. For long working distance air objectives, transparent, warmed bottom plates are available. However, this does not work with high NA oil objectives, which are thermally coupled to the coverslip by the immersion oil. The objective acting as a heat sink will especially cool the spot that is currently under observation, generating temperature gradients of unknown magnitude. To alleviate this problem, stage-top incubators are often used in combination with heated collars around the objective. A troubling aspect of this design is temperature fluctuations and gradients generated in the objective. Heating the front lens to 37°C will require a much higher temperature of the heating collar. In addition, glass and metal have very different thermal expansion coefficients, and it is easy to imagine how temperature gradients and repeated heating and cooling will negatively affect optical performance and lifetime of these very expensive, precision-engineered compound objective lenses.
Full microscope enclosures: For dedicated live cell imaging systems, we therefore find that a full heated enclosure that contains specimen, stage, objective turret and parts of the microscope body to be the preferred solution. Such enclosures are mostly made from acrylic glass. One disadvantage is that these need to be custom-designed to fit around specific imaging systems and allow for sufficient space for cameras, filter wheels and other peripheral devices. This may also make it more difficult to change components at a later time. Advantages are that full enclosures allow the best thermal equilibration. We always leave our system set to 37°C because it will take several hours for full temperature equilibration. This also reduces focus drift and minimizes thermal cycling experienced by optical and mechanical components. In addition, the microscope stage remains free and there are no limitations to the type of sample chambers that can be used.
3.2. Media Composition and pH
Most tissue culture medium is buffered to physiological pH by sodium bicarbonate and 5% CO2. Thus, pH in an open tissue culture dish on a microscope stage will rapidly increase and leave the physiological range within minutes as CO2 outgases into the atmosphere. One common approach to control pH is the use of CO2-independent media, or addition of 10–25 mM HEPES to increase buffering capacity. However, HEPES alone will not completely control long-term alkalization of the bicarbonate-buffered media, and it is also important to test how different imaging media or HEPES addition affect the process under investigation. For example, bicarbonate transporters are principal regulators of intracellular pH in animal cells, which will not function correctly in bicarbonate-free media or buffers such as PBS. A better, but technically more involved approach is to control CO2 concentration such that regular tissue culture medium can be used, which can be especially important for long term imaging experiments. Most high-end stage incubators have the option of active CO2 control in which a sensor ideally placed near the specimen measures CO2 concentration providing a feedback signal to an active gas mixer. However, active CO2 controllers are unreasonably expensive and not without trouble, and a malfunctioning sensor can offer a false sense of security.
Alternatively, passive control of CO2 concentration can be achieved by using premixed 5% CO2 (often sold as biological atmosphere) or produced by passive volumetric mixing. 5% CO2 can then be streamed slowly onto the specimen covered for example with a very low cost inverted plastic dish to minimize gas leakage. In our hands this has been effective to control pH for days. Of note, it is not feasible and likely dangerous to fill a full microscope enclosure with 5% CO2. For live imaging of tissue slices, more complex gas control including increased oxygen supply may be necessary (e.g. 40% oxygen, 5% CO2, 55% N2) (Attardo, Calegari, Haubensak, Wilsch-Brauninger, & Huttner, 2008). In any case, to prevent evaporation of cell culture medium during imaging, it is necessary to humidify the gas mixture. This can be achieved by bubbling gas through a bottle with sterile deionized water or by channeling through semi-permeable tubing immersed in water. The latter solution is preferable as it introduces less vibration and results in better humidification. To maintain sample temperature, humidifiers should either be warmed or placed inside the heated microscope enclosure. If necessary, additional humidification can be provided by placing wet tissue paper inside the imaging chamber.
Of note, most tissue culture medium contains fluorescent compounds such as for example phenol red. In our hands, background fluorescence of regular DMEM or similar tissue culture media is negligible on our spinning disk confocal setup at 488 nm or 561 nm excitation with optimized filter sets. However, phenol red is highly fluorescent when excited at 440 nm, and phenol red free media has to be used for imaging of cyan FPs.
