Abstract
Migration of macrophages is a key process for a variety of physiological functions, such as pathogen clearance or tissue homeostasis. However, it can also be part of pathological scenarios, as in the case of tumor-associated macrophages. This review presents an overview of the different migration modes macrophages can adopt, depending on the physical and chemical properties of specific environments, and the constraints they impose upon cells. We discuss the importance of these environmental and also of cellular parameters, as well as their relative impact on macrophage migration and on the formation of matrix-lytic podosomes in 2D and 3D. Moreover, we present an overview of routinely used and also newly developed assays for the study of macrophage migration in both 2D and 3D contexts, their respective advantages and limitations, and also their potential to reliably mimic in vivo situations.
Keywords: cell migration, extracellular matrix, macrophages, MMP, podosomes, proteases
Macrophage Migration
Macrophages are an important part of the innate immune system and can be found throughout the human body. As tissue resident macrophages, they constantly survey their immediate surroundings,1 and as professional phagocytes, they remove apoptotic cells and cellular debris, participate in the host response to infectious diseases, and perform tissue remodeling after injury.2-4 The ability of macrophages to migrate through most tissues in the human body is a prerequisite to fulfill their various functions. However, movement of macrophages through tissues can also be detrimental, as in the case of tumor-associated macrophages. These cells can be found in close proximity to cancer cells5,6 and are associated with poor prognosis for cancer patients,7 which is based on the promotion of tumor growth and cancer cell metastasis.8 Moreover, macrophage tissue infiltration plays a critical role in several pathological conditions such as neurodegenerative diseases and chronic inflammation.9
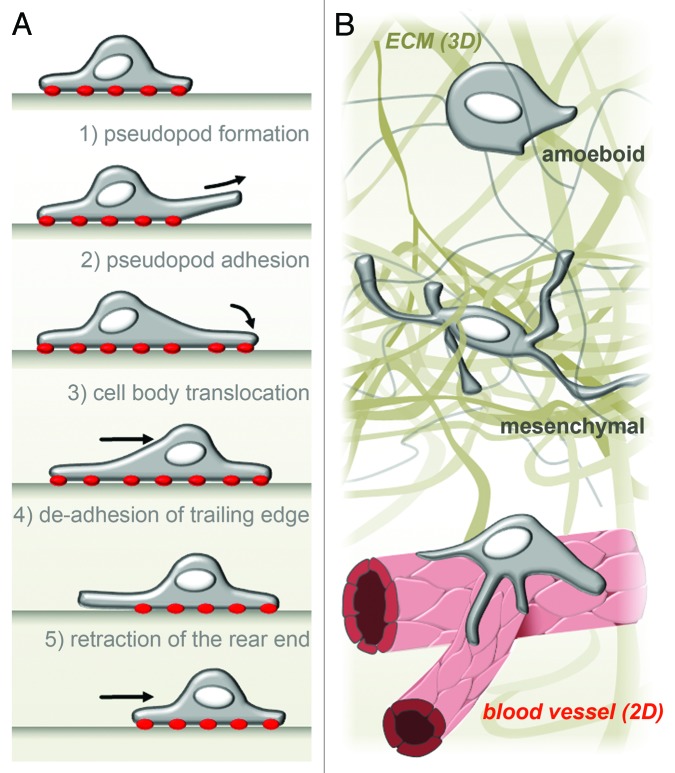
According to the classical cycling five-step model of cell migration (Fig. 1A), movement of cells on 2D surfaces comprises (1) the formation of a leading pseudopod; (2) the adhesion to matrix contacts; (3) translocation of the cell body; (4) the release of the rear edge; and (5) the retraction and recycling of membrane and receptors from the rear to the front of the cell.10 Indeed, macrophages encounter 2D surfaces in vivo, such as the endothelial monolayers of blood vessels11 (Fig. 1B) or extracellular matrix (ECM) barriers, such as basement membranes.12 Macrophage locomotion occurs also in the context of three-dimensional (3D) interstitial matrices, which exhibit a variety of different mechanical and biochemical compositions. Comparable to cancer cells,13-15 macrophages can apply at least two distinct migration modes while moving through or infiltrating into three-dimensional matrices: amoeboid or mesenchymal migration (Fig. 1B).16-18 The amoeboid migration mode is characterized by a spherical cell shape with a small number of short protrusions and a relatively high velocity (~0.7 µm/min). Amoeboid migration strongly depends on the Rho/ROCK pathway, which regulates actomyosin contractility, but not on proteolytic activity.17,19 In contrast, in the mesenchymal mode, cells display an elongated morphology with multiple long protrusions and display low migration speed (~0.2 µm/min). Importantly, mesenchymal migration strongly depends on proteolytic degradation of matrix material.17,19 A further difference between the two migration modes is that amoeboid migration shows only weak or no dependency on integrin-mediated adhesion,20 whereas mesenchymal migration is strongly dependent on adhesion to the ECM.15,21 However, during the last years, studies on cancer cells proposed at least a third mode, corresponding to an intermediate state between amoeboid and mesenchymal migration. In this pseudopodial amoeboid migration, cells utilize the deformability of the nucleus to enable migration through narrow spaces of the ECM without degradation of the surrounding ECM. Cells using this non-proteolytic process strongly depend on adhesion to the ECM via integrins, as in mesenchymal migration, but also on cell contractility mediated by Rho/ROCK, as in amoeboid migration.19,22 Due to the variable restrictions posed by a changing environment, cells seem to quickly adapt the necessary abilities such as adhesion, nucleus deformation, or matrix proteolysis to achieve maximal locomotion, which may result in a continuum of migration modes, in which amoeboid or mesenchymal migration constitute only the most extreme variants.19
Figure 1. Macrophage migration in 2D and 3D. (A) In vitro, on 2D surfaces, human macrophages adopt a rounded, flat cell shape and follow a classical five-step model of cell migration. Adhesion to the substrate is marked by red dots. (B) In vivo, macrophages are confronted with both 2D (B, bottom) and 3D (B, middle and top) situations. For example, during vascularization, macrophages can be found on vascular junctions attached to a 2D endothelial wall. Depending on the extracellular environment, macrophages migrating through interstitial space can adopt a rounded cell shape and use the amoeboid, non-proteolytic migration mode (B top) or a prolonged protrusion-rich morphology and use mesenchymal, proteolysis-dependent migration (B middle).
Podosomes and Proteolytic Migration
Podosomes in 2D
In macrophages, adhesion and proteolytic degradation of extracellular matrix (ECM) material are closely linked processes. Central for both functions are podosomes, which belong to the group of invasion-mediating adhesions (invadosomes) that exhibit the ability for focal degradation of the ECM by matrix–lytic proteases.23,24 Podosomes are constitutively formed in cells of the monocytic lineage such as macrophages,25 dendritic cells,26 and osteoclasts,27 but can also be found or induced in a variety of other cell types, including endothelial28 and smooth muscle cells.29
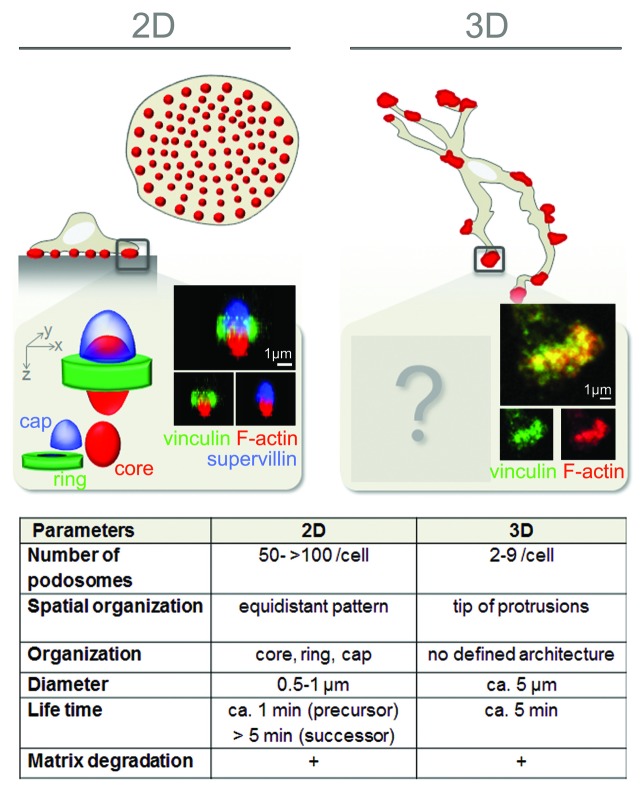
On 2D surfaces, primary human macrophages display an evenly spaced pattern of constitutively formed podosomes, ranging in numbers from 10 to several hundred per cell.30-32 Macrophage podosomes typically appear in a dot-like shape and exhibit a tripartite architecture (Fig. 2). They consist of a densely packed F-actin-rich core, which also contains actin-associated proteins (e.g., Arp2/3 complex, cortactin, and gelsolin), and a ring structure surrounding the core, which contains adhesion plaque proteins, such as vinculin and talin.33,34 Strikingly, the application of high-resolution imaging revealed that the podosome ring shows a discontinuous organization and in fact consists of individual clusters that surround the core.35-37 Recent work also revealed the existence of a third substructure at podosomes, the so-called “cap,” which localizes on top of the podosome and partially overlaps with core and ring structures. To date, only a few cap proteins have been identified, including the myosin-binding protein supervillin32 and the formin FMNL-1.38 Collectively, these substructures add up to a podosome diameter of approximately 0.5–1 µm, a typical height of 0.2–0.4 µm, and a stiffness of 44 kPa, parameters which remain constant independently of the ECM composition.39,40 Besides these dot-like individual podosomes, also podosome super-structures have been described in macrophages, the so-called podosome rosettes.41 These assemblies are more likely to be formed in macrophages activated by M-CSF42 or LPS +/− IFNγ41 and form an area of ECM degradation, which is larger than that of individual podosomes but still focused.
Figure 2. Macrophage podosomes in 2D and 3D. Schematic overview over podosome structure and ultrastructure in human macrophages attached to a 2D surface (left) or embedded in a 3D matrix (right). In 2D, macrophages form numerous dot-like podosomes (red dots) on the substrate-attached cell site. Podosome components are organized in core, ring, and cap structures. Reconstructed z-stacks of confocal micrographs of a single podosome show typical members of the different substructures (F-actin, vinculin, and supervillin) (adapted with permission from Bhuwania et al.32). In dense 3D environments, human macrophages form 3D podosomes at protrusions. So far, no substructures have been described, and components of the core and ring of 2D podosomes mostly co-localize (see confocal micrographs of F-actin and vinculin; adapted with permission from van Goethem et al.57). The table lists several parameters typical of podosomes in 2D and 3D, respectively.
Podosomes are highly dynamic organelles, which undergo constant rearrangement. Several studies on the lifespan of podosomes have described an average period of 2–12 min from the point of assembly of the structure to its disassembly.27,30-32,43 In addition to dissolution, also fission or fusion with neighboring podosomes has been observed.31,43,44 A recent study has revealed a succession series of some typical podosome marker proteins in murine osteoclast-like cells,45 although the precise sequence of recruitment or turnover of many podosome components is still unknown.
In monocyte-derived cells, at least two subgroups of podosomes occur, which can be defined by their lifetime, size, and dynamic behavior: so-called “precursor” podosomes are larger structures, preferentially localized at the cell periphery (Fig. 2) or the leading edge of polarized cells. They are characterized by their high rates of fission and fusion.43,46 In contrast, “successor” podosomes localize more centrally and are also more stable.32,43 As the name suggests, successor podosomes are not only localized behind precursors, relative to the cell edges, but a substantial part of the successor subpopulation also develops through fission of precursors.32,43,46 Little is known about differences in composition or molecular mechanisms that regulate these subpopulations. So far, supervillin was found to localize preferentially to successor podosomes and to recruit myosin-dependent contractility especially to this podosome subset.32
Work on the function of podosomes has progressed steadily during the last decade. It is now well-accepted that they act as (1) adhesive structures,37,46 (2) mechanosensors and—transducers,37,47 and (3) are able to degrade extracellular matrix.42,49 Evidence for these functions is based on (1) the presence of integrins and other ECM receptors, such as the hyaluronic acid receptor CD44 at podosomes,47,50-52 and (2) the ability of podosome components to act as mechanotransducers by converting mechanical cues into chemical signals. Talin, for example, has been shown to undergo force‐induced conformational changes in in vitro experiments on adhesion structures, resulting in the exposure of a vinculin binding site,53,54 and it may undergo a similar transition in situ at podosomes. Moreover, podosomes can also sense the topography of the substratum, as shown for dendritic cells.40,55,56 (3) The presence of proteases with lytic ability toward components of the ECM indicates that macrophage podosomes are also functional in matrix degradation.24,39 For detailed information, see section Podosomes and matrix degradation.
Podosomes in 3D
Compared with 2D situations, as described above, 3D systems exhibit additional physiochemical signals, which can have significant influence on both inter- and intracellular processes. Accordingly, 3D in vitro models that mimic the scenario of cells in a tissue context have received increased attention during the last years. Recent work on monocyte‐derived macrophages migrating through gelled collagen I demonstrated the existence of actin-rich structures at the end of long cell protrusions, which resemble podosomes in composition and function and are termed here “3D podosomes” (Fig. 2).17,57-59 In good agreement with podosomes as local sites of matrix degradation, 3D podosomes are only formed during protease-dependent mesenchymal migration, but not during protease-independent amoeboid migration.17,57
Comparable to podosomes in 2D, 3D podosomes are F-actin accumulations that show an enrichment of several classical podosome marker proteins, such as vinculin or cortactin.47,58 However, the typical tripartite architecture of 2D podosomes (core, ring, cap) is not apparent in the 3D context. This might, in part, be due to the general difficulty of visualizing subcellular structures such as adhesions structures in a 3D context.60 In addition, it might also point to a different architecture of 3D podosomes, compared with their 2D counterparts. For example, podosome ring proteins, such as vinculin and paxillin, do not surround the F-actin core of 3D podosomes but are found mostly at their distal ends, at the tip of the podosome-forming protrusion.57 Also, in contrast to the high numbers of podosomes formed on 2D surfaces (> 100 podosomes/cell), 3D podosomes show strongly decreased numbers (2–9 3D podosomes/cell). Concomitantly, 3D podosomes are bigger in size (~5 µm in diameter), while their lifetime is in the range of regular podosomes (4.9 ± 0.5 min).57 Conversely, not all F-actin-enriched accumulations at protrusions are positive for the typical podosome components.57 This could be based on a variety of non-exclusive reasons: (1) some F-actin accumulations have no relation at all to podosomes; (2) similar to different stages of podosome formation45 or invadopodia maturation,61 3D podosomes might exhibit different maturation steps; (3) alternatively, this could also reflect the directional nature of mesenchymal migration, with 3D podosomes only forming at the migration front.
The primary functions of podosomes in 2D have been defined as adhesion to the underlying matrix, as mechanosensing and the generation of traction forces as well as the degradation of extracellular matrix.39 It is likely that 3D podosomes exhibit adhesive properties, which is based on the presence of ECM receptors, such as β1 integrins and CD44.57 Much less is known about the role of 3D podosomes in the transduction of forces inside the cell and from the cell to the matrix. Still, force generation by migrating primary human macrophages on 3D fibrillar collagen has been observed (C. Wiesner et al., unpublished data).62 To date, degradation of extracellular matrix is the best studied function of 3D podosomes,17,57,59,62 as described in the following sections.
Collectively, these results indicate that 3D podosomes observed in collagen and MatrigelTM networks indeed constitute the 3D counterparts of the classical podosomes found in 2D. This is based on the typical molecular composition, although the components appear to be arranged in a slightly different architecture, and also on the co-localization of 3D podosomes with sites of matrix degradation.57,62
Podosomes and matrix degradation
Efficient ECM degradation by podosomes requires both the structural integrity of podosomes, as well as tightly regulated trafficking of proteases to podosomes.30-32,49,62 For example, deletion of proteins involved in the stability and organization of podosomes, such as filamin A and Hck, results in altered podosome-mediated matrix degradation in 2D and also in deficient 3D mesenchymal migration.16,18 The degradation of gelatin (a hydrolyzed form of collagen) by macrophages depends on the formation of podosomes and the recruitment of proteolytic enzymes to these structures.17,49 So far, the actual process of protease secretion or surface exposure at podosomes has not been demonstrated. However, the enrichment of matrix–lytic enzymes at podosomes63-65 and the dot-like pattern of degraded matrix underneath podosomes clearly indicates that podosomes are sites of focalized ECM degradation.47,49 Comparable to their 2D counterparts, 3D podosomes formed within collagen gels have been shown to be targeted by proteases and to represent sites of focal degradation in 3D environments.57,59,62 In addition to focalized matrix degradation, 3D podosomes probably also participate in the formation of tunnels in the matrix, as observed when macrophages migrate into 3D MatrigelTM.57
To date, several types of proteases involved in the breakdown of ECM components have been identified,65 including metalloproteases such as matrix metalloproteases (MMPs) and ADAMs (a disintegrin and metalloprotease),67 as well as serine proteases and their associated receptors, the plasmin activation (PA) system,68,69 and cysteine proteases (e.g., cysteine cathepsins [Cts]).70 Depending on the presence of membrane-anchoring motifs, such as glycosylphosphatidylinositol (GPI) anchors or a transmembrane domain, ECM–lytic proteases can be either exposed at the cell surface for focal degradation or secreted into the extracellular space (diffused proteolysis).58
In humans, there are 24 known members of the matrix–metalloprotease family, which are either secreted or associated with the plasma membrane via transmembrane domains (membrane type-MMPs [MT-MMPs]) or GPI anchors.71 From this group, the membrane-type metalloproteinase MT1-MMP (MMP-14) has emerged as a “master-switch” protease, as it cleaves a variety of matrix proteins and matrix receptors, and also proteolytically activates other MMPs such as MMP-2.72-74 Vesicles containing MT1-MMP have been shown to be recruited to podosomes via podosome-contacting microtubules.43,49,62 Accordingly, MT1-MMP has been detected at podosomes65,75,76 and its activity was shown to be important for podosomal matrix degradation.49,65 Consistently, overexpression of MT1-MMP results in an enhanced invasive ability of cells, while its silencing leads to an almost complete inhibition of cell migration and infiltration.62,77 By contrast, the potential connection between cysteine cathepsins and podosomes in 2D is still unclear. Cathepsins belong to the family of cysteine proteases and include 11 members in humans.78 They are mostly sorted into lysosomal compartments, but can also be secreted, either as active enzymes or inactive proenzymes.79 Cathepsin B and S have been reported to be involved in the 2D migration of tumor-associated macrophages.80 However, cathepsin activity in macrophages has been linked to the trailing end of cells, whereas podosomes are usually found at the leading edge of migrating macrophages.46,57 In fact, a cocktail of protease inhibitors targeting mostly lysosomal proteases does not inhibit gelatin degradation by podosomes, while the general MMP inhibitor GM6001 does.17
In contrast to cathepsins, plasmin has been described to be localized at the leading edge of migrating macrophages.81 However, similar to cathepsins, also plasminogen activation seems to be necessary for matrix remodeling on 2D surfaces.82,83 Concomitantly, the urokinase plasmin activator uPA has been reported to be required for pericellular matrix remodeling, as peritoneal macrophages from uPA-knockout (uPA−/−) mice are not able to degrade the ECM.81
Adding additional complexity, activation and regulation events overlap between the different groups of proteases. In the case of extracellular proteolysis, for example, active cathepsin B in the pericellular space can promote the activation of uPA, which subsequently activates plasmin, which in turn, can activate specific MMPs.84,85 Interestingly, in MatrigelTM, optimal inhibition of macrophage mesenchymal migration is only obtained by a mix of MMP-, cysteine-, serine-, and aspartic-protease inhibitors, suggesting that all of these protease families could be involved.17
The relationship between matrix degradation and podosome activity on 2D surfaces is well explored. By contrast, much less is known about the formation of podosomes in 3D and the potential recruitment of lytic enzymes to these structures. This may in part be due to the fact that the experimental analysis of macrophages migrating through 3D matrices poses a particular challenge. So far, accumulations of surface-localized MT1-MMP have been described at 3D podosomes in macrophages.62 Consistently, also focal degradation of ECM components has been demonstrated at these structures.57,62 In dense ECM material, such as MatrigelTM or gelled collagen I, macrophage migration shows a strong dependency on proteolytic activity.16,17 Consistently, inference with the activity of individual MMP isoforms (such as MT1-MMP, MMP-10, MMP-13) has resulted in a prominent decrease in the proteolytic migration of macrophages in vitro.62,86,87 By contrast, other studies have reported that broad inhibition of MMPs or the loss of MMP-2 and MMP-9, respectively, showed no influence on macrophage infiltration into MatrigelTM.17,88,89 Therefore, the link between MMPs and 3D podosomes with mesenchymal migration of macrophages is still poorly defined and needs to be further investigated.
In the case of cysteine cathepsins, various isoforms (CtsB, -H, -L, -S, -X) have been shown to localize at 3D podosomes in macrophages embedded in MatrigelTM. Jevnikar et al.59 described finger- and cup-like structures of cathepsins at 3D podosomes of primary human macrophages, which may hint at the existence of a more structured architecture of 3D podosomes.59 Cathepsins have also been associated with macrophages in mesenchymal migration through MatrigelTM, linking CtsB, -L, and -S with extracellular digestion and CtsB and –L with ingestion of dense collagen material.59 Furthermore, the serine protease plasmin has also been linked with migration of murine macrophages.90 In line with this study, murine uPA- and uPAR-knockout macrophages also show severe migration defects through MatrigelTM.91 However, the existence of a potential link to 3D podosomes is currently unexplored.
In conclusion, comparable to the ECM lytic activity of 2D podosomes, 3D podosomes have been associated with degradation of ECM components and mesenchymal migration of macrophages through dense ECM material. However, due to sometimes contradictory results from different studies, the relative impact of specific classes of proteases on 3D podosome-dependent matrix degradation is currently unclear. A possible hypothesis is that macrophages use distinct sets of proteases depending on the type of 3D matrix they infiltrate.
Determinants of proteolytic migration
The addition of a third dimension in cell migration experiments adds further complexity to the studied system. Most of the studies on migration and degradation in 2D use thin layers of matrix material on a carrier such as glass. Due to the small height of the matrix in these experiments, the most important parameters are the stiffness of the glass coverslip and the molecular composition of the matrix, rather than its architecture or viscoelastic properties. In a 3D context, physical properties of the matrix, as well as cellular determinants, such as the deformability of the nucleus,22 have crucial roles for the migration of cells through the matrix (Fig. 3). The following section provides an overview over cell- and matrix-dependent parameters, which determine the eventual migration mode.
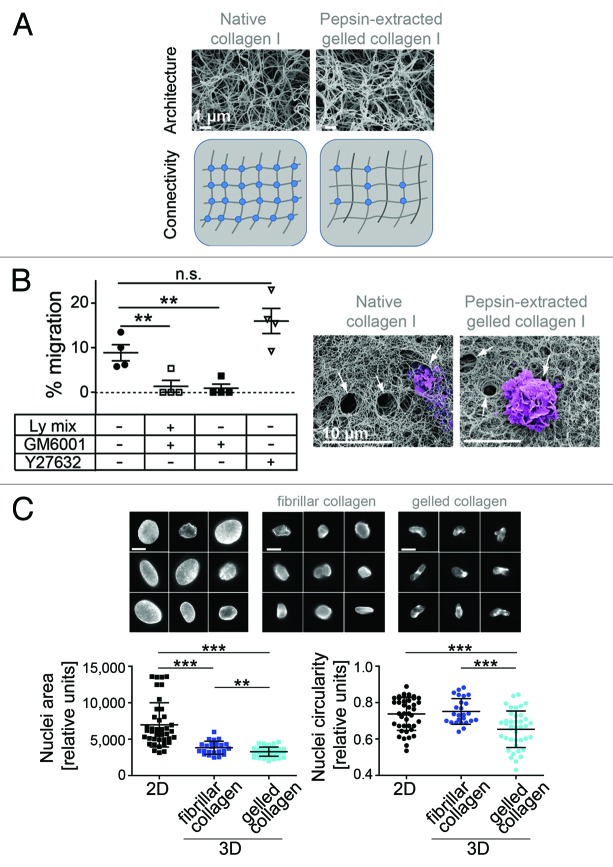
Figure 3. Mesenchymal migration involves distinct proteases and nuclear constriction. (A) SEM micrographs (with permission from Renaud Poincloux, Tri Imaging Toulouse France; bars: 1 µm) of native collagen I and pepsin-extracted gelled collagen I show similar architecture and schematic presentation of distinct inter-fiber connectivity. (B) The percentage of macrophages migrating through native collagen I in the absence or presence of Ly-mix (lysosomal protease inhibitors) and/or GM6001 (MMP inhibitor), or Y27632 (ROCK inhibitor) was quantified. Macrophage migration through native collagen is not inhibited by Y27632 and is abolished by GM6001, indicating that cells use the mesenchymal migration mode, but not the amoeboid migration. hMDMs isolated from four independent healthy donors (n = 4). Statistics: two-tailed unpaired Student t test; **P < 0.005; n.s. P = 0.165. Macrophages penetrating matrices were examined by scanning electron microscopy (for methods, see van Goethem et al.17). Note the presence of holes made by cells (purple) in the matrix of native collagen I and pepsin-extracted gelled collagen I (arrows). Representative pictures are shown. Bars: 10 µm. (C) Reduced nuclear circularity, a signature of mesenchymal migration. Galleries showing confocal micrographs of macrophage nuclei stained with DAPI 72 h post-seeding (bars: 1 µm). Macrophages on 2D surfaces (upper left) mostly adopt the “fried egg” morphology, and nuclei are flattened, as shown by the higher nuclear area compared with nuclear area of cells in 3D matrices (upper middle and right). 3D matrices were prepared using pepsin-treated fibrillar collagen I. In gelled collagen I (upper right), cells use the mesenchymal migration mode and display a higher deformation of the nucleus compared with cells in fibrillar collagen, which use the amoeboid mode (upper middle). Both nuclear area (lower left graph) and nuclear circularity (lower right graph) were measured. Statistics: two-tailed unpaired Student t test. ***P < 0.0001, **P = 0.0056. Nuclear area and circularity were measured in a total of 25 (for fibrillar collagen) and 40 (for gelled collagen and 2D) macrophages from each time three independent donors.
Besides cellular parameters, migration modes are determined by mechanical, structural and biochemical properties of the surrounding matrix. Accordingly, even slight variations in a single parameter can result in great differences in outcome.15 Careful comparison of different studies is thus indispensable for correct interpretation of results. Most differences are caused by the choice of matrix material. For example, different types of collagens (e.g., type I or IV) or MatrigelTM are used in eukaryotic cell-based migration assays. MatrigelTM is a multi-component gelatinous matrix, derived from Engelbreth-Holm-Swarm (EHS) mouse sarcoma. It is composed of structural proteins, such as laminin, entactin, and collagen, as well as several growth factors.92 Due to its multi-component nature, this matrix is often believed to be closer to the physiological situation, compared to use of a single specific matrix component. However, MatrigelTM might not be the perfect option for experiments, which focus on the influence of specific matrix components. Still, even use of a single component system, such as type I collagen matrices, does not ensure direct comparability of experiments. This is due to the fact that collagen I can form different microstructures, such as a gel or various fibrillar states with multiple fibril organizations that vary in density or thickness, and also in the resulting pore size. These situations can be induced by changing the temperature, the acidity, ionic strength, ion stoichiometry, or monomer concentration during matrix polymerization.93-95 Moreover, also viscoelastic properties of the material contribute to the elasticity of the matrix, which is based on intra- and intermolecular crosslinks.96 While intramolecular connections are regulated by the specific nature of the molecule (e.g., collagens, laminin, fibronectin), intermolecular connectivity can be considerably modified due to diverse treatments of the material prior to or during polymerization.15,17,97 Currently, a number of collagen substrates are commercially available, which differ in their origin (e.g., rat, bovine), tissue source (e.g., skin, tendon, placenta), and isolation conditions (e.g., different degrees of salinity and acidity, application of proteolytic enzymes [pepsin or pronase] or enzyme-free preparations).15,97,98 Perhaps the most pronounced differences have been demonstrated for collagens treated with pepsin during the isolation procedure, compared with collagens isolated without use of enzymes.15,97,99 Pepsin-treated collagen generates networks of lower connectivity (lower number of intermolecular cross-links), and consequently, of a higher viscosity. However, compared with enzyme-free collagen (e.g., native collagen), it provides equal characteristics in pore size and density of fibrils, thus similar architecture.15,97 When macrophage migration is studied in fibrillar pepsin-treated collagen I, which forms large pores and poorly connected fibers, they use the amoeboid mode.17 In contrast, they use the mesenchymal mode in both pepsin-treated gelled collagen I (small pores, poorly interconnected fibers) and enzyme-free collagen I (small pores, highly interconnected fibers) polymerized at neutral pH. These results further support our previous data showing the importance of the matrix architecture in the choice of the migration mode.17 Of note, protease-dependent migration in enzyme-free collagen requires only MMP activity (Fig. 3B), whereas this is not essential for mesenchymal migration in pepsin-treated collagen I.17 The difference between these two matrices lies in the presence of intermolecular cross-links between enzyme-free collagen fibers15 (Fig. 3A). Whether it also affects the type of enzyme required for macrophage migration deserves further investigation.
For a comparison of the described experimental conditions with in vivo situations, it has to be considered that collagen-rich connective tissues are heterogeneous, with both dense and loose areas. In vivo approaches like confocal reflection microscopy or two-photon excited second or third harmonic generation imaging do not provide a complete picture of the tissue architecture. However, the pore diameter of human dermis, a loose tissue, is similar to the pore diameter of fibrillar collagen polymerized in vitro.100 In addition, the viscosity and elasticity values of the matrices polymerized in vitro are in the range of 100 Pa,17 a value also found for brain tissue.101
Moreover, matrices exert physical constraints on migrating cells. Nuclear deformation has been described in tumor cells.22 In macrophages, when they use the amoeboid mode in fibrillar collagen, we observed that the circularity of the nucleus was not affected, whereas it was markedly reduced (Fig. 3C) when cells use the mesenchymal mode in gelled collagen I and dig narrow tunnels to move.57
In summary, the migration of macrophages is determined by cellular and matrix-based parameters such as abilities for cellular, mostly nuclear, and matrix deformation. Porous matrices characterized by physical deformability, induce moderate nuclear contraction and non-proteolytic amoeboid migration. Dense matrices characterized by a high level of connectivity also necessitate nuclear deformation, proteolytic mesenchymal migration, and thus the formation of 3D podosomes.
Migration Assays in 2D and 3D
General remarks
The ability to infiltrate tissues is crucial for macrophages to fulfill their physiological role. To correctly mimic this process in vitro, and in order to gain reproducible values, it is important to limit the number of variables, such as physical or chemical parameters, and to choose the appropriate dimensionality of the assay. This results mostly in a choice between assays investigating random or directional migration in 2D vs. 3D environments,102,103 either of single cell types or of co-cultures.104,105 In particular, different kinds of 3D migration assays have been established for studies of macrophages and other cell types,102 including ex vivo explants (such as dermal explants),106 tumor cell spheroids,6 and matrices polymerized in vitro (e.g., MatrigelTM, fibrillar/gelled type I collagen, or native basement membrane).17,55,58,59,107 However, each type of assay presents intrinsic disadvantages, for example, limited access for imaging processes in the case of thick 3D matrices. It is therefore important to consider both benefits and disadvantages before choosing the correct assay for a specific study. Some of the most commonly used assays, their relation to the study of (macrophage) migration and invasion, as well as their respective advantages and limitations, are discussed in the following section.
Migration assays in 2D and 3D
Until recently, 2D migration assays, such as scratch wound or Boyden chamber assays, have been the most utilized set ups for studying cell migration.108 While the molecular and cellular mechanisms involved in leukocyte 2D migration have been extensively studied, three-dimensional migration of macrophages is only beginning to be investigated. To date, a number of different matrix materials are commercially available, such as multi-component basement membrane extracts (BME) (e.g., MatrigelTM [BD Biosciences]; Geltrex® [life technologies]; MaxGelTM [Sigma-Aldrich]), single component ECM extracts either produced in vitro by cell lines (e.g., laminin), or isolated from diverse animal sources (e.g., type I collagen [from rat tail], fibronectin [from bovine plasma]), as well as hydrogel-based synthetic scaffolds (e.g., HydroMaxTM [Sigma-Aldrich], MAPTrixTM [Kollodis Biosciences]). Furthermore, tumor cell spheroids, which are organotypic-like structures comprising cells and ECM proteins6 and ex vivo preparations (such as dermal or bone explants) have been used in these assays.106,109 However, it has to be stressed that the often applied reasoning “2D = in vitro,” and “3D = in vivo,” is not stringent. For example, 2D situations of cell migration also exist in vivo, such as movement of leukocytes along vessel walls or along the lumen of organs.39 Moreover, simply embedding cells in 3D matrices, without knowledge of their exact structural and physical properties, may not lead to results that reflect the specific situation within certain tissues.
Primary human macrophages are usually cultured on stiff plastic or glass surfaces, without matrix coatings.110 This is appropriate for 2D random migration assays, where quantifiable parameters include speed, distance over time, the number of migrating cells, and the directionality of movement (meandering index; Fig. 4A).111 The addition of a chemoattractant (e.g., M-CSF,112 MCP-1,113 or MIP-1β114) for gradient formation enables the study of directional migration (Fig. 4B).115,116 Moreover, gap closure assays, commonly used for monolayer-forming cells such as primary endothelial or tumor cells, can also be utilized in set ups with macrophages. The creation of a gap by scratching the cell population induces directional movement of the cells without the addition of a chemoattractant (Fig. 4C).117 This method is cost-effective and requires no specific equipment. However, the mostly manually performed scratch (using a needle or pipette tip) often results in an uneven cell-free area. Additionally, application of a scratch causes changes in the chemical environment, due to the release of components from damaged cells. To avoid this, gaps can be generated by seeding cells around a barrier (for example silicon stoppers), which are subsequently removed (Fig. 4D). This results in uniform borders of the cell-free area, mostly without cellular damage. Silicon stoppers are commercially available in different shapes (linear or round) and are mostly designed for 96-well plates. Collectively, these 2D migration assays are easily adaptable for real-time imaging and available software packages (e.g., Volocity® 3D Image Analysis Software [Perkin Elmer] or Imaris Track [Bitplane])118 and enable high-throughput analysis. In addition, the carrier surface of these assays can be coated with a thin layer of specific matrix. However, it has to be considered that thin matrix layers on rigid substrates, such as glass or plastic, display a higher rigidity compared with thick layers or three-dimensional gels of the same matrix material.97,119 Furthermore, scratching or removal of an insert may damage the matrix coating, thus affecting the movement of cells.
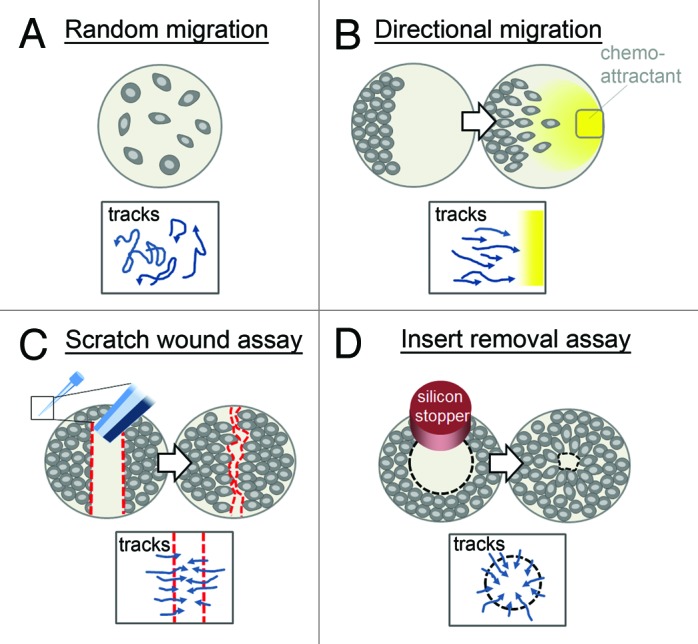
Figure 4. Assays for the study of macrophage migration in 2D. In vitro, migration of macrophages on 2D surfaces can be analyzed by monitoring (A) the random movement of spontaneously migrating cells, (B) the directional movement of cells along a chemoattractant gradient (yellow; e.g., macrophage-colony stimulating factor), or (C and D) the directional movement of cells into a gap, which can be generated by the application of (C) a wound (red dotted line; based on a scratch by a pipette tip or needle) or (D) the removal of a silicon stopper. Directions of possible cell movement are indicated in boxes below. In all applications, cells can be imaged over time by standard microscopy methods and tracked/quantified with imaging software.
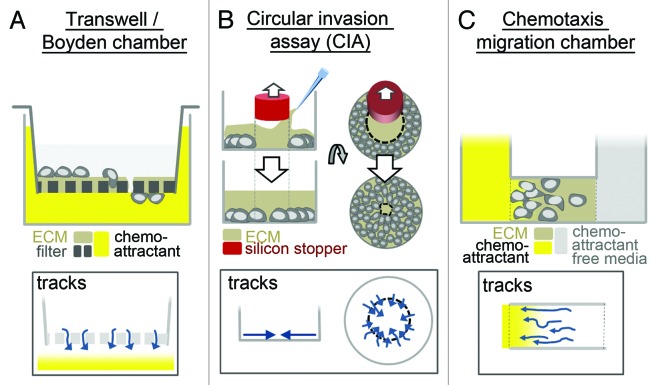
In another type of assay, the transmembrane/Boyden chamber, cells in a chamber of attractant-free medium are separated by a filter from a chamber containing media with an attractant. The cells can transmigrate through the pores of the filter120 (Fig. 5A), with subsequent quantification of the transmigrated cells. Modification of this system by the addition of ECM gels on top and within the pores of the filter allows the study of invasive cell migration,121 and multi-well systems (96-well plates) are available for respective high-throughput screenings. However, actual tracking of individual cells is not possible, which limits this assay to an end-point measurement of the number of transmigrated cells. A further way of including 3D gels in an otherwise 2D system is performing gap closure assays in the presence of an ECM gel placed on top of the cells and within the gap122 (Fig. 5B). This particular setup also enables high-throughput approaches, with the added advantage of an option for live cell imaging. However, cells still move along a relatively stiff surface (a pre-coated thin layer of ECM), which results in a not completely three-dimensional context.
Figure 5. Migration assays for macrophages on 2D surfaces embedded in 3D matrices. Several assays enable the study of macrophage migration through 3D matrix (such as MatrigelTM or type I collagen). However, cells still mostly adhere to 2D surfaces within these assays. (A) Standard Boyden chamber, with a thin layer of matrix added on the filter and within the pores, (B) gap closure assay (circular invasion assay), with a thick layer of matrix added on top, (C) chemotaxis chamber, with cells the seeded in 3D matrix. In Boyden/Chemotaxis chambers, cells migrate along a chemoattractant gradient (yellow), whereas in gap closure assays, cells migrate into the generated gap without additional stimulation. Directions of possible cell movement are indicated in boxes below. In all applications, cells can be imaged by standard microscopy methods. Note that Boyden chambers do not allow monitoring over time, as gap closure or chemotaxis chamber assays do, but enable easy access to the cells (e.g., for staining procedures).
Insertion of three-dimensional matrix gels into a chemotaxis migration chamber enables the study of chemotactic cell migration within a true three-dimensional context.123 Several systems are commercially available, which all share the same principle: cells are embedded in a 3D matrix, which is placed between chemoattractant-containing medium on the one side and chemoattractant-free medium on the other. Over time, a chemoattractant gradient develops within the matrix, inducing chemotactic migration of cells (Fig. 5C). These systems enable live cell imaging and long-term observations. A drawback lies in the mostly small chamber size (e.g., 1 mm in length and 70 µm in height, µ-slides Chemotaxis3D [ibidi]), which often results in adherence of a considerable amount of cells to the chamber walls. Additionally, once it is set up, there is mostly limited scope for manipulation, such as addition of inhibitors.
The development of genuine 3D migration assays, in which cells are surrounded exclusively by matrix material, has opened the door to a new field of cellular in vitro experiments.103 One of the first described 3D invasion assays was the vertical gel invasion assay, in which lymphocytes penetrate into a thick gel of native collagen.124 Since then, multiple assays based on this model have been developed. Usually, a thick (1 to 2 mm) gel of ECM (such as MatrigelTM or collagen I) is polymerized, cells are placed on top of it, and subsequently infiltrate into the gel. Invasive cells can be easily distinguished from non-invasive cells, which remain on the surface of the gel (Fig. 6A). This assay allows quantification of migrating cells vs. non-motile cells, cell imaging by bright field microscopy, and further biochemical/genetic analysis following their isolation out of the gel.17 High-throughput screens use numerous units of gel that can be polymerized in 96-well plates.125 For the incorporation of a chemoattractant gradient in this system, ECM gel needs to be polymerized on top of the filter/membrane of a Boyden chamber. The cells are then placed on top of the gel and migrate through it along the gradient and can be imaged or during that process.6 In this assay, cells have to switch from a 2D situation on top of the gel to a 3D migratory mode in order to penetrate into the matrix. It can be a useful model for cells that have to cross tissue barriers. In addition, self-prepared matrices present the advantage to modify matrix parameters such as biochemical composition, architecture, and viscoelasticity17 to mimic the diversity of interstitial tissues.126-129
Figure 6. Migration assays for macrophages in 3D. Macrophage infiltration into or migration through 3D matrices can be studied by vertical invasion, spherical invasion, or spheroid gel invasion assays. (A) In a vertical invasion assay, cells are placed on top of a thick matrix. Cells start to infiltrate into the gel (top right, red dots) and can be distinguished from non-invasive cells (black dots), which remain on the gel surface. The proportion of migrating cells, their morphology, and the distance of migration can be quantified over time from bright field microscopy image stacks. Cells can also be imaged by confocal microscopy, as shown by the z-stack reconstruction (bottom right) of human macrophages overexpressing creating a tunnel…white line; overexpressed mCherry-Lifeact (red) is used to stain F-actin. Bar: 20 µm (adapted, with permission, from van Goethem et al.57 and Guiet et al.6). (B) Spherical invasion assay (SIA). Cells are embedded in a dense plug of type I collagen, which is surrounded by slightly less dense collagen I, containing a chemoattractant. Cells invade into the surrounding matrix and can be monitored by confocal microscopy over time (right panel, bright field images of human macrophages, time of image acquisition after start of experiment is indicated; bar: 10 µm [adapted, with permission from Wiesner et al.62]). The number of infiltrating cells, as well as distance from the plug border, can be quantified. (C) Spheroid gel invasion assay. Macrophages co-cultured for 3 d with spheroids of cancer cells (diameter: 0.5 mm) penetrate into spheroids. Spheroids infiltrated by macrophages are subsequently embedded in MatrigelTM. Both macrophages and tumor cells leave the spheroid and invade the surrounding matrix. Cells are imaged by multi-photon microscopy (merged micrographs of human macrophages stained with cell tracker [red]; bar: 10 µm). Number of cells moving out of the spheroid (red triangles; bottom scheme) and area of cell invasion (green dotted line) can be quantified (adapted with permission from Guiet et al.6). Schemes at the bottom indicate direction of cell movement within each assay.
Cells can be also embedded in a 3D matrix already at the start of the experiment (spherical invasion assay; Fig. 6B). Cells are seeded in a dense (2.5 mg/ml) fibrillar collagen I plug, which is then transferred into a bigger chamber, and the surrounding space filled with slightly less dense fibrillar collagen I (2 mg/ml) containing a chemoattractant. These modifications are helpful to induce migration from the inner plug into the outer shell. Infiltration of the surrounding shell can be quantified by measuring parameters, such as the number of infiltrated cells, migratory displacement over time, or velocity of migration.62,130 The advantages of this method are the absence of 2D surfaces and the possibility of (live cell) imaging, even by bright field illumination. Chemical agents (such as chemoattractants or inhibitors) can be added at the step of polymerization to the gels, but not during the run of the experiment.
These latter assays can also be used for co-culture and -migration studies of several cell types. For example, small tumors and their surrounding tumor-associated macrophages (TAMs) can be mimicked in vitro by embedding cancer cell spheroids in a 3D matrix6,131 (such as MatrigelTM) and co-embedding of surrounding macrophages6 (Fig. 6C). Invading cells can be detected by optical methods, and parameters such as the infiltrated area, migration distance over time, or the relative amount of tumor cells, which migrate in association to macrophages vs. single cells can be acquired.6 As in the case of the spherical invasion assays, inhibitors or other agents can be applied prior to the polymerization of the matrix. A difficulty common to all 3D gel assays is to image cells that have migrated deep inside the matrix. The use of bright field microscopy and small magnification but large numerical aperture objectives allows to easily image cells within the matrix from the top to the bottom of the 3D matrix, but with a limited resolution. Analysis of intracellular protein intracellular localization by immunofluorescence can be enabled by confocal or multi-photon confocal microscopy.17,57,132 These approaches require cell fixation, which results in shrinking of matrices. In addition, it should be considered that not all used matrices enable immunofluorescence, as, for example, MatrigelTM generates a high fluorescence background.
To date, only a few studies have been published on 3D podosomes in macrophages seeded in a 3D matrix context. These studies used vertical invasion assays,6,16,17,57,59 spheroid gel invasion assays,6 and spherical invasion assays62 to investigate migration processes, the appearance of 3D podosomes, and matrix degradation of macrophages. As matrices, mostly MatrigelTM and type I collagen were selected,6,16,17,57,59,62 although the specific influence of different types of 3D matrices on the formation of 3D podosomes is currently unclear. Especially considering that not all matrices appear to support podosome formation even in 2D to an equal extent,40 this aspect should be addressed carefully in future work also in 3D contexts.
In sum, both 2D and 3D in vitro models are required to elucidate the molecular and cellular mechanisms of migration used by macrophages. The recent developments of 3D environments for the study of macrophage migration are a welcome and indispensable addition that enables us to draw a more complete picture of macrophage motility in the body. However, considering the variability arising from 2D migration assays on glass, plastic, or matrix protein-coated culture device or on devices with distinct stiffness, as well as the variability in 3D migration assays arising from biochemical and physical parameters of the matrix, its connectivity, or elasticity, care should be taken in comparing the results of these assays to specific in vivo situations. Moreover, the dimensionality of the assay should also reflect the specific situation of the studied cells in vivo, such as infiltration within a 3D tissue or crossing of a 2D interface.
All of the assays discussed present their unique combination of advantages and drawbacks, and the ultimate choice of assay depends on the particular scientific question. For the study of 2D podosomes and their dynamics, the directional migration assay (Fig. 4B) is a good choice, as it is easy to set up, and the continued formation and turnover of numerous podosomes at the leading edge of cells allows quantitative analyses. To study macrophage invasion and 3D migration, both transwell (Fig. 6A) and spherical invasion assays (Fig. 6B) are recommended, as they allow easy visualization of cells and podosomes and also quantification of invasion. To study the interplay between macrophages, 3D podosomes, and tumor cells, the spheroid gel invasion assay (Fig. 6C) is particularly appropriate.
Future Perspectives
Macrophages cultured in vitro on stiff 2D surfaces, such as glass or plastic, exhibit a typical “fried egg” morphology. The ventral surface of these cells is covered with numerous podosomes that present a well-defined substructure. Transfer into a softer but dense 3D matrix, such as collagen I or MatrigelTM, results in an altered morphology or cells that form numerous protrusions that dynamically extend from a spindle-like cell body. At the tips of many of these protrusions, F-actin-rich accumulations are formed, which contain many classical podosome marker proteins and proteases, although with no clearly defined substructure. These structures co-localize with sites of matrix degradation and constitute thus most probably the equivalents of podosomes in 3D.
3D podosomes are predominantly formed in macrophages, which migrate through dense ECM material, and thus, adopt the proteolytic mesenchymal migration mode.16,17,57 Accordingly, several cysteine cathepsins have been localized at 3D podosomes,59 and the matrix metalloprotease MT1-MMP has been shown to localize at membrane sites at the tips of cell protrusions, where matrix degradation takes place.62 Although the existence of 3D podosomes has been clearly demonstrated, a variety of questions regarding the structure and function of these organelles are still unanswered. These include the potential existence of substructures, which may depend on the nature or viscoelastic properties of the surrounding matrix, and the potential recruitment and release of other proteases, such as secreted MMP isoforms or the urokinase plasminogen activator system. Furthermore, it is unclear whether other functions of 2D podosomes, such as mechanosensing and transduction,37,48 are also fulfilled by 3D podosomes. Moreover, different maturation and activation states of macrophages might affect the formation, structure, and function of 3D podosomes, as it has been shown for podosomes in 2D.133
It is also an intriguing speculation that podosomes and invadopodia, which present as similar but distinct structures in 2D systems,39 might show convergence in their morphology and properties in cells that are present in a 3D context. The more invadopodia-like morphology and localization of podosomes in 3D, as well as the relevance of 3D podosomes and invadopodia for 3D infiltration of cells,16,56,134,135 should provide a sufficient basis to stimulate respective research.
To address all these questions, 3D in vitro and ex vivo experimental systems will be indispensable. Already, the introduction of these systems has proven helpful in reproducing conditions that macrophages are likely to encounter within a tissue context. However, the wide variety of tissues which all exhibit their unique combination of physical, structural, and chemical properties still makes the recreation of in vivo situations challenging. A further task for the future will thus also be the development of fine-tuned 3D systems, which reliably mimic the physical and chemical properties of specific tissues.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Research in the Linder S lab has received funding from Deutsche Forschungsgemeinschaft (LI925/2-1, LI925/3-1), Wilhelm Sander-Stiftung (2007.0202.02), and the European Union’s Seventh Framework Program (FP7/2007-2013) under grant agreement number FP7-237946. Research in the Maridonneau-Parini I lab has been supported by ANR 2010 Midi 01301,FRM DEQ 20110421312 and the European Community’s Seventh Framework Program (FP7/2007-2013) under grant agreement HEALTH-F4-2011-282095. We thank SJ Weiss for providing us with native collagen I and Andrea Mordhorst for excellent technical assistance. We apologize to all authors whose work was not mentioned owing to space limitations.
Glossary
Abbreviations:
- 2D
two-dimensional
- 3D
three-dimensional
- ADAM
a disintegrin and metalloprotease
- BME
basement membrane extracts
- CD44
cluster of differentiation 44
- CIA
circular invasion assay
- Cts
cysteine cathepsins
- DAPI
4′,6-Diamidin-2-phenylindol
- ECM
extracellular matrix
- EHS
Engelbreth-Holm-Swarm
- FMNL-1
Formin-like protein 1
- GPI
glycosylphophatidylinositol
- hMDMs
human monocyte derived macrophages
- IFNγ
interferon gamma
- kPa
kilopascal
- LPS
lipopolysaccharide
- M-CSF
macrophage-colony stimulating factor
- µm
micrometer
- mg
milligram
- min
minutes
- ml
milliliter
- mm
millimeter
- MMP
matrix metalloprotease
- MT-MMP
membrane type-MMP
- n.s.
not significant
- Rho
Ras homologue
- ROCK
Rho-associated kinase
- SIA
spherical invasion assay
- uPA
urokinase plasmin activator
- uPAR
urokinase plasmin activator receptor
References
- 1.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–37. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erwig LP, Henson PM. Immunological consequences of apoptotic cell phagocytosis. Am J Pathol. 2007;171:2–8. doi: 10.2353/ajpath.2007.070135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Twigg HL., 3rd Macrophages in innate and acquired immunity. Semin Respir Crit Care Med. 2004;25:21–31. doi: 10.1055/s-2004-822302. [DOI] [PubMed] [Google Scholar]
- 4.Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol. 2013;229:176–85. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- 5.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–56. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 6.Guiet R, Van Goethem E, Cougoule C, Balor S, Valette A, Al Saati T, Lowell CA, Le Cabec V, Maridonneau-Parini I. The process of macrophage migration promotes matrix metalloproteinase-independent invasion by tumor cells. J Immunol. 2011;187:3806–14. doi: 10.4049/jimmunol.1101245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196:254–65. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 8.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–55. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheetz MP, Felsenfeld D, Galbraith CG, Choquet D. Cell migration as a five-step cycle. Biochem Soc Symp. 1999;65:233–43. [PubMed] [Google Scholar]
- 11.Fantin A, Vieira JM, Gestri G, Denti L, Schwarz Q, Prykhozhij S, Peri F, Wilson SW, Ruhrberg C. Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood. 2010;116:829–40. doi: 10.1182/blood-2009-12-257832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boehncke WH, Wortmann S, Kaufmann R, Mielke V, Sterry W. A subset of macrophages located along the basement membrane (“lining cells”) is a characteristic histopathological feature of psoriasis. Am J Dermatopathol. 1995;17:139–44. doi: 10.1097/00000372-199504000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Friedl P, Zänker KS, Bröcker EB. Cell migration strategies in 3-D extracellular matrix: differences in morphology, cell matrix interactions, and integrin function. Microsc Res Tech. 1998;43:369–78. doi: 10.1002/(SICI)1097-0029(19981201)43:5<369::AID-JEMT3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 14.Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Bröcker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–77. doi: 10.1083/jcb.200209006. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol. 2009;185:11–9. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cougoule C, Le Cabec V, Poincloux R, Al Saati T, Mège JL, Tabouret G, Lowell CA, Laviolette-Malirat N, Maridonneau-Parini I. Three-dimensional migration of macrophages requires Hck for podosome organization and extracellular matrix proteolysis. Blood. 2010;115:1444–52. doi: 10.1182/blood-2009-04-218735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Goethem E, Poincloux R, Gauffre F, Maridonneau-Parini I, Le Cabec V. Matrix architecture dictates three-dimensional migration modes of human macrophages: differential involvement of proteases and podosome-like structures. J Immunol. 2010;184:1049–61. doi: 10.4049/jimmunol.0902223. [DOI] [PubMed] [Google Scholar]
- 18.Guiet R, Vérollet C, Lamsoul I, Cougoule C, Poincloux R, Labrousse A, Calderwood DA, Glogauer M, Lutz PG, Maridonneau-Parini I. Macrophage mesenchymal migration requires podosome stabilization by filamin A. J Biol Chem. 2012;287:13051–62. doi: 10.1074/jbc.M111.307124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–9. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lämmermann T, Sixt M. Mechanical modes of ‘amoeboid’ cell migration. Curr Opin Cell Biol. 2009;21:636–44. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Wolf K, Müller R, Borgmann S, Bröcker EB, Friedl P. Amoeboid shape change and contact guidance: T-lymphocyte crawling through fibrillar collagen is independent of matrix remodeling by MMPs and other proteases. Blood. 2003;102:3262–9. doi: 10.1182/blood-2002-12-3791. b. [DOI] [PubMed] [Google Scholar]
- 22.Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201:1069–84. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gimona M, Buccione R. Adhesions that mediate invasion. Int J Biochem Cell Biol. 2006;38:1875–92. doi: 10.1016/j.biocel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 2007;17:107–17. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Linder S, Nelson D, Weiss M, Aepfelbacher M. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc Natl Acad Sci U S A. 1999;96:9648–53. doi: 10.1073/pnas.96.17.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burns S, Thrasher AJ, Blundell MP, Machesky L, Jones GE. Configuration of human dendritic cell cytoskeleton by Rho GTPases, the WAS protein, and differentiation. Blood. 2001;98:1142–9. doi: 10.1182/blood.V98.4.1142. [DOI] [PubMed] [Google Scholar]
- 27.Destaing O, Saltel F, Géminard JC, Jurdic P, Bard F. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol Biol Cell. 2003;14:407–16. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreau V, Tatin F, Varon C, Génot E. Actin can reorganize into podosomes in aortic endothelial cells, a process controlled by Cdc42 and RhoA. Mol Cell Biol. 2003;23:6809–22. doi: 10.1128/MCB.23.19.6809-6822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgstaller G, Gimona M. Actin cytoskeleton remodelling via local inhibition of contractility at discrete microdomains. J Cell Sci. 2004;117:223–31. doi: 10.1242/jcs.00839. [DOI] [PubMed] [Google Scholar]
- 30.Cornfine S, Himmel M, Kopp P, El Azzouzi K, Wiesner C, Krüger M, Rudel T, Linder S. The kinesin KIF9 and reggie/flotillin proteins regulate matrix degradation by macrophage podosomes. Mol Biol Cell. 2011;22:202–15. doi: 10.1091/mbc.E10-05-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stölting M, Wiesner C, van Vliet V, Butt E, Pavenstädt H, Linder S, Kremerskothen J. Lasp-1 regulates podosome function. PLoS One. 2012;7:e35340. doi: 10.1371/journal.pone.0035340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhuwania R, Cornfine S, Fang Z, Krüger M, Luna EJ, Linder S. Supervillin couples myosin-dependent contractility to podosomes and enables their turnover. J Cell Sci. 2012;125:2300–14. doi: 10.1242/jcs.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linder S, Aepfelbacher M. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 2003;13:376–85. doi: 10.1016/S0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 34.Linder S, Higgs H, Hüfner K, Schwarz K, Pannicke U, Aepfelbacher M. The polarization defect of Wiskott-Aldrich syndrome macrophages is linked to dislocalization of the Arp2/3 complex. J Immunol. 2000;165:221–5. doi: 10.4049/jimmunol.165.1.221. [DOI] [PubMed] [Google Scholar]
- 35.Cox S, Rosten E, Monypenny J, Jovanovic-Talisman T, Burnette DT, Lippincott-Schwartz J, Jones GE, Heintzmann R. Bayesian localization microscopy reveals nanoscale podosome dynamics. Nat Methods. 2011;9:195–200. doi: 10.1038/nmeth.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent C, Siddiqui TA, Schlichter LC. Podosomes in migrating microglia: components and matrix degradation. J Neuroinflammation. 2012;9:190. doi: 10.1186/1742-2094-9-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van den Dries K, Schwartz SL, Byars J, Meddens MB, Bolomini-Vittori M, Lidke DS, Figdor CG, Lidke KA, Cambi A. Dual-color superresolution microscopy reveals nanoscale organization of mechanosensory podosomes. Mol Biol Cell. 2013;24:2112–23. doi: 10.1091/mbc.E12-12-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mersich AT, Miller MR, Chkourko H, Blystone SD. The formin FRL1 (FMNL1) is an essential component of macrophage podosomes. Cytoskeleton (Hoboken) 2010;67:573–85. doi: 10.1002/cm.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linder S, Wiesner C, Himmel M. Degrading devices: invadosomes in proteolytic cell invasion. Annu Rev Cell Dev Biol. 2011;27:185–211. doi: 10.1146/annurev-cellbio-092910-154216. [DOI] [PubMed] [Google Scholar]
- 40.Labernadie A, Thibault C, Vieu C, Maridonneau-Parini I, Charrière GM. Dynamics of podosome stiffness revealed by atomic force microscopy. Proc Natl Acad Sci U S A. 2010;107:21016–21. doi: 10.1073/pnas.1007835107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poincloux R, Vincent C, Labrousse A, Castandet J, Rigo M, Cougoule C, Bordier C, Le Cabec V, Maridonneau-Parini I. Re-arrangements of podosome structures are observed when Hck is activated in myeloid cells. Eur J Cell Biol. 2006;85:327–32. doi: 10.1016/j.ejcb.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi H, Pixley F, Condeelis J. Invadopodia and podosomes in tumor invasion. Eur J Cell Biol. 2006;85:213–8. doi: 10.1016/j.ejcb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 43.Kopp P, Lammers R, Aepfelbacher M, Woehlke G, Rudel T, Machuy N, Steffen W, Linder S. The kinesin KIF1C and microtubule plus ends regulate podosome dynamics in macrophages. Mol Biol Cell. 2006;17:2811–23. doi: 10.1091/mbc.E05-11-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cervero P, Panzer L, Linder S. Podosome reformation in macrophages: assays and analysis. Methods Mol Biol. 2013;1046:97–121. doi: 10.1007/978-1-62703-538-5_6. [DOI] [PubMed] [Google Scholar]
- 45.Luxenburg C, Winograd-Katz S, Addadi L, Geiger B. Involvement of actin polymerization in podosome dynamics. J Cell Sci. 2012;125:1666–72. doi: 10.1242/jcs.075903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans JG, Correia I, Krasavina O, Watson N, Matsudaira P. Macrophage podosomes assemble at the leading lamella by growth and fragmentation. J Cell Biol. 2003;161:697–705. doi: 10.1083/jcb.200212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linder S, Kopp P. Podosomes at a glance. J Cell Sci. 2005;118:2079–82. doi: 10.1242/jcs.02390. [DOI] [PubMed] [Google Scholar]
- 48.Collin O, Na S, Chowdhury F, Hong M, Shin ME, Wang F, Wang N. Self-organized podosomes are dynamic mechanosensors. Curr Biol. 2008;18:1288–94. doi: 10.1016/j.cub.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiesner C, Faix J, Himmel M, Bentzien F, Linder S. KIF5B and KIF3A/KIF3B kinesins drive MT1-MMP surface exposure, CD44 shedding, and extracellular matrix degradation in primary macrophages. Blood. 2010;116:1559–69. doi: 10.1182/blood-2009-12-257089. [DOI] [PubMed] [Google Scholar]
- 50.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 51.Clark K, Langeslag M, van Leeuwen B, Ran L, Ryazanov AG, Figdor CG, Moolenaar WH, Jalink K, van Leeuwen FN. TRPM7, a novel regulator of actomyosin contractility and cell adhesion. EMBO J. 2006;25:290–301. doi: 10.1038/sj.emboj.7600931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varon C, Basoni C, Reuzeau E, Moreau V, Kramer IJ, Génot E. TGFbeta1-induced aortic endothelial morphogenesis requires signaling by small GTPases Rac1 and RhoA. Exp Cell Res. 2006;312:3604–19. doi: 10.1016/j.yexcr.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 53.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–41. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Margadant F, Chew LL, Hu X, Yu H, Bate N, Zhang X, Sheetz M. Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol. 2011;9:e1001223. doi: 10.1371/journal.pbio.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van den Dries K, van Helden SF, te Riet J, Diez-Ahedo R, Manzo C, Oud MM, van Leeuwen FN, Brock R, Garcia-Parajo MF, Cambi A, et al. Geometry sensing by dendritic cells dictates spatial organization and PGE(2)-induced dissolution of podosomes. Cell Mol Life Sci. 2012;69:1889–901. doi: 10.1007/s00018-011-0908-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderegg F, Geblinger D, Horvath P, Charnley M, Textor M, Addadi L, Geiger B. Substrate adhesion regulates sealing zone architecture and dynamics in cultured osteoclasts. PLoS One. 2011;6:e28583. doi: 10.1371/journal.pone.0028583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Goethem E, Guiet R, Balor S, Charrière GM, Poincloux R, Labrousse A, Maridonneau-Parini I, Le Cabec V. Macrophage podosomes go 3D. Eur J Cell Biol. 2011;90:224–36. doi: 10.1016/j.ejcb.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 58.Vérollet C, Charrière GM, Labrousse A, Cougoule C, Le Cabec V, Maridonneau-Parini I. Extracellular proteolysis in macrophage migration: losing grip for a breakthrough. Eur J Immunol. 2011;41:2805–13. doi: 10.1002/eji.201141538. [DOI] [PubMed] [Google Scholar]
- 59.Jevnikar Z, Mirković B, Fonović UP, Zidar N, Švajger U, Kos J. Three-dimensional invasion of macrophages is mediated by cysteine cathepsins in protrusive podosomes. Eur J Immunol. 2012;42:3429–41. doi: 10.1002/eji.201242610. [DOI] [PubMed] [Google Scholar]
- 60.Kubow KE, Horwitz AR. Reducing background fluorescence reveals adhesions in 3D matrices. Nat Cell Biol. 2011; 13(1):3-5; author reply 5-7 Erratum in. Nat Cell Biol. 2012;14:1344. doi: 10.1038/ncb2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega-Larson N, Tyska MJ, Weaver AM. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013;5:1159–68. doi: 10.1016/j.celrep.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiesner C, El Azzouzi K, Linder S. A specific subset of RabGTPases controls cell surface exposure of MT1-MMP, extracellular matrix degradation and three-dimensional invasion of macrophages. J Cell Sci. 2013;126:2820–33. doi: 10.1242/jcs.122358. [DOI] [PubMed] [Google Scholar]
- 63.Delaissé JM, Engsig MT, Everts V, del Carmen Ovejero M, Ferreras M, Lund L, Vu TH, Werb Z, Winding B, Lochter A, et al. Proteinases in bone resorption: obvious and less obvious roles. Clin Chim Acta. 2000;291:223–34. doi: 10.1016/S0009-8981(99)00230-2. [DOI] [PubMed] [Google Scholar]
- 64.Tatin F, Varon C, Génot E, Moreau V. A signalling cascade involving PKC, Src and Cdc42 regulates podosome assembly in cultured endothelial cells in response to phorbol ester. J Cell Sci. 2006;119:769–81. doi: 10.1242/jcs.02787. [DOI] [PubMed] [Google Scholar]
- 65.Nusblat LM, Dovas A, Cox D. The non-redundant role of N-WASP in podosome-mediated matrix degradation in macrophages. Eur J Cell Biol. 2011;90:205–12. doi: 10.1016/j.ejcb.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.López-Otín C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800–8. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- 67.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hildenbrand R, Allgayer H, Marx A, Stroebel P. Modulators of the urokinase-type plasminogen activation system for cancer. Expert Opin Investig Drugs. 2010;19:641–52. doi: 10.1517/13543781003767400. [DOI] [PubMed] [Google Scholar]
- 69.Deryugina EI, Quigley JP. Cell surface remodeling by plasmin: a new function for an old enzyme. J Biomed Biotechnol. 2012;2012:564259. doi: 10.1155/2012/564259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–75. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 71.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–33. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deryugina EI, Ratnikov B, Monosov E, Postnova TI, DiScipio R, Smith JW, Strongin AY. MT1-MMP initiates activation of pro-MMP-2 and integrin alphavbeta3 promotes maturation of MMP-2 in breast carcinoma cells. Exp Cell Res. 2001;263:209–23. doi: 10.1006/excr.2000.5118. [DOI] [PubMed] [Google Scholar]
- 73.Itoh Y, Seiki M. MT1-MMP: a potent modifier of pericellular microenvironment. J Cell Physiol. 2006;206:1–8. doi: 10.1002/jcp.20431. [DOI] [PubMed] [Google Scholar]
- 74.van Hinsbergh VW, Engelse MA, Quax PH. Pericellular proteases in angiogenesis and vasculogenesis. Arterioscler Thromb Vasc Biol. 2006;26:716–28. doi: 10.1161/01.ATV.0000209518.58252.17. [DOI] [PubMed] [Google Scholar]
- 75.Sato T, del Carmen Ovejero M, Hou P, Heegaard AM, Kumegawa M, Foged NT, Delaissé JM. Identification of the membrane-type matrix metalloproteinase MT1-MMP in osteoclasts. J Cell Sci. 1997;110:589–96. doi: 10.1242/jcs.110.5.589. [DOI] [PubMed] [Google Scholar]
- 76.Osiak AE, Zenner G, Linder S. Subconfluent endothelial cells form podosomes downstream of cytokine and RhoGTPase signaling. Exp Cell Res. 2005;307:342–53. doi: 10.1016/j.yexcr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 77.Itoh Y. MT1-MMP: a key regulator of cell migration in tissue. IUBMB Life. 2006;58:589–96. doi: 10.1080/15216540600962818. [DOI] [PubMed] [Google Scholar]
- 78.Rossi A, Deveraux Q, Turk B, Sali A. Comprehensive search for cysteine cathepsins in the human genome. Biol Chem. 2004;385:363–72. doi: 10.1515/BC.2004.040. [DOI] [PubMed] [Google Scholar]
- 79.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, Berman T, Joyce JA. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–55. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kindzelskii AL, Amhad I, Keller D, Zhou MJ, Haugland RP, Garni-Wagner BA, Gyetko MR, Todd RF, 3rd, Petty HR. Pericellular proteolysis by leukocytes and tumor cells on substrates: focal activation and the role of urokinase-type plasminogen activator. Histochem Cell Biol. 2004;121:299–310. doi: 10.1007/s00418-004-0639-3. [DOI] [PubMed] [Google Scholar]
- 82.Falcone DJ, Borth W, Khan KM, Hajjar KA. Plasminogen-mediated matrix invasion and degradation by macrophages is dependent on surface expression of annexin II. Blood. 2001;97:777–84. doi: 10.1182/blood.V97.3.777. [DOI] [PubMed] [Google Scholar]
- 83.Brownstein C, Deora AB, Jacovina AT, Weintraub R, Gertler M, Khan KM, Falcone DJ, Hajjar KA. Annexin II mediates plasminogen-dependent matrix invasion by human monocytes: enhanced expression by macrophages. Blood. 2004;103:317–24. doi: 10.1182/blood-2003-04-1304. [DOI] [PubMed] [Google Scholar]
- 84.Koblinski JE, Ahram M, Sloane BF. Unraveling the role of proteases in cancer. Clin Chim Acta. 2000;291:113–35. doi: 10.1016/S0009-8981(99)00224-7. [DOI] [PubMed] [Google Scholar]
- 85.Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 86.Shipley JM, Wesselschmidt RL, Kobayashi DK, Ley TJ, Shapiro SD. Metalloelastase is required for macrophage-mediated proteolysis and matrix invasion in mice. Proc Natl Acad Sci U S A. 1996;93:3942–6. doi: 10.1073/pnas.93.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murray MY, Birkland TP, Howe JD, Rowan AD, Fidock M, Parks WC, Gavrilovic J. Macrophage migration and invasion is regulated by MMP10 expression. PLoS One. 2013;8:e63555. doi: 10.1371/journal.pone.0063555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matsumura S, Iwanaga S, Mochizuki S, Okamoto H, Ogawa S, Okada Y. Targeted deletion or pharmacological inhibition of MMP-2 prevents cardiac rupture after myocardial infarction in mice. J Clin Invest. 2005;115:599–609. doi: 10.1172/JCI22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gong Y, Hart E, Shchurin A, Hoover-Plow J. Inflammatory macrophage migration requires MMP-9 activation by plasminogen in mice. J Clin Invest. 2008;118:3012–24. doi: 10.1172/JCI32750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O’Connell PA, Surette AP, Liwski RS, Svenningsson P, Waisman DM. S100A10 regulates plasminogen-dependent macrophage invasion. Blood. 2010;116:1136–46. doi: 10.1182/blood-2010-01-264754. [DOI] [PubMed] [Google Scholar]
- 91.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008; Chapter 14: Unit 14.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–90. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 93.Roeder BA, Kokini K, Sturgis JE, Robinson JP, Voytik-Harbin SL. Tensile mechanical properties of three-dimensional type I collagen extracellular matrices with varied microstructure. J Biomech Eng. 2002;124:214–22. doi: 10.1115/1.1449904. [DOI] [PubMed] [Google Scholar]
- 94.Raub CB, Suresh V, Krasieva T, Lyubovitsky J, Mih JD, Putnam AJ, Tromberg BJ, George SC. Noninvasive assessment of collagen gel microstructure and mechanics using multiphoton microscopy. Biophys J. 2007;92:2212–22. doi: 10.1529/biophysj.106.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Raub CB, Unruh J, Suresh V, Krasieva T, Lindmo T, Gratton E, Tromberg BJ, George SC. Image correlation spectroscopy of multiphoton images correlates with collagen mechanical properties. Biophys J. 2008;94:2361–73. doi: 10.1529/biophysj.107.120006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vesentini S, Redaelli A, Gautieri A. Nanomechanics of collagen microfibrils. Muscles Ligaments Tendons J. 2013;3:23–34. doi: 10.11138/mltj/2013.3.1.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kreger ST, Bell BJ, Bailey J, Stites E, Kuske J, Waisner B, Voytik-Harbin SL. Polymerization and matrix physical properties as important design considerations for soluble collagen formulations. Biopolymers. 2010;93:690–707. doi: 10.1002/bip.21431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chandrakasan G, Torchia DA, Piez KA. Preparation of intact monomeric collagen from rat tail tendon and skin and the structure of the nonhelical ends in solution. J Biol Chem. 1976;251:6062–7. [PubMed] [Google Scholar]
- 99.Demou ZN, Awad M, McKee T, Perentes JY, Wang X, Munn LL, Jain RK, Boucher Y. Lack of telopeptides in fibrillar collagen I promotes the invasion of a metastatic breast tumor cell line. Cancer Res. 2005;65:5674–82. doi: 10.1158/0008-5472.CAN-04-1682. [DOI] [PubMed] [Google Scholar]
- 100.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20:931–41. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9:108–22. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hulkower KI, Herber RL. Cell migration and invasion assays as tools for drug discovery. Pharmaceutics. 2011;3:107–24. doi: 10.3390/pharmaceutics3010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kramer N, Walzl A, Unger C, Rosner M, Krupitza G, Hengstschläger M, Dolznig H. In vitro cell migration and invasion assays. Mutat Res. 2013;752:10–24. doi: 10.1016/j.mrrev.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 104.Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V, Kamm RD. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip. 2009;9:269–75. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 105.Holt DJ, Chamberlain LM, Grainger DW. Cell-cell signaling in co-cultures of macrophages and fibroblasts. Biomaterials. 2010;31:9382–94. doi: 10.1016/j.biomaterials.2010.07.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Petrie RJ, Gavara N, Chadwick RS, Yamada KM. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J Cell Biol. 2012;197:439–55. doi: 10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schoumacher M, Louvard D, Vignjevic D. Cytoskeleton networks in basement membrane transmigration. Eur J Cell Biol. 2011;90:93–9. doi: 10.1016/j.ejcb.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 108.Comley J. Cell Migration Assay Trends Report, published by HTStec Limited, Cambridge, UK 2012. [Google Scholar]
- 109.Chan ME, Lu XL, Huo B, Baik AD, Chiang V, Guldberg RE, Lu HH, Guo XE. A Trabecular Bone Explant Model of Osteocyte-Osteoblast Co-Culture for Bone Mechanobiology. Cell Mol Bioeng. 2009;2:405–15. doi: 10.1007/s12195-009-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Davies JQ, Gordon S. Isolation and culture of human macrophages. Methods Mol Biol. 2005;290:105–16. doi: 10.1385/1-59259-838-2:105. [DOI] [PubMed] [Google Scholar]
- 111.Beltman JB, Marée AF, de Boer RJ. Analysing immune cell migration. Nat Rev Immunol. 2009;9:789–98. doi: 10.1038/nri2638. [DOI] [PubMed] [Google Scholar]
- 112.Webb SE, Pollard JW, Jones GE. Direct observation and quantification of macrophage chemoattraction to the growth factor CSF-1. J Cell Sci. 1996;109:793–803. doi: 10.1242/jcs.109.4.793. [DOI] [PubMed] [Google Scholar]
- 113.Yoshimura T, Yuhki N, Moore SK, Appella E, Lerman MI, Leonard EJ. Human monocyte chemoattractant protein-1 (MCP-1). Full-length cDNA cloning, expression in mitogen-stimulated blood mononuclear leukocytes, and sequence similarity to mouse competence gene JE. FEBS Lett. 1989;244:487–93. doi: 10.1016/0014-5793(89)80590-3. [DOI] [PubMed] [Google Scholar]
- 114.Cheung R, Malik M, Ravyn V, Tomkowicz B, Ptasznik A, Collman RG. An arrestin-dependent multi-kinase signaling complex mediates MIP-1beta/CCL4 signaling and chemotaxis of primary human macrophages. J Leukoc Biol. 2009;86:833–45. doi: 10.1189/jlb.0908551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.van Roosmalen W, Le Dévédec SE, Zovko S, de Bont H, van de Water B. Functional screening with a live cell imaging-based random cell migration assay. Methods Mol Biol. 2011;769:435–48. doi: 10.1007/978-1-61779-207-6_29. [DOI] [PubMed] [Google Scholar]
- 116.Pixley FJ. Macrophage Migration and Its Regulation by CSF-1. Int J Cell Biol. 2012;2012:501962. doi: 10.1155/2012/501962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cory G. Scratch-wound assay. Methods Mol Biol. 2011;769:25–30. doi: 10.1007/978-1-61779-207-6_2. [DOI] [PubMed] [Google Scholar]
- 118.Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 119.Maloney JM, Walton EB, Bruce CM, Van Vliet KJ. Influence of finite thickness and stiffness on cellular adhesion-induced deformation of compliant substrata. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;78:041923. doi: 10.1103/PhysRevE.78.041923. [DOI] [PubMed] [Google Scholar]
- 120.Chen HC. Boyden chamber assay. Methods Mol Biol. 2005;294:15–22. doi: 10.1385/1-59259-860-9:015. [DOI] [PubMed] [Google Scholar]
- 121.Simon N, Noël A, Foidart JM. Evaluation of in vitro reconstituted basement membrane assay to assess the invasiveness of tumor cells. Invasion Metastasis. 1992;12:156–67. [PubMed] [Google Scholar]
- 122.Kam Y, Guess C, Estrada L, Weidow B, Quaranta V. A novel circular invasion assay mimics in vivo invasive behavior of cancer cell lines and distinguishes single-cell motility in vitro. BMC Cancer. 2008;8:198. doi: 10.1186/1471-2407-8-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zengel P, Nguyen-Hoang A, Schildhammer C, Zantl R, Kahl V, Horn E. μ-Slide Chemotaxis: a new chamber for long-term chemotaxis studies. BMC Cell Biol. 2011;12:21. doi: 10.1186/1471-2121-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schor SL, Allen TD, Winn B. Lymphocyte migration into three-dimensional collagen matrices: a quantitative study. J Cell Biol. 1983;96:1089–96. doi: 10.1083/jcb.96.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Burgstaller G, Oehrle B, Koch I, Lindner M, Eickelberg O. Multiplex profiling of cellular invasion in 3D cell culture models. PLoS One. 2013;8:e63121. doi: 10.1371/journal.pone.0063121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nat Rev Mol Cell Biol. 2011;12:308–19. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Friedl P, Weigelin B. Interstitial leukocyte migration and immune function. Nat Immunol. 2008;9:960–9. doi: 10.1038/ni.f.212. [DOI] [PubMed] [Google Scholar]
- 128.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer. 2003;3:422–33. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 129.Schindler M, Nur-E-Kamal A, Ahmed I, Kamal J, Liu HY, Amor N, Ponery AS, Crockett DP, Grafe TH, Chung HY, et al. Living in three dimensions: 3D nanostructured environments for cell culture and regenerative medicine. Cell Biochem Biophys. 2006;45:215–27. doi: 10.1385/CBB:45:2:215. [DOI] [PubMed] [Google Scholar]
- 130.Montagnac G, Meas-Yedid V, Irondelle M, Castro-Castro A, Franco M, Shida T, Nachury MV, Benmerah A, Olivo-Marin JC, Chavrier P. αTAT1 catalyses microtubule acetylation at clathrin-coated pits. Nature. 2013;502:567–70. doi: 10.1038/nature12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sutherland RM, McCredie JA, Inch WR. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J Natl Cancer Inst. 1971;46:113–20. [PubMed] [Google Scholar]
- 132.Centonze VE, White JG. Multiphoton excitation provides optical sections from deeper within scattering specimens than confocal imaging. Biophys J. 1998;75:2015–24. doi: 10.1016/S0006-3495(98)77643-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cougoule C, Van Goethem E, Le Cabec V, Lafouresse F, Dupré L, Mehraj V, Mège JL, Lastrucci C, Maridonneau-Parini I. Blood leukocytes and macrophages of various phenotypes have distinct abilities to form podosomes and to migrate in 3D environments. Eur J Cell Biol. 2012;91:938–49. doi: 10.1016/j.ejcb.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 134.Tolde O, Rösel D, Veselý P, Folk P, Brábek J. The structure of invadopodia in a complex 3D environment. Eur J Cell Biol. 2010;89:674–80. doi: 10.1016/j.ejcb.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 135.Yu X, Machesky LM. Cells assemble invadopodia-like structures and invade into matrigel in a matrix metalloprotease dependent manner in the circular invasion assay. PLoS One. 2012;7:e30605. doi: 10.1371/journal.pone.0030605. [DOI] [PMC free article] [PubMed] [Google Scholar]