Abstract
Podosomes are small, circular adhesions formed by cells such as osteoclasts, macrophages, dendritic cells, and endothelial cells. They comprise a protrusive actin core module and an adhesive ring module composed of integrins and cytoskeletal adaptor proteins such as vinculin and talin. Furthermore, podosomes are associated with an actin network and often organize into large clusters. Recent results from our laboratory and others have shed new light on podosome structure and dynamics, suggesting a revision of the classical “core-ring” model. Also, these studies demonstrate that the adhesive and protrusive module are functionally linked by the actin network likely facilitating mechanotransduction as well as providing feedback between these two modules. In this commentary, we briefly summarize these recent advances with respect to the knowledge on podosome structure and discuss force distribution mechanisms within podosomes and their emerging role in mechanotransduction.
Keywords: actin, cell adhesion, invadopodia, mechanotransduction, podosome, tension
The Podosome Modules
Although the earliest observations date back to 1980,1 the first official report on podosomes appeared in 1985 when Marchisio and colleagues described the localization of actin, vinculin, and pTyr in distinct punctae in Rous sarcoma virus-transformed fibroblasts, which they called podosomes (i.e., little feet), referring to their localization at the ventral plasma membrane.2 Subsequently, studies involving interference reflection microscopy (IRM), fluorescence microscopy, and electron microscopy contributed to the identification of additional podosome components such as integrins and talin.2-6 More recently, two distinct pools of polymerizing actin have been distinguished, the parallel branched actin of the core and the anti-parallel filamentous actin of the ring, the latter also known as the actin cloud or actin network.7-9 Currently, podosomes are defined as highly dynamic dot-shaped adhesion complexes, approximately one micron in size, comprising an adhesive ring and a protrusive core.10 Podosomes are often arranged into higher-ordered structures, such as large clusters in macrophages and DCs, rosettes in endothelial cells, or circular belts in osteoclasts.11-14
Recent super-resolution microscopy studies have revealed details of podosome organization at the nanoscale challenging the traditional concept of a defined ring surrounding the core. By Bayesian localization microscopy, it was shown that podosome rings often have a polygonal shape and that vinculin, in contrast to talin, localizes in short strands that radiate from the podosome edge.15 More recently, by dual-color Stochastic Optical Reconstruction Microscopy (dSTORM), we revealed that vinculin is preferentially enriched at the rim of the core and along the radiating actin filaments, whereas islets of αMβ2 integrin and talin are homogeneously distributed in core-free areas within the podosome cluster.16 Together, these observations demonstrated that each of the ring proteins has a specific localization within the podosome cluster most likely related to its distinct role within podosomes. More specifically, vinculin localization seems guided by the actin network, while talin distribution seems dictated by the organization of the integrin molecules. The super-resolution data also highlight that podosome clusters should be regarded as one multifunctional zone consisting of three main modules: actin-dense cores as protrusion module, integrin islets differentially populated by adaptor proteins acting as adhesion module, and a well-organized network of radiating actin filaments that interconnects podosomes, possibly working as mechanotransduction module. Importantly, it should also be mentioned that, similarly to invadopodia formed by invading cancer cells, podosomes have the capacity to degrade extracellular matrix.17,18 Although several reports documented the existence of polarized secretory pathways responsible for matrix metalloprotease delivery and extracellular release at podosome sites,19-24 their nanoscale organization as well as their molecular and temporal connection with the podosome modules are still poorly defined.
Interestingly, the organization of each of the podosome modules can be altered under specific conditions. In macrophages, reduced or enhanced core actin polymerization by altered PAK4 kinase activity has been shown to lead to smaller or larger podosome cores, respectively.25 Defective ring formation is observed in bone marrow macrophages lacking Rac126 and bone marrow dendritic cells (DCs) lacking phospholipase C gamma2.27 Furthermore, osteoclasts lacking kindlin-3 are unable to form the actin network resulting in small, unstable podosome cores.9 More recently, it has been shown that podosome size and composition are also controlled by substrate properties. When DCs are seeded on collagen impregnated filters with 1 micron sized pores, podosomes are enlarged and recruit transmembrane metalloproteases and c-type lectins.20,28 Proteomic analysis of podosome fractions isolated from primary human macrophages has revealed about 170 potential novel components.29 The next challenge is to determine their spatiotemporal organization and role in preserving the integrity of each of the podosome modules. Development of image analysis software dedicated to specifically identify podosomes has allowed quantitative analysis of large numbers of podosomes, thus enabling robust statistics to determine subtle changes in podosome composition.30-32 We envisage that bioimaging techniques allowing 3D reconstruction and multicolor labeling will soon allow detailed characterization of the structural arrangement of the podosome modules shedding new light on the molecular mechanisms regulating podosome composition and function.
Balancing Forces within Podosomes
Podosomes, in sharp contrast to focal adhesions (FAs), have classically been associated with cytoskeletal relaxation.33,34 However, evidence is emerging that mechanical forces and cytoskeletal tension also play an important role in podosomes, albeit differently from FAs.
For cytoskeletal tension to be generated, two opposing forces are required. In the case of FAs, forces are actively generated by the actomyosin apparatus and opposed by the binding of activated integrins to immobilized extracellular matrix proteins. The resulting tension within the adhesion complex drives the recruitment of tension-sensitive molecules vinculin and zyxin creating stable, FAs. Importantly, in the absence of internal forces, such as upon myosin inhibition35 and actin filament disruption,36 or when external forces are weak (e.g., on compliant substrates37 or in the absence of ligand38), FAs do not stabilize and rapidly turnover.
Interestingly, podosomes appear to form and grow in the absence of the classical forces described for FAs. Actomyosin contractility is not required for podosome stability or the presence and recruitment of the tension-sensitive molecules vinculin and zyxin.31 Also, integrin ligation appears redundant for podosome formation, as podosomes rapidly form on uncoated substrates and in the absence of ligand-containing serum in Src-transformed fibroblasts and monocyte-derived DCs.2,39 Moreover, REF52 fibroblasts have recently been shown to form podosomes on fluid Arg-Gly-Asp (RGD) peptide-lipid surfaces, demonstrating that podosomes indeed form in the absence of traction forces.40
How do podosomes generate tension within the adhesion scaffold to drive the recruitment of tension-sensitive molecules in the absence of classical traction forces? One interesting possibility is that this distinguishing feature is facilitated by the intrinsic ability of podosomes to account for two opposing forces that are balanced within the podosome structure (Fig. 1). Whereas FAs are anchored to the ventral plasma membrane by a tangential-shaped cluster of integrins (Fig. 1, top view), thus requiring integrin–ECM ligation as a counter force to generate tension (Fig. 1, side view), podosomes are anchored to the plasma membrane through many different integrin clusters that are organized around the actin core and are connected to it through the F-actin network. This unique architecture ensures that the combination of actin polymerization in the core and myosin contractility in the radiating F-actin filaments is apparently sufficient to create two opposing forces within the podosome structure, thereby creating tension on the F-actin filaments (Fig. 1, side view). Once a podosome core is formed, it will continuously grow and shrink, vertically oscillating over time before it dissolves again. Our model implies that the tension on the filamentous actin network will successively increase and decrease during podosome oscillations. This is supported by our observation that the levels of the tension-sensitive components vinculin and zyxin oscillate in harmony with the actin core, while the levels of the tension-insensitive components are relatively stable.31 Future studies should directly determine whether tension within the filamentous actin is indeed built up during podosome core growth or myosin IIA-mediated contraction. To fully support this model, the presence of a linkage between the central protruding core and the surrounding adhesive module is essential. To date, the exact molecular nature of this connection remains unknown.
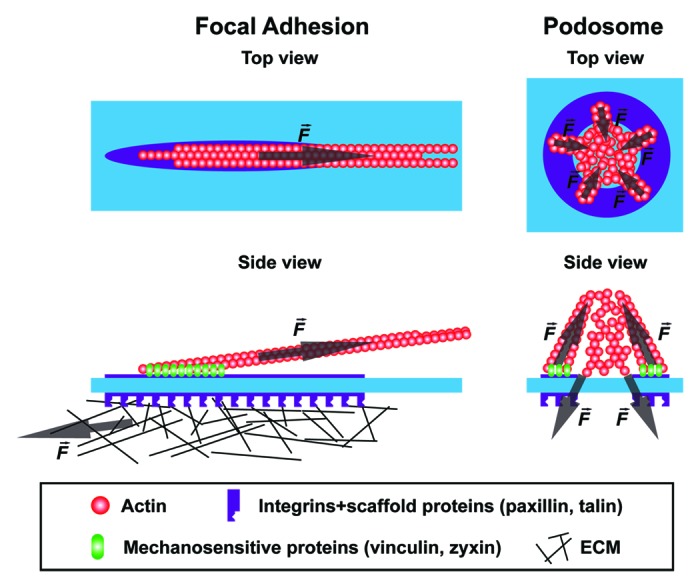
Figure 1. Force distribution within podosomes and focal adhesions. Schematic representation of the top and side view of a focal adhesion and a podosome. The forces generated within these adhesions are indicated with an arrow. The growth of focal adhesions and podosomes is both tension-mediated but since opposite forces are generated within a podosome, force derived from integrin-mediated ECM ligation is not necessary to facilitate the growth of podosomes.
A recent elegant study by the Waterman’s group using interferometric photoactivated localization microscopy (iPALM) showed for the first time the composite multilaminar protein architecture of FAs, which share similar components with podosomes.41 In the future, similar 3D super-resolution techniques should be applied to understand whether podosome adhesive modules have a similar composite multilaminar structure responsible for translating the tension within the ring into intracellular signals.
Podosomes as Mechanotransducers
Mechanotransduction is the process by which cells translate mechanical cues such as matrix elasticity and geometric constraints into chemical signals, directing key processes such as adhesion, growth, and differentiation.42-44 By controlling both the perception of mechanical cues and the execution of the cellular response, integrin-based adhesions are thought to play a crucial role in mechanotransduction. Indeed, FAs have been shown to respond to substrate mechanical properties, thereby regulating cellular processes. For example, FA size and dynamics are critically regulated by substrate elasticity,37,45 thereby regulating the migration of cells on a rigidity gradient toward higher rigidity.45,46 Although much less is known about the role of podosomes in mechanotransduction, clear evidence is emerging that they also respond to substrate mechanical properties such as substrate texture and rigidity.
Substrate texture has been shown to influence podosomes in osteoclasts and DCs. In osteoclasts, podosomes organize into small unstable sealing zones on smooth surfaces and large stable actin rings on rough surfaces.47 In DCs, we have shown that podosomes respond to 3-D geometric cues by specifically aligning on the edges of micropatterned substrates.39 Although the mechanisms that regulate podosome behavior on textured substrates are not well understood, it is tempting to speculate that the surface induces membrane curvatures, thereby recruiting specific lipids and proteins to the ventral plasma membrane, in turn influencing the dynamics and spatial organization of podosomes. In this respect, it is interesting to note that lipids associated with membrane curvature, such as phosphoinositides (PtdIns), especially PtdIns(4,5)P2 and PtdIns(3,4,5)P3, were reported to regulate the recruitment and binding of many podosome components during podosome initiation but also in later stages of podosome assembly.48 Also, membrane curvature-sensing proteins have been shown to be present within podosomes in macrophages.49 How the complex interplay between membrane proteins and lipids regulates podosome texture sensing remains to be elucidated.
Podosome organization is also regulated by substrate rigidity in various cell types. Substrate rigidity positively correlates with the lifetime and stability of individual podosomes and podosome rosettes in 3T3 fibroblasts.50 Furthermore, traction stress, exhibited by rings of podosomes in rous sarcoma virus-transformed baby hamster kidney cells, increases with substrate rigidity.51 More recently, it was demonstrated that podosomes in DCs seeded on filters with 1 µm pores specifically form on soft spots of low physical resistance (i.e., the pores).20,28 Once a podosome is formed on such a soft spot, it becomes increasingly invasive and, when possible, protrudes into the substrate. Substrate stiffness sensing by podosomes is likely to be regulated by the balance between myosin IIA activity and actin polymerization that control the tension on the actin network. The tension on the network could subsequently be sensed by structural proteins such as cofilin52 or formin and profilin53 to control podosome oscillations and the recruitment of mechanosensitive proteins. As podosome cores have been shown to undergo periodic stiffness oscillations,54 another possibility is that substrate stiffness sensing is orchestrated directly by the stiffness of the podosome core itself. Altogether, it would be interesting to investigate whether actin network tension, podosome oscillations, and/or podosome core stiffness are differentially regulated on substrates with variable stiffness. Also, future work should be directed toward unraveling the signaling pathways and feedback mechanisms that are responsible for translating podosome stiffness sensing into cellular decisions, such as cell protrusion and migration within tissues.
Outlook and Perspectives
Podosomes have recently been identified in cells that are seeded within 3D matrices,55-57 indicating that podosomes are not just artificial adhesion structures formed by cells seeded on 2D substrates. Future challenges will include the characterization of the functional role of podosomes in 3D migration and how podosomes and FAs, through their different functions and mechanosensing abilities, together orchestrate the migration of cells. The availability of novel 3D imaging setups58 and 3D biomimetic migration models59 finally enables us to address these exciting questions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We apologize to our colleagues whose work could not be cited due to space limitations. van den Dries K was supported by the European Commission’s Seventh Framework Programme (FP7-ICT-2011-7) under grant agreement no. 288263 (NanoVista) and Bolomini-Vittori M is supported by a program grant from the Human Frontier Science Program (HFSP) (RGP0027/2012) awarded to Cambi A. Cambi A is the recipient of a Meervoud grant (836.09.002) from The Netherlands Organisation for Scientific Research (NWO).
References
- 1.David-Pfeuty T, Singer SJ. Altered distributions of the cytoskeletal proteins vinculin and alpha-actinin in cultured fibroblasts transformed by Rous sarcoma virus. Proc Natl Acad Sci U S A. 1980;77:6687–91. doi: 10.1073/pnas.77.11.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tarone G, Cirillo D, Giancotti FG, Comoglio PM, Marchisio PC. Rous sarcoma virus-transformed fibroblasts adhere primarily at discrete protrusions of the ventral membrane called podosomes. Exp Cell Res. 1985;159:141–57. doi: 10.1016/S0014-4827(85)80044-6. [DOI] [PubMed] [Google Scholar]
- 3.Marchisio PC, Capasso O, Nitsch L, Cancedda R, Gionti E. Cytoskeleton and adhesion patterns of cultured chick embryo chondrocytes during cell spreading and Rous sarcoma virus transformation. Exp Cell Res. 1984;151:332–43. doi: 10.1016/0014-4827(84)90384-7. [DOI] [PubMed] [Google Scholar]
- 4.Shriver K, Rohrschneider L. Organization of pp60src and selected cytoskeletal proteins within adhesion plaques and junctions of Rous sarcoma virus-transformed rat cells. J Cell Biol. 1981;89:525–35. doi: 10.1083/jcb.89.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchisio PC, Bergui L, Corbascio GC, Cremona O, D’Urso N, Schena M, Tesio L, Caligaris-Cappio F. Vinculin, talin, and integrins are localized at specific adhesion sites of malignant B lymphocytes. Blood. 1988;72:830–3. [PubMed] [Google Scholar]
- 6.Gavazzi I, Nermut MV, Marchisio PC. Ultrastructure and gold-immunolabelling of cell-substratum adhesions (podosomes) in RSV-transformed BHK cells. J Cell Sci. 1989;94:85–99. doi: 10.1242/jcs.94.1.85. [DOI] [PubMed] [Google Scholar]
- 7.Destaing O, Saltel F, Géminard JC, Jurdic P, Bard F. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol Biol Cell. 2003;14:407–16. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luxenburg C, Geblinger D, Klein E, Anderson K, Hanein D, Geiger B, Addadi L. The architecture of the adhesive apparatus of cultured osteoclasts: from podosome formation to sealing zone assembly. PLoS One. 2007;2:e179. doi: 10.1371/journal.pone.0000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt S, Nakchbandi I, Ruppert R, Kawelke N, Hess MW, Pfaller K, Jurdic P, Fässler R, Moser M. Kindlin-3-mediated signaling from multiple integrin classes is required for osteoclast-mediated bone resorption. J Cell Biol. 2011;192:883–97. doi: 10.1083/jcb.201007141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linder S, Aepfelbacher M. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 2003;13:376–85. doi: 10.1016/S0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 11.Lakkakorpi P, Tuukkanen J, Hentunen T, Järvelin K, Väänänen K. Organization of osteoclast microfilaments during the attachment to bone surface in vitro. J Bone Miner Res. 1989;4:817–25. doi: 10.1002/jbmr.5650040605. [DOI] [PubMed] [Google Scholar]
- 12.Evans JG, Correia I, Krasavina O, Watson N, Matsudaira P. Macrophage podosomes assemble at the leading lamella by growth and fragmentation. J Cell Biol. 2003;161:697–705. doi: 10.1083/jcb.200212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns S, Hardy SJ, Buddle J, Yong KL, Jones GE, Thrasher AJ. Maturation of DC is associated with changes in motile characteristics and adherence. Cell Motil Cytoskeleton. 2004;57:118–32. doi: 10.1002/cm.10163. [DOI] [PubMed] [Google Scholar]
- 14.Tatin F, Varon C, Génot E, Moreau V. A signalling cascade involving PKC, Src and Cdc42 regulates podosome assembly in cultured endothelial cells in response to phorbol ester. J Cell Sci. 2006;119:769–81. doi: 10.1242/jcs.02787. [DOI] [PubMed] [Google Scholar]
- 15.Cox S, Rosten E, Monypenny J, Jovanovic-Talisman T, Burnette DT, Lippincott-Schwartz J, Jones GE, Heintzmann R. Bayesian localization microscopy reveals nanoscale podosome dynamics. Nat Methods. 2012;9:195–200. doi: 10.1038/nmeth.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van den Dries K, Schwartz SL, Byars J, Meddens MB, Bolomini-Vittori M, Lidke DS, Figdor CG, Lidke KA, Cambi A. Dual-color superresolution microscopy reveals nanoscale organization of mechanosensory podosomes. Mol Biol Cell. 2013;24:2112–23. doi: 10.1091/mbc.E12-12-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schachtner H, Calaminus SD, Thomas SG, Machesky LM. Podosomes in adhesion, migration, mechanosensing and matrix remodeling. Cytoskeleton (Hoboken) 2013;70:572–89. doi: 10.1002/cm.21119. [DOI] [PubMed] [Google Scholar]
- 18.Buccione R, Orth JD, McNiven MA. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol. 2004;5:647–57. doi: 10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- 19.Daubon T, Buccione R, Génot E. The Aarskog-Scott syndrome protein Fgd1 regulates podosome formation and extracellular matrix remodeling in transforming growth factor β-stimulated aortic endothelial cells. Mol Cell Biol. 2011;31:4430–41. doi: 10.1128/MCB.05474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gawden-Bone C, Zhou Z, King E, Prescott A, Watts C, Lucocq J. Dendritic cell podosomes are protrusive and invade the extracellular matrix using metalloproteinase MMP-14. J Cell Sci. 2010;123:1427–37. doi: 10.1242/jcs.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schachtner H, Calaminus SD, Sinclair A, Monypenny J, Blundell MP, Leon C, Holyoake TL, Thrasher AJ, Michie AM, Vukovic M, et al. Megakaryocytes assemble podosomes that degrade matrix and protrude through basement membrane. Blood. 2013;121:2542–52. doi: 10.1182/blood-2012-07-443457. [DOI] [PubMed] [Google Scholar]
- 22.Xiao H, Bai XH, Kapus A, Lu WY, Mak AS, Liu M. The protein kinase C cascade regulates recruitment of matrix metalloprotease 9 to podosomes and its release and activation. Mol Cell Biol. 2010;30:5545–61. doi: 10.1128/MCB.00382-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornfine S, Himmel M, Kopp P, El Azzouzi K, Wiesner C, Krüger M, Rudel T, Linder S. The kinesin KIF9 and reggie/flotillin proteins regulate matrix degradation by macrophage podosomes. Mol Biol Cell. 2011;22:202–15. doi: 10.1091/mbc.E10-05-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiesner C, Faix J, Himmel M, Bentzien F, Linder S. KIF5B and KIF3A/KIF3B kinesins drive MT1-MMP surface exposure, CD44 shedding, and extracellular matrix degradation in primary macrophages. Blood. 2010;116:1559–69. doi: 10.1182/blood-2009-12-257089. [DOI] [PubMed] [Google Scholar]
- 25.Gringel A, Walz D, Rosenberger G, Minden A, Kutsche K, Kopp P, Linder S. PAK4 and alphaPIX determine podosome size and number in macrophages through localized actin regulation. J Cell Physiol. 2006;209:568–79. doi: 10.1002/jcp.20777. [DOI] [PubMed] [Google Scholar]
- 26.Wheeler AP, Wells CM, Smith SD, Vega FM, Henderson RB, Tybulewicz VL, Ridley AJ. Rac1 and Rac2 regulate macrophage morphology but are not essential for migration. J Cell Sci. 2006;119:2749–57. doi: 10.1242/jcs.03024. [DOI] [PubMed] [Google Scholar]
- 27.Cremasco V, Benasciutti E, Cella M, Kisseleva M, Croke M, Faccio R. Phospholipase C gamma 2 is critical for development of a murine model of inflammatory arthritis by affecting actin dynamics in dendritic cells. PLoS One. 2010;5:e8909. doi: 10.1371/journal.pone.0008909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baranov M, Ter Beest M, Reinieren-Beeren I, Cambi A, Figdor CG, van den Bogaart G. Podosomes of dendritic cells facilitate antigen sampling. J Cell Sci. 2014 doi: 10.1242/jcs.141226. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cervero P, Himmel M, Krüger M, Linder S. Proteomic analysