Abstract
To investigate the role of stored and neosynthesized mRNAs in seed germination, we examined the effect of α-amanitin, a transcriptional inhibitor targeting RNA polymerase II, on the germination of nondormant Arabidopsis seeds. We used transparent testa mutants, of which seed coat is highly permeable, to better ascertain that the drug can reach the embryo during seed imbibition. Even with the most permeable mutant (tt2-1), germination (radicle protrusion) occurred in the absence of transcription, while subsequent seedling growth was blocked. In contrast, germination was abolished in the presence of the translational inhibitor cycloheximide. Taken together, the results highlight the role of stored proteins and mRNAs for germination in Arabidopsis and show that in this species the potential for germination is largely programmed during the seed maturation process. The α-amanitin-resistant germination exhibited characteristic features. First, this germination was strongly slowed down, indicating that de novo transcription normally allows the synthesis of factor(s) activating the germination rate. Second, the sensitivity of germination to gibberellic acid was reduced 15-fold, confirming the role of this phytohormone in germination. Third, de novo synthesis of enzymes involved in reserve mobilization and resumption of metabolic activity was repressed, thus accounting for the failure in seedling establishment. Fourth, germinating seeds can recapitulate at least part of the seed maturation program, being capable of using mRNAs stored during development. Thus, commitment to germination and plant growth requires transcription of genes allowing the imbibed seed to discriminate between mRNAs to be utilized in germination and those to be destroyed.
Seed germination is a complex, multistage process that can be divided into three phases—imbibition, increased metabolic activity, and initiation of growth—which loosely parallel the triphasic water uptake of dry mature seeds. Morphologically, initiation of growth corresponds to radicle emergence; subsequent growth is generally defined as seedling growth. By definition, germination sensu stricto incorporates those events that start with the uptake of water by the nondormant quiescent dry seed and terminate with the protrusion of the radicle and the elongation of the embryonic axis (Bewley and Black, 1994; Bewley, 1997; Koornneef et al., 2002). Upon imbibition, the quiescent dry seed rapidly resumes metabolic activity.
While the processes and genes regulating embryogenesis and maturation are being identified (Errampalli et al., 1991; Girke et al., 2000; Brocard-Gifford et al., 2003; Gallardo et al., 2003; Tzafrir et al., 2003), only very recent studies addressed the question of the exact requirements for germination, particularly in terms of de novo RNA and protein syntheses (Bradford et al., 2000; Bove et al., 2001; Gallardo et al., 2001, 2002a; Koornneef et al., 2002; Potokina et al., 2002; van der Geest, 2002; Duque and Chua, 2003; Jabrin et al., 2003; Ogawa et al., 2003). Although it is widely documented that, in addition to the storage proteins, mature dry seeds accumulate during maturation a number of proteins that are involved in various cellular processes such as DNA replication and transcription, translation, metabolism, or defense mechanisms (e.g. see Bewley and Black, 1994; Gallardo et al., 2001, 2002a, 2002b), their role and functionality has not yet been thoroughly documented. It is also known that mature dry seeds contain mRNAs stored during maturation, also called long-lived mRNAs to indicate that they can survive desiccation and remain active in dry quiescent embryos. Early studies based on the use of metabolic inhibitors (cycloheximide, actinomycin D) had supported the view that protein synthesis occurs on such long-lived templates during the early stages of seed germination and that de novo transcription is not necessary (Dure and Waters, 1965; Waters and Dure, 1966; for review, see Raghavan, 2000). Different results were obtained for wheat embryos for which germination appeared to be strongly impeded in the presence of the transcriptional inhibitor α-amanitin (Jendrisak, 1980).
We wish to reinvestigate these questions by using the model plant Arabidopsis. The widespread acceptance of Arabidopsis as a model plant is based on the genetic and genomic methods and resources that are available for it, which have facilitated the investigation of a range of biological problems (Arabidopsis Genome Initiative, 2000; for review, see Somerville and Koornneef, 2002). It is anticipated that a better knowledge of the role played by the mRNAs stored in mature seeds of this model species can help facilitate the interpretation of microarray analyses of the seed germination process. In the present work we analyzed the effect of α-amanitin on Arabidopsis seed germination. This amatoxin, which is produced by the toadstool Amanita phalloides, specifically inhibits RNA synthesis catalyzed by DNA-dependent RNA polymerase II both in vivo (Jendrisak, 1980) and in vitro (Guilfoyle and Jendrisak, 1978; de Mercoyrol et al., 1989; Bushnell et al., 2002). However α-amanitin is a relatively large molecule (Mr 919.0), and it might therefore have restricted access to the embryos that are covered by a seed coat composed of five cell layers forming the outer and inner integuments (Debeaujon et al., 2000). To circumvent this problem, we used in the present work the transparent testa (tt) mutants of Arabidopsis described by Debeaujon et al. (2000) and Debeaujon and Koornneef (2000), of which some (e.g. tt2-1) possess highly permeable seed coats.
Our general finding is that Arabidopsis seed germination resists inhibition by α-amanitin, even in the presence of an excess (500 μm) of the fungal toxin. The rate and uniformity of seed germination are, however, considerably affected, indicating that new transcripts must be synthesized during imbibition to enhance seed vigor. In marked contrast, seed germination proved to be totally inhibited in the presence of cycloheximide, indicating a requirement for protein synthesis in germinating embryos. These combined results strongly suggest that germination-specific proteins are made from the long-lived stored mRNAs. Furthermore, the proteome of seeds germinated on α-amanitin, as characterized through the incorporation of [35S]Met into newly synthesized proteins, was markedly different from that of seeds germinated on water, being more reminiscent of a proteome characteristic of the seed maturation phase than of germination. Thus, factors must be synthesized from new transcripts to avoid the use of some of the long-lived stored mRNAs during normal germination, promoting the shift from a developmental to a germination program.
RESULTS AND DISCUSSION
Sensitivity of Wild-Type and tt-Mutant Arabidopsis Seed Germination to α-Amanitin
In initial experiments, the effect of α-amanitin on Arabidopsis seed germination was assessed by using nondormant wild-type seeds from the Landsberg erecta (Ler) ecotype. In all eukaryotes, this fungal toxin specifically inhibits transcription by DNA-dependent RNA polymerase II (Kd = 10 nm) by impeding the progression of the enzyme along the DNA template during transcription elongation (de Mercoyrol et al., 1989). On water and under optimal conditions (25°C), radicle protrusion started at 1.5 d of imbibition and it took almost 1.8 d (T50) for 50% of the seeds to reach this phase (Fig. 1A). Under the same conditions, but in the presence of 500 μm of α-amanitin, a retardation in radicle emergence was constantly observed (T50 ≈ 2.2 d; Fig. 1A). However, seeds were still able to germinate, that is, all seeds germinated after 2.9 d (data not shown). In contrast, plant growth was totally repressed following radicle protrusion (data not shown). This suggests that new mRNA synthesis is not required for germination, although it is for plantlet growth.
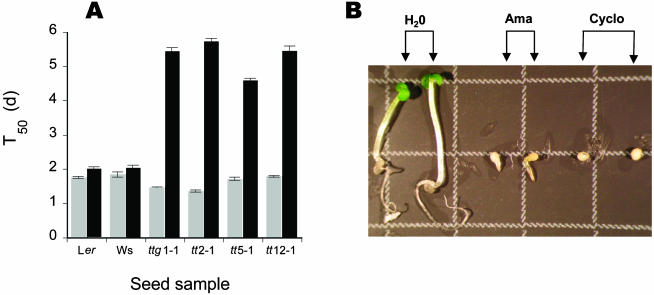
Figure 1.
Influence of α-amanitin and cycloheximide on seed germination and seedling establishment of several Arabidopsis ecotypes (Ler and Ws) and tt mutants. A, Influence of α-amanitin on germination. Seeds were germinated as described in “Materials and Methods,” in the absence (gray bars) or presence (black bars) of 500 μm α-amanitin. T50 (time to reach 50% germination) values were calculated from experiments run in triplicate. B, Phenotypes observed with the tt2-1 mutant 8 d after sowing the seeds on water (H2O), 500 μm α-amanitin (Ama) or 100 μm cycloheximide (Cyclo). Square, 3 × 3 mm.
However, the question arises as to whether the observed effects could be due to a lack of penetration of α-amanitin through the seed coat. Germination experiments were conducted to answer this question by using seeds of tt mutants from Arabidopsis, which are affected in flavonoid pigmentation (Shirley et al., 1995). As a consequence of these mutations, the seed coat of tt mutants is much more permeable than that of the corresponding wild-type ecotype, as demonstrated by uptake assays of tetrazolium salts by the embryo (Debeaujon et al., 2000). Figure 1A summarizes the germination data obtained with several of the tt mutants incubated either in water or in the presence of 500 μm α-amanitin. Upon incubation in water, nondormant tt and wild-type seeds exhibited kinetics of germination globally similar, with T50 values in the range of 1.37 to 1.93 d. In these conditions, tt2-1 and transparent testa glabra ttg1-1 mutant seeds germinated at a slightly faster rate than the other seeds (T50 in the order of 1.4 d). In the presence of 500 μm α-amanitin, impediment of germination brought about by the fungal toxin was substantially more pronounced with the tt mutant than with wild-type seeds, as indicated by T50 value determinations (Fig. 1A). However, for all tt-mutants seed germination resisted α-amanitin inhibition. As for the wild-type seeds, further post-germinative growth from all tt-mutant seeds was totally repressed by α-amanitin (Fig. 1B).
Among tt mutants, seed germination from the tt2-1 and ttg1-1 mutants proved to be the most affected by α-amanitin (Fig. 1A). TT2 is specifically involved in tannin biosynthesis being only expressed in the endothelium, whereas TTG1 is also involved in the formation of testa mucilage and trichomes and in root development (Koornneef, 1990; Masucci and Schiefelbein, 1996; Debeaujon et al., 2000). Hence, the tt2-1 mutant was used in the following experiments.
Germination of tt2-1 seeds was scored at different times in the absence or presence of various concentrations of α-amanitin (Fig. 2A). Confirming that seed germination can resist inhibition by this compound, plots of T10 (time to reach 10% germination), T50, and T90 (time to reach 90% germination) versus [α-amanitin] exhibited saturation behavior with an IC50 (concentration of α-amanitin yielding 50% inhibitory effect) in the order of 50 μm (Fig. 2B). However, the sensitivity of radicle protrusion to α-amanitin inhibition depended on the time at which germination was scored. Figure 2C shows that the shorter the time at which germination was scored the higher the apparent sensitivity to α-amanitin. This occurs because the germination curves (as expressed by cumulative germination frequency versus time of imbibition) intrinsically follow saturation kinetics.
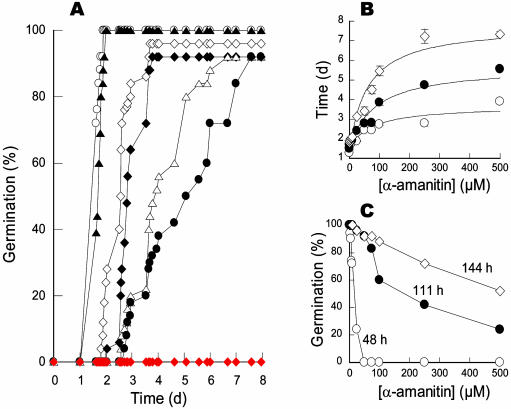
Figure 2.
Analysis of the effect of α-amanitin and cycloheximide on the time courses of seed germination of the tt2-1 Arabidopsis mutant. A, Influence of α-amanitin and cycloheximide on the germination profiles. Seeds were germinated as described in “Materials and Methods,” in the absence (○) or presence of α-amanitin (▴, 1 μm; ⋄, 25 μm; ♦, 50 μm; ▵, 100 μm; •, 250 μm) or 100 μm cycloheximide (♦). The figure shows a representative experiment carried out three times in triplicate. B, Influence of α-amanitin concentration on the kinetic parameters of germination curves for the tt2-1 mutant. T10 (○), T50 (•), and T90 (⋄) values, corresponding to the times (d) required for 10%, 50%, and 90% germination, respectively, were calculated from germination curves as shown in A. C, Influence of α-amanitin concentration on germination frequency monitored 48 h (○), 111 h (•), and 144 h (⋄) after sowing. The figure shows a representative experiment carried out three times in triplicate.
The present data demonstrate that in Arabidopsis α-amanitin does not prevent seed germination (radicle protrusion), but instead considerably slows down the process. Thus, proteins and mRNAs stored in the dry mature seeds are sufficient for germination sensu stricto as well as starting of root growth. At first sight our results seem at variance with those reported for the germination of wheat embryos that appeared to be strongly inhibited by low concentrations of α-amanitin (Jendrisak, 1980). However, two points warrant comment. (1) These previous experiments measured embryonic axis elongation as determined by fresh weight analysis from isolated wheat embryos (Jendrisak, 1980) and therefore cannot address important features of normal seed germination such as the piercing of the radicle through the seed coat. (2) The sensitivity of isolated wheat embryos to α-amanitin was evaluated at a single time point, that is at 36 h after imbibition (Jendrisak, 1980), and there were no data reported on the effect of the fungal toxin for longer times of germination. When considering only the early stages of seed germination (e.g. 48 h imbibition in Fig. 2C) our data agree well with those of Jendrisak (1980), since the germination of both intact tt2-1 Arabidopsis seeds (this study) and isolated wheat embryos (Jendrisak, 1980) appears to respond to low concentrations of α-amanitin, in the μm range.
That germination was not blocked by α-amanitin indicates that in Arabidopsis the potential for seed germination is largely programmed during the seed maturation phase. In other words, this potential relies only on the stored mRNAs and/or stored proteins. However, the fact that the rate of germination was substantially slowed down by the fungal toxin disclosed an important role for de novo RNA synthesis in the control of the rate and uniformity of seed germination. Thus, quantitative factors governing seed vigor (that is, the potential to establish seedling growth) must be synthesized de novo soon after imbibition takes place.
α-Amanitin Prevents UMP Incorporation into TCA-Precipitable RNAs
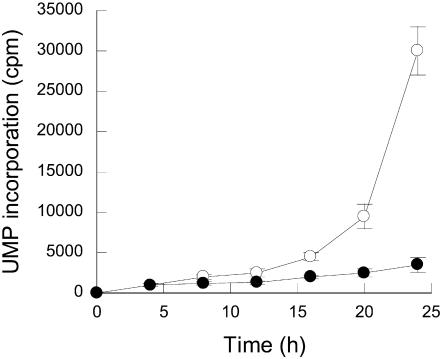
To address more directly the question of whether the fungal toxin can penetrate through the seed coat, we assessed the effect of α-amanitin on in vivo RNA synthesis during germination. Labeled RNA was synthesized in vivo by tt2-1 seeds imbibed on water in the presence of [α-33P]UTP and in the presence or absence of 500 μm of α-amanitin. At time intervals, the extent of radioactivity incorporated into TCA precipitable material was quantitated as described in “Materials and Methods.” For the seeds incubated in water only, Figure 3 shows that transcriptional activity was rather low during the first 16 h of seed imbibition and then considerably increased at 24 h postimbibition corresponding to the completion of germination sensu stricto, just before radicle protrusion through the seed coat (all seeds germinated after 48-h imbibition; Fig. 2). Figure 3 also shows a considerable impediment of such an incorporation from seeds incubated in the presence of α-amanitin compared to that from seeds incubated in its absence. Thus, the drug can readily enter the embryo tissues and exert a strong inhibitory effect on transcriptional activity mediated by DNA-dependent RNA polymerase II.
Figure 3.
Influence of α-amanitin on the time courses of UMP incorporation into RNA with the tt2-1 Arabidopsis mutant. Seeds were germinated as described in “Materials and Methods,” in the presence of [α-33P]UTP and in the absence (○) or presence of 500 μm α-amanitin (•). Following incubation, RNA synthesis was scored at time intervals as indicated in “Materials and Methods.”
Cycloheximide Inhibits Arabidopsis Seed Germination
To directly test if protein synthesis is required for seed germination, seeds were germinated in the presence of the translational inhibitor cycloheximide. This compound was found to totally inhibit seed germination from both wild-type (data not shown) and tt2-1 seeds (Fig. 2A). The results are similar to those obtained for isolated wheat embryos (Jendrisak, 1980).
A comparison of the effects on tt2-1 seed germination brought about by α-amanitin and cycloheximide indicates that although de novo mRNA synthesis is not required for radicle protrusion in germinating Arabidopsis embryos, de novo protein synthesis must occur. Likewise these comparative data suggest that protein synthesis required for germination proceeds from translation of the stored mRNA pool. The present data highlight the importance and significance of stored mRNAs in the dry seed and its role in germination.
It is worth noting that, contrary to translational inhibitors, transcriptional inhibitors do not inhibit the germination of spores of Aspergillus nidulans (Osherov and May, 2000), nor do they inhibit pollen grain germination in conifers (Pinus monticola, P. aristata, P. contorta, P. griffithii, P. lambertiana, Cedrus deodara, Picea orientalis, Abies amabilis, Picea stichensis) (Fernando et al., 2001). All these studies are consistent with the view that some form of translational control involving stored mRNAs operates during the transition from a quiescent to a metabolically active state in different systems.
Influence of GAs
The central role of GAs in promoting seed germination is well established (Chrispeels and Varner, 1966; Davies, 1995; Olszewski et al., 2002) and confirmed by the identification of GA-deficient mutants of Arabidopsis and tomato (Lycopersicon esculentum Mill.), seeds of which will not germinate unless exogenously supplied with GAs (Koornneef and van der Veen, 1980; Groot and Karssen, 1987). Furthermore, the fact that inhibitors of GA biosynthesis such as paclobutrazol (PAC), uniconazol, and tetcyclacis prevent germination (Karssen et al., 1989; Nambara et al., 1991; Jacobsen and Olszewski, 1993) indicated the requirement for de novo synthesis of GAs in germination.
On this basis, it might be hypothesized that α-amanitin inhibited Arabidopsis seed germination because, under these assay conditions, the capacity of the seeds to produce GAs was lowered. To investigate this possibility, germination experiments were performed in the absence or presence of α-amanitin, PAC and/or GA. In the absence of α-amanitin, and as found previously (Debeaujon and Koornneef, 2000), PAC blocked tt2-1 seed germination, while addition of GA4+7 to the PAC-containing germination medium strongly promoted germination, up to the frequency observed in the absence of PAC (Fig. 4A). This repeatedly shows that the dry mature seeds of the tt2-1 mutant do not contain sufficient amounts of GAs for germination and exhibit an absolute requirement of GAs for germination. In the presence of α-amanitin, tt2-1 seeds retained a strict requirement of GAs for germination, as shown by a complete blocking of residual seed germination in the presence of PAC (Fig. 4A). Yet, as judged by the different inhibitory behaviors observed with PAC (complete block of germination; Fig. 4A) and α-amanitin (germination was only delayed and slowed down; Fig. 2), it appears that the GA-biosynthetic pathway can function during germination even in the presence of an excess of the transcriptional inhibitor. This finding suggests strongly that some of the basic enzymes in this pathway are already present in the dry mature seeds while others are being synthesized from the stored mRNAs.
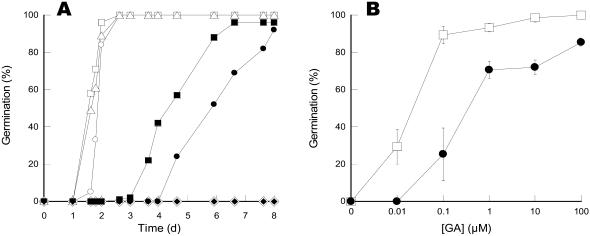
Figure 4.
Influence of GAs on seed germination of the tt2-1 Arabidopsis mutant in the presence or absence of α-amanitin. A, Germination curves. Seeds were incubated on water (○), water plus 100 μm GA4+7 (▵), water plus 100 μm PAC (⋄), water plus 100 μm PAC and 100 μm GA4+7 (□), 500 μm α-amanitin (•), 500 μm α-amanitin plus 100 μm GA4+7 (▪), and 500 μm α-amanitin plus 100 μm PAC (♦), and germination was scored by the frequency of radicle protrusion through the seed coat. The figure shows a typical experiment repeated three times in triplicate; errors bars are of the size of the symbols. B, Influence of α-amanitin on the sensitivity to GAs of tt2-1 seed germination. Seeds of the tt2-1 mutant were incubated in the presence of 100 μm PAC, various concentrations of GA4+7 as indicated on the plot, and in the presence (•) or absence (□) of 500 μm α-amanitin. Germination was scored 43 and 127 h after sowing for the seeds incubated in the absence or presence of α-amanitin, respectively. The figure shows a typical experiment carried out two times in triplicate.
Adding GA4+7 to the α-amanitin-containing medium reversed substantially the inhibitory effect of α-amanitin on seed germination (Fig. 4A). Thus, the GA-biosynthetic pathway was not functioning at full rate in the presence of the transcriptional inhibitor, suggesting that, in addition to the stored proteins and the pool of stored mRNAs, de novo transcription also contributes to the GA-biosynthetic pathway. Consistent with this hypothesis, microarray analyses indicated that several GA biosynthesis genes [e.g. genes encoding an ent-kaurene oxidase (AtKO1), a GA 20-oxidase (AtGA20ox3), and a GA 3-oxidase (AtGA3ox1)] are up-regulated during Arabidopsis seed germination (Ogawa et al., 2003). Other genes controlling the levels of GAs in plants have been described. For example, RSG (for repression of shoot growth) is a transcriptional activator involved in the control of endogenous GA levels that binds and activates the promoter of the gene GA3, which encodes ent-kaurene oxidase in the GA biosynthetic pathway (Fukazawa et al., 2000). Transgenic tobacco plants expressing a dominant-negative form of RSG exhibit reduced germination frequency (Fukazawa et al., 2000), showing that RSG does play a role in seed germination. Recent work also documented the role played by thioredoxins, which are small Mr proteins containing a redox-active disulfide group (Yano et al., 2002), in seed germination. This study showed that overexpression of wheat thioredoxin h in barley was associated with enhanced germination and increased GA levels in the transgenic germinating seeds (Wong et al., 2002), suggesting that thioredoxins can somehow regulate some of the enzymes in the GA biosynthetic pathway and/or factors modulating the rate and extent of GA biosynthesis.
It must be stressed, however, that, contrary to what is observed with PAC, exogenous GA4+7 only partly reversed the impediment in seed germination brought about by α-amanitin (Fig. 4A). Furthermore, we observed that exogenous GAs (100 μm) did not alleviate the complete blocking of seed germination imposed by cycloheximide (100 μm), at least up to 8-d imbibition (data not shown). These findings raise the question of whether α-amanitin can alter the sensitivity of germination to GAs. A series of germination experiments was conducted to answer this question, in which different concentrations of GAs were supplied to the seeds in the presence of an inhibitory amount of PAC (100 μm), in the presence or absence of the transcriptional inhibitor. Figure 4B demonstrates that the sensitivity of the tt2-1 seeds to exogenous GAs differed markedly in the presence of α-amanitin compared to that in its absence. To reach 50% germination, a concentration of GAs in the order of 0.5 μm was needed in the presence of α-amanitin, which is about 15-fold higher than the concentration needed for the seeds incubated in the absence of the fungal toxin (Fig. 4B). There are some leads in the literature to the molecular mechanisms governing the sensitivity of seed germination to GAs. For example, an Arabidopsis mutant line with a T-DNA insertion in gene DAG2 encoding a Dof zinc finger protein involved in the control of seed germination showed a 10-fold reduction in sensitivity to external GAs compared to wild type (Gualberti et al., 2002). Also, Arabidopsis seeds that carry a protein null mutation in the gene encoding the G protein α-subunit GPA1 are 100-fold less responsive to GAs, whereas seeds ectopically expressing GPA1 are at least a millionfold more responsive to GA, yet still requiring this phytohormone for germination (Ullah et al., 2002). Further, DELLA proteins, which are transcription factors inhibiting GA response, have been described, and models of GA regulation response through the degradation of these negative regulators have been proposed (Silverstone et al., 1997, 1998; Dill et al., 2001; Wen and Chang, 2002). For example, in Arabidopsis, RGL2 (a DELLA protein) behaves as an imbibition-induced negative regulator that is switched off by GAs to allow germination completion (Lee et al., 2002; Peng and Harberd, 2002). We note that the GA-deficient ga1-3 rgl2 double mutant of Arabidopsis is capable of germination (Lee et al., 2002), indicating that in the absence of RGL2 germination occurs in the absence of GAs. Because germination in the presence of α-amanitin still retained PAC sensitivity (Fig. 4A), albeit with reduced response to exogenous GAs (Fig. 4B), our results suggest that the RGL2 protein is already present in the mature seeds and/or can be translated from the stored mRNA pool. It is believed that active GA signaling inhibits the repressor action of the DELLA proteins by destabilizing them (Dill et al., 2001). However, the molecular mechanisms by which GAs affect the stability of the DELLA proteins have not yet been characterized (Itoh et al., 2003).
In summary, our present data showing that the requirement of GAs for seeds to germinate is strongly dependent on de novo transcription during imbibition suggest the hypothesis that if the genes encoding some of the modulators of GA level, sensitivity and/or action described above were to be controlled at the transcriptional level during germination, it would provide an explanation for the presently observed effect of α-amanitin. The possibility that additional factors controlling the rate of seed germination are synthesized through de novo transcription and act independently of GAs cannot be excluded. However, in view of the total block of seed germination imposed by PAC (Fig. 4), in such a case, the action of such factors could only occur after the germination program had been triggered by the GA-dependent mechanisms.
Proteomic Analyses
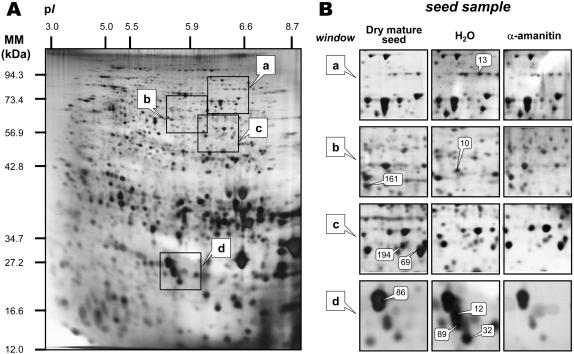
We have initiated a global study of Arabidopsis seed germination by proteomics (Gallardo et al., 2001, 2002a, 2002b; http://seed.proteome.free.fr). In the present work, we wished to characterize those proteins for which accumulation level can be affected by α-amanitin during germination, thereby allowing the identification of genes that are under transcriptional control during this process. Toward this goal, total soluble proteins were extracted from tt2-1 seeds (dry mature seeds, seeds imbibed for 1 d on water or in the presence of 500 μm of α-amanitin). This stage corresponded to germination sensu stricto for the tt2-1 seeds incubated on water since none of the seeds showed radicle protrusion during this period (Fig. 2). The protein extracts were then separated by two-dimensional (2D) gel electrophoresis, and, following silver nitrate coloration, protein patterns were analyzed by image analysis (see “Materials and Methods”). Typical gels are presented in Figure 5.
Figure 5.
Influence of α-amanitin on the proteome of tt2-1 Arabidopsis mutant seeds 24 h after sowing. An equal amount (200 μg) of total protein extracts was loaded in each gel. A, Silver-stained 2D gel of total proteins from tt2-1 seeds incubated for 24 h in water. The indicated portions of the gel, a, b, c, and d, are reproduced in B. B, enlarged windows (a–d) of 2D gels as shown in A for tt2-1 dry mature seeds (left), tt2-1 dry mature seeds incubated in water for 24 h (middle) and tt2-1 dry mature seeds incubated in 500 μm α-amanitin for 24 h (right). The nine labeled protein spots (protein nos. 13, 161, 10, 194, 69, 86, 89, 12, and 32) were identified by comparison with Arabidopsis seed protein reference maps (Gallardo et al., 2001, 2002a, http://seed.proteome.free.fr; see Table II). Protein spot quantitation was carried out as described in “Materials and Methods,” from at least three gels for each seed sample.
Because our previous protein maps were obtained from seeds of the wild-type Ler ecotype (Gallardo et al., 2001, 2002a), we first investigated the effect of the tt2-1 mutation on the mature dry seed proteome. Only a few protein spots showed reproducible variations in their accumulation level when comparing the tt2-1 seed proteome presently analyzed with that of the corresponding wild-type Ler seed proteome previously described. They are listed in Table I.
Table I.
Arabidopsis proteins whose abundance was significantly different in dry mature Ler seeds than in dry mature seeds from the tt2-1 mutant
| No.a | Exp. MM | Exp. PI | Arabidopsis Protein Nameb | Cov. | Theo. MM | Theo. PI | AGI No. | Relative Abundancec |
|---|---|---|---|---|---|---|---|---|
| kD | % | kD | ||||||
| 17 | 72.98 | 6.57 | Phosphoenolpyruvate carboxykinase | 17 | 73.40 | 6.61 | At4g37870 | 1.6 ± 0.06 |
| 25 | 23.32 | 6.89 | β-Cruciferin 12S seed storage protein | 43 | 21.20 | 6.19 | At4g28520 | 3.8 ± 0.05 |
| 69 | 43.67 | 6.29 | 12S Cruciferin precursor | 18 | 50.56 | 6.53 | At1g03880 | 2.0 ± 0.08 |
| 160 | 82.27 | 5.94 | Met synthase | 12 | 84.36 | 6.09 | At5g17920 | 3.0 ± 0.06 |
Cov., coverage; Exp., experimental; Theo., theoretical.
Protein numbering following Gallardo et al. (2001, 2002a) and Arabidopsis seed protein reference maps (http://seed.proteome.free.fr).
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a).
Data obtained from densitometric analysis of individual spots from 2D gels colored with silver nitrate (see Fig. 5 for an example of 2D gel): normalized spot volume in the mature dry tt2-1 mutant seeds divided by the normalized spot volume in the wild-type Ler dry mature seeds ±sd, from three different gels and independent extractions.
The effect of α-amanitin on the seed proteome was then assessed during germination sensu stricto of the tt2-1 seeds. Globally, the protein profiles obtained from tt2-1 seeds germinated on water for 24 h in the presence or absence of α-amanitin looked highly similar. However, out of 1,193 protein spots detected on reproducible gels, 8 showed reproducible variations in their accumulation level upon incubation in the presence of α-amanitin (Fig. 5; Table II). For four of these proteins (spot nos. 12, 13, 32, and 89) the fungal toxin caused an inhibition in their accumulation (Fig. 5, Table II). For three other proteins (spot nos. 10, 69, and 161) the reverse was observed; that is, the drug prevented their disappearance during germination. Finally, one protein spot (no. 194) showed accelerated disappearance in the presence of α-amanitin (Fig. 5, Table II). Out of these eight spots, four (protein nos. 12, 32, 69, and 89) corresponded to 12S cruciferin (globulins), which are the major seed protein reserves in Arabidopsis seeds. As found previously for the wild-type Ler seeds (Gallardo et al., 2001, 2002a), in the case of the tt2-1 mutant protein no. 69, which is a precursor form of the 12S cruciferin, readily disappeared during germination, while spot nos. 12, 13, and 89, which correspond to proteolytic fragments of 12S cruciferins, transiently accumulated during germination (Fig. 5, Table II). The observed effect of α-amanitin on the accumulation level of these spots indicates that the expression of some genes involved in the initial mobilization of 12S globulins during early germination is under transcriptional control. These data further support the finding that the drug can readily enter the embryonic tissues during early steps of germination.