3.3. Imaging Chambers
For live cell microscopy, cells are typically grown on coverslips and viewed with an inverted microscope from below. Most microscope objectives are designed for No. 1.5 coverslips (0.17 mm thick), and use of coverslips of a different thickness will result in spherical aberration. However, practical considerations can sometimes demand the use of thinner coverslips. For example, we have used No. 1 coverslips for high resolution imaging of epithelial organoids embedded in a 3D extracellular matrix to increase the useable working distance of high NA oil objectives (Gierke & Wittmann, 2012). Coverslips should be cleaned well before seeding cells, for example by using the ‘squeaky clean’ coverslip cleaning protocol outlined in Chapter 20. It may also be necessary to coat coverslips with appropriate extracellular matrix molecules to improve adhesion and promote normal cell dynamics.
Stage-top incubators often come with dedicated live cell imaging and perfusion chambers for mounting coverslips, but it is important to consider whether a complicated imaging chamber is necessary for a particular experiment. Imaging chambers can be arbitrarily complex requiring assembly of multiple O-rings, spacers and screws, which increases the risk of leakage, contamination or drying of the specimen. It is also important to note that imaging chambers with bottom parts thicker than the working distance of a high NA oil objective, which is typically in the range of 0.1–0.2 mm, have to be used very carefully to avoid damage to the objective front lens. Chambers with thick bottom assemblies can also significantly limit the observable coverslip area.
Alternative solutions are disposable cell culture dishes in which a coverslip is glued to the bottom of a plastic tissue culture dish. Several variations of this theme are commercially available such as 35 mm round dishes (MaTek, Cat. No. P35G-1.5-14-C; In Vivo Scientific, Cat. No. D35-10-1.5-N) or rectangular formats (LabTek, Nunc Cat. No.155361). Although supplied in sterile packaging, these commercial dishes are often not as clean as one would like them to be for live-cell imaging. Glass bottom dishes can be self-made by drilling a hole into a plastic tissue-culture dish and gluing matching coverslips onto the bottom by using UV-hardened glue. If the necessary equipment is available, this can be a good costeffective alternative to commercial products. In an open tissue culture dish at 37°C, a significant amount of water will evaporate within minutes, which increases the osmolarity of the tissue culture medium much more rapid than one might anticipate and may negatively affect cell dynamics in the absence of an apparent change in medium volume. Thus, even in relatively short experiments glass-bottom dishes should be sealed. This can be done by running a bead of silicon grease around the inner edge of the lid before closing the dish. If drugs need to be added during the experiment, a small glass plate on top of the dish will also help to reduce evaporation. Alternatively, a layer of mineral oil can be overlaid on top of the medium.
In experiments during which cells do not need to be accessed during imaging, we have been quite successful using simple, reusable anodized aluminum slides that can be easily custom-made (e.g. online at www.emachineshop.com). These slides have a hole and counterbore on each side (Fig. 2A). A clean 15 mm round coverslip (Warner, CS-15R15, Cat. No. 64-0713) and one with the cultured cells are attached to the ledge of the counterbore on either side with silicon grease (Dow Corning, High Vacuum Grease), which creates a seal that eliminates evaporation and restricts gas exchange. After assembly, it is important to clean the outside of the coverslip carrying the cells with ethanol to remove any trace of silicon grease that will contaminate the immersion oil and deteriorate image quality. Despite the small media volume (~200 µl), cell health in these chambers is usually excellent, and we have, for example, imaged F-actin dynamics during epithelial-to-mesenchymal transition for up to 48 hours using these slides (Haynes et al., 2011). After use we clean the slides by several washes and short sonication in soapy water, deionized water and ethanol. For special applications such as chemotaxis assays, different designs of disposable microfluidic chambers are available (e.g. from Ibidi). To allow high resolution imaging, these chambers use plastic coverslips with a refractive index that matches glass. It is important to note that organic solvents in different types of immersion oils may dissolve this plastic, and it is thus important to test compatibility.
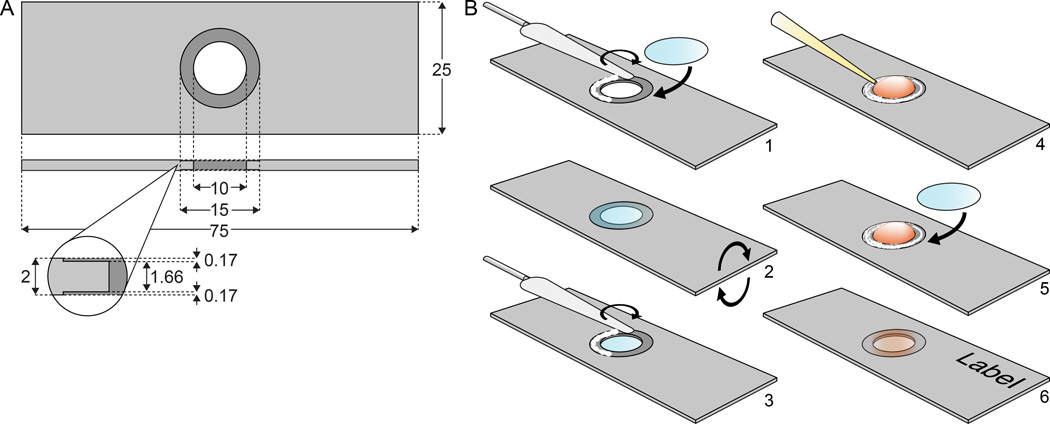
Figure 2. Diagram of reusable aluminum slides.
(A) Example dimensions for a slide made to fit 15 mm round coverslips. All dimensions are in mm, and can be adapted to fit different microscope stages. (B) Assembly of metal slides: A bead of silicon grease is distributed on one side of the slide with a small spatula and a clean cover slip is attached (1); after turning the slide (2), a thin layer of silicon grease is likewise spread on the other side (3), a drop of cell culture medium is added (4) and a cover glass with cells is mounted with the cells facing inside (5). Before imaging, the outside of the coverslip needs to be cleaned thoroughly to avoid contamination of the immersion oil.
3. Fluorescent Proteins
The strength of live-cell fluorescence imaging is in the specificity with which proteins and cellular structures can be labelled, imaged and analyzed. Genetically encoded fluorescent protein (FP) tags have revolutionized the analysis of intracellular dynamics and are now the most commonly used. In the last decades, dozens of different types of FPs have been cloned, engineered and optimized, and Chapter 6 provides an overview of recent FP developments. Here, we briefly summarize important considerations that should enter the design of live cell imaging experiments using FPs:
Matching FP spectra and fluorescence filter sets: Excitation and emission spectra of modern FP variants cover almost the entire visible spectrum. However, in reality, the usefulness of specific FPs will be determined largely by the available excitation and emission bands in a live cell imaging system. To deliver the correct wavelength of light and collect as much signal as possible, excitation and emission filter sets need to be optimized. While epifluorescence filter sets can easily be adapted for different wavelengths, spinning disk confocal microscopes are much less flexible. Excitation and emission bands in a spinning disk confocal are largely determined by the dichroic beamsplitter in the spinning disk head. Because these beamsplitters operate in reverse (the emission light is reflected) compared with beamsplitters in epifluorescence filter cubes (in which the excitation light is reflected) fewer choices are available. In addition, excitation wavelengths in spinning disk confocals and other laser-based systems are limited to a relatively small number of available laser lines with sufficient power. For example, while 488 nm is almost optimal for EGFP excitation, it is not for red-shifted brighter variants such as Clover (Lam et al., 2012). Similarly, to optimize the yield of emitted fluorescence, it is essential to match emission filters to the FP emission spectrum. In general, FP emission spectra are broad. Thus, in single wavelengths experiments, longpass emission filters should be used to collect as many emitted photons as possible. Ideally, bandpass emission filters should only be used in experiments with multiple FPs, and bandpass filters with the widest permissible transmission bands to avoid crosstalk between channels should be selected. Narrower emission bands may be required in specimens with high autofluorescence, and optimized filters sets should be determined empirically. If emitted light is detected inefficiently, higher excitation light intensity will be needed to achieve the same signal-to-noise ratio, which increases photobleaching and phototoxicity.
Other factors influencing FP choice: While fluorescence properties are obviously important for successful live cell imaging, in choosing a specific FP, one should also consider its performance in a specific biological system. This includes the FPs tendency to dimerize or aggregate, which will depend on multiple parameters such as temperature, and an FP that works well in yeast may aggregate in mammalian cells. Especially for newer FP variants where reports from the literature may be scarce, it is advisable to test cells for toxic effects and unspecific aggregation when expressing fusion constructs. Finally, maturation time of the fluorophore varies widely between FPs, and has to be considered together with FP stability when FP reporters are used to assess protein turnover or rapid gene expression changes.
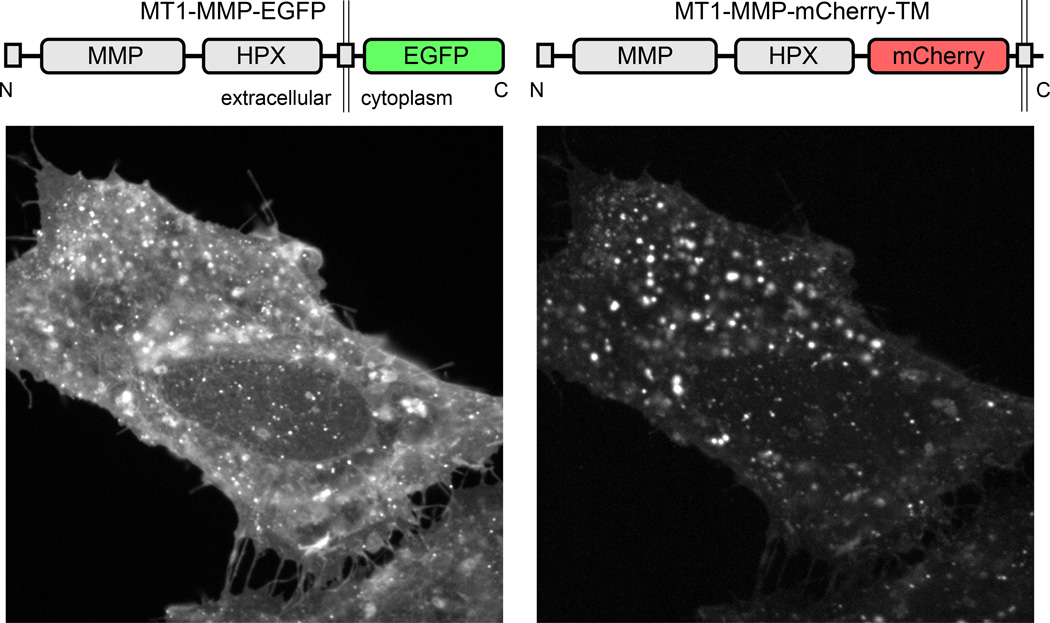
Placement of the FP tag: We will not cover details of cloning of FP-constructs, but it is important to think where the relatively large FP moiety will interfere least with endogenous protein function. FP tags are most commonly placed at the N- or C-termini of the protein of interest, but can also be inserted in intrinsically disordered protein regions between folded domains. Immunofluorescence of the endogenous protein as well as functional assays should be used to test to what extent addition of the FP tag interferes with protein localization and function. It is also important to note that different FPs can have very different effects on different proteins of interest. Fig. 3 shows an example of two FP-tagged constructs of the membrane-bound matrix metalloprotease MT1-MMP that have both been used in a number of publications (Wang & McNiven, 2012; Sakurai-Yageta et al., 2008), and these two constructs label almost entirely different intracellular compartments. Intracellular MT1-MMP transport is complex, and it is possible that differential inhibition of targeting signals enriches FP-tagged MT1-MMP in different transport compartments. This example underscores the importance of testing different constructs and understanding the underlying biology. If a protein tolerates FP fusion, we have been successful in doubling up EGFP tags to increase brightness and avoid overexpression artifacts. For example, the microtubule plus end tracking protein EB1 tends to bind along microtubules at high expression levels, which can be minimized by using two tandem EGFP tags resulting in increased signal-to-noise ratio at lower expression levels and lower excitation light intensity (Gierke et al., 2012).
FP photobleaching: Unfortunately, photobleaching is unavoidable, and is often the limiting factor that determines how many images can be acquired. FP photobleaching results from a complex combination of mechanisms specific to different FPs, and currently there is no simple solution to eliminate photobleaching. Although recent reports propose that FP photobleaching in biological media is reduced in the absence of vitamins that can act as electron acceptors (Bogdanov, Bogdanova, Chudakov, Gorodnicheva, Lukyanov, & Lukyanov, 2009), in our hands the use of vitamin-free media did not significantly improve EGFP photostability. The origin of this variability is unclear, but it is possible that intracellular concentrations of relevant compounds are different in different cell types. In addition, the characteristics of the imaging system, i.e. the specifics of excitation and emission bands, as well as intracellular environment will influence the apparent brightness, and thus apparent photobleaching of specific FPs.
Figure 3. Influence of tagging strategy on the apparent localization of FP-tagged MT1-MMP.
HeLa cells were co-transfected with the constructs indicated. The two constructs localize to mostly distinct intracellular compartments.
3.1 Protocol for analyzing FP photobleaching
Spinning disk confocal microscopy is the most prominent method of high resolution live cell imaging of intracellular dynamics using FPs. Yet, the performance of different FPs is rarely examined by spinning disk confocal microscopy. Although FP photobleaching dynamics are complex and discussed in more detail in Chapter 6, we included a specific protocol and results of comparing a variety of different FPs on our spinning disk microscope setup that can be adapted to other microscopes:
Transfect cells with FP expression constructs. We used a number of different green and red FPs in the same vector backbone based on Clontech’s original EGFP-N1 plasmid transfected into immortalized mouse embryonic fibroblasts on 35 mm glass bottom dishes using Fugene 6 (Promega) following the manufacturer’s instructions.
Image cells the following day. Locate expressing cells by epifluorescence illumination taking care to minimize light exposure, and let cells recover in the dark for a few minutes before imaging. We imaged cells in 2 ml DMEM containing 20 mM HEPES in dishes sealed with vacuum grease at 37°C using a Nikon 40× CFI S Plan Fluor N.A. 0.6 air objective to mimic realistic light exposure during a live cell imaging experiment as close as possible. Spinning disk confocal images were taken every 500 ms using a 300 ms exposure with continuous illumination (i.e. shutter remained open between exposures) for several minutes. For the GFP channel, the excitation wavelength was 488 nm and images were collected through a 505 nm longpass filter. For RFP variants, excitation was 561 nm and a 575 nm longpass emission filter.
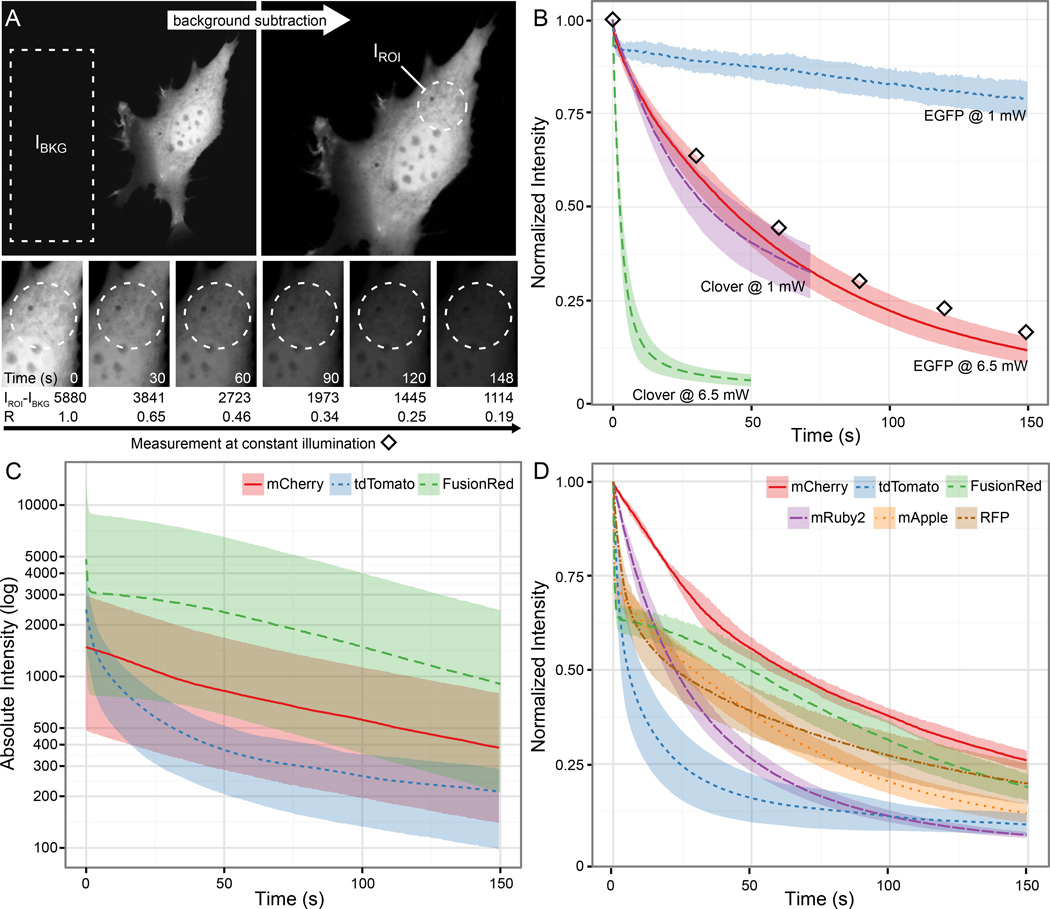
Calculate normalized photobleaching kinetics R by dividing the mean background-corrected fluorescence intensity I in an intracellular region of interest (ROI) in each frame by the intensity in the first frame (Fig. 4a): R = (IROI(n) − IBKG)/(IROI(1) − IBKG). It is important to calculate such normalized photobleaching curves because FP expression levels in different cells will vary, thus precluding direct comparison of absolute fluorescence of different constructs (Fig 4c). To perform this analysis in open source image analysis software such as ImageJ or Fiji (http://fiji.sc/Fiji) (Schindelin et al., 2012), one can use the ‘Multi Measure’ function that is accessed via the ‘ROI Manager’ tool, which will measure the average intensity and other parameters in a selected area for every image, and the output can be saved as text file and analyzed further as described above.
Figure 4. FP photobleaching characterization on a spinning disk confocal microscope.
(A) Example time-lapse sequence of an EGFP-expressing cell illustrating calculation of normalized photobleaching curves. See text for details. (B) Normalized photobleaching curves for Clover and EGFP with continuous 488 nm excitation at two different light power settings. Diamonds correspond to panels and values in (A). (C) Absolute background-corrected fluorescence intensities measured for three different red FPs at the same ~8 mW 561 nm excitation, illustrating the large variation of absolute intensities in different cells, which is likely due to different expression levels. (D) Normalized photobleaching curves showing widely different photobleaching kinetics for five different red FPs. The normalized curves for mCherry, tdTomato and FusionRed photobleaching correspond to the absolute intensities shown in (C). Solid and dotted lines in (B–D) represent mean values of five cells. Shaded areas in (B) and (D) are 95 % confidence intervals; shaded areas in (C) represent the range of measurements.
This experiment had the somewhat surprising outcome that both old-fashioned EGFP and mCherry performed better compared with all of the newer FP variants tested. Although initially brighter, Clover fluorescence decreased significantly more rapid compared with EGFP at both excitation power levels tested (Fig. 4b). Similarly, mCherry was not the brightest red FP (Fig. 4c), but displayed the slowest photobleaching compared with five other red and orange FPs tested (Fig. 4d). This is not easily explained and does not completely reflect what one would expect from reported fluorescence spectra, extinction coefficients, and quantum efficiencies of purified FPs in vitro. tdTomato (Peak excitation: λEx = 554 nm/Peak emission: λEm = 581 nm) is not a good match for a typical spinning disk illumination setup (Shaner, Campbell, Steinbach, Giepmans, Palmer, & Tsien, 2004). A significant part of the emission spectrum is cut off by the 575 nm longpass filter. Accordingly, tdTomato fluorescence is not imaged efficiently and as expected displays a very high apparent photobleaching rate. In contrast, mRuby (λEx = 558 nm/λEm = 605 nm) (Kredel et al., 2009) is theoretically a much better match to our illumination setup compared with mCherry (λEx = 587 nm/λEm = 610 nm) (Shaner et al., 2004), but still bleaches much more rapidly. This comparison further highlights the importance of testing FP performance in the experimental system and imaging setup to be used. It also reaffirms our more qualitative impression that EGFP and mCherry is possibly still the best FP protein pair for spinning disk confocal imaging with commonly available filter sets although we have certainly sampled only a small set of the available range of green and red FPs.
4. Other Fluorescent Probes
Although genetically encoded FP tags are the most popular and straightforward choice, other methods to label intracellular structures or specific proteins with chemical fluorophors exist. Many modern fluorescent dyes, such as the series of Alexa dyes, have evolved through derivatization of earlier fluorescent molecules such as fluorescein or rhodamine. These modern dyes have advantages such as better quantum efficiency and brightness compared with FPs, which can be essential for certain techniques. For example, fluorescent speckle microscopy relies on a high signal-to-noise ratio of clusters of few fluorescent molecules in F-actin networks or microtubules, and works best with fluorophor-conjugated actin or tubulin (Mendoza, Besson, & Danuser, 2012). A disadvantage of direct chemical fluorophor conjugation is the need for significantly more complicated experimental protocols including purification, fluorescent labeling and microinjection of the labeled protein that in many cases may not be feasible. Hybrid systems that combine genetic encoding and chemistry such as for example the SNAP-, CLIP- and Halo-tags (Stagge, Mitronova, Belov, Wurm, & Jakobs, 2013) are growing in popularity and utilize non-fluorescent genetically encoded enzyme tags that can specifically react with cell-permeable fluorogenic molecules. However, potential background fluorescence and unspecific sticking of hydrophobic fluorogenic molecules to cell membranes may limit the usefulness of these systems for live cell imaging, although almost no direct comparison of SNAP-, CLIP- or Halo-tags with FP-tagged proteins is available. In contrast to FPs, oxidative photobleaching of many fluorescent dyes can be reduced by oxygen depletion of the imaging medium, for example by the addition of Oxyrase (Oxyrase Inc.) a proprietary enzyme system to the imaging medium that oxidizes lactate and other organic acids (Wittmann, Bokoch, & Waterman-Storer, 2003). This only works in well-sealed imaging chambers that effectively prevent gas exchange with atmospheric oxygen. However, photobleaching of chemical fluorescent dyes also tends to be more cytotoxic possibly because the cytoplasm is less protected from reactive free radical breakdown products compared with FPs in which the fluorophor is isolated from the cytoplasm by the FP beta barrel structure.
Finally, a variety of cell-permeable fluorescent molecules are available that more or less specifically label intracellular organelles. These include intercalating DNA dyes such DAPI, molecules that become trapped in specific organelles after chemical modification such as MitoTracker (Invitrogen), which is oxidized and retained in mitochondria, or fluorescent molecules that preferentially bind to specific membrane compositions. Although many of these probes are marketed as live cell imaging reagents, it is important to note that imaging of these molecules often rapidly produces highly cytotoxic breakdown products. This light-induced cytotoxicity is easily observed in MitoTracker-labeled cells in which mitochondria swell rapidly during illumination. Of note, many small molecule inhibitors used in cell biology research are also highly fluorescent and in addition to increasing background fluorescence can generate extremely cytotoxic breakdown products. Thus, special care should be taken to ensure cell health when using any of these vital dye fluorescent probes or potentially fluorescent small molecule compounds.
5. Conclusion
Photobleaching is a fact of life. Even the best fluorescent molecule converts only a fraction of absorbed energy into fluorescence. Some of this energy will inevitably be channeled into chemical reactions that result in fluorophor breakdown. Thus, in addition to maintaining a physiological environment and confirming specific labeling and function of the protein or organelle of interest, the single most important factor to successful live cell imaging and meaningful data is to limit excitation light as much as possible. This requires an in-depth understanding of the microscope used and optimization of the components that control exposure, wavelengths selection and collection of emitted photons. Similarly important is a good practice in setting up experiments. Wait for dark adaptation of the visual system before attempting to find a dim sample. Modern cameras are much more sensitive than the human eye, and it is often better to take quick snapshots to locate areas of interest rather than staring through the eyepiece for an extended period of time. However, never use live camera modes to find your sample. Every photon is sacred.
Acknowledgements
This work was supported by National Institutes of Health grants R01 GM079139, R01 GM094819 and Shared Equipment Grant S10 RR26758.
Footnotes
Based on the American Society for Testing and Materials (ASTM) Terrestrial Reference Spectra
References
- Attardo A, Calegari F, Haubensak W, Wilsch-Brauninger M, Huttner WB. Live imaging at the onset of cortical neurogenesis reveals differential appearance of the neuronal phenotype in apical versus basal progenitor progeny. PLoS. One. 2008;3:e2388. doi: 10.1371/journal.pone.0002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov AM, Bogdanova EA, Chudakov DM, Gorodnicheva TV, Lukyanov S, Lukyanov KA. Cell culture medium affects GFP photostability: a solution. Nat. Methods. 2009;6:859–860. doi: 10.1038/nmeth1209-859. [DOI] [PubMed] [Google Scholar]
- Gao L, Shao L, Higgins CD, Poulton JS, Peifer M, Davidson MW, et al. Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell. 2012;151:1370–1385. doi: 10.1016/j.cell.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierke S, Wittmann T. EB1-recruited microtubule +TIP complexes coordinate protrusion dynamics during 3D epithelial remodeling. Curr. Biol. 2012;22:753–762. doi: 10.1016/j.cub.2012.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes J, Srivastava J, Madson N, Wittmann T, Barber DL. Dynamic actin remodeling during epithelial-mesenchymal transition depends on increased moesin expression. Mol. Biol. Cell. 2011;22:4750–4764. doi: 10.1091/mbc.E11-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K, Rajfur Z, Vitriol E, Hahn K. Chromophore-assisted laser inactivation in cell biology. Trends Cell Biol. 2008;18:443–450. doi: 10.1016/j.tcb.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredel S, Oswald F, Nienhaus K, Deuschle K, Rocker C, Wolff M, et al. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS. One. 2009;4:e4391. doi: 10.1371/journal.pone.0004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalkens B, Testa I, Willig KI, Hell SW. MRT letter: Nanoscopy of protein colocalization in living cells by STED and GSDIM. Microsc. Res. Tech. 2012;75:1–6. doi: 10.1002/jemt.21026. [DOI] [PubMed] [Google Scholar]
- Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza MC, Besson S, Danuser G. Quantitative fluorescent speckle microscopy (QFSM) to measure actin dynamics. Curr. Protoc. Cytom. 2012;Chapter 2(Unit2) doi: 10.1002/0471142956.cy0218s62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai-Yageta M, Recchi C, Le DG, Sibarita JB, Daviet L, Camonis J, et al. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J. Cell Biol. 2008;181:985–998. doi: 10.1083/jcb.200709076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Stagge F, Mitronova GY, Belov VN, Wurm CA, Jakobs S. SNAP-, CLIP- and Halo-tag labelling of budding yeast cells. PLoS. One. 2013;8:e78745. doi: 10.1371/journal.pone.0078745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S, Pemble H, Murrow L, Wittmann T. Imaging intracellular protein dynamics by spinning disk confocal microscopy. Methods Enzymol. 2012;504:293–313. doi: 10.1016/B978-0-12-391857-4.00015-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Babbey CM, Dunn KW. Performance comparison between the high-speed Yokogawa spinning disc confocal system and single-point scanning confocal systems. J. Microsc. 2005;218:148–159. doi: 10.1111/j.1365-2818.2005.01473.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, McNiven MA. Invasive matrix degradation at focal adhesions occurs via protease recruitment by a FAK-p130Cas complex. J. Cell Biol. 2012;196:375–385. doi: 10.1083/jcb.201105153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J. Cell Biol. 2003;161:845–851. doi: 10.1083/jcb.200303082. [DOI] [PMC free article] [PubMed] [Google Scholar]