Key Points
AML cells resistant to cytarabine are more susceptible to NK-mediated cell lysis.
c-Myc regulates ULBP1/2/3 expression and interferes with NK cell susceptibility in primary cytarabine resistant AML blasts.
Abstract
Cytarabine (cytosine arabinoside) is one of the most effective drugs for the treatment of patients diagnosed with acute myeloid leukemia (AML). Despite its efficiency against AML cells, the emergence of drug resistance due to prolonged chemotherapy in most patients is still a major obstacle. Several studies have shown that drug resistance mechanisms alter the sensitivity of leukemia cells to immune system effector cells. To investigate this phenomenon, parental acute myeloid cell lines, HL-60 and KG-1, were continuously exposed to increasing doses of cytarabine in order to establish equivalent resistant cell lines, HL-60(R) and KG-1(R). Our data indicate that cytarabine-resistant cells are more susceptible to natural killer (NK)-mediated cell lysis as compared with parental cytarabine-sensitive cells. The increased susceptibility correlates with the induction of UL-16 binding proteins (ULBP) 1/2/3 and NK group 2, member D (NKG2D) ligands on target cells by a mechanism involving c-Myc induction. More importantly, chromatin immunoprecipitation assay revealed that ULBP1/3 are direct targets of c-Myc. Using drug-resistant primary AML blasts as target cells, inhibition of c-Myc resulted in decreased expression of NKG2D ligands and the subsequent impairment of NK cell lysis. This study provides for the first time, the c-Myc dependent regulation of NKG2D ligands in AML.
Introduction
Acute myeloid leukemia (AML) is a hematologic malignancy characterized by proliferation of malignant precursors of the myeloid lineage coupled with impaired differentiation of normal hematopoietic progenitors.1 Chemotherapy is the first line treatment against most leukemia disorders, and cytarabine (cytosine arabinoside) has been one of the most widely used chemotherapy agents against AML blasts for more than 30 years.2-6 Although cytarabine is an efficient antileukemic agent for AML and other leukemias,7 emergence of drug resistance due to prolonged chemotherapy in most patients is a major obstacle.8,9 Accumulating evidence indicates that the acquisition of drug resistance enhances the sensitivity of leukemic blasts to cytotoxic cells of the immune system. However, other reports indicate decreased susceptibility of leukemic cells to cytotoxic cells.10-18
Allogeneic bone marrow transplantation is the only curative treatment of many intermediate and high-risk leukemias. Recent studies suggest that immunotherapy may continue to be an effective approach for patients with leukemia,19-21 and emerging strategies are currently under investigation based on adoptive transfer of natural killer (NK) cells. NK cells are a component of an innate immune system that play important roles as first line-defenders in the host response to tumors and infections, as well as in transplant rejection and in the development of tolerance.22-27 Due to their strong ability to target tumor cells, NK cells have been described as promising effectors for adoptive immunotherapy of cancer.28 It is well established that NK cell activity is regulated by a balance between inhibitory and stimulatory signals that are transmitted by cell-surface receptors after interaction with their respective ligands on target cells.29,30 NK group 2, member D (NKG2D) is one of the activating receptors expressed by NK cells, γ/δ T cells, and activated CD8+ T cells in humans.31-33 Several ligands for this receptor have been identified in humans, including major histocompatibility complex (MHC) class I-related chain A (MICA), MICB, and UL16-binding proteins (ULBP) 1/2/3/4/5. These ligands are abundantly expressed by tumor cells, rendering these cells susceptible to NK-cell–mediated cytotoxicity.32,34-36
While the functional role of NKG2D is well established,37 the regulation of its ligands (NKG2DL) remains only partially understood. Various molecular pathways, including extracellular signal-regulated kinase (ERK), AKT, p53, and signal transducer and activator of transcription 3 have been reported to play a regulatory, both at the transcriptional or posttranscriptional level.38-48
In this study, we investigated the molecular basis of cytarabine resistance in AML cells. We found that these cells exhibited increased susceptibility to NK lysis that correlates with an increase in c-Myc induction and the subsequent upregulation of ULBPs. Therefore, this study reveals a new regulatory mechanism of ULBPs in AML involving the c-Myc pathway. This knowledge could help predict the efficacy and response to NK-cell–based therapy, and allow for better designing of NK-based immunotherapy.
Methods
Culture of cell lines and resistant cell line establishment
Human AML cell lines (KG-1 and HL-60) were grown in RPMI 1640 medium supplemented with 10% fetal calf serum (Seromed) and 1% penicillin-streptomycin. Human NK cell lines were grown in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% penicillin-streptomycin, and 300 IU/mL IL-2. Cytarabine-resistant KG-1 and HL-60 cell sub lines were established by exposing parental cells to increasing concentrations of the drug. All experiments were performed using cytarabine-resistant cells subcultured at 7 day intervals without further addition of the drug. Approval for these studies was obtained from the Gustave Roussy Cancer Campus Institutional Review Board. Informed consent was provided in accordance with the Declaration of Helsinki.
Antibodies (Abs), reagents, and inhibitors
Monoclonal antibodies (mAbs) directed against ULBP1/2/3 were purchased from R&D Systems. Anti-CD107a coupled to CyChrome mAb was provided by Becton Dickinson. mAbs-recognizing MICA, MICB, and MHC-I was obtained from BioLegend. Small-molecular-weight c-Myc inhibitor (10058-F4; [Z,E]-5-[4- ethylbenzylidine]-2-thioxothiazolidin-4-one) was purchased from Calbiochem (San Diego, CA). The ERK inhibitor (PD98059) was obtained from GIBCO (Invitrogen, Cergy Pontoise, France) and the AKT inhibitor (MK-2206) was from Selleck Chemicals.
Abs against c-Myc was purchased from Santa Cruz Biotechnology. Abs against total ERK, phospho-ERK (Thr202/Tyr204), total AKT, and phospho-AKT (ser-473) were from Cell Signaling Technology. Abs against β-actin was from Sigma-Aldrich.
RNA silencing
Specific c-Myc small interfering RNA (siRNA) (sc-29226) and negative control siRNA (sc-37007) were purchased from Santa Cruz Biotechnology. siRNA was transfected into HL-60 or KG-1 cells by electroporation (Amaxa, Gaithersburg, MD) following the manufacturer’s instructions. Briefly, 2 × 106 cells were electroporated in 100 μL nucleofector solution (Amaxa Reagent V for HL-60 cells and Amaxa Reagent R for KG-1 cells) containing 50 pmol of each siRNA using the preselected Amaxa program T-019 for HL-60 cells and Amaxa program V-001 for KG-1 cells. siRNA transfected cells were plated in a 6-well plate with 5 mL supplemented RPMI 1640 medium for 72 hours and harvested for further experiments.
RNA isolation and real-time quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from cell samples with TRIzol solution (Invitrogen). DNase I-treated 1 μg of total RNA was converted into complementary DNA by using TaqMan Reverse Transcription Reagent (Applied Biosystems), and messenger RNA (mRNA) levels were quantified by SYBR Green qPCR method (Applied Biosystems). Relative expression was calculated by using the comparative Ct method (2-ΔCt). Primer sequences are available upon request.
Determination of cell viability
Leukemia cells seeded onto flat-bottom 96-well plates were exposed to different concentrations of the drug for 48 hours. Viability of cell was measured with a modified 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay (Sigma-Aldrich, Saint-Quentin, France). The percentage of cell viability was calculated as follows: percentage of viability = (A1/A0) × 100, where A1 and A0 represent absorbance obtained, respectively, for treated and untreated cells.
Western blot
Tumor cells were washed in phosphate-buffered saline and lysed in plates with lysis buffer (62.5 mM Tris-HCl [pH 6.8], 2% weight/volume sodium dodecyl sulfate, 10% glycerol, 1 mM sodium orthovanadate, 2 mM phenylmethylsulfonyl fluoride, 25 mM leupeptin, 5 mM benzamidine, 1 mM pepstatin, and 25 mM aprotinin). Lysates were sonicated on ice, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (60 mg/lane), and transferred onto nitrocellulose membranes. After incubation in blocking buffer, the membranes were probed overnight at 4°C with the indicated Abs. The labeling was visualized using peroxidase-conjugated secondary Abs and an enhanced chemiluminescence kit (Amersham International). Blots were scanned and processed by Adobe Photoshop 7.0 software.
Cytotoxicity assay
The cytotoxic activity of the NK cells was measured by a conventional 4-hour 51Cr-release assay by using triplicate cultures in round-bottom 96-well plates. Different effector to target (E:T) ratios were used as previously described.49
NK degranulation assay
NK cells were cocultured with laser-assisted microdissection target cells for 4 hours at 2:1 E:T ratio. Degranulation of NK cells was analyzed by flow cytometric analysis of CD107a expression. For c-Myc inhibition experiments, the leukemia cells were treated with c-Myc inhibitor overnight and then assayed by flow cytometry.
Flow cytometry analysis
Flow cytometry analysis was performed using a FACSCalibur and LSRII flow cytometer. Data were processed using CellQuest software (BD Biosciences).
Conjugate formation assay
NK cells and leukemia cells were incubated for 1 hour, respectively, with red cell tracer and green cell tracer. Cells were then washed and put in coculture (at 2:1 E:T ratio) for 1 hour, and then immediately analyzed by flow cytometry.
Chromatin immunoprecipitation (ChIP) assay
For ChIP experiments, 3.107 untreated or treated KG-1 cells and primary blasts were harvested and processed with the EpiTect ChiP OneDay Kit (Qiagen) according to the manufacturer’s instructions. The amount of input chromatin per ChIP was 10 µg. For c-Myc–specific ChIP, chromatin was immunoprecipitated with 2 µg monoclonal antibody (sc-40; Santa Cruz Biotechnology) or 2 µg of a respective isotype control mAb (sc-2031; Santa Cruz Biotechnology). The relative amounts of chromatin immunoprecipitated by either isotype control or c-Myc mAb was determined by real-time PCR with specific primers for c-Myc binding site (c-Myc BS) in ULBP1/3 gene promoter, respectively (TGCTATGTC CATGGGACCTG, GCAGGCACCACGTGCCATAGC) purchased from Qiagen.
Statistical analyses
Data were analyzed with GraphPad Prism. A Student t test was used for single comparisons. A P value of .05 was considered statistically significant.
Results
The acquisition of resistance to cytarabine correlates with an increased susceptibility to NK-mediated cell lysis of resistant AML cell lines
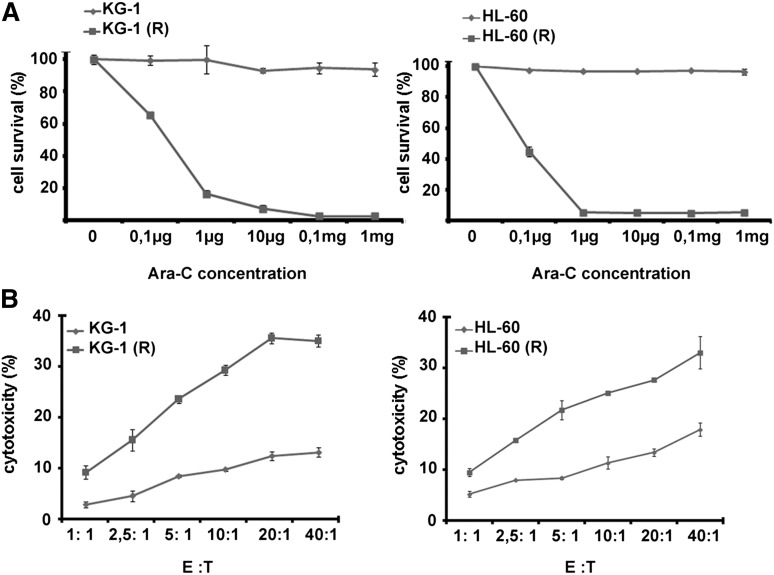
The cytarabine-resistant AML cell lines, KG-1 and HL-60 cell sublines, were established by exposing parental cells to increasing concentrations of the drug. Results depicted in Figure 1A show that cell viability is correlated to drug concentration in sensitive cell lines. When cytarabine was used at the different indicated concentrations, it had no effect on the viability of resistant cells. To examine the susceptibility of cytarabine-resistant KG-1(R) and HL-60(R) cells to NK-mediated cytotoxicity, we used the NK-cell line (NKL) that selectively expresses NKG2D but does not express most other receptors. Cytotoxicity by NK cells was determined by a conventional 4-hour 51Cr release assay at different E:T ratios in resistant and parental cells. As shown in Figure 1B, both cytarabine-resistant cell lines were more susceptible to NK cell lysis compared with parental (ie, cytarabine-sensitive) cells, in all tested ratios.
Figure 1.
Effects of cytarabine on leukemia cells. (A) Dose-dependent antiproliferative effect of ara-C on HL-60 and KG-1 resistant and sensitive cell lines using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay. (B) NK-cell cytotoxicity of sensitive and resistant leukemic cells. Target cells were co-incubated with NKL cells at the indicated E:T ratios for 4 hours at 37°C.
Increased NKG2D ligand expression on cytarabine resistant cells
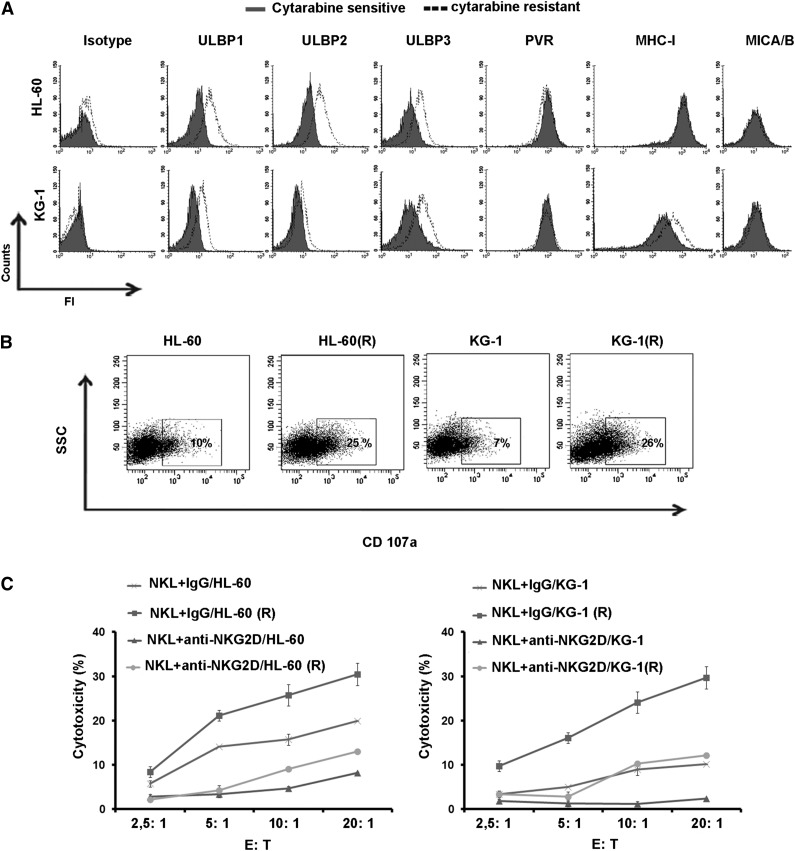
To elucidate the mechanism underlying the increased susceptibility to NK cells by cytarabine resistant targets, we examined the expression pattern of NK cell receptor ligands by parental and cytarabine-resistant cell lines. Our results showed that the expression of ULBP1, 2, and 3 was up-regulated on the surface of KG-1(R) and HL-60 (R) cell lines, while the expression of MICA/B, poliovirus receptor, and MHC-I was not affected (Figure 2A). We next performed the degranulation assay by measuring CD107a expression on NK cells after 4-hour coculture with leukemia cells at 2:1 effector/target ratio. The results showed that the expression of CD107a increased on NK cells after coculture with resistant cells as compared to sensitive cells (Figure 2B). To examine the role of NKG2D and its ligands in the acquisition of leukemic cell resistance to cytarabine, we first incubated leukemic cells with anti-NKG2D mAb. Data revealed that blocking this receptor resulted in the inhibition of the cytotoxic activity of NK cells (Figure 2C). Taken together, these results suggest that induction of NKG2D ligand expression on cytarabine-resistant AML cells renders them susceptible to NK-cell–mediated killing.
Figure 2.
Expression of ligands for NK-cell–activating receptors in leukemic cells. (A) Flow cytometric analysis for the expression of ligands in KG-1 and KG-1(R) cells, and HL-60 and HL-60(R) cells. One representative of at least 3 separate experiments is shown. (B) Flow cytometric analysis of CD107a expression NK cells cocultured for 4 hours with candidate tumor cells at E:T ratio of 2:1 (representative blot of n = 3). (C) Sensitive and resistant leukemia cells incubated with anti-NKG2D mAb or isotype as negative control. The lytic activity of the NKL cells toward these target cells was assessed in a standard 4-hour chromium release assay at different indicated E:T ratios.
The upregulation of ULBP1/2/3 on cytarabine resistant cells is c-Myc dependent
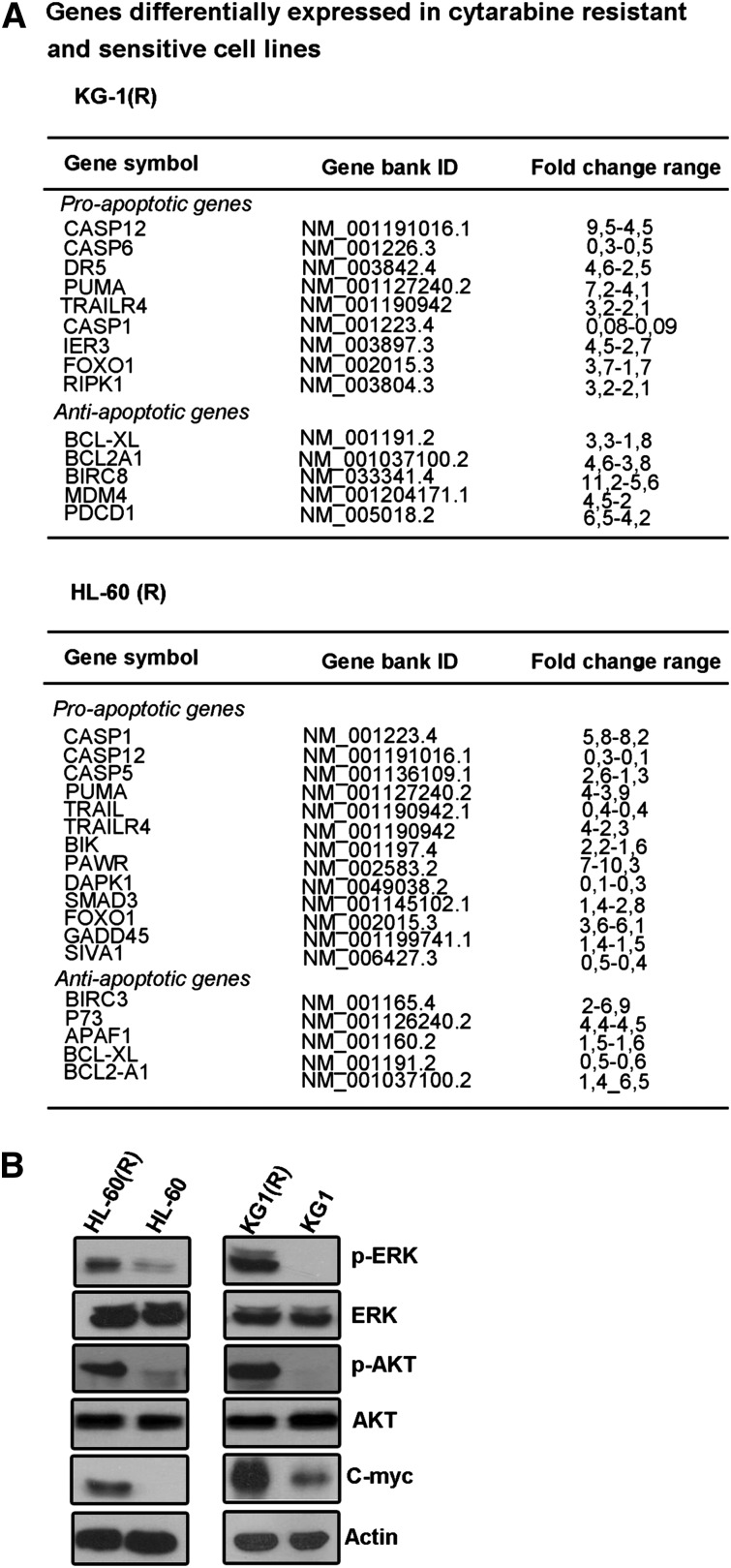
To investigate the molecular mechanisms associated with the enhancement of target cell susceptibility to NK cells following acquisition of cytarabine resistance, we analyzed the expression of a series of antiapoptotic and proapoptotic genes using an apoptosis-related array of 94 genes. In the course of these studies, the gene differentiation analysis was performed using noncloned cell lines in order to be closer to what is observed in patient blasts. The results revealed no common expression changes in proapoptotic or antiapoptotic genes between cytarabine-resistant cell lines and their sensitive equivalents (Figure 3A).
Figure 3.
Expression of pro- and antiapoptotic genes, c-Myc, AKT, and ERK in cytarabine-resistant cells. (A) Pro- and antiapoptotic gene transcript quantification by qPCR using a 94-gene apoptosis dedicated array. Results are representative of 2 independent experiments. (B) Western blot analysis for c-Myc and constitutive AKT and ERK activation was assessed using antibodies that recognize AKT phosphorylated at ser-473, and ERK1/2 phosphorylated at Thr202/Tyr204 on whole cell extracts. Actin was used as a protein loading (40 mg per lane; representative experiment of n = 3).
It has been shown that ERK and AKT pathways can modulate NKG2D ligand expression. Furthermore, as cytarabine induces apoptosis in AML cells through cell-cycle arrest, we investigated the putative involvement of c-Myc, which is a major cell-cycle modulator in leukemia disorders. Therefore, we assessed the expression of AKT, ERK, and c-Myc. Western blot analysis showed a stronger phosphorylated ERK and AKT, and induced expression of c-Myc in KG-1(R) and HL-60(R) cells as compared with KG-1 and HL-60 cells (Figure 3B).
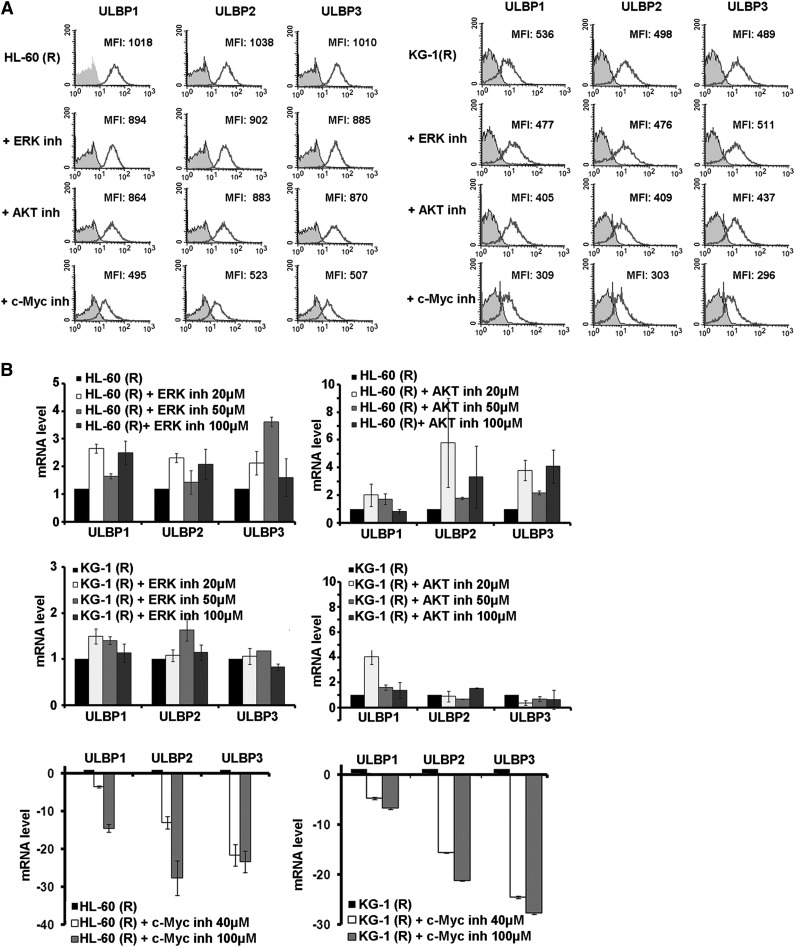
Next, we asked whether the inhibition of the expression of ERK, AKT, and c-Myc affects the expression of ULBP1/2/3. The expression at the transcriptional level by real-time qPCR and also at the cell surface by flow cytometry was examined. Results depicted in Figure 4A-B revealed that inhibition of c-Myc by chemical inhibitor reduced the mRNA level and cell-surface expression of ULBP1/2/3, while inhibition ERK or AKT by pharmacologic inhibitors had no effect. These results indicate that c-Myc is involved in the regulation of ULBP1/2/3 in cancer cells.
Figure 4.
Effect of c-Myc inhibition on NKG2D ligands expression. (A) Flow cytometric analysis for the expression of ULBP1/2/3 for resistant leukemia cells after specific inhibition of c-Myc using 10058-F4, and inhibition of ERK and AKT activation using PD98059 and MK-2206, respectively, after 24 hours of treatment. Data are representative of at least 3 experiments. (B) Real-time quantitative reverse-transcription PCR for the mRNA expression levels of ULBP1/2/3, transcripts in leukemia cells after 24 hours of treatment with c-Myc, ERK, or AKT inhibitor.
c-Myc targeting in cytarabine-resistant cells is accompanied by a decrease in NK-mediated lysis
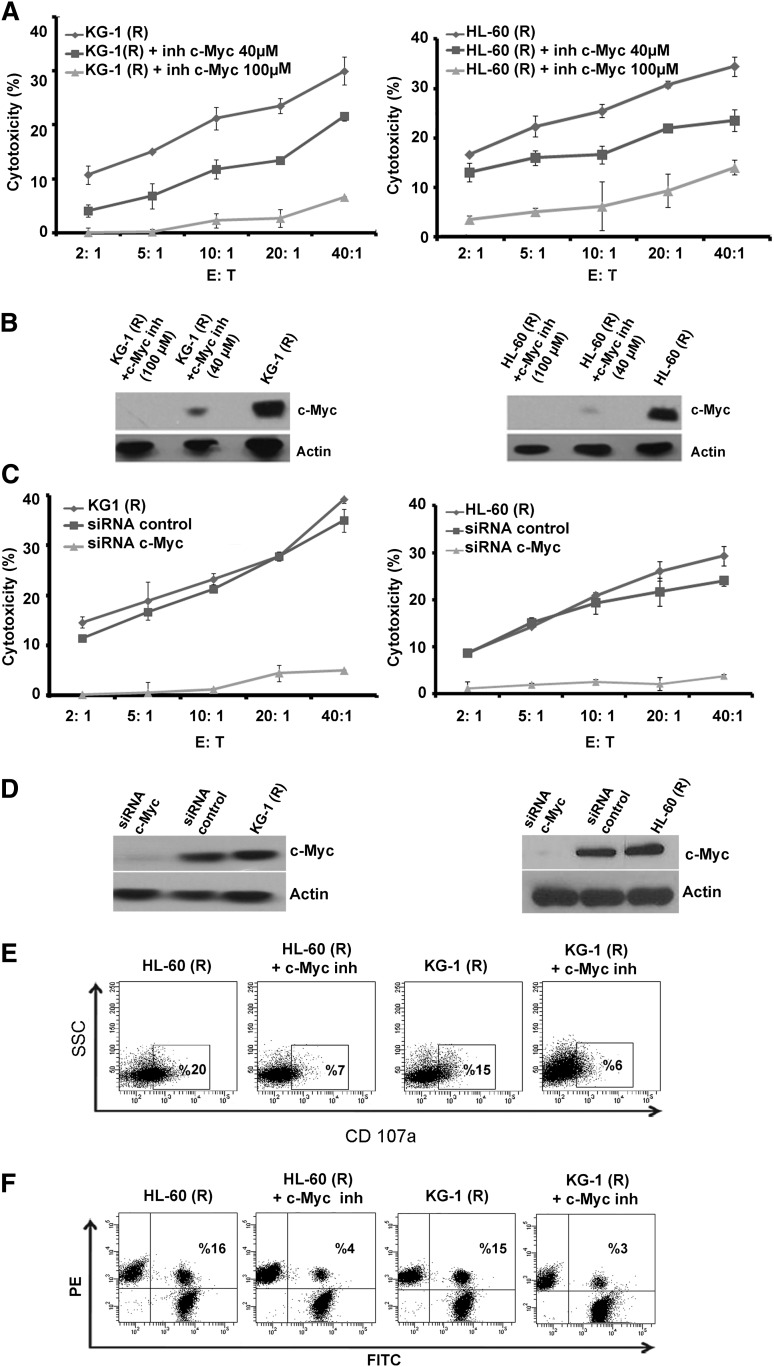
To delineate the role of c-Myc in NK-cell susceptibility of cytarabine-resistant cells, we investigated the effect of c-Myc inhibition in KG-1(R) and HL-60(R) cells using specific siRNA or a c-Myc pharmacologic inhibitor. c-Myc neutralization, whether by siRNA or pharmacological inhibitor, was associated with sensitization to NK-mediated lysis (Figure 5A,C). The inhibition was confirmed by western blot assay, which showed that there was no effect on c-Myc protein level when luciferase siRNA was used as a control (Figure 5B,D). To examine whether c-Myc inhibition in cytarabine-resistant cells would alter the stimulation of cytolytic activity of NK cells, CD107a assay was performed (as described earlier with NK cells) in the presence or absence of c-Myc inhibitor. The results shown in Figure 5E demonstrate that CD107a expression on NK cells decreased dramatically when cocultured with leukemic cells treated with c-Myc inhibitor as compared with nontreated cells (Figure 5E). We also performed the conjugate formation assay to analyze the capacity of NK cells to form immunologic synapse with leukemia cells treated or untreated with c-Myc inhibitor. The results showed that the c-Myc treated KG-1(R) and HL-60(R) cells form much less conjugates with NK cells than with nontreated cells (Figure 5F).
Figure 5.
Effect of c-Myc inhibition on NK-cell–mediated killing of cytarabine-resistant cells. (A) NK-cell cytotoxicity of KG-1(R) and HL-60(R) cells after incubation with 10058-F4 (40 µM and 100 µM) for 24 hours. (B) Experimental values were determined by western blot analysis using anti–c-Myc antibody. Actin was used as the protein level control. (C) NK-cell cytotoxicity of KG-1(R) and Hl-60(R) cells after inhibition of c-Myc by specific siRNA or a negative control siRNA (Luc). (D) Transfection efficiency was determined by western blot analysis using anti–c-Myc antibody. Actin was used as the protein level control. (E) Flow cytometric analysis of CD107a expression on NK cells cocultured for 4 hours with KG-1(R) and HL-60(R) with or without 10058-F4 (100 µM for 24 hours) at E:T ratio of 2:1 (representative blot of n = 3). (F) Conjugate formation assay between NK cells and KG-1(R) and HL-60(R) with or without 10058-F4 (100 µM for 24 hours) at E:T ratio of 2:1 (representative blot of n = 3).
c-Myc regulates ULBP1/2/3 expression and interferes with NK cell susceptibility in primary AML blasts
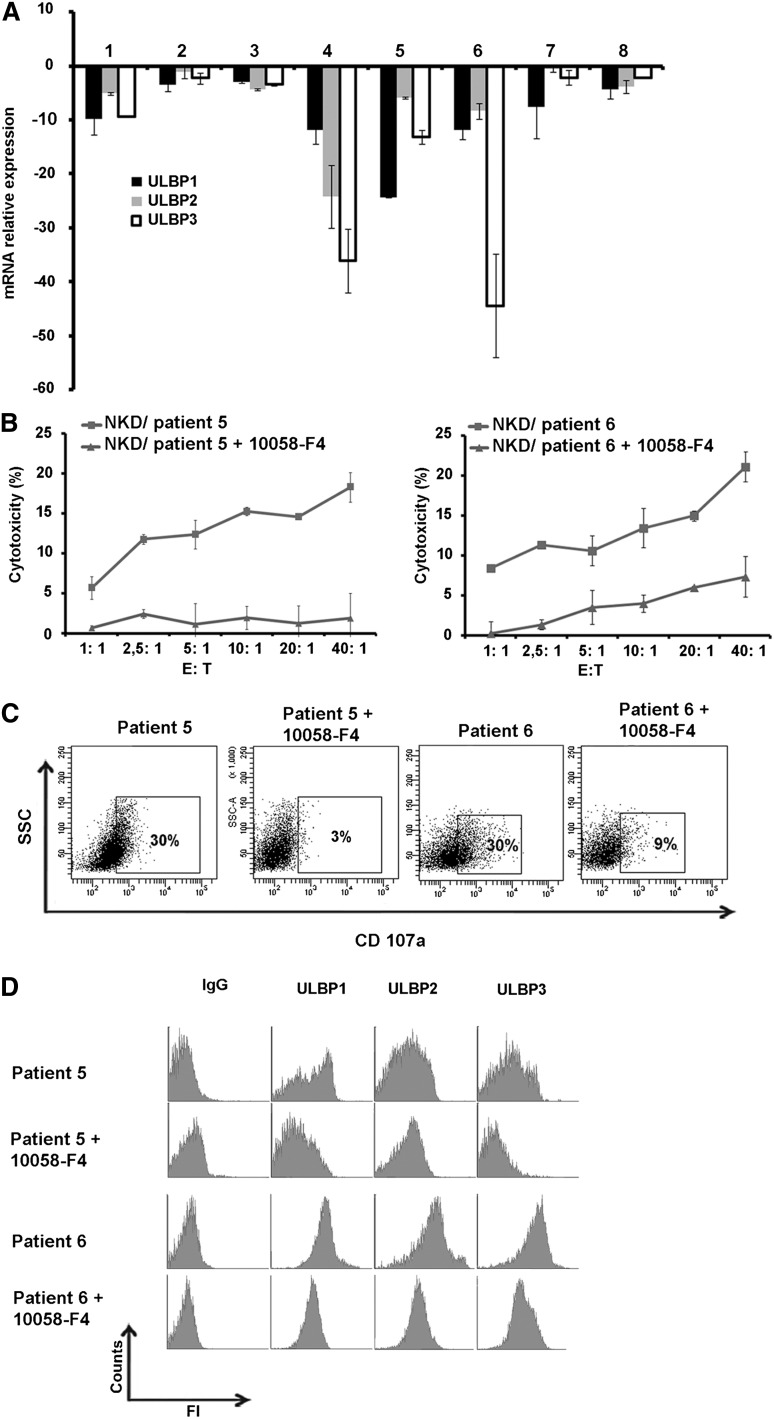
To validate the role of c-Myc in regulation of NKG2D ligands expression in primary AML cells, blasts from 9 different patients in a refractory stage (resistant to chemotherapy) were isolated and treated with c-Myc inhibitor for 24 hours. ULBP expression was measured at the transcriptional level using real-time qPCR and also examined at the cell surface by flow cytometry. The results indicate that ULBP1/2/3 mRNA level in blasts treated with c-Myc inhibitor was decreased in all patients, with varying degrees of down-regulations (Figure 6A).
Figure 6.
Effect of inhibition of c-Myc in primary cells on NKG2D ligand expression and NK-cell–mediated cytotoxicity. (A) Blasts from chemotherapy-resistant patients were isolated and incubated with c-Myc inhibitor for 24 hours and relative ULBP1/2/3 mRNA expression by real-time qPCR was measured and compared with nontreated blasts. (B) NK-cell–cytotoxicity of blasts with or without treatment with 10058-F4 at indicated E:T ratios. (C) Flow cytometric analysis of CD107a expression on NK cells cocultured for 4 hours in refractory blasts with or without inhibition of c-Myc with 10058-F4 (treatment of 24 hours, 100 µM) at E:T ratio of 10:1 (representative blot of n = 3). (D) Flow cytometric analysis for the expression of ULBP1/2/3 of AML refractory patients after treatment with c-Myc inhibitor for 24 hours.
To further confirm the role of c-Myc in the regulation of primary AML susceptibility to NK-mediated cell lysis, the blasts were incubated with c-Myc inhibitor for 24 hours at 100 µM concentration. Surprisingly, we observed that the refractory blasts of all of the tested patients were resistant to lysis mediated by NKL cells. Since NKL cells express a much lower level of granzyme B as compared with healthy donor NK cells, as well as not expressing most of the stimulatory receptors involved in recognition and elimination of primary blasts, we hypothesized that these NK cells are not cytotoxic enough to eliminate leukemia cells. However, when we used peripheral NK cells of a healthy donor (activated with IL-2, 300 U/mL for 2 days), we observed that NK-cell cytotoxic activity against AML blasts, which had been treated with c-Myc inhibitor, was abolished as compared with nontreated cells (Figure 6B). We also observed that CD107a expression on NK cells decreased dramatically when cocultured with cells treated with c-Myc inhibitor as compared with nontreated cells (Figure 6C). The analysis of ULBP expression at the cell surface revealed that c-Myc inhibition resulted in a dramatic decrease in expression of ULBP1/2/3 (Figure 6D).
ULBP1/3 are direct c-Myc target genes in AML cells
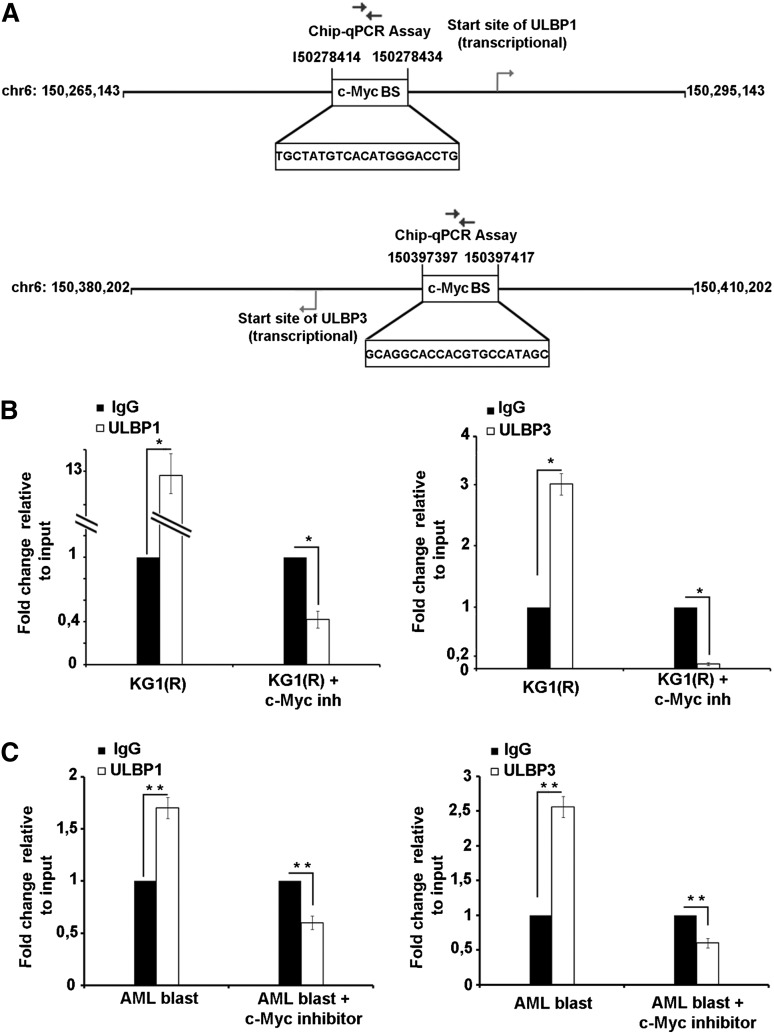
To get more insight into the mechanism of ULBP gene regulation by c-Myc, we asked whether the transcriptional upregulation of ULBP1/2/3 involved direct c-Myc binding to c-Myc BS in the promoter region of ULBP1/2/3 genes (based on the SABiosciences database). Data depicted in Figure 7A indicate that potential c-Myc BS was only found for the ULBP1/3 genes. ChIP with a c-Myc–specific mAb revealed the binding of c-Myc to c-Myc BS of the ULBP1/3 in KG-1(R) cells (Figure 7B). We also observed that the binding of c-Myc to c-Myc BS of ULBP1/3 genes was decreased dramatically when cells were treated with c-Myc inhibitor (Figure 7B). In addition, we also performed a ChIP experiment with AML drug-resistant primary blasts, and similar results were obtained (Figure 7C). Together, these data indicate that c-Myc modulates ULBP1/3 expression directly by interacting with c-Myc BS at promoter region of ULBP1/3 genes.
Figure 7.
c-Myc is a direct target of ULBP1 and ULBP3. (A) Schematic picture of the potential c-Myc BS in the ULBP1/3 genes. (B-C) Binding of c-Myc to the potential c-Myc BS in the ULBP1/3 gene promoters was analyzed in KG-1(R), and primary blasts were or were not treated with c-Myc inhibitor (10058-F4) by ChIP assay. The amount of immunoprecipitated chromatin bound by either isotype control or c-Myc mAb was quantified by real-time PCR with specific primers. Specific signals were set relative to signals obtained for the input chromatin. One representative experiment of 2 ChIP independently conducted experiments is shown. Data were analyzed with GraphPad Prism. Two-tailed Student t test was used for single comparisons. Statistically significant differences are indicated by asterisks (*P < .05; **P < .005; ***P < .0005).
Discussion
There is renewed interest in exploiting the antitumor effect of NK cells due to data from preclinical and clinical work on the potential of alloreactive NK cells to kill tumor cells. Potential approaches include not only enhancing the generation of NK cells, but also manipulating the susceptibility of blast cells to NK-mediated killing. Pursuing these approaches will require a more thorough understanding of the different mechanisms of resistance and their relationship with susceptibility to NK-mediated killing. For example, following allogeneic hematopoietic stem-cell transplantation, activated donor NK cells can kill residual AML target cells in the recipient and contribute to protection from relapse. In this regard, manipulating NK-cell alloreactivity might improve outcomes after hematopoietic stem-cell transplantation; however, cross-resistance among blasts remains a drawback. In this study, we attempted to elucidate how acquisition of drug resistance in AML cells influences NK-cell recognition and the killing of drug-resistant blasts. We showed that the in vitro acquisition of AML cell resistance to cytarabine resulted in an increase in their susceptibility to NK-mediated cell cytotoxicity. Meanwhile, some studies report decreased sensitivity of drug-resistant leukemic cells to cellular cytotoxicity; others showed that chemotherapeutic drugs, including ara-C, sensitize acute lymphoblastic leukemia cells for NK-cell–mediated apoptosis.10-18
Even if the susceptibility of blasts in refractory patients is increased, it is essential to take into account the alteration of autologous NK cells in these patients. In fact, leukemic cells may escape from NK-cell surveillance through different mechanisms, including NK-cell qualitative deficiency (due to expression of HLA class I molecules, impaired activation, downregulation of ligands relevant in natural cytotoxicity receptors, and impaired differentiation signaling), as well as NK quantitative defects. Given the impairment of NK-cell cytotoxicity in almost hematologic malignancies, the restoration of normal NK function or adoptive NK cell therapy remains an attractive option to treat leukemic patients. In this context, NK-based cell therapy approaches should be based on the administration of highly cytotoxic NK cells in the context of target susceptibility to lysis.
The in vitro acquisition of resistance of AML cells to cytarabine was associated with an enhanced expression of NKG2D ligands by a mechanism involving c-Myc induction. More importantly, using AML blasts isolated from patients at refractory stage, c-Myc inhibitor was found to be efficient in downregulating NKG2D ligands expression. Future experiments will focus on the elucidation of c-Myc level, as well as NKG2DL expression in AML patients before and after drug resistance acquisition. In previous reports, it has been shown that ERK or AKT pathways could modulate NKG2DL expression; however, in our experimental model, despite the induction of these pathways, the pharmacologic inhibition of these proteins did not result in downregulation of ULBP1/2/3. We hypothesized that these pathways are switched on in a response to a hostile cell culture condition, which allows cells to survive and overcome drug-induced apoptosis.50,51 Nevertheless, while inhibition of AKT or ERK had no effect on ULBP1/2/3, inhibition of c-Myc resulted in a decrease in the transcriptional level expression of ULBP1/2/3 and inhibition of NK cytotoxic response. It is assumed that this transcriptional factor plays a critical role in cell growth, proliferation, and apoptosis, by its capacity to orchestrate the expression of more than 15% of all cellular genes. c-Myc is frequently activated in AML and plays an important role in the induction of leukemogenesis, regulating several genes both positively and negatively. Unni et al have provided evidence that NKG2D ligands are induced on spontaneously arising tumors in a murine model of lymphomagenesis and that c-Myc is involved in their regulation.52
We attempted therefore, to shed light on the correlation between those 2 important genes, NKG2D ligands and c-Myc, respectively, involved in immune response and leukemogenesis in the context of a drug resistance.
ChIP studies revealed a direct correlation between c-Myc activation and ULBP1/3 expression, although no c-Myc BS for ULBP2 was identified. This indicates not only that c-Myc can regulate ULBP expression directly, but also that it may modify its expression indirectly. These data would suggest that quantification of c-Myc in AML blast cells may be useful as a prognostic indicator in AML for NK-cell response in bone marrow transplantation cell therapy trials.
Recently, it has been reported that p53 regulates ULBP1/2 expression at the transcriptional level.44 This mechanism is ruled out in our cell models since the one we used displays a mutated p53. Recent studies also indicate that DNA damage could regulate NKG2D ligand expression.39 This mechanism is also unlikely since no modification for MICA/B or poliovirus receptor expressions, which are regulated by DNA damage pathway, was observed. In addition, it is well established that leukemic stem cells play a central role in the relapse of acute AML and the resistance to NK-cell–mediated cytotoxicity.53
Nevertheless, under our experimental conditions, the acquisition of resistance cells to cytarabine did not correlate with the induction of stem-cell markers (not shown).
Collectively, these studies suggest that the acquisition of AML resistance to cytarabine does not confer cross-resistance to NK-mediated cell lysis, and that the increased susceptibility resulting from the acquisition of drug resistance may show promise in NK-cell therapy following allogeneic stem-cell transplantation for drug refractory patients with AML.
Acknowledgments
This work was supported by the Institut National du Cancer, Association Laurette Fugain, and the Qatar Foundation.
Footnotes
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.N., C.P., and A.M. performed the experiments; S.C. designed the study; A.N. and S.C. wrote the manuscript; and all authors critically reviewed the manuscript and gave their final approval.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Salem Chouaib, U753 INSERM, Institut Gustave Roussy, 114 rue Edouard Vaillant, 94805 Villejuif cedex, France; e-mail: chouaib@igr.fr.
References
- 1.Shipley JL, Butera JN. Acute myelogenous leukemia. Exp Hematol. 2009;37(6):649–658. doi: 10.1016/j.exphem.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Rustum YM, Preisler HD. Correlation between leukemic cell retention of 1-beta-D-arabinofuranosylcytosine 5′-triphosphate and response to therapy. Cancer Res. 1979;39(1):42–49. [PubMed] [Google Scholar]
- 3.Estey E. Treatment of refractory AML. Leukemia. 1996;10(6):932–936. [PubMed] [Google Scholar]
- 4.Willemze R, Suciu S, Archimbaud E, et al. A randomized phase II study on the effects of 5-Aza-2′-deoxycytidine combined with either amsacrine or idarubicin in patients with relapsed acute leukemia: an EORTC Leukemia Cooperative Group phase II study (06893). Leukemia. 1997;11(suppl 1):S24–S27. [PubMed] [Google Scholar]
- 5.Keating MJ, McCredie KB, Bodey GP, Smith TL, Gehan E, Freireich EJ. Improved prospects for long-term survival in adults with acute myelogenous leukemia. JAMA. 1982;248(19):2481–2486. [PubMed] [Google Scholar]
- 6.Löwenberg B. Sense and nonsense of high-dose cytarabine for acute myeloid leukemia. Blood. 2013;121(1):26–28. doi: 10.1182/blood-2012-07-444851. [DOI] [PubMed] [Google Scholar]
- 7.Wang WS, Tzeng CH, Chiou TJ, et al. High-dose cytarabine and mitoxantrone as salvage therapy for refractory non-Hodgkin’s lymphoma. Jpn J Clin Oncol. 1997;27(3):154–157. doi: 10.1093/jjco/27.3.154. [DOI] [PubMed] [Google Scholar]
- 8.Shaffer BC, Gillet JP, Patel C, Baer MR, Bates SE, Gottesman MM. Drug resistance: still a daunting challenge to the successful treatment of AML. Drug Resist Updat. 2012;15(1-2):62–69. doi: 10.1016/j.drup.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiller GJ. Treatment of resistant disease. Leukemia. 1998;12(suppl 1):S20–S24. [PubMed] [Google Scholar]
- 10.Komada Y, Zhou YW, Zhang XL, et al. Fas/APO-1 (CD95)-mediated cytotoxicity is responsible for the apoptotic cell death of leukaemic cells induced by interleukin-2-activated T cells. Br J Haematol. 1997;96(1):147–157. doi: 10.1046/j.1365-2141.1997.8742505.x. [DOI] [PubMed] [Google Scholar]
- 11.Debatin KM. Cytotoxic drugs, programmed cell death, and the immune system: defining new roles in an old play. J Natl Cancer Inst. 1997;89(11):750–751. doi: 10.1093/jnci/89.11.750. [DOI] [PubMed] [Google Scholar]
- 12.Micheau O, Solary E, Hammann A, Martin F, Dimanche-Boitrel MT. Sensitization of cancer cells treated with cytotoxic drugs to fas-mediated cytotoxicity. J Natl Cancer Inst. 1997;89(11):783–789. doi: 10.1093/jnci/89.11.783. [DOI] [PubMed] [Google Scholar]
- 13.Renvoizé C, Roger R, Moulian N, Bertoglio J, Bréard J. Bcl-2 expression in target cells leads to functional inhibition of caspase-3 protease family in human NK and lymphokine-activated killer cell granule-mediated apoptosis. J Immunol. 1997;159(1):126–134. [PubMed] [Google Scholar]
- 14.Friesen C, Fulda S, Debatin KM. Deficient activation of the CD95 (APO-1/Fas) system in drug-resistant cells. Leukemia. 1997;11(11):1833–1841. doi: 10.1038/sj.leu.2400827. [DOI] [PubMed] [Google Scholar]
- 15.Posovszky C, Friesen C, Herr I, Debatin KM. Chemotherapeutic drugs sensitize pre-B ALL cells for CD95- and cytotoxic T-lymphocyte-mediated apoptosis. Leukemia. 1999;13(3):400–409. doi: 10.1038/sj.leu.2401327. [DOI] [PubMed] [Google Scholar]
- 16.Classen CF, Fulda S, Friesen C, Debatin KM. Decreased sensitivity of drug-resistant cells towards T cell cytotoxicity. Leukemia. 1999;13(3):410–418. doi: 10.1038/sj.leu.2401335. [DOI] [PubMed] [Google Scholar]
- 17.Classen CF, Falk CS, Friesen C, Fulda S, Herr I, Debatin KM. Natural killer resistance of a drug-resistant leukemia cell line, mediated by up-regulation of HLA class I expression. Haematologica. 2003;88(5):509–521. [PubMed] [Google Scholar]
- 18.Treichel RS, Bunuan M, Hahn N, Wee K. Altered conjugate formation and altered apoptosis of multidrug-resistant human leukemia cell line affects susceptibility to killing by activated natural killer (NK) cells. Int J Cancer. 2004;108(1):78–85. doi: 10.1002/ijc.11555. [DOI] [PubMed] [Google Scholar]
- 19.Bornhäuser M, Thiede C, Platzbecker U, et al. Prophylactic transfer of BCR-ABL-, PR1-, and WT1-reactive donor T cells after T cell-depleted allogeneic hematopoietic cell transplantation in patients with chronic myeloid leukemia. Blood. 2011;117(26):7174–7184. doi: 10.1182/blood-2010-09-308569. [DOI] [PubMed] [Google Scholar]
- 20.Rubnitz JE, Inaba H, Ribeiro RC, et al. NKAML: a pilot study to determine the safety and feasibility of haploidentical natural killer cell transplantation in childhood acute myeloid leukemia. J Clin Oncol. 2010;28(6):955–959. doi: 10.1200/JCO.2009.24.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller JS, Soignier Y, Panoskaltsis-Mortari A, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. 2005;105(8):3051–3057. doi: 10.1182/blood-2004-07-2974. [DOI] [PubMed] [Google Scholar]
- 22.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9(5):503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 23.Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol. 2006;6(10):715–727. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
- 24.Iannello A, Debbeche O, Samarani S, Ahmad A. Antiviral NK cell responses in HIV infection: I. NK cell receptor genes as determinants of HIV resistance and progression to AIDS. J Leukoc Biol. 2008;84(1):1–26. doi: 10.1189/jlb.0907650. [DOI] [PubMed] [Google Scholar]
- 25.Arina A, Murillo O, Dubrot J, et al. Cellular liaisons of natural killer lymphocytes in immunology and immunotherapy of cancer. Expert Opin Biol Ther. 2007;7(5):599–615. doi: 10.1517/14712598.7.5.599. [DOI] [PubMed] [Google Scholar]
- 26.Manilay JO, Sykes M. Natural killer cells and their role in graft rejection. Curr Opin Immunol. 1998;10(5):532–538. doi: 10.1016/s0952-7915(98)80219-7. [DOI] [PubMed] [Google Scholar]
- 27.Kroemer A, Edtinger K, Li XC. The innate natural killer cells in transplant rejection and tolerance induction. Curr Opin Organ Transplant. 2008;13(4):339–343. doi: 10.1097/MOT.0b013e3283061115. [DOI] [PubMed] [Google Scholar]
- 28.Ljunggren HG, Malmberg KJ. Prospects for the use of NK cells in immunotherapy of human cancer. Nat Rev Immunol. 2007;7(5):329–339. doi: 10.1038/nri2073. [DOI] [PubMed] [Google Scholar]
- 29.Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol. 2006;6(7):520–531. doi: 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- 30.Farag SS, Caligiuri MA. Human natural killer cell development and biology. Blood Rev. 2006;20(3):123–137. doi: 10.1016/j.blre.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Bauer S, Groh V, Wu J, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285(5428):727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 32.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA. 1996;93(22):12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verneris MR, Karami M, Baker J, Jayaswal A, Negrin RS. Role of NKG2D signaling in the cytotoxicity of activated and expanded CD8+ T cells. Blood. 2004;103(8):3065–3072. doi: 10.1182/blood-2003-06-2125. [DOI] [PubMed] [Google Scholar]
- 34.Textor S, Dürst M, Jansen L, et al. Activating NK cell receptor ligands are differentially expressed during progression to cervical cancer. Int J Cancer. 2008;123(10):2343–2353. doi: 10.1002/ijc.23733. [DOI] [PubMed] [Google Scholar]
- 35.Pende D, Rivera P, Marcenaro S, et al. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 2002;62(21):6178–6186. [PubMed] [Google Scholar]
- 36.Pende D, Cantoni C, Rivera P, et al. Role of NKG2D in tumor cell lysis mediated by human NK cells: cooperation with natural cytotoxicity receptors and capability of recognizing tumors of nonepithelial origin. Eur J Immunol. 2001;31(4):1076–1086. doi: 10.1002/1521-4141(200104)31:4<1076::aid-immu1076>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 37.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3(10):781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 38.Ogbomo H, Michaelis M, Klassert D, Doerr HW, Cinatl J., Jr Resistance to cytarabine induces the up-regulation of NKG2D ligands and enhances natural killer cell lysis of leukemic cells. Neoplasia. 2008;10(12):1402–1410. doi: 10.1593/neo.08972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soriani A, Zingoni A, Cerboni C, et al. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood. 2009;113(15):3503–3511. doi: 10.1182/blood-2008-08-173914. [DOI] [PubMed] [Google Scholar]
- 40.Butler JE, Moore MB, Presnell SR, Chan HW, Chalupny NJ, Lutz CT. Proteasome regulation of ULBP1 transcription. J Immunol. 2009;182(10):6600–6609. doi: 10.4049/jimmunol.0801214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerboni C, Zingoni A, Cippitelli M, Piccoli M, Frati L, Santoni A. Antigen-activated human T lymphocytes express cell-surface NKG2D ligands via an ATM/ATR-dependent mechanism and become susceptible to autologous NK- cell lysis. Blood. 2007;110(2):606–615. doi: 10.1182/blood-2006-10-052720. [DOI] [PubMed] [Google Scholar]
- 42.López-Soto A, Quiñones-Lombraña A, López-Arbesú R, López-Larrea C, González S. Transcriptional regulation of ULBP1, a human ligand of the NKG2D receptor. J Biol Chem. 2006;281(41):30419–30430. doi: 10.1074/jbc.M604868200. [DOI] [PubMed] [Google Scholar]
- 43.Wu X, Tao Y, Hou J, Meng X, Shi J. Valproic acid upregulates NKG2D ligand expression through an ERK-dependent mechanism and potentially enhances NK cell-mediated lysis of myeloma. Neoplasia. 2012;14(12):1178–1189. doi: 10.1593/neo.121236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Textor S, Fiegler N, Arnold A, Porgador A, Hofmann TG, Cerwenka A. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res. 2011;71(18):5998–6009. doi: 10.1158/0008-5472.CAN-10-3211. [DOI] [PubMed] [Google Scholar]
- 45.Himmelreich H, Mathys A, Wodnar-Filipowicz A, Kalberer CP. Post-transcriptional regulation of ULBP1 ligand for the activating immunoreceptor NKG2D involves 3′ untranslated region. Hum Immunol. 2011;72(6):470–478. doi: 10.1016/j.humimm.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 46.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436(7054):1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bortul R, Tazzari PL, Billi AM, et al. Deguelin, a PI3K/AKT inhibitor, enhances chemosensitivity of leukaemia cells with an active PI3K/AKT pathway. Br J Haematol. 2005;129(5):677–686. doi: 10.1111/j.1365-2141.2005.05504.x. [DOI] [PubMed] [Google Scholar]
- 48.Bedel R, Thiery-Vuillemin A, Grandclement C, et al. Novel role for STAT3 in transcriptional regulation of NK immune cell targeting receptor MICA on cancer cells. Cancer Res. 2011;71(5):1615–1626. doi: 10.1158/0008-5472.CAN-09-4540. [DOI] [PubMed] [Google Scholar]
- 49.Noman MZ, Buart S, Van Pelt J, et al. The cooperative induction of hypoxia-inducible factor-1 alpha and STAT3 during hypoxia induced an impairment of tumor susceptibility to CTL-mediated cell lysis. J Immunol. 2009;182(6):3510–3521. doi: 10.4049/jimmunol.0800854. [DOI] [PubMed] [Google Scholar]
- 50.Grandage VL, Gale RE, Linch DC, Khwaja A. PI3-kinase/Akt is constitutively active in primary acute myeloid leukaemia cells and regulates survival and chemoresistance via NF-kappaB, Mapkinase and p53 pathways. Leukemia. 2005;19(4):586–594. doi: 10.1038/sj.leu.2403653. [DOI] [PubMed] [Google Scholar]
- 51.Yin B, Morgan K, Hasz DE, Mao Z, Largaespada DA. Nfl gene inactivation in acute myeloid leukemia cells confers cytarabine resistance through MAPK and mTOR pathways. Leukemia. 2006;20(1):151–154. doi: 10.1038/sj.leu.2404033. [DOI] [PubMed] [Google Scholar]
- 52.Unni AM, Bondar T, Medzhitov R. Intrinsic sensor of oncogenic transformation induces a signal for innate immunosurveillance. Proc Natl Acad Sci USA. 2008;105(5):1686–1691. doi: 10.1073/pnas.0701675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.She M, Niu X, Chen X, et al. Resistance of leukemic stem-like cells in AML cell line KG1a to natural killer cell-mediated cytotoxicity. Cancer Lett. 2012;318(2):173–179. doi: 10.1016/j.canlet.2011.12.017. [DOI] [PubMed] [Google Scholar]