Table II.
Arabidopsis proteins whose abundance was altered by α-amanitin during 1-d imbibition
| Relative Abundancec
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Noa. | Exp. MM | Exp. PI | Arabidopsis Protein Nameb | Cov. | Theo. MM | Theo. PI | AGI No. | H2O/DS | Ama/DS |
| kD | % | kD | |||||||
| 10 | 57.36 | 5.77 | Ribulose bisphosphate carboxylase large chain (precursor) | 28 | 53.47 | 6.25 | AtCg00490 | 0.25 ± 0.08 | 0.86 ± 0.06 |
| 161 | 52.11 | 5.71 | Dihydrolipoamide S-acetyltransferase (mitochondrial) | 10 | 45.07 | 5.93 | At3g25860 | 0.05 ± 0.05 | 0.54 ± 0.05 |
| 194 | 45.50 | 6.15 | 3-Oxoacyl[Acyl-carrier-protein] synthase I | 19 | 50.41 | 8.29 | At5g46290 | 0.46 ± 0.07 | 0.17 ± 0.09 |
| 13 | 73.27 | 6.32 | MLO-like-protein | 15 | 67.23 | 9.55 | At1g61560 | 1.67 ± 0.09 | 1.17 ± 0.08 |
| 32 | 15.16 | 5.75 | β-Subunit of 12S-2 seed storage protein (fragment) | 18 | 20.80 | 7.03 | At1g03880 | 4.31 ± 0.12 | 0.30 ± 0.08 |
| 12 | 16.14 | 5.71 | β-Subunit of 12S-2 seed storage protein (fragment) | 30 | 20.80 | 7.03 | At1g03880 | 3.60 ± 0.14 | 0.24 ± 0.03 |
| 89 | 16.14 | 5.71 | β-Subunit of 12S-2 seed storage protein (fragment) | 36 | 20.80 | 7.03 | At1g03880 | 2.1 ± 0.15 | 0.35 ± 0.05 |
| 69 | 43.67 | 6.29 | 12S seed storage protein precursor | 18 | 50.56 | 6.53 | At1g03880 | 0.16 ± 0.04 | 0.4 ± 0.07 |
DS, tt2-1 dry mature seeds; H2O, tt2-1 seeds incubated for 1 d in water; Ama, α-amanitin, and tt2-1 seeds incubated for 1 d in 500 μm α-amanitin as described in “Materials and Methods”; Cov., coverage; Exp., experimental; Theo., theoretical.
Protein numbering following Gallardo et al. (2001, 2002a) and Arabidopsis seed protein reference maps (http://seed.proteome.free.fr).
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a).
Data obtained from densitometric analysis of individual spots from 2D gels colored with silver nitrate (see Fig. 5 for an example of 2D gel): normalized spot volume in the tt2-1 mutant seeds incubated for 1 d in water divided by the normalized spot volume in the tt2-1 dry mature seeds (H2O/DS), and in the tt2-1 seeds incubated for 1 d in 500 μm α-amanitin divided by the normalized spot volume in the tt2-1 dry mature seeds (Ama/DS) ±sd, from three different gels and independent extractions.
De Novo Protein Patterns
To get direct insight on protein synthesis during germination sensu stricto, labeled proteins were synthesized in vivo by tt2-1 seeds imbibed on water for 1 d in the presence of [35S]Met and in the presence or absence of α-amanitin. Then protein extracts were prepared and submitted to 2D-PAGE. Figure 6 shows the results obtained following revelation of the radioactively labeled proteins as described in “Materials and Methods.”
Figure 6.
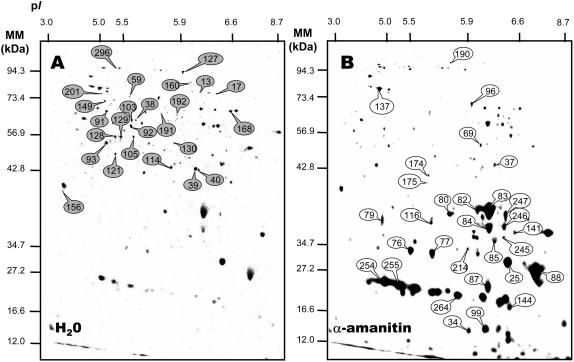
Influence of α-amanitin on de novo protein synthesis during 24-h germination of the tt2-1 Arabidopsis mutant. Seeds were incubated for 24 h in the presence of [35S]Met, in the absence (A) or presence (B) of 500 μm α-amanitin. Proteins were extracted, submitted to 2D gel electrophoresis, and the radiolabeled proteins revealed as described in “Materials and Methods.” The labeled protein spots (gray circles, proteins that showed reduced accumulation levels in the α-amanitin-incubated seeds; white circles, proteins that showed increased accumulation levels in the α-amanitin-incubated seeds) were identified either by MALDI-TOF analysis, by Edman protein sequencing, or by comparison with Arabidopsis seed protein reference maps (Gallardo et al., 2001, 2002a, http://seed.proteome.free.fr; see Tables III and IV). A number of labeled spots exhibiting contrasting specific expression are not annotated because they had no match with identified proteins from the reference maps; that is, these spots correspond most presumably to low abundance unidentified proteins. Protein spot quantitation was carried out as described in “Materials and Materials” and, from at least three gels for each seed sample.
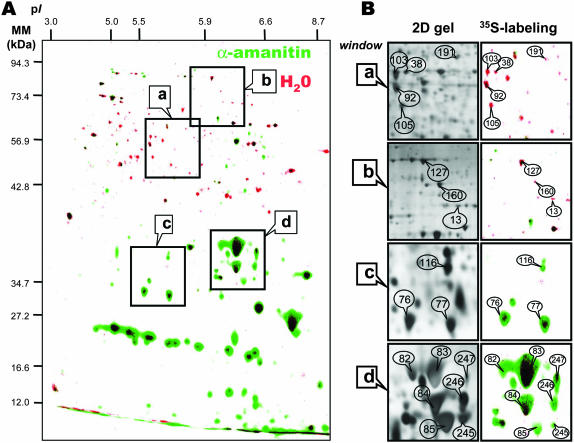
The two radioactive protein patterns differed remarkably. Upon incubation for 24 h on water, seeds synthesized a number of proteins of molecular mass larger than 30 kD and of pI < 7.3. Under the same conditions but in the presence of 500 μm α-amanitin a large number of these protein spots were no more detectable or were only present at a much lower level than in the water control. The radiolabeled proteome in Figure 6A was compared with the total proteome obtained after silver staining of the same gels (see Fig. 7) and also with the reference maps established for the Arabidopsis seed proteome (Gallardo et al., 2001, 2002a, 2002b). These comparisons can provide a means to tentatively assign some of the [35S]Met-labeled protein spots to proteins previously characterized by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) analysis, although we cannot rule out the possibility that at least some of the [35S]Met-labeled spots corresponded to very low abundant proteins that co-migrated with more abundant proteins. However, as shown in Figure 7B, in many cases there was quite a good match between protein patterns revealed by coloration with silver nitrate and those revealed through labeling with the [35S]Met precursor. Under these assumptions, the proteins for which de novo synthesis was repressed by α–amanitin are listed in Table III. Several of them are involved in the mobilization of seed reserves and reactivation of metabolic activity during germination. Thus, seven enzymes that are involved in the pathways of triacylglycerol metabolism and hexose assimilation (spot nos. 17, 39, 40, 59, 103, 114, 121, and 168) exhibit decreased de novo synthesis in the presence of α-amanitin, indicating that the accumulation level of theses enzymes is regulated at the transcription level during germination. In this context, it is worth mentioning recent data demonstrating that icl mutants, which are deficient in the glyoxysomal enzyme isocitrate lyase, show a significant decrease in the frequency of seedling establishment upon germination in the dark (Eastmond et al., 2000; Eastmond and Graham, 2001). Further, analysis of the ped1 mutant, which is deficient in the β-oxidation enzyme 3-ketoacyl-CoA thiolase, also demonstrated that fatty acid breakdown is essential for seedling establishment, because ped1 seedling growth arrests shortly after germination (Hayashi et al., 1998). From image analysis of silver nitrate-colored 2D gels (Gallardo et al., 2001, 2002a), out of the seven protein spots discussed above, four (protein nos. 17, 39, 40, and 168, which correspond to phosphoenolpyruvate carboxykinase, two forms of cytosolic glyceraldehyde-3-phosphate dehydrogenase, and glyoxysomal malate synthase, respectively) have previously been shown to accumulate during 1-d imbibition of Arabidopsis seeds. The fact that these three protein spots can be labeled by incorporation of [35S]Met (Fig. 6A; Table III) is therefore in agreement with this finding. In contrast, the other three spots (spot nos. 103, 114, and 121, which correspond to enolase, malate dehydrogenase, and phosphoglycerate kinase, respectively) were detected by image analysis of silver nitrate-colored 2D gels in the dry mature seeds and shown to retain constant accumulation levels up to radicle protrusion (Gallardo et al., 2001, 2002a). Here, the fact that these protein spots could be labeled by [35S]Met (Fig. 6A; Table III) indicates the occurrence of protein turnover during germination sensu stricto. This finding therefore reveals the existence of regulatory mechanisms to maintain constant the accumulation levels of such enzymes during the germination process, necessary for proper functioning of metabolic pathways. More generally, these data illustrate the power of combining classical proteomics with de novo protein biosynthesis in the interpretation of protein accumulation patterns.
Figure 7.
Comparison of de novo protein synthesis patterns during 24-h germination of the tt2-1 Arabidopsis mutant in the presence or absence of α-amanitin. The data are from Figure 6. Seeds were incubated for 24 h in the presence of [35S]Met, in the absence or presence of 500 μm α-amanitin. Proteins were extracted, submitted to 2D gel electrophoresis, and the radiolabeled proteins revealed as described in “Materials and Methods.” A, Superimposition of the two labeled protein patterns shown in Figure 6A and B. The data are represented in false colors: Red, incubation in water (Fig. 6A); Green, incubation in α-amanitin (Fig. 6B). The indicated portions of the gel, a, b, c, and d, are reproduced in B. B (right; 35S-labeling), Enlarged windows a, b, c, and d of 2D gels as shown in A for tt2-1 dry mature seeds incubated in water for 24 h (red spots) or in the presence of 500 μm α-amanitin (green spots). B (left; 2D gel), Same enlarged windows (a–d) of a 2D gel of proteins extracted from tt2-1 dry mature seeds incubated in water for 24 h and revealed by silver nitrate coloration. The labeled protein spots were identified by comparison with Arabidopsis seed protein reference maps (Gallardo et al., 2001, 2002a, http://seed.proteome.free.fr). They are listed in Tables III and IV. The following spots are shown in the four selected windows (protein no., protein name). Window (a): 103, enolase; 38, Leu aminopeptidase; 92, myrosinase-binding protein; 105, elongation factor1-γ2; 191, mitochondrial processing protease. Window (b): 127, elongation factor EF-2; 160, Met synthase; 13, MLO-like protein. Window (c): 116, P1 clone:MRA19, strong similarity to unknown protein T05029; 76 and 77, α-cruciferin 12S seed storage protein (fragments). Window (d): 82, 83, 84, 85 and 245, α-cruciferin 12S seed storage protein (fragments); 246, globulin seed storage protein precursor (fragment); 247, hydrolase.
Table III.
| No. | Exp. MM | Exp. PI | Arabidopsis Protein Name | Cov. | Theo. MM | Theo. PI | AGI No. | Relative Abundancec |
|---|---|---|---|---|---|---|---|---|
| kD | % | kD | Ama/H2O | |||||
| 114a | 39.56 | 5.87 | Cytosolic malate dehydrogenase | 21 | 35.57 | 6.11 | At1g04410 | 0.20 ± 0.01 |
| 17a | 72.98 | 6.57 | Phosphoenolpyruvate carboxykinase | 17 | 73.40 | 6.61 | At4g37870 | 0.16 ± 0.01 |
| 168a | 62.91 | 7.08 | Malate synthase | 10 | 63.89 | 8.02 | At5g03860 | 0.13 ± 0.00 |
| 40a | 38.55 | 6.30 | Cytosolic GAPDH | 26 | 36.91 | 6.62 | At3g04120 | 0.03 ± 0.00 |
| 39a | 38.52 | 6.27 | Cytosolic GAPDH | 34 | 36.91 | 6.62 | At3g04120 | 0.02 ± 0.00 |
| 59b | 67.31 | 5.61 | Succinate dehydrogenase | 16 | 69.66 | 5.86 | At2g18450 or At5g66760 | ≤0.01 |
| 103a | 57.95 | 5.59 | Enolase | 13 | 47.72 | 5.54 | At2g36530 | ≤0.01 |
| 121a | 40.92 | 5.45 | Cytosolic phosphoglycerate kinase | 16 | 42.13 | 5.49 | At1g79550 | ≤0.01 |
| 130b | 47.75 | 5.84 | Isocitrate dehydrogenase | 16 | 45.75 | 6.13 | At1g65930 | ≤0.01 |
| 160a | 82.27 | 5.94 | Met synthase | 12 | 84.36 | 6.09 | At5g17920 | ≤0.01 |
| 191a | 61.11 | 5.81 | Mitochondrial processing peptidase | 17 | 59.16 | 6.30 | At3g02090 | ≤0.01 |
| 192a | 60.17 | 5.88 | ATP synthase α-chain (mitochondrial) or expressed protein | 14 | 55.04 | 6.23 | AtMg01190 or At2g07698 | ≤0.01 |
| 201a | 70.29 | 5.11 | Vacuolar ATP synthase catalytic subunit A | 28 | 68.81 | 5.11 | At1g78900 | ≤0.01 |
| 296b | 94.30 | 5.32 | Pyruvate, orthophosphate dikinase | 5 | 98.24 | 5.55 | At4g15530 | ≤0.01 |
| 38b | 57.92 | 5.62 | Leucine aminopeptidase | 27 | 54.51 | 5.66 | At2g24200 | ≤0.01 |
| 128b | 47.45 | 5.45 | Elongation factor 1B-γ | 13 | 46.66 | 5.36 | At1g09640 | 0.18 ± 0.02 |
| 127a | 93.92 | 5.89 | Elongation factor EF-2 | 31 | 94.25 | 5.89 | At1g56070 | 0.07 ± 0.00 |
| 105a | 48.63 | 5.61 | Elongation factor 1-γ 2 | 38 | 46.40 | 5.55 | At1g57720 | ≤0.01 |
| 156b | 36.04 | 3.99 | Elongation factor 1B α-subunit | 15 | 24.20 | 4.42 | At5g19510 | ≤0.01 |
| 129a | 47.20 | 5.5 | Eukaryotic initiation factor 4A-1 | 28 | 46.70 | 5.47 | At3g13920 | ≤0.01 |
| 93a | 42.55 | 5.27 | Actin 7 | 20 | 41.73 | 5.31 | At5g09810 | 0.30 ± 0.02 |
| 92a | 53.10 | 5.59 | Myrosinase-binding protein | 30 | 51.21 | 5.50 | At2g33070 | 0.26 ± 0.02 |
| 149a | 64.96 | 5.18 | HSP60 | 15 | 61.28 | 5.66 | At3g23990 | 0.19 ± 0.02 |
| 91a | 61.88 | 5.06 | LEA protein-like | 13 | 52.08 | 5.29 | At3g53040 | 0.02 ± 0.00 |
| 13a | 73.27 | 6.32 | MLO-like protein | 15 | 67.23 | 9.55 | At1g61560 | ≤0.01 |
H2O, tt2-1 seeds incubated for 1 d in water and [35S]Met; Ama, α-amanitin, and tt2-1 seeds incubated for 1 d in 500 μm α-amanitin and [35S]Met as described in “Materials and Methods”; Cov., coverage; Exp., experimental; Theo., theoretical.
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a).
Listed proteins correspond to proteins identified during this work.
Data obtained from densitometric analysis of individual spots from 2D gels revealed by PhosphorImager analysis (see Fig. 6 for examples of in vivo protein synthesis of seeds labeled with [35S]Met): normalized spot volume in the tt2-1 mutant seeds incubated for 1 d in 500 μm α-amanitin divided by the normalized spot volume in the tt2-1 mutant seeds incubated for 1 d in water ±sd, from three different gels and independent extractions; ≤0.01 means that the accumulation level of the corresponding protein in seeds incubated for 1 d in the presence of 500 μm α-amanitin was close to background.
Another metabolic enzyme whose de novo synthesis was repressed is the cobalamin-independent Met synthase (Table III, spot no. 160), which corresponds to the last enzyme in the Met biosynthetic pathway (Ravanel et al., 1998). This metabolic pathway is of paramount importance for Arabidopsis seed germination and seedling growth since seed germination was shown to be strongly delayed in the presence of DL-propargylglycine, a specific inhibitor of Met synthesis, while this compound totally inhibited seedling growth (Gallardo et al., 2002b).
Besides these metabolic enzymes, five other protein spots (nos. 105, 127, 128, 129, and 156) for which de novo synthesis was strongly depressed by α-amanitin during germination (Fig. 6, Table III) corresponded to translation elongation and initiation factors. In agreement with the cycloheximide data in Figure 2, this finding points out the importance of the translational process for germination and seedling establishment in Arabidopsis. Such translational control of seed germination has also been exemplified in maize (Sánchez de Jiménez and Aguilar, 1984; Dinkova and Sánchez de Jiménez, 1999).
Finally, it is worth noting the inhibitory effect of α-amanitin on de novo accumulation of enzymes that play important roles in mitochondrial bioenergetics. The one is the mitochondrial processing peptidase (Table III, spot no. 191). This processing peptidase is a fascinating enzyme that catalyzes the specific cleavage of the diverse presequence peptides from hundreds of the nuclear-encoded mitochondrial precursor proteins that are synthesized in the cytosol and imported into the mitochondrion. Contrary to other sources, the plant enzyme is integrated into the mitochondrial cytochrome bc1 complex (also called complex III) of the respiratory chain, which renders the bc1 complex in plants bifunctional, being involved both in electron transport and in protein processing (Glaser and Dessi, 1999). Knowing the central role of mitochondria in seed germination and seedling establishment (Bewley and Black, 1994; Bewley et al., 2000), the possibility of controlling this important enzyme during germination would provide a simple and very efficient means to functionally control these essential organelles. Consistent with this hypothesis, developmental regulation of protein import in plant mitochondria has been documented (Murcha et al., 1999). Another control of the efficiency of mitochondria could occur at the level of succinate dehydrogenase (Table III, spot no. 59) owing to the role of this enzyme in mitochondrial bioenergetics. Succinate is a major substrate of seed mitochondria in a number of species (Bewley and Black, 1994), and a tuning of succinate dehydrogenase might exert a dramatic control upon energy and carbon metabolism, especially in lipid-storing seeds such as Arabidopsis.
Very surprisingly, Figure 6 also shows that in the presence of α-amanitin the seeds massively synthesized proteins of molecular mass smaller than 40 kD, which were barely detected from the radiolabeled proteome obtained in the absence of the fungal toxin (Figs. 6 and 7). However, it is worth noting that some of them could also be labeled, albeit to a lower extent, in the absence of α-amanitin (see, for example, spot nos. 76, 77, 83, 84, and 247 in Fig. 7B), thus reinforcing the good matching between the protein patterns revealed by coloration with silver nitrate (total proteome) and through the incorporation of radiolabeled Met into newly synthesized proteins (de novo proteome). Several of the protein spots whose de novo synthesis increased in the presence of α-amanitin could be assigned to known proteins from reference maps of Arabidopsis seed proteins (Gallardo et al., 2001, 2002a, 2002b). They are listed in Table IV. The majority of these proteins corresponded to proteins normally synthesized during seed development, such as 12S globulin subunits and members of the dehydrin family (group 2 late embryogenesis abundant [LEA] proteins; Finkelstein, 1993; Bewley and Black, 1994; Cuming, 1999). In other words, it seemed that transcriptional inhibition by α-amanitin somehow allowed the germinating seeds to recapitulate part of the maturation program, by favoring the use of stored mRNAs encoding maturation proteins. This finding is not without precedent. Thus, Lopez-Molina et al. (2002) showed that Arabidopsis seed germination comprises a short window of time after the start of imbibition during which the seed can still recruit late maturation programs (e.g. the expression of genes encoding maturation proteins such as the LEA proteins can be reinduced) in order to remain osmo-tolerant. This abscisic acid (ABA)-mediated developmental check point, which requires the bZIP transcription factor ABI5, has physiological significance since control of this developmental transition to auxotrophic growth may enable seeds to monitor environmental water status and to mount appropriate adaptive responses (Lopez-Molina et al., 2002). This behavior is consistent with the demonstration that lea gene expression can be reinduced by ABA, albeit at low level, in 8-d-old Arabidopsis seedlings (Finkelstein, 1993). Also, pkl mutants of Arabidopsis, which carry mutations in the PICKLE (PKL) gene encoding an essential chromatin remodeling protein, retain during germination and seedling growth characteristics of embryonic tissue (Ogas et al., 1997; Dean Rider et al., 2003). Thus, PKL is a component of a developmental switch that functions during germination to prevent reexpression of the embryonic developmental stage. It must be stressed, however, that our data showing an apparent reinduction of the embryonic maturation program during germination were obtained in the presence of α-amanitin. This situation therefore does not favor mechanisms in which seed maturation proteins accumulated through reexpression of their genes. Rather our data indicate that the fungal toxin repressed the synthesis of factor(s) preventing the use of some of the stored mRNAs during germination. In maize axes, different types of translation initiation factors differentially accumulate during germination (Dinkova and Sánchez de Jiménez, 1999; Dinkova et al., 2000, 2003), accounting at least in part for the observation that stored mRNAs are selectively translated during germination (Sánchez de Jiménez and Aguilar, 1984).
Table IV.
| No. | Exp. MM | Exp. PI | Arabidopsis Protein Name | Cov. | Theo. MM | Theo. PI | AGI No. | Relative Abundanced |
|---|---|---|---|---|---|---|---|---|
| kD | % | kD | Ama/H2O | |||||
| 69a | 43.67 | 6.29 | 12S seed storage protein precursor | 18 | 50.56 | 6.53 | At1g03880 | ≥100 |
| 87a | 18.43 | 6.36 | β-Cruciferin 12S seed storage protein | 33 | 20.72 | 6.36 | At1g03880 | 34.5 ± 0.22 |
| 141a | 29.35 | 7.42 | α-Cruciferin 12S seed storage protein | 21 | 29.23 | 6.49 | At5g44120 | 31.4 ± 0.58 |
| 214b | 27.14 | 6.10 | α-Cruciferin 12S seed storage protein | 23 | 29.86 | 6.60 | At1g03880 | 18.6 ± 0.28 |
| 82a | 33.89 | 6.24 | α-Cruciferin 12S seed storage protein (fragment) | 42 | 34.68 | 6.42 | At4g28520 | 18.2 ± 0.62 |
| 246b | 32.75 | 6.54 | Globulin seed storage protein precursor (fragment) | 14 | 55.06 | 6.64 | At3g22640 | 10.7 ± 0.51 |
| 85a | 27.20 | 6.50 | α-Cruciferin 12S seed storage protein | 32 | 29.86 | 6.60 | At1g03880 | 8.7 ± 0.31 |
| 245b | 29.95 | 6.53 | α-Cruciferin 12S seed storage protein | 16 | 29.86 | 6.60 | At1g03880 | 4.1 ± 0.49 |
| 76a | 23.94 | 5.58 | α-Cruciferin 12S seed storage protein (fragment) | 27 | 34.68 | 6.42 | At4g28520 | 2.7 ± 0.51 |
| 84a | 30.46 | 6.61 | α-Cruciferin 12S seed storage protein | 42 | 29.23 | 6.49 | At5g44120 | 2.7 ± 0.02 |
| 25a | 23.32 | 6.89 | β-Cruciferin 12S seed storage protein (fragment) | 43 | 21.20 | 6.19 | At4g28520 | 2.6 ± 0.21 |
| 77a | 25.42 | 5.78 | α-Cruciferin 12S seed storage protein (fragment) | 30 | 34.68 | 6.42 | At4g28520 | 2.4 ± 0.33 |
| 80a | 32.64 | 5.85 | α-Cruciferin 12S seed storage protein (fragment) | 33 | 34.68 | 6.42 | At4g28520 | 2.0 ± 0.49 |
| 83a | 34.35 | 6.42 | α-Cruciferin 12S seed storage protein | 33 | 34.68 | 6.42 | At4g28520 | 2.0 ± 0.15 |
| 88a | 22.82 | 8.68 | α-Cruciferin 12S seed storage protein (fragment) | 44 | 34.68 | 6.42 | At4g28520 | 2.0 ± 0.13 |
| 255b | 21.65 | 5.18 | Dehydrin RAB18-related protein | Seqc | 18.46 | 7.10 | At5g66400 | 2.4 ± 0.19 |
| 254b | 22.10 | 4.96 | Dehydrin RAB18-related protein | Seqc | 18.46 | 7.10 | At5g66400 | 2.0 ± 0.06 |
| 190a | 101.60 | 5.82 | HSP101 | 13 | 101.29 | 5.81 | At1g74310 | ≥100 |
| 137a | 76.06 | 5.07 | HSP70 | 8 | 71.10 | 5.14 | At3g12580 | 2.0 ± 0.11 |
| 34b | 13.32 | 6.02 | Major latex protein | 37 | 18.06 | 6.88 | At1g14950 | ≥100 |
| 99a | 13.32 | 6.28 | Major latex protein | 59 | 18.06 | 6.88 | At1g14950 | 21.0 ± 0.47 |
| 37a | 40.29 | 6.49 | Cytosolic GAPDH | 26 | 36.91 | 6.62 | At3g04120 | ≥100 |
| 79a | 27.24 | 5.20 | F4I1.20 protein | 26 | 23.67 | 4.86 | At2g44390 | ≥100 |
| 96a | 64.62 | 6.08 | β-Glucosidase | 15 | 60.01 | 6.20 | At3g21370 | ≥100 |
| 116a | 30.24 | 5.77 | P1 clone:MRA19, Strong similarity to unknown protein T05029 | 33 | 28.78 | 5.92 | At5g45690 | 74.1 ± 0.28 |
| 144a | 15.87 | 6.82 | Peptidyl-prolyl cis-trans isomerase | 48 | 18.37 | 7.68 | At4g38740 | 6.8 ± 0.48 |
| 174b | 38.70 | 5.73 | Cys synthase or O-acetylserine-thiol-lyase | 9 | 33.80 | 5.90 | At4g14880 | 6.1 ± 0.30 |
| 175a | 37.85 | 5.71 | Cys synthase or O-acetylserine-thiol-lyase | 30 | 33.80 | 5.90 | At4g14880 | ≥100 |
| 247b | 34.90 | 6.56 | Hydrolase | 19 | 37.64 | 8.90 | At4g39955 | 2.8 ± 0.26 |
| 264b | 18.70 | 6.05 | β-Cruciferin 12S seed storage protein | 29 | 20.72 | 6.36 | At1g03880 | 11.6 ± 0.44 |
H2O, tt2-1 seeds incubated for 1 d in water; Ama, α-amanitin, and tt2-1 seeds incubated for 1 d in 500 μm α-amanitin as described in “Materials and Methods”; Cov., coverage; Exp., experimental; Seq, protein identified by Edman sequencing; Theo., theoretical.
Listed proteins correspond to previously identified proteins (Gallardo et al., 2001, 2002a).
Listed proteins correspond to proteins identified during this work.
From Edman sequencing, the sequence QNRPGGQATE was found for both protein nos. 254 and 255; BLAST analysis showed that this sequence is present in the Arabidopsis dehydrin related RAB18 protein At5g66400. Note that although these two proteins correspond to abundant proteins in 2D gels colored with the GelCode blue stain reagent (see “Materials and Methods”), they are poorly stained by silver nitrate.
Data obtained from densitometric analysis of individual spots from 2D gels revealed by PhosphorImager analysis (see Fig. 6 for examples of in vivo protein synthesis of seeds labeled with [35S]Met): normalized spot volume in the tt2-1 mutant seeds incubated for 1 d in 500 μm α-amanitin divided by the normalized spot volume in the tt2-1 mutant seeds incubated for 1 d in water ±sd, from three different gels and independent extractions; ≥100 means that the accumulation level of the corresponding protein in seeds incubated 1 d on water was close to background.
CONCLUSIONS
In summary, the use of α-amanitin has allowed a better characterization of the distinct roles played by the stored and neosynthesized mRNA pools in Arabidopsis seed germination and plantlet growth. We show that in this species seed germination, as monitored by radicle protrusion through the seed coat, can occur even in the presence of an excess of the transcriptional inhibitor. Under the same experimental conditions seedling establishment was totally blocked, showing that beyond radicle protrusion, further development requires the participation of de novo transcription. In contrast, seed germination was totally blocked by the translational inhibitor cycloheximide. Taken together, these results enlighten the importance of protein synthesis from the pool of stored mRNAs for radicle protrusion. The present data would suggest that commitment to seedling growth occurs after radicle protrusion. However, this issue might be complicated by the fact that under the nonphysiological conditions presently used (germination in the presence of α-amanitin) the usual sequence of events (exit from phase 2 of germination, loss of desiccation tolerance, and commitment to seedling growth) is distorted. Further experiments are required to characterize better the different developmental phases in germination.
The present data document that proteins and mRNAs stored in the dry mature seeds are sufficient for germination sensu stricto as well as starting of root growth, thus showing that the potential for seed germination is largely programmed during the seed maturation phase. In agreement with this proposal, transcriptional activity appeared to be very weak during the first 16 h of germination (Fig. 3).
Compared to normal germination, germination in the presence of α-amanitin exhibited four specific features. First, the germination process was strongly delayed and slowed down, pointing out the importance of regulators of the rate of seed germination that are under transcriptional control, none of which have been characterized at present. Second, the sensitivity of seed germination to GAs was reduced 15-fold, confirming the role of this phytohormone in seed germination. Third, de novo synthesis of several enzymes involved in reserve mobilization and resumption of metabolic activity was strongly repressed, which can be related to the germination and seedling establishment defects described for Arabidopsis mutants altered in storage lipids mobilization (Hayashi et al., 1998; Eastmond et al., 2000; Eastmond and Graham, 2001). Also in agreement with the importance of metabolic control of seed germination and seedling growth is the fact that α-amanitin prevented de novo accumulation of housekeeping enzymes, such as Met synthase and essential mitochondrial enzymes. Fourth, germinating seeds seemed to recapitulate at least part of the maturation program of seed development. This peculiar feature has recently been shown to naturally occur in Arabidopsis, with the demonstration that lea gene expression can be re-induced during early stages of seed germination (Lopez-Molina et al., 2002). Our results reveal a different mechanism, involving the use of mRNAs stored during maturation. If such a use of stored mRNAs encoding maturation proteins also occurred during normal germination, particularly under unfavorable germination conditions, it might have physiological implication, because, as stressed by Lopez-Molina et al. (2002), it would allow germinating seeds to mount adaptive responses to environmental water stress. That stored mRNAs encoding maturation proteins (e.g. cruciferin subunits) can be translated during normal germination is indicated by the data in Figure 7B showing that common protein spots are detected in the novo protein patterns obtained in the absence or presence of α-amanitin. This last finding also raises an old, yet hitherto unanswered, question (Bewley and Black, 1994) that commitment to germination and plant growth requires transcription of genes allowing the imbibed seed to discriminate between mRNAs to be utilized in germination and those to be destroyed. Microarray analyses of seed germination in the absence or presence of α-amanitin will allow better characterization of the fate of these mRNA pools during germination.
MATERIALS AND METHODS
Plant Material and Germination Experiments
Nondormant seeds of Arabidopsis, ecotype Landsberg erecta (Ler), are referred to as wild-type seeds in this work. The isolation of the transparent testa (tt) mutants in the Ler and Wassilewskija (Ws) backgrounds was described by Koornneef and van der Veen (1980), Koornneef (1990), and Debeaujon et al. (2001), and their physiological properties were analyzed by Debeaujon et al. (2000) and Debeaujon and Koornneef (2000). Seeds of mutants tt2-1, tt5-1, ttg1-1 (Ler background), and tt12-1 (Ws background) have been used here.
Germination assays were carried out on three replicates of 75 seeds and independent experiments. Seeds were incubated at 25°C, with 8 h light daily, on three sheets of absorbent paper (Roundfilter paper circles, Ø 45 mm, Schleicher & Schuell, Dassel, Germany) and a black membrane filter with a white grid (ME 25/31, Ø 45 mm, Schleicher & Schuell) wetted with 1.3 mL of distilled water, in covered plastic boxes (Ø 50 mm). Assays were carried out in the presence or absence of various concentrations of α-amanitin (Sigma, St Quentin Fallavier, France), and/or 100 μm cycloheximide (Sigma), and/or 100 μm PAC (Greyhound Chromatography and Allied Chemicals, Birkenhead, Merseyside, UK), and/or 100 μm GA4+7 (Plant Protection, Fernhurst, UK). A seed was regarded as germinated when the radicle protruded through the seed coat. The Seed Calculator software (Plant Research International B.V., Wageningen, The Netherlands) was used in curve-fitting analyses to estimate the germination parameters from the germination curves. The following values were calculated: T1, T10, T50, and T90, corresponding to the time to reach 1%, 10%, 50%, or 90% germination, respectively, and Gmax corresponding to the maximal germination frequency, along with their sd values.
Synthesis of Radiolabeled RNA
Labeled RNA was synthesized in vivo by tt2-1 seeds imbibed on water as above in the presence of [α-33P]-UTP (1.48 MBq; ICN Biomedicals, S.A.R.L., Orsay, France) and in the presence or absence of 500 μm of α-amanitin. Following incubation for various times, total RNA was extracted according to the protocol of Vicient and Delseny (1999). Then, RNA synthesis was measured by TCA precipitation of aliquots of reaction mixtures spotted on Whatmann GF/C filters; after eight washing steps in cold 5% TCA and 0.04 m sodium pyrophosphate and two washing steps in absolute methanol, filters were dried and counted for radioactivity in a liquid scintillation counter (Dietrich et al., 1985).
Preparation of Total Protein Extracts
Total protein extracts were prepared from dry mature seeds, and from seeds at different stages of germination. Following grinding of seeds using mortar and pestle (150 mg representing approximately 6,500 tt2-1 seeds) in liquid nitrogen, total proteins were extracted at 2°C in 1.2 mL of thiourea/urea lysis buffer (Harder et al., 1999) containing 7 m urea (Amersham Biosciences, Orsay, France), 2 m thiourea (Merck, Lyon, France), 4% (w/v) CHAPS (Amersham Biosciences), and 1% (v/v) Pharmalyte pH 3 to 10 carrier ampholytes (Amersham Biosciences). This extraction buffer also contained 18 mm Tris-HCl (Trizma HCl; Sigma); 14 mm Trizma base (Sigma); the protease inhibitor cocktail, complete Mini from Roche Diagnostics (Mannheim, Germany); 53 units/mL DNAse I (Roche Diagnostics); 4.9 Kunitz units/mL RNAse A (Sigma); and 0.2% (v/v) Triton X-100. After 10 min at 4°C, 14 mm dithiothreitol (DTT; Amersham Biosciences) was added and the protein extracts were stirred for 20 min at 4°C, then centrifuged (35,000g, 10 min) at 4°C. The supernatant was submitted to a second clarifying centrifugation as above. The final supernatant corresponded to the total protein extract. Protein concentrations in the various extracts were measured according to Bradford (1976). Bovine serum albumin was used as a standard.
Two-Dimensional Electrophoresis
Proteins were first separated by electrophoresis according to charge. Isoelectric focusing was carried out with protein samples with an equivalent to an extract of 110 seeds, corresponding to about 200 μg protein for all samples. Proteins from the various extracts were separated using gel strips forming an immobilized nonlinear pH gradient from 3 to 10 (Immobiline DryStrip pH 3–10 NL, 18 cm; Amersham Biosciences). Strips were rehydrated for 14 h at 22°C with the thiourea/urea lysis buffer containing 2% (v/v) Triton X-100, 20 mm DTT, and the protein extracts. Isoelectric focusing was performed at 22°C in the Multiphor II system (Amersham Biosciences) for 1 h at 300 V and 7 h at 3,500 V. Proteins were then separated according to size. Prior to the second dimension, the gel strips were equilibrated for 2 × 20 min in 2 × 100 mL equilibration solution containing 6 m urea, 30% (v/v) glycerol, 2.5% (w/v) SDS, 0.15 m BisTris, and 0.1 m HCl (Görg et al., 1987; Harder et al., 1999). DTT (50 mm) was added to the first equilibration solution, and iodoacetamide [4% (w/v)] to the second (Harder et al., 1999). Equilibrated gel strips were placed on top of vertical polyacrylamide gels [10% (v/v) acrylamide, 0.33% (w/v) piperazine diacrylamide, 0.18 m Trizma base, 0.166 m HCl, 0.07% (w/v) ammonium persulfate, 0.035% (v/v) Temed]. A denaturing solution [1% (w/v) low-melting agarose (Gibco BRL), 0.4% (w/v) SDS, 0.15 m BisTris, and 0.1 m HCl] was loaded on gel strips. After agarose solidification, electrophoresis was performed at 10°C in a buffer (pH 8.3) containing 25 mm Trizma base, 200 mm taurine, and 0.1% (w/v) SDS, for 1 h at 35 V and 14 h at 110 V. Ten gels (200 × 250 × 1.0 mm) were run in parallel (Isodalt system from Amersham Biosciences). For each condition analyzed, 2D gels were made in triplicate and from two independent protein extractions.
Protein Staining and Analysis of 2D Gels
Gels were stained with either silver nitrate according to a modified procedure of Blum et al. (1987) or the GelCode blue stain reagent from Pierce (Rockford, IL), using the Hoefer Automated Gel Stainer apparatus from Amersham Biosciences. Silver-stained gels were scanned with the Sharp JX-330 scanner equipped with the Labscan version 3.00 from Amersham Biosciences. Image analysis was carried out with the ImageMaster 2-D Elite version 3.01 software (Amersham Biosciences), according to the instruction booklet ImageMaster 2D Elite from Amersham Biosciences. After spot detection and background substraction (mode: average on boundary), 2D gels were aligned, matched, and the quantitative determination of the spot volumes was performed (mode: total spot volume normalization). For each analysis, statistical data showed a high level of reproducibility between normalized spot volumes of gels produced in triplicate from the two independent protein extractions.
Protein Identification by Mass Spectrometry and Edman Sequencing
Some of the new proteins characterized in this work were identified by MALDI-TOF analysis. Spots of interest were excised from GelCode-stained 2D gels and digested by sequence grade trypsin (Promega Biotec, Madison, WI). After digestion, the supernatant containing peptides was concentrated by batch adsorption on POROS 50 R2 beads (Roche Molecular Biochemicals, Basel), and used for MALDI-mass spectrometry analysis on a Bruker Reflex II MALDI-TOF spectrometer after on-target desorption with matrix solution (Gevaert et al., 1998). Before each analysis, the instrument was externally calibrated using two synthetic peptides spotted as near as possible to the biological sample. Proteins were identified by peptide mass fingerprinting. As far as possible, individual peptide ions were selected and subjected to postsource decay analysis, using the reflectron made of the instrument. This fragmentation information yielded further confirmation to the protein's identity. Searches were carried out with the SwissProt and NRDB sequence databases using MASCOT (http://www.matrixscience.com). Theoretical masses and pIs of identified proteins were predicted by entering the sequence at http://www.expasy.org/tools/peptide-mass.html. To denote a protein as unambiguously identified, the following criteria were used: coverage of the protein by the matching peptides must reach a minimum of 10%, and at least four independent peptides should match within a stringent 10-ppm maximum deviation of mass accuracy.
The search for sequence homology was carried out at http://www.arabidopsis.org/Blast.
De Novo Protein Synthesis
Labeled proteins were synthesized in vivo by tt2-1 seeds imbibed on water for 24 h as above in the presence of [35S]Met (1.85 MBq; ICN Biomedicals, S.A.R.L.) and in the presence or absence of 500 μm of α-amanitin. Following incubation, protein extracts were prepared and submitted to 2D gel electrophoresis as described above. Proteins on the 2D gels were colored by silver nitrate (see above). Then, colored 2D gels were dried for 1 week at room temperature in a sandwich composed of, from bottom to top, one sheet of cellophane model Gel Dryer (Bio-Rad, Marnes la Coquette, France), 2D gel, one sheet of Saran wrap (VWR international SAS, Strasbourg, France), and one sheet of cellophane model Gel Dryer (Bio-Rad). After drying, the upper sheet of cellophane and the Saran sheet were peeled and gels were submitted to PhosphorImager analysis (Molecular Dynamics Storm 840 phosphorimager, Amersham Biosciences). Labeled 2D protein patterns were scanned as described above for the silver-nitrate colored gels. Proteins of interest were identified by MALDI-TOF analysis or by comparison with the reference protein maps for Arabidopsis seed proteome available at http://seed.proteome.free.fr. MALDI-TOF analysis failed to identify spot nos. 254 and 255 (Table IV). Here, further identification was carried out by amino acid sequencing performed by Dr. Jacques d'Alayer (Institut Pasteur, Paris) by automated Edman degradation of the peptides with a PE Applied Biosystem sequencer. By this technique, the two spots were found to correspond to dehydrin RAB18 (see Table IV).
Acknowledgments
We thank Magda Puype and Hans Demol for assistance in the protein identification procedures.
The PhD thesis of L.R. is supported by Bayer CropScience and the French Ministry of Industry. J.V. was supported by the Concerted Research Actions of the Flemish Community (Belgium).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036293.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bewley JD (1997) Seed germination and plant dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Black M (1994) Seeds. Physiology of Development and Germination. Plenum Press, New York
- Bewley D, Hempel FD, McCormick S, Zambryski P (2000) Reproductive development. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry & Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 988–1043
- Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93–99 [Google Scholar]
- Bove J, Jullien M, Grappin P (2001) Functional genomics in the study of seed germination. Genome Biol 3: 1002.1–1002.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bradford KJ, Chen F, Cooley MB, Dahal P, Downie B, Fukunaga KK, Gee OH, Gurusinghe S, Mella RA, Nonogaki H, et al. (2000) Gene expression prior to radicle emergence in imbibed tomato seeds. In M Black, KJ Bradford, J Vázquez-Ramos, eds, Seed biology: advances and applications. CAB International, Wallingford, UK, pp 231–251
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR (2003) Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol 131: 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell DA, Cramer P, Kornberg RD (2002) Structural basis of transcription: α-amanitin-RNA polymerase II cocrystal at 2.8 Å resolution. Proc Natl Acad Sci USA 99: 1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrispeels M, Varner JE (1966) Inhibition of gibberellic acid induced formation of α-amylase by abscisin II. Nature 212: 1066–1067 [Google Scholar]
- Cuming AC (1999) LEA proteins. In PR Shewry, R Casey, eds, Seed Proteins. Kluwer Academic Press, Dordrecht, The Netherlands, pp 753–780
- Davies PJ (1995) Plant Hormones: Physiology, Biochemistry and Molecular Biology. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Dean Rider S, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J (2003) Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J 35: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Koornneef M (2000) Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol 122: 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, Koornneef M (2001) The TRANSPARENT TESTA 12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13: 853–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mercoyrol L, Job C, Job D (1989) Studies on the inhibition by alpha-amanitin of single-step addition reactions and productive RNA synthesis catalysed by wheat germ RNA polymerase II. Biochem J 258: 165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich J, Teissère M, Job C, Job D (1985) Poly(dAT) dependent trinucleotide synthesis catalysed by wheat-germ RNA polymerase II. Effects of nucleotide substrates and cordycepin triphosphate. Nucleic Acids Res 13: 6155–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Jung H-S, Sun T-P (2001) The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova TD, Aguilar R, Sánchez de Jiménez E (2000) Expression of maize initiation factor (eIF) iso4E is regulated at translational level. Biochem J 351: 825–831 [PMC free article] [PubMed] [Google Scholar]
- Dinkova TD, Aguilar R, Sánchez de Jiménez E (2003) Translational control by differential CAP-dependency in selected sub-populations of maize-stored mRNAs. In G Nicolás, KJ Bradford, D Côme, HW Pritchard, eds, The Biology of Seeds: Recent Research Advances. CAB International, Wallingford, UK, pp 181–189
- Dinkova TD, Sánchez de Jiménez E (1999) Differential expression and regulation of translation initiation factors −4E and −iso4E during maize germination. Physiol Plant 107: 419–425 [Google Scholar]
- Duque P, Chua NH (2003) IMB1, a bromodomain protein induced during seed imbibition, regulates ABA- and phyA-mediated responses of germination in Arabidopsis. Plant J 35: 787–799 [DOI] [PubMed] [Google Scholar]
- Dure LS III, Waters LC (1965) Long-lived messenger RNA: evidence from cotton seed germination. Science 147: 410–412 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA (2000) Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA 97: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Graham IA (2001) Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci 6: 72–77 [DOI] [PubMed] [Google Scholar]
- Errampalli D, Patton D, Castle L, Mickelson L, Hansen K, Schnall J, Feldmann K, Meinke D (1991) Embryonic lethals and T-DNA insertional mutagenesis in Arabidopsis. Plant Cell 3: 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando DD, Owens JN, Yu X, Ekramoddoullah AKM (2001) RNA and protein synthesis during in vitro pollen germination and tube elongation in Pinus monticola and other conifers. Sex Plant Reprod 13: 259–264 [Google Scholar]
- Finkelstein RR (1993) Abscisic acid-insensitive mutations provide evidence for stage-specific signal pathways regulating expression of an Arabidopsis late embryogenesis-abundant (lea) gene. Mol Gen Genet 238: 401–408 [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y (2000) Repression of SHOOT GROWTH, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol 126: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2002. a) Proteomics analysis of Arabidopsis seed germination. A comparative study of wild-type and GA-deficient seeds. Plant Physiol 129: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2002. b) Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol Plant 116: 238–247 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Le Signor C, Vandekerckhove J, Thompson R, Bustin J (2003) Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol 133: 664–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert K, Demol H, Sklyarova T, Vandekerckhove J, Houthaeve T (1998) A peptide concentration and purification method for protein characterization in the subpicomole range using matrix assisted laser desorption/ionisation-postsource decay (MALDI-MS) sequencing. Electrophoresis 19: 909–917 [DOI] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser E, Dessi P (1999) Integration of the mitochondrial-processing peptidase into the cytochrome bc1 complex in plants. J Bioenerg Biomembr 31: 259–274 [DOI] [PubMed] [Google Scholar]
- Görg A, Postel W, Weser J, Günther S, Strahler JR, Hanash SM, Somerlot L (1987) Elimination of point streaking on silver stained two-dimensional gels by addition of iodoacetamide to the equilibration buffer. Electrophoresis 8: 122–124 [Google Scholar]
- Groot SPC, Karssen CM (1987) Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta 171: 525–531 [DOI] [PubMed] [Google Scholar]
- Gualberti G, Papi M, Belluci L, Ricci I, Bouchez D, Camilleri C, Costantino P, Vittorioso P (2002) Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14: 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Jendrisak JJ (1978) Plant DNA-dependent RNA polymerases: subunit structures and enzymatic properties of the class II enzymes from quiescent and proliferating tissues. Biochemistry 17: 1860–1866 [DOI] [PubMed] [Google Scholar]
- Harder A, Wildgruber R, Nawrocki A, Fey SJ, Larsen PM, Görg A (1999) Comparison of yeast cell protein solubilization procedures for two-dimensional electrophoresis. Electrophoresis 20: 826–829 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M (1998) 2,4-dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Matsuoka M, Steber CM (2003) A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends Plant Sci 8: 492–497 [DOI] [PubMed] [Google Scholar]
- Jabrin S, Ravanel S, Gambonnet B, Douce R, Rebeille F (2003) One-carbon metabolism in plants. Regulation of tetrahydrofolate synthesis during germination and seedling development. Plant Physiol 131: 1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrisak JJ (1980) The use of α-amanitin to inhibit in vivo RNA synthesis and germination in wheat embryos. J Biol Chem 255: 8529–8533 [PubMed] [Google Scholar]
- Karssen CM, Zagorski S, Kepczynski J, Groot SPC (1989) Key role for endogenous gibberellins in the control of seed germination. Ann Bot (Lond) 63: 71–80 [Google Scholar]
- Koornneef M (1990) Mutations affecting the testa colour in Arabidopsis. Arabidopsis Inf Serv 27: 1–4 [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 51: 33–36 [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang H, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua N-H (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW (1996) Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha MW, Huang T, Whelan J (1999) Import of precursor proteins into mitochondria from soybean tissues during development. FEBS Lett 464: 53–59 [DOI] [PubMed] [Google Scholar]
- Nambara E, Akazawa T, McCourt P (1991) Effects of the gibberellin biosynthesis inhibitor uniconazol on mutants of Arabidopsis. Plant Physiol 97: 736–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Cheng J-C, Sug R, Somerville C (1997) Cellular differentiation regulated by gibberellins in the Arabidopsis thaliana pickle mutant. Science 277: 91–94 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kumiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun T-P, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherov N, May G (2000) Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 155: 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Harberd NP (2002) The role of GA-mediated signaling in the control of seed germination. Curr Opin Plant Biol 5: 376–381 [DOI] [PubMed] [Google Scholar]
- Potokina E, Srevinasulu N, Altschmied L, Michalek W, Graner A (2002) Differential gene expression during seed germination in barley (Hordeum vulgare L.). Funct Integr Genomics 2: 28–39 [DOI] [PubMed] [Google Scholar]
- Raghavan B (2000) Seed germination. In Developmental Biology of Flowering Plants. Springer-Verlag, New York, pp 7–24
- Ravanel S, Gakière B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95: 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez de Jiménez E, Aguilar R (1984) Protein synthesis patterns: relevance of old and new messenger RNA in germinating maize embryos. Plant Physiol 75: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun TP (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Martinez EC, Sun TP (1997) The RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Koornneef M (2002) A fortunate choice: the history of Arabidopsis as a model plant. Nat Rev Genet 3: 883–889 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D (2003) The Arabidopsis SeedGenes Project. Nucleic Acids Res 31: 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen J-G, Wang S, Jones AM (2002) Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol 129: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Geest AHM (2002) Seed genomics: germinating opportunities. Seed Sci Res 12: 145–153 [Google Scholar]
- Vicient CM, Delseny M (1999) Isolation of total RNA from Arabidopsis thaliana seeds. Anal Biochem 268: 412–413 [DOI] [PubMed] [Google Scholar]
- Waters LC, Dure LS III (1966) Ribonucleic acid synthesis in germinating cotton seeds. J Mol Biol 19: 1–27 [DOI] [PubMed] [Google Scholar]
- Wen C-K, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JH, Kim Y-B, Ren P-H, Cai N, Cho M-J, Hedden P, Lemaux PG, Buchanan BB (2002) Transgenic barley overexpressing thioredoxin shows evidence that the starchy endosperm communicates with the embryo and the aleurone. Proc Natl Acad Sci USA 99: 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano H, Kuroda S, Buchanan BB (2002) Disulfide proteome in the analysis of protein function and structure. Proteomics 2: 1090–1096 [DOI] [PubMed] [Google Scholar]