Abstract
In mammals, TUBBY-like proteins play an important role in maintenance and function of neuronal cells during postdifferentiation and development. We have identified a TUBBY-like protein gene family with 11 members in Arabidopsis, named AtTLP1-11. Although seven of the AtTLP genes are located on chromosome I, no local tandem repeats or gene clusters are identified. Except for AtTLP4, reverse transcription-PCR analysis indicates that all these genes are expressed in various organs in 6-week-old Arabidopsis. AtTLP1, 2, 3, 6, 7, 9, 10, and 11 are expressed ubiquitously in all the organs tested, but the expression of AtTLP5 and 8 shows dramatic organ specificity. These 11 family members share 30% to 80% amino acid similarities across their conserved C-terminal tubby domains. Unlike the highly diverse N-terminal region of animal TUBBY-like proteins, all AtTLP members except AtTLP8 contain a conserved F-box domain (51–57 residues). The interaction between AtTLP9 and ASK1 (Arabidopsis Skp1-like 1) is confirmed via yeast (Saccharomyces cerevisiae) two-hybrid assays. Abscisic acid (ABA)-insensitive phenotypes are observed for two independent AtTLP9 mutant lines, whereas transgenic plants overexpressing AtTLP9 are hypersensitive to ABA. These results suggest that AtTLP9 may participate in the ABA signaling pathway.
The TUBBY gene was first identified from obese mice via positional cloning (Kleyn et al., 1996; Noben-Trauth et al., 1996). Mutation in the tubby gene causes maturity-onset obesity, insulin resistance, retinal degeneration, and neurosensory hearing loss (Coleman and Eicher, 1990; Heckenlively et al., 1995; Ohlemiller et al., 1995). In addition to TUBBY, four TUBBY related proteins, including TUBBY-like proteins (TULPs) 1 through 3 and TUBBY superfamily protein (TUSP), have since been identified in mammals (North et al., 1997; Nishina et al., 1998; Li et al., 2001). These proteins all feature an approximately 270-amino acid tubby domain at their C-terminal region, but their N-terminal sequences are more divergent both in length and in amino acid sequences. Mutations in this highly conserved C-terminal portion of mouse TUBBY and TULP1 lead to different deteriorated phenotypes (Kleyn et al., 1996; Noben-Trauth et al., 1996; North et al., 1997). Currently, members of this family have been identified in various multicellular organisms but not in single-celled organisms (North et al., 1997). The highly conserved tubby domain in different species suggests that these proteins must have fundamental biological functions in multicellular organisms.
Though target genes have not been identified, reports suggest that the TUBBY protein is a bipartite transcription regulator (Boggon et al., 1999). Santagata et al. (2001) showed that mouse TUBBY is transported from the plasma membrane to the nucleus after phospholipase C-β activation of a subclass of G proteins called Gαq. Unlike the highly diverse N-terminal sequence of animal TULPs, a conserved F-box-containing domain is present in plant TUBBY-like protein (TLP) members (Gagne et al., 2002; Risseeuw et al., 2003). The F-box domain was first described as a sequence motif found in cyclin F that interacts with the protein S-phase kinase-associated protein 1 (SKP1) (Bai et al., 1996). Experimental data from work in yeast (Saccharomyces cerevisiae) indicate that SKP1 interacts with the CDC53 (Cullin) proteins and F-box proteins to form designated SKP1/Cullin/F-Box (SCF) complexes (E3 ubiquitin ligase complex; Krek, 1998; Patton et al., 1998).
F-box proteins can interact with SKP1-related proteins via an N-terminal F-box domain and with the substrates via a protein-protein interaction domain in C termini (Xiao and Jang, 2000). The review of the Arabidopsis F-box proteins suggests that most of the F-box domains are followed by specific amino acid sequences (such as the WD40 repeat, kelch repeats, and the Leu-rich repeat), which serve as protein-protein interacting domains for recruiting specific proteins and targeting them for ubiquitin mediated proteolysis (Patton et al., 1998; Xiao and Jang, 2000). Yeast F-box protein Grrlp of SCFGrrl recruits phosphorylated Cln1 and Cln2 via Leu-rich repeats (Skowyra et al., 1999), and mammalian F-box protein β-TrCP of SCFβ-TrCP recruits phosphorylated IkBd and β-catenin through WD40 repeats (Winston et al., 1999).
F-box proteins regulate diverse cellular processes, including cell cycle transition, transcriptional regulation, and signal transduction. Examples of F-box proteins regulating plant growth and development include TIR1 in auxin response (Rugger et al., 1998), UFO in floral organ identity determination (Samach et al., 1999), COI1 in jasmonic acid-regulated defense response (Xie et al., 1998), and ZTL and FKF1 in the control of the circadian clock (Somers et al., 2000; Nelson et al., 2000).
The existence of multiple TLPs implies their vital function in plants, though in-depth studies of plant TLPs have yet to be done. To reveal the potential functions of TLPs in plants, extensive bioinformatics and molecular characterization of TLPs in Arabidopsis were performed in this report. Using the yeast two-hybrid assays, we have confirmed that AtTLP9 is an F-box protein that can interact with ASK1 (Arabidopsis Skp1-like 1). At least one of the AtTLPs functions in the abscisic acid (ABA)-regulated pathway, based on the observation that transgenic plants with suppressed expression and overexpression of AtTLP9 show aberrant sensitivity to ABA during seed germination and early seedling development.
RESULTS
Identification of the AtTLP Family
In the attempt to identify plant tubby domain-containing proteins, we used the tubby consensus sequence (Pfam PF01167) to search the Arabidopsis expressed sequence tag (EST) database and the completed Arabidopsis genome sequence. Our search revealed eleven TUBBY-like protein genes, termed AtTLP1 to AtTLP11, in the Arabidopsis genome (Table I). For each of the 11 AtTLP genes, the corresponding bacterial artificial chromosome (BAC) locus (The Arabidopsis Information Resource), Tentative Consensus (TC) group, Arabidopsis Genome Initiative (AGI) gene code, cDNA GenBank accession number, and predicted protein length (no. of amino acid) are assembled in Table I.
Table I.
Characteristics of AtTLP family members
| Gene Family Name | BAC Locus | TC Group | AGI Gene Code | cDNA GenBank Accession No. | Predicted Protein Lengtha |
|---|---|---|---|---|---|
| AtTLP1 | F22K20.1 | TC95487 | At1g76900 | AF487267 | 455 |
| AtTLP2 | T30D6.21 | TC86308 | At2g18280 | AY045773 | 394 |
| AtTLP3 | F17A22.29 | TC86633 | At2g47900 | AY045774 | 406 |
| AtTLP4 | F8K4.13 | — | At1g61940 | — | 265 |
| AtTLP5 | T10P12.9 | TC102456 | At1g43640 | AY046921 | 429 |
| AtTLP6 | F8G22.1 | TC90700 | At1g47270 | AF487268 | 388 |
| AtTLP7 | F12M16.22 | TC88599 | At1g53320 | AY092403 | 379 |
| AtTLP8 | T24D18.17 | — | At1g16070 | AF487269 | 397 |
| AtTLP9 | F24P17.15 | TC102624 | At3g06380 | AF487270 | 380 |
| AtTLP10 | F4F7.13. | TC101291 | At1g25280 | AF487271 | 445 |
| AtTLP11 | T1A4.60 | − | At5g18680 | AY046922 | 380 |
All the cDNAs of AtTLP genes, except AtTLP4, were cloned and sequenced to experimentally determine mRNA sequence; cDNAs were isolated and sequenced, and the encoded protein sequence was deduced from cDNA sequence.
Based on the sequence of predicted open reading frame and the corresponding EST in the database, gene-specific 5′ and 3′ primers were designed. When reverse transcription (RT)-PCR was performed with total RNA from 2-week-old Arabidopsis seedlings, 10 AtTLP cDNAs (AtTLP1 to 3 and AtTLP5 to 11) were successfully amplified. Except for AtTLP2 and AtTLP11, the amino acid sequences deduced from the AtTLP1, AtTLP3, AtTLP5, AtTLP6, AtTLP7, AtTLP8, AtTLP9, and AtTLP10 cDNA sequences are identical to the predicted open reading frames in the database. Our AtTLP2 cDNA sequence showed that intron 3 of AtTLP2 was located between 708 and 781 bp, whereas the predicted splicing sites for this intron were 663 and 766 bp. Our AtTLP11 cDNA sequence showed that intron 2 and intron 4 were located at 669 to 803 bp and 1,334 to 1,575 bp, respectively, whereas the computer predicted intron 2 was at 621 to 803 bp, and there is no predicted intron 4. cDNA sequences obtained from this study were submitted to GenBank (see Table I for accession nos.).
Sequence Analysis of the AtTLP Proteins
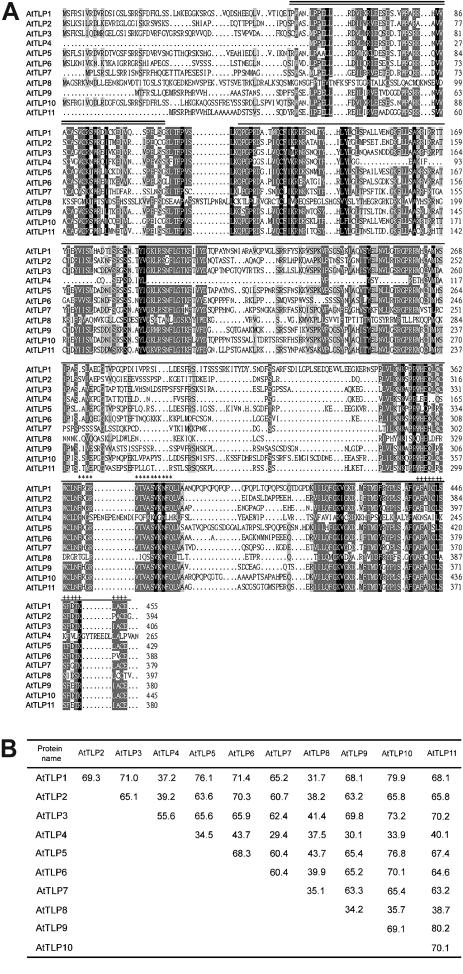
With the exception of AtTLP4 and 8, AtTLPs have a well-conserved tubby domain at their C termini. Unlike the highly diverse N-terminal sequence of animal TLUPs, a conserved F-box (51–57 residues)-containing domain (Pfam PF00646) is present in all AtTLP members except AtTLP8 (Fig. 1A). Among the AtTLP proteins, pairwise comparisons reveal that the tubby domains share 30% to 80% amino acid similarities (Fig. 1B). When analyzing the tubby domain in detail, we identified two PROSITE signature patterns as TUB1 (Prosite accession no. PS01200) and TUB2 motif (Prosite accession no. PS01201). The TUB1 and TUB2 motifs are located at the C terminus of AtTLPs and contain 14 and 16 amino acid residues, respectively. These two TUB motifs are highly conserved among the TLPs from various organisms. Though AtTLP4 and 8 do not have obvious signatures for TUB1 and TUB2 motifs, their C-terminal tubby domains are recognizable by MOTIF SCANNING (N score > 15; Pagni et al., 2001).
Figure 1.
Sequence comparisons of the deduced proteins from AtTLPs. A, Sequence alignment of AtTLPs. Identical and similar amino acid residues are shaded in black and gray, respectively. The locations of the F-box domain and tubby domain are indicated with double and single solid lines above the sequences, respectively. Asterisks and crosses shown above the sequences represent TUB1 and TUB2 motifs. Two PROSITE signature patterns are F-[KRHQ]-GR-V-[ST]-x-A-S-V-K-N-F-Q for TUB1 motif and A-F-[AG]-I-[GSAC]-[LIVM]-[ST]-S-F-x-[GST]-K-x-A-C-E for TUB2 motif. The amino acid sequences of AtTLP1-3 and AtTLP5-11 are deduced from the cDNAs reported here. The amino acid sequence of AtTLP4 is deduced from annotated At1g61940. The alignment is generated by the ClustalW program. The search for all known motifs in the deduced amino acid sequences was achieved by the MOTIF SCANNING (Pagni et al., 2001). B, Amino acid similarities among AtTLP C-terminal tubby domain.
Location and Gene Structure Comparison of the AtTLP Gene Family
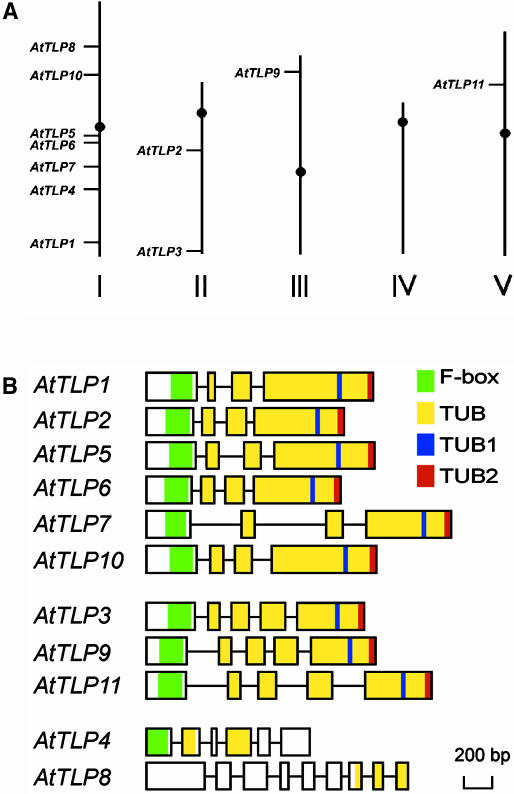
From the data in Figure 2A, the genes are obviously not evenly distributed in chromosomes I, II, III, or V. Seven genes (AtTLP1, 4, 5, 6, 7, 8, and 10) are located on chromosome I, whereas two genes (AtTLP2 and 3), one gene (AtTLP9), and one gene (AtTLP11) are located on chromosomes II, III, and V, respectively. Although most of the AtTLP genes are located on chromosome I, no local tandem repeats or gene clusters are identified.
Figure 2.
Location and gene structure comparison of the AtTLP gene family. A, Chromosomal locations of the AtTLP genes. The relative sizes of five Arabidopsis chromosomes are derived from the National Center for Biotechnology Information database. B, The intron-exon structure of the AtTLP gene family. Except for AtTLP4, the positions of the exons/introns of each individual AtTLP gene were confirmed by comparison of the cDNAs with their corresponding genomic DNA sequences. Exons and introns are represented by boxes and lines, respectively. The green and yellow shaded boxes indicate approximate positions encoding the F-box and tubby domain of the AtTLPs. Two PROSITE signature patterns (TUB1 and TUB2 motif) in tubby domain are shown in blue and red shaded boxes, respectively. The scale shown below is in base pairs.
By comparing the sequences of the RT-PCR products and the Arabidopsis genome, except for AtTLP4, the corrected exon-intron organization of the AtTLP genes is presented in Figure 2B. Gene structure indicates that the N-terminal leading sequences, the F-box, and the nine-residue spacer between the F-box and tubby domain are all located in exon 1, indicating that they might have arisen from the same ancestral gene. The C-terminal tubby domain sequences are distributed in exons interrupted by either two or three introns (Fig. 2B). On the basis of their exon and intron composition, the AtTLP genes can be classified into three groups. The genes of the first group (AtTLP1, 2, 5, 6, 7, and 10) contain three introns. Members of the second group, AtTLP3, 9, and 11, contain an additional intron in the C-terminal region of the tubby domain. The third and most distinct group is composed of AtTLP4 and 8, containing five and eight introns, respectively.
Expression of AtTLP Genes
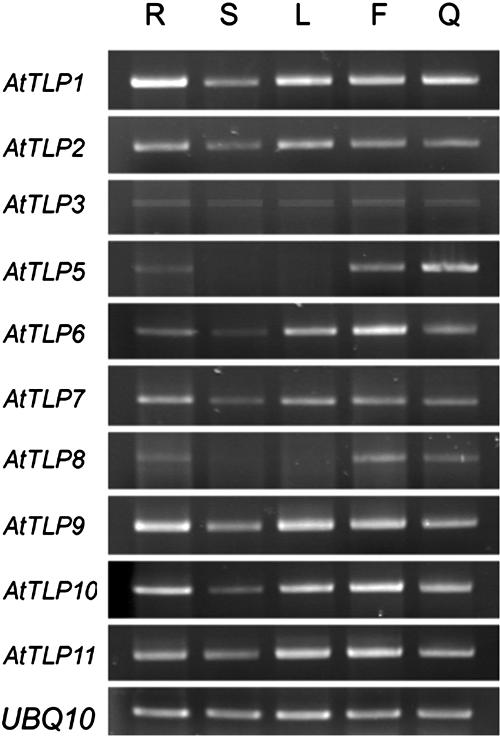
To understand how the expression of AtTLP gene family was regulated in different organs, total RNA from root, main and lateral stems, rosette leaves, flower clusters, and green siliques of soil-grown Arabidopsis were isolated for organ-specificity expression analyses of AtTLPs. Because the basal expression levels of AtTLPs were too low to be detected by RNA gel blotting with total RNA, we used coupled RT and PCR for the analysis. Results demonstrate that AtTLP1, 2, 3, 6, 7, 9, 10, and 11 are expressed in all the organs tested, with slight variations in mRNA accumulation (Fig. 3). By contrast, AtTLP5 and AtTLP8 primarily express in root, flower, and silique. The organ-specific expression of AtTLP5 and AtTLP8 may reflect their specific roles in particular organs.
Figure 3.
RT-PCR analyses of AtTLP gene expression patterns. Root (R), main and lateral stems (S), rosette leaves (L), flower clusters (F), and green siliques (Q) were harvested from 42-d-old plants grown in a growth chamber as described in “Materials and Methods.” Expression of each AtTLP gene was analyzed by RT-PCR using specific primers as described in “Materials and Methods.” The expression level of a UBQ10 gene was used as an internal control.
Although the expression of AtTLP1, 2, 3, 6, 7, 9, 10, and 11 is omnipresent in tissues tested in this study, the possibility that these genes are expressed with cell-type specificity could not be excluded. It is also possible that differential expression of these AtTLP genes could only be observed when internal developmental programming was altered or specific environmental stimuli were applied to the plants. To test this hypothesis, we took advantage of the public Arabidopsis gene expression database at the Stanford Microarray Database, where hundreds of expression data sets based on cDNA microarray (http://genome-www5.stanford.edu/MicroArray/SMD/) are available. A search of the Arabidopsis EST database and the clone list used to generate Arabidopsis cDNA microarray revealed representative DNA elements corresponding to AtTLP2, 7, 9, and 10. This allowed us to analyze the global gene expression pattern of these four genes by querying the microarray database. With the 2-fold expression difference cutoff, the expression profiles of these four genes are summarized in Table II. Results show factors like hormone fluctuation and environmental stimuli could modulate the expression of these four AtTLP genes.
Table II.
AtTLP gene expression analysis via searching microarray expression databasea
| Ch2/Ch1 Normalized (Mean)bc
|
||||||
|---|---|---|---|---|---|---|
| Experiment | Channel 1 Description | Channel 2 Description | AtTLP2 | AtTLP7 | AtTLP9 | AtTLP10 |
| Hormone Effect | ||||||
| Auxin response | msgd seedlings, untreated | msg seedlings; 10 μm indole-3-acetic acid for 30 min | 0.32 | 2.21 | ||
| Auxin induction | Mock-treated Col-0 roots | Naphthylacetic acid-treated Col-0 roots | 0.46 | 2.22 | ||
| Cytokinin response | Control | 15 min of cytokinin treatment | 2.19 | |||
| Abscisic acid insensitive 1 | Wild-type control | Abscisic acid insensitive 1 mutant | 0.49 | 2.6 | 0.35 | |
| edr1 mutant | Wild-type leaves | edr1 mutant leaves | 2.33 | |||
| Downstream genes of KN1 | Control | Overexpression of KN1-GR in Col-0 background | 0.43 | 2.78 | 4.73 | |
| Stress | ||||||
| Effects of elevated atmospheric CO2 | Col-0 leaves 360 ppm CO2 | Col-0 leaves 1,000 ppm CO2 | 0.33 | |||
| Genes involved in chilling tolerance | Cold-treated Col-0 wild-type tissue | Cold-treated cls8 mutant tissue | 0.22 | 0.15 | ||
| Genes involved in potassium nutrition | [K+] = 120 μm | [K+] = 2 mm | 0.34 | 0.2 | ||
| Cadmium | Control | 10 μm cadmium-treated plant | 2.72 | |||
These data are obtained from http://genome-www5.stanford.edu.
All data are corresponding with fluorescence intensities greater than 500 in both channels and ch2:ch1 normalized ratio ≧2.0 or ≦0.5.
When searching dbEST with BLASTN, we find Arabidopsis EST corresponding to four of our AtTLPs represented on microarray data generated by AFGC. AtTLP2 is corresponding to the EST clones 289B10T7 and 173K22T7. AtTLP7, AtTLP9, and AtTLP10 are corresponding to the EST clones 173G1T7, 201E19T7, and F3E6T7, respectively.
It is the mutant of massugu gene, which has meanwhile been officially renamed to NPH4 (nonphototropic hypocotyl 4) gene.
The four AtTLP genes examined showed differential responses to treatments of various hormones. AtTLP2 gene expression instantaneously increases more than 2-fold with cytokinin treatment but decreases to one-third when treated with indole-3-acetic acid. Cytokinin and auxin may play antagonistic roles in regulating AtTLP2 gene expression. Another cytokinin-related experiment was aimed at identifying downstream genes of KN1. KN1-like protein is a homeobox transcription factor, the overexpression of which up-regulates cytokinin production and leads to delayed senescence (Vollbrecht et al., 1991). The expression of AtTLP7 and 10 is up-regulated in KN1 overexpression transgenic plant, while AtTLP2 is down-regulated. The opposite response of AtTLP7 from AtTLP2 and AtTLP9 to ABA treatment is also worth noticing. In abscisic acid insensitive 1 mutant (Pei et al., 1997), the expression of AtTLP2 and AtTLP10 decreases 2- to 3-fold, but AtTLP7 expression increases more than 2-fold. Interestingly, AtTLP2 and AtTLP7 respond oppositely to auxin treatment, abscisic acid insensitive 1 mutant, and KN1 overexpression transgenic plant. These two AtTLP genes may function antagonistically in regulating phytohormone signaling pathways. The expression level of AtTLP2 rises in the edr1 (enhanced disease resistance 1) mutant leaves. EDR1 gene encodes a putative MAP kinase similar to CTR1, a negative regulator of ethylene response in Arabidopsis (Frye et al., 2001). The edr1 mutation of Arabidopsis also confers resistance to powdery mildew disease (Frye and Innes, 1998). The regulation of AtTLP2 gene expression is possibly associated with salicylic acid-inducible and ethylene defense mechanism.
Environmental stresses also impose influences on the expression of AtTLP genes. For example, similar to the cold treatment on cls8 mutant, elevated concentration of CO2 inhibits the expression of AtTLP2. K+ deficiency augments the expression of AtTLP7 and AtTLP10 3-fold and 5-fold, respectively. Heavy metal cadmium treatment also stimulates the expression of AtTLP9 expression.
In conclusion, the expression data of these four AtTLP genes imply their potential involvement in phytohormone and environmental stress signaling. A comprehensive survey of AtTLP gene expression regulation upon hormone and stress treatment is expected to shed more light on this line of research but is beyond the scope of current studies.
AtTLP9 Can Interact with ASK1 Protein
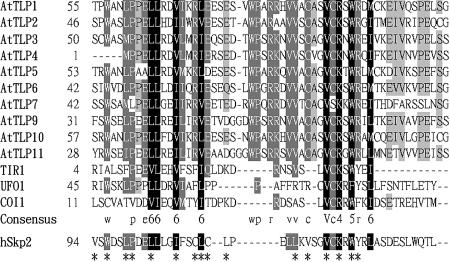
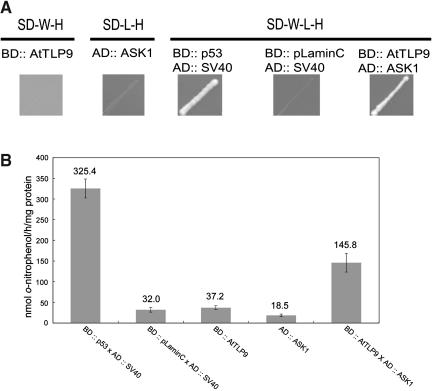
Homology searches in the public databases reveal that TULPs are also present in multiple plant species (Lemna paucicostata, Oryza sativa, Cicer arietinum, Zea mays, and Arabidopsis). Unlike the highly diverse N-terminal sequence of animal TULPs, a conserved F-box-containing domain is present in plant TLP members. Sequence alignment of the F-box cores from AtTLP, TIR, UFO, COI1, and the human F-box protein Skp2 revealed conserved islands separated by regions with weak homology (Fig. 4), but many of the conserved residues correspond with those known by x-ray crystallographic analysis to be important for Skp association (Schulman et al., 2000; Zheng et al., 2002). When members of AtTLP were aligned individually, the N-terminal F-box domain revealed highly conserved region (Fig. 1A). To test whether AtTLP could interact with the ASK1, one member of AtTLPs (AtTLP9) was chosen to test this hypothesis by yeast two-hybrid analysis. As shown in Figure 5, A and B, the acquiring of His biosynthesis and the presence of β-galactosidase activity suggested that AtTLP9 has physical interaction with ASK1.
Figure 4.
Sequence alignment of the core F-box sequences from AtTLPs, TIR1, UFO, COI1, and the human F-box protein Skp2 were aligned by ClustalW. The amino acid sequence of AtTLP4 is deduced from annotated At1g61940. Identical and similar amino acid residues are shaded with black and gray, respectively. Dots denote gaps. Amino acid substitution groups were numbered according to the Blosum 35 matrix: (1) ED; (2) NQ; (3) ST; (4) KR; (5) FYW; and (6) LIVM. Asterisks indicate the amino acids positions important for the Skp/F-box interactions between human Skp1 and Skp2 (Schulman et al., 2000; Zheng et al., 2002).
Figure 5.
The interaction between AtTLP9 and ASK1 via yeast two-hybrid assays. p53-SV40 (SV40 T-antigen) and LaminC-SV40 represent known interacting and noninteracting protein partners, respectively. A, Interaction of AtTLP9 and ASK1 with yeast two-hybrid His auxotrophic growth. Yeast cells transformed with the plasmid pairs were cultured on synthetic dropout (SD) medium lacking selected amino acids. SD-W-H, SD media without Trp and His; SD-L-H, SD media without Leu and His; SD-W-L-H, SD media without Trp, Leu, and His. B, β-galactosidase activities of the interaction pairs. Values are the means ± sd of assays from least three independent transformants indicated. β-galactosidase activity is expressed in nmol o-nitrophenol h−1 mg−1 yeast protein.
Effect of ABA on the Seed Germination of Attlp9 Mutants and Overexpression Lines
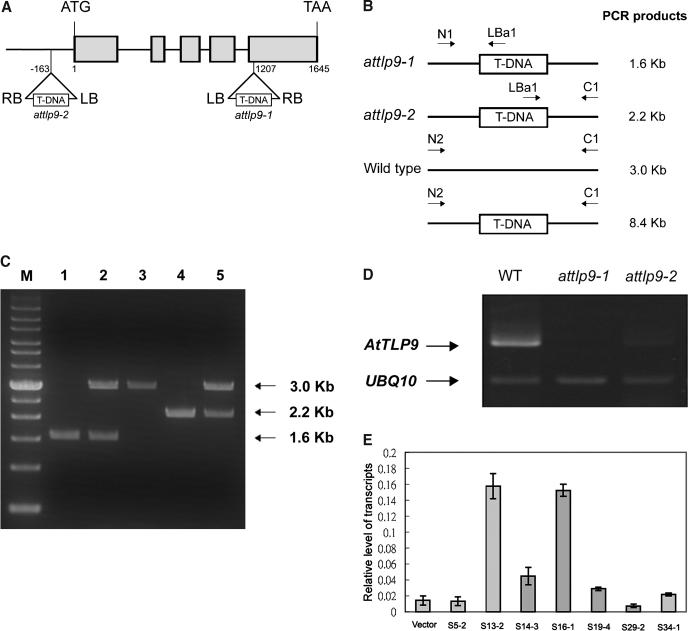
Based on the above observation that the expression of AtTLP9 is altered in abscisic acid insensitive 1 mutant and AtTLP9 could interact with ASK1, AtTLP9 was chosen in this study to investigate the putative in vivo functions of AtTLPs. Both loss-of-function and overexpression approaches were taken to address the biological role of AtTLP9. We obtained two AtTLP9 T-DNA insertion lines from the Arabidopsis Biological Resource Center (ABRC; SALK_016678 and 051138) and designated attlp9-1 and attlp9-2, respectively. The insertion site of attlp9-1 and attlp9-2 were confirmed and are illustrated in Figure 6A. In brief, the mutant attlp9-1 has a T-DNA insertion in the coding sequence at codon 705, whereas attlp9-2 has an insertion in the 5′ distal region of this gene (Fig. 6A). The T-DNA insertion site of attlp9-1 is identical to that originally described in SIGnAL; however, the T-DNA insertion site of attlp9-2 is in the promoter region instead of exon 1 predicted in SIGnAL (the latter is supported by a potential full-length cDNA corresponding to At3g06380 generated in RIKEN; accession no. BT004092). Southern-blot analysis probed with nptII gene suggested one and three T-DNA insertion events in the T4 attlp9-1and attlp9-2 T-DNA insertion mutants, respectively (data not shown). Genotypes of attlp9-1 and attlp9-2 mutant lines were characterized both by their kanamycin resistance and genomic PCR with gene-specific primers and T-DNA left-border primer (Fig. 6, B and C). RT-PCR analyses of T4 homozygous attlp9-1 and attlp9-2 plants showed that attlp9-1 is a null allele, whereas attlp9-2 is somewhat leaky (Fig. 6D).
Figure 6.
Molecular characterization of AtTLP9 T-DNA insertion lines. A, Schematic presentation of the structure of the AtTLP9 genomic structure. Start ATG and termination codons are indicated. The five exons are represented by gray boxes. The T-DNA location for each of the two attlp9 knockout lines, attlp9-1 and attlp9-2, are indicated. B, Schematic diagrams of the attlp9-1 and attlp9-2 genotyping: N1, forward primer of AtTULP9; LBa1, left-border primer of T-DNA; and C1, reverse primer of AtTLP9. The N1 and LBa1 primers produced the 1.6-kb PCR fragment from the attlp9-1 genomic DNA, and C1 and LBa1 primers produced the 2.2-kb PCR fragment from the attlp9-2 T-DNA genomic DNA; N2 and C1 primers produced a 3.0-kb PCR product from the wild-type DNA, while no PCR band was obtained from the attlp9-1 and attlp9-2 genomic DNA because the expected fragment was too large to amplify. C, Genotype determination for attlp9-1 and attlp9-2. Lane 1, homozygous attlp9-1; lane 2, heterozygous attlp9-1; lane 3, wild type; lane 4, homozygous attlp9-2; and lane 5, heterozygous attlp9-2. M, Mr marker. Size of major PCR products was indicated in kilobase pairs. D, Examination of AtTLP9 transcript in attlp9-1 and attlp9-2 plants. RT-PCR was performed with total RNA extracted from 14-d-old seedlings of attlp9-1, attlp9-2, and the wild type (Col-0) as described in “Materials and Methods.” UBQ10-specific primers were also included as an internal control. E, mRNA level for AtTLP9 gene in 14-d-old seedlings of AtTLP9 sense transgenic plants. Total RNA was extracted from 14-d-old seedlings of vector control and seven independent T3 sense homozygous transgenic plants. Poly(A+) mRNA was isolated, converted to cDNA, and subjected to real-time PCR analysis as described in “Materials and Methods.” Transcript levels are given as relative values to UBQ10 (the value of 1). Each data point represents the mean of triplicate experiments. The small bars represent se. Vector represents transgenic plants carrying the empty vector.
We also generated transgenic plants with additional copy of AtTLP9 cDNA driven by 35S promoter of cauliflower mosaic virus. From the transformation, we obtained 38 independent lines of transgenic plants (T0 generation). Among them, we analyzed AtTLP9 expression of seven independent homozygous lines from the T3 sense transgenic plants in detail, as they contained a single copy of the transgene (data not shown). Two independent transgenic lines (S13-2 and S16-1), which showed dramatic increases in the endogenous level of AtTLP9 transcript levels (Fig. 6E), were selected for more detailed analysis. As controls, a number of transgenic lines were generated by transforming with Agrobacterium tumefaciens with PBI121 vector alone.
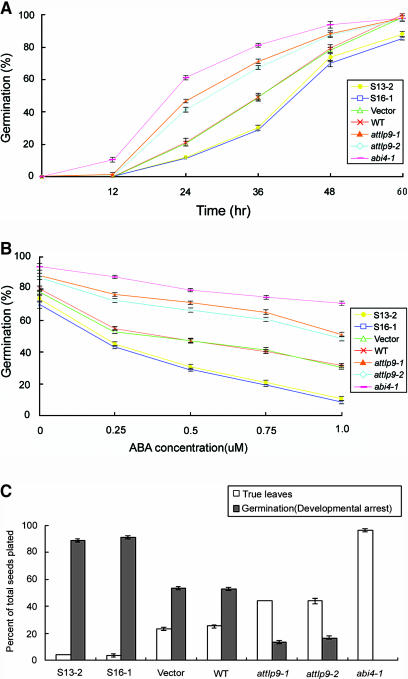
The general development and growth phenotypes of the attlp9-1 and attlp9-2 knockout plants appear to be similar to those of the wild-type plants. However, when seeds were plated on nutrient agar media, the germination time of mutant attlp9-1 and attlp9-2 seeds was advanced several hours compared with that of the wild-type plants, whereas the selected sense line seeds germinated slightly later than vector control seeds (Fig. 7A). Seed germination is the outcome of an integration of many signals, coordinated by the interactions of stage-specific developmental regulators and the competing effects of hormonal signals (Finkelstein and Gibson, 2002). The most critical hormone promoting embryo maturation and preventing germination is ABA. To determine whether the transgenic plants display altered ABA responses, AtTLP9 transgenic plants were germinated on media containing various concentrations of ABA. In the presence of 1 μm ABA, the germination of sense line seeds was further delayed, and the germination rate was reduced to less than 10% (Fig. 7B). By contrast, the germination rate of attlp9-1 and attlp9-2 mutant seeds nearly reached 50%, and about 30% of wild-type seeds were able to germinate in the presence of 1 μm ABA. This suggests that the disruption of the AtTLP9 gene affects the sensitivity of seeds to exogenous ABA. In addition to a reduced rate of seed germination, ABA also inhibited the growth and the greening process in cotyledons of the sense transgenic lines. In MS agar medium containing 1 μm ABA and 1% Suc, 90% of the 10-d-old seedlings showed developmental arrest, although the radicles of most sense lines seeds emerged (Fig. 7C). Under the same conditions, attlp9-1 and attlp9-2 plants continue to grow, and about 45% of the seedlings continue to develop true leaves, although at slower rates than abi4-1 mutant does. These results indicate that the alteration of AtTLP9 modulate plant's sensitivity to ABA during seed germination and early seedling development.
Figure 7.
AtTLP9 can modulate plant's sensitivity to ABA during seed germination and early seedling development. WT, Columbia wild type; Vector, transgenic plants carrying the empty vector; abi4-1, ABA perception mutant. Plant material was prepared as described in “Materials and Methods.” Each data point represents the mean of triplicate experiments (n = 50 each). The small bars represent se. A, AtTLP9 can modulate the seed germination rate. Seeds of WT, abi4-1, attlp9-1, attlp9-2, and sense transgenic plants S13-2 and S16-1 were plated on ABA-free medium, and germination was scored at time points indicate. B, ABA dose response of seed germination. Seeds were plated on media containing various concentrations of ABA and grown for 2 d. Seedlings with fully emerged radicles were counted. C, Sensitivity of seedling development to ABA. Shown are the percentages of 10-d-old seedlings with developmental arrest and true leaves over total number of seeds planted on Murashige and Skoog media supplemented with 1 μm ABA and 1% Suc.
AtTLP9 Expression Is Transiently Up-Regulated during Imbibition of Seeds
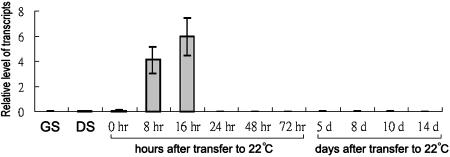
Real-time PCR experiments were used to quantify AtTLP9 transcript levels at seed maturation, seed germination, and early development stage. During seed maturation and seed imbibition at 4°C for 72 h, AtTLP9 transcripts remained at a relatively low level (Fig. 8). When the seeds were transferred to 22°C for further incubation, AtTLP9 transcript levels rose in seeds after 8 h, reached the highest level at 16 h, and fell rapidly after 24 h, when the radicle emerged (Fig. 8). The AtTLP9 transcripts were barely detectable afterward. We have shown that AtTLP9 is expressed early during germination and especially in the preemergent radicle.
Figure 8.
Real-time PCR analysis of AtTLP9 expression during seed maturation, germination, and early development. Wild-type (Col-0) seeds were kept in darkness at 4°C for 3 d, and then seeds for germination test were floated on liquid Murashige and Skoog medium and incubated at 22°C under a 16-h-light/8-h-dark photoperiod for 8, 16, 24, 48, and 72 h (0 indicates the time immediately following transfer), while seeds for early development test were plated on solid medium containing 0.7% phytoagar for 5, 8, 10, and 14 d. Poly(A+) mRNA was isolated, converted to cDNA, and subjected to real-time PCR analysis as described in “Materials and Methods.” Transcript levels are given as relative values to UBQ10 (the value of 1). Each data point represents the mean of triplicate experiments. The small bars represent se. GS, green siliques; DS, dry seeds.
DISCUSSION
In this study, we have defined and characterized a novel TUBBY-like protein gene family in Arabidopsis with a conserved tubby domain in their C-terminal region. Except for AtTLP4, the cDNAs of 10 AtTLP genes are successfully obtained via RT-PCR. Our attempt to amplify the corresponding cDNA for AtTLP4 via RT-PCR has been futile. The EST TC group for AtTLP4 is also absent (Table I). It is possible that AtTLP4 gene is not expressed or its transcript is too low to be detected with the experimental approaches undertaken to date. More work is needed to determine whether AtTLP4 is indeed expressed or is a pseudogene.
An obvious feature of AtTLPs is the tubby domain. This highly conserved domain in different species suggests that these proteins must have fundamental biological functions in multicellular organisms. Three positively charged amino acid residues, R332, R363, and K330, are thought to be crucial for phosphatidylinositol 4,5-bisphosphate [PI (4,5) P2] binding in the tubby domain of mouse TUBBY protein (Santagata et al., 2001). Except for AtTLP4 and 8, a sequence alignment of AtTLP tubby domain with mouse TUBBY protein reveals a putative PI (4,5) P2 binding domain (data not shown). The presence of a conserved PI (4,5) P2 binding domain in AtTLPs suggests that they may similarly bind to PI (4,5) P2. Whether AtTLP can interact with PI (4,5) P2, as has been observed in mammals, will be further investigated.
Experimental data show that TUBBY protein is a bipartite transcription regulator; the tubby domain of mouse TUBBY protein exhibits double-stranded DNA binding activity, and the N-terminal segment seems to modulate transcription (Boggon et al., 1999). In plants, the N-terminal region of TLPs is quite different from that in mammal TULPs. Unlike TUBBY and TULP1 in mouse, AtTLP9 fails to activate transcription from a GAL4 promoter when fused to the GAL4 DNA binding domain in a heterologous system (Fig. 5, A and B). However, further studies are needed to find out whether TLP plays a role as transcription factor in Arabidopsis.
Unlike the highly diverse N-terminal sequence of animal TULPs, all AtTLP members except AtTLP8 contain a conserved putative F-box domain (Fig. 4). Our data from yeast two-hybrid analysis showed that AtTLP9 can interact with ASK1 (Fig. 5, A and B). The F-box domain was first described as a sequence motif found in cyclin F that interacts with the protein SKP1 (Bai et al., 1996). Experimental data in yeast indicate that SKP1 interacts with the Cdc53 (Cullin) proteins and F-box proteins to form designated SCF complex (Krek, 1998; Patton et al., 1998). Analysis of the completed Arabidopsis genome sequence revealed 21 Skp1-like ASK genes that exhibit different expression patterns (Farrás et al., 2001; Zhao et al., 2003). Other studies demonstrated that ASK proteins exhibit differences in their association with both cullins and F-box proteins (Gagne et al., 2002; Risseeuw et al., 2003). ASK1, one of the 21 Skp proteins, is involved in both vegetative growth and reproductive development (Yang et al., 1999; Zhao et al., 1999, 2003). ASK1 was able to interact with one of several F-box proteins in yeast two-hybrid or in vitro-binding assays (del Pozo and Estelle , 1999; Dieterle et al., 2001; Gray et al., 2001; Woo et al., 2001; del Pozo et al., 2002; Gagne et al., 2002; Xu et al., 2002; Risseeuw et al., 2003). But the F-box domain alignment from the F-box proteins interacting with ASK1 showed poor conservation in an earlier (Risseeuw et al., 2003) and this study (Fig. 4). The best-studied role of F-box protein is a component of SCF complexes, which acts as a factor for substrate recognition (Pickart, 2001). In Arabidopsis, several SCF complexes have been characterized, such as SCFTIR1, SCFCOI1, and SCFAtSKP2 (Gray et al., 1999; del Pozo et al., 2002; Xu et al., 2002). In Caenorhabditis elegans and yeast, it has been shown that SKP1 may form a complex lacking Cullin (Yamanaka et al., 2002; Stemmann et al., 2002). Whether AtTLP can form SCF complex will be further investigated by the coimmunoprecipitation experiment of AtTLP9 with CUL1 or RBX1.
To investigate the in vivo functions of AtTLPs, a representative member of the group II (AtTLP9) was selected for physiological study. Unlike C. elegans, Drosophila melanogaster, and mammals, plants appear to have relatively large numbers of TLPs (Nishina et al., 1998). According to the results in Table I, TLPs in Arabidopsis belong to a multigene family with 11 members. Functional redundancy within this family may mask the effects of an individual loss-of-function allele. To address this question, we also used gain-of-function approaches to generate overexpressed transgenic plants. Our analyses of these transgenic plants showed that AtTLP9 overexpression and suppression affect sensitivity to ABA during seed germination and early seedling development. In Arabidopsis, ABA-insensitive mutations (e.g. abi1, abi2, abi3, abi4, and abi5) reduce seed dormancy and allow germination at ABA concentrations that are normally inhibitory to wild-type germination (Koornneef et al., 1984). Compared with abi4-1, our results indicate that the germination of attulp9-1 knockout plants is weakly insensitive to ABA (Fig. 7, B and C). Seed germination can be divided into three phases, imbibition, increased metabolic activity, and initiation of growth, which loosely parallel the triphasic water uptake of dry mature seeds (Bewley, 1997). The transient expression of AtTLP9 indicated that AtTLP9 may function at stage II of seed germination as a checkpoint before radicle protrusion (Fig. 8). The exact mechanism of AtTLP9 function during seed germination will be further studied.
MATERIALS AND METHODS
Database Screening and Isolation AtTLP Genes cDNA Clones
Searching of the Arabidopsis database (The Institute of Genome Research) was performed with multiple BLAST algorithms to locate all the sequences sharing significant similarities with the tubby domain (Kleyn et al., 1996; Noben-Trauth et al., 1996; P value less than 0.0085). RT-PCR with gene-specific primers was performed to obtain cDNA clones for each AtTLP gene. To achieve this, total RNA from Arabidopsis tissues was isolated with Trizol reagent (Invitrogen, Carlsbad, CA) according the manufacturer's suggestion. Poly(A+) mRNA was isolated with oligo(dT)-coated magnetic beads and the PolyATract system (Promega, Madison, WI). First-strand cDNA was synthesized from 0.5 μg of poly(A+) mRNA using SuperScript II RNase H RT (Invitrogen) according to the protocol of the supplier. For full-length cDNA amplification, the primer pairs used are as follows: AtTLP1-5′ (5′-ATGTCGTTCCGTAGCATAGTTCGT-3′), AtTLP1-3′ (5′-TTATTCGCAAGCAAGTTTTGTGTCG-3′), AtTLP2-5′ (5′-ATGTCTTTGAAAAGCATCCTTCGTGATC-3′), AtTLP2-3′ (5′-TTACCCTTCACATGCCGGTTTGGTGTCA-3′), AtTLP3-5′ (5′-ATGTCCTTCAAGAGTCTCATTCAG-3′), AtTLP3-3′ (5′-TCATTCACATGCTATCTTGGTGTC-3′), AtTLP5-5′ (5′-ATGTCGTTTCTGAGTATTGTTCG-3′), AtTLP5-3′ (5′-TTATTCACATGCCAATTTAGTAT-3′), AtTLP6-5′ (5′-ATGTCATTGAAGAACATAGTGAA-3′), AtTLP6-3′ (5′-TCATTCGCAGACTGGCTTCGTGT-3′), AtTLP7-5′ (5′-ATGCCTTTGTCACGGTCCCTC-3′), AtTLP7-3′ (5′-TCACTCGCAGGCAAGTTTAGTG-3′), AtTLP8-5′ (5′-ATGGCTGGTTCGAGAAAAGTGAA-3′), AtTLP8-3′ (5′-TCAAACAGTACAACAAAGCTTGG-3′), AtTLP9-5′ (5′-ATGACGTTCCGAAGTTTACTCCA-3′), AtTLP9-3′ (5′-TTATTCACAGGCAATTCTGGTTT-3′), AtTLP10-5′ (5′-ATGTCGTTTCGAGGCATTGTTCA-3′), AtTLP10-3′ (5′-CTATTCACAAGCAAGCTTGGTGT-3′), AtTLP11-5′ (5′-ATGTCGTTTCTGAGTATTGTTCG-3′), and AtTLP11-3′ (5′-TTATTCACATGCCAATTTAGTAT-3′).
Except for AtTLP4 gene, gene-specific primer pairs were used for amplifying cDNA of each AtTLP gene from first-strand cDNA via PCR. PCR conditions were as follows: 3 min at 94°C followed by 25 cycles each of 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 1 min 30 s of extension at 72°C. The PCR products were purified with the QIAquick PCR purification kit (Qiagen, Valencia, CA) and subcloned into T-Easy vector (Promega). Each of these clones was sequence verified.
Bioinformatics
Chromosome localizations of the different AtTLP genes were analyzed by MapViewer (http://www.arabidopsis.org/servlets/mapper; Huala et al., 2001). The search for all known motifs in the deduced amino acid sequences was achieved by MOTIF SCANNING (Pagni et al., 2001). Multiple sequence alignment was performed with ClustalW (Thompson et al., 1994).
Expression Study
Spatial expression patterns of AtTLP genes were studied using a RT-PCR-based method. Total RNA was isolated from 42-d-old roots, main and lateral stems, rosette leaves, flower clusters, and green siliques. For each predicted gene or cDNA, a pair of gene-specific primers was chosen, and PCR amplifications were carried out using 15 ng of first-strand cDNA synthesized as described above. Primers of ubiquitin gene UBQ10 (5′-ATTTCTCAAAATCTTAAAAACTT-3′ and 5′-TGATAGTTTTCC CAGTCAAC-3′) were used for an internal loading standard (Norris et al., 1993). Microarray database analysis was performed by searching the Arabidopsis Functional Genomics Consortium (AFGC) microarray expression database located at the Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD/; Wu et al., 2001).
Yeast Two-Hybrid Analysis
The yeast two-hybrid vectors pAD-GAL4-2.1 and pBD-GAL4 Cam (Stratagene, La Jolla, CA), were used for C-terminal GAL4 AD and BD fusion constructions. A 1.1-kb SalI-PstI fragment containing the entire coding region of AtTLP9 was cloned into the SalI-PstI site of the pBD-GAL4 Cam vector. A 480-bp EcoRI-PstI fragment containing the entire coding region of ASK1 (At1g75950) was cloned into the EcoRI-PstI site of the pAD-GAL4-2.1 vector. The yeast strain YRG-2 [MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3 112 gal4-542 gal80-538 LYS2::UASGAL1-TATAGAL1-HIS3 URA3::UASGAL4 17mers(×3)-TATACYC1-lacZ] was used in this study. The Y2H analysis was performed according to the manufacturer's recommendations (Stratagene).
Transgenic Lines
To identify attlp9 T-DNA insertion mutant, we used AtTLP9 (At3g06380) to search the T-DNA Express database (http://signal.salk.edu/cgi-bin/tdnaexpress). Two putative attlp9 T-DNA insertion mutants (ABRC seed stock SALK_016678 and 051138) were identified and designated attlp9-1 and attlp9-2. T3 seeds of attlp9-1 and attlp9-2 were obtained from the ABRC (Ohio State University, Columbus, OH). The position of the T-DNA within the AtTLP9 was reconfirmed by sequencing the PCR-amplified fragment using pairs of primers corresponding to the T-DNA left borders and the AtTLP9 gene-specific primer. The following primer pairs were used for attlp9-1 and attlp9-2 specific amplification: for attlp9-1, N1 (5′-ATGACGTTCCGAAGTTTACTC-3′) and LBa1 (5′-TGGTTCACGTAGTGGGCCATC-3′); for attlp9-2, C1 (5′-TTATTCACAGGCAATTCTGGT-3′) and LBa1 (5′-TGGTTCACGTAGTGGGCCATC-3′). The T-DNA of attlp9-1 and attlp9-2 carries a gene leading to resistance to kanamycin. Southern blot was probed with the nptII marker gene to determine the T-DNA insertion number in attlp9-1 knockout mutant. Homozygous analyses of attlp9-1 and attlp9-2 plants were carried out by kanamycin selection and PCR method as described in Figure 5B. For the phenotype investigation, attlp9-1 and attlp9-2 T4 homozygous lines were used for detailed analysis.
For the 35S::AtTLP9 sense construct, a gel-purified XbaI-SmaI fragment of AtTLP9 including the entire coding regions was inserted into an XbaI-SmaI site of the pBI121 Ti-vector (CLONTECH, Palo Alto, CA). The constructs were introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation and transformed into wild-type plants by the floral dip method (Clough and Bent, 1998). For the phenotypic investigation, T3 homozygous lines were used.
Plant Growth Conditions
The Arabidopsis ecotype Columbia-0 (Col-0) was used in this study. abi4-1 was obtained from Dr. Wan-Hsing Cheng (Institute of Botany, Academia Sinica, Taipei, Taiwan). The phenotypes of abi4-1 were confirmed as described (Söderman et al., 2000) prior to use. Seeds were surface sterilized with 70% ethanol for 30 s and then with 6% household bleach for 5 min before being washed five times with sterile water. For aseptic growth, they were then plated on solid medium containing Murashige and Skoog salts (Invitrogen), vitamins (Duchefa, Haarlem, The Netherlands), 0.7% phytoagar (Invitrogen), and 1% Suc and transferred to a tissue culture room. For soil growth, seedlings were transferred into individual pots 8 to 10 d after germination and maintained in the growth chamber. Plants were grown at 22°C under a 16-h-light/8-h-dark photoperiod aseptically or on soil.
To determine the sensitivity of germination to ABA, seeds collected at the same or similar times were used. After surface sterilization, sterile seeds were suspended in 0.15% agarose and kept in the dark at 4°C for 3 d to break residual dormancy. Then seeds were plated on agar plates in six replicates containing no ABA or 0.25, 0.5, 0.75, or 1.0 μm ABA in 12-cm plastic petri dishes. Each agar plate was divided into seven halves, and 50 seeds of wild-type and AtTLP9 transgenic seeds were plated on each part. A seed was regarded as germinated when the radicle protruded through the seed coat.
Real-Time PCR
The relative expression of AtTLP9 was measured using real-time PCR on the ABI PRISM 7700 sequence detection system (Applied Biosystems, Scoresby, Victoria, Australia). UBQ10 was used as the endogenous control (Norris et al., 1993).
Primers were designed using Primer Express 1.0 software (Applied Biosystems, Foster City, CA). The primers used were: AtTLP9 forward primer, 5′-TAGGCCACACCGTGTAGTTCA-3′; AtTLP9 reverse primer, 5′-CGTCAACAGTCTCAACCCTAATCA-3′; UBQ10 forward primer, 5′-AGAAGTTCAATGTTTCGTTTCATGTAA-3′; and UBQ10 reverse primer, 5′-GAACGGAAACATAGTAGAACACTTATTCA-3′.
The real-time PCR was performed in 50 μL of reaction mixture containing 500 ng first-strand cDNA, 2.5 μm each primer, and 1× SYBR Green PCR Master Mix (Applied Biosystems). PCR cycling was 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The UBQ10 mRNA quantity was set at 1, and AtTLP9 expression was determined relative to control samples. Threshold cycles were determined using Sequence Detection System version 1.7a software (Applied Biosystems) for all results.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AF487267, AY045773, AY045774, AY046921, AF487268, AY092403, AF487269, AF487270, AF487271, and AY046922.
Acknowledgments
We thank Miss May-Chih Fen, Institute of Botany, Academia Sinica, for DNA sequencing. We also thank Professor Anthony Huang of University of California, Riverside; Professor Tuan-hua David Ho of Academia Sinica; and Professor Teh-hui Kao of Penn State University for their critical reading of this manuscript. We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and ABRC for providing seeds of attlp9-1 and attlp9-2.
This work was supported by a grant from the Genomic Program, Academia Sinica, Taipei, Taiwan.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.037820.
References
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 [DOI] [PubMed] [Google Scholar]
- Bewley JD (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro SL (1999) Implication of tubby proteins as transcription factors by structure-based functional analysis. Science 286: 2119–2125 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coleman DL, Eicher EM (1990) Fat (fat) and tubby (tub): two autosomal recessive mutations causing obesity syndromes in the mouse. J Hered 81: 424–427 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M (1999) The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc Natl Acad Sci USA 96: 15342–15347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Zhou YC, Schafer E, Funk M, Kretsch T (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrás R, Ferrando A, Jasik J, Kleinow T, Okresz L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C (2001) SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J 20: 2742–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gibson SI (2002) ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr Opin Plant Biol 5: 26–32 [DOI] [PubMed] [Google Scholar]
- Frye CA, Innes RW (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 98: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Heckenlively JR, Chang B, Erway LC, Peng C, Hawes NL, Hageman GS, Roderick TH (1995) Mouse model for usher syndrome: linkage mapping suggests homology to usher type I reported at human chromosome 11p15. Proc Natl Acad Sci USA 92: 11100–11104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29: 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleyn PW, Fan W, Kovats SG, Lee JJ, Pulido JC, Wu Y, Berkemeier LR, Misumi DJ, Holmgren L, Charlat O, et al (1996) Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell 85: 281–290 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Krek W (1998) Proteolysis and the G1-S transition: the SCF connection. Curr Opin Genet Dev 8: 36–42 [DOI] [PubMed] [Google Scholar]
- Li QZ, Wang CY, Shi JD, Ruan QG, Eckenrode S, Davoodi-Semiromi A, Kukar T, Gu Y, Lian W, Wu D, et al (2001) Molecular cloning and characterization of the mouse and human TUSP gene, a novel member of the tubby superfamily. Gene 273: 275–284 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340 [DOI] [PubMed] [Google Scholar]
- Nishina PM, North MA, Ikeda A, Yan Y, Naggert JK (1998) Molecular characterization of a novel tubby gene family member, TULP3, in mouse and humans. Genomics 54: 215–220 [DOI] [PubMed] [Google Scholar]
- Noben-Trauth K, Naggert JK, North MA, Nishina PM (1996) A candidate gene for the mouse mutation tubby. Nature 380: 534–538 [DOI] [PubMed] [Google Scholar]
- North MA, Naggert JK, Yan Y, Noben-Trauth K, Nishina PM (1997) Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc Natl Acad Sci USA 94: 3128–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SR, Meyer SE, Callis J (1993) The intron of polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol Biol 21: 895–906 [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK, Hughes RM, Mosinger-Ogilvie J, Speck JD, Grosof DH, Silverman MS (1995) Cochlear and retinal degeneration in the tubby mouse. Neuroreport 6: 845–849 [DOI] [PubMed] [Google Scholar]
- Pagni M, Iseli C, Junier T, Falquet L, Jongeneel V, Bucher P (2001) TrEST, trGEN and Hits: access to databases of predicted protein sequences. Nucleic Acids Res 29: 148–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M (1998) Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet 14: 236–243 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI (1997) Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9: 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J 34: 753–767 [DOI] [PubMed] [Google Scholar]
- Rugger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Llenz JE, Kohalmi SE, Risseeuw E, Haughn GW, Grosby WL (1999) The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J 20: 433–445 [DOI] [PubMed] [Google Scholar]
- Santagata S, Boggon TJ, Baird CL, Gomez JA, Zhao J, Shan WS, Myszka DG, Shapiro SL (2001) G-protein signaling through tubby proteins. Science 292: 2041–2050 [DOI] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408: 381–386 [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW (1999) Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284: 662–665 [DOI] [PubMed] [Google Scholar]
- Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR (2000) Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124: 1752–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Stemmann O, Neidig A, Kocher T, Wilm M, Lechner J (2002) Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore. Proc Natl Acad Sci USA 99: 8585–8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S (1991) The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350: 241–243 [DOI] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW (1999) The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev 13: 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SH, Ramonell K, Gollub J, Somerville SC (2001) Plant gene expression profiling with DNA microarrays. Plant Physiol Biochem 39: 917–926 [Google Scholar]
- Xiao W, Jang J (2000) F-box proteins in Arabidopsis. Trends Plant Sci 5: 454–457 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D (2002) The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka A, Yada M, Imaki H, Koga M, Ohshima Y, Nakayama KI (2002) Multiple Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with Cullins and F-Box proteins. Curr Biol 12: 267–275 [DOI] [PubMed] [Google Scholar]
- Yang M, Hu Y, Lodhi M, McCombie WR, Ma H (1999) The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc Natl Acad Sci USA 96: 11416–11421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Ni W, Feng B, Han T, Petrasek MG, Ma H (2003) Members of the Arabidopsis-SKP1-like gene family exhibit a variety of expression patterns and may play diverse roles in Arabidopsis. Plant Physiol 133: 203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Yang M, Solava J, Ma H (1999) The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev Genet 25: 209–223 [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al (2002) Structure of the Cul1-Rbx1-Skp1-F box Skp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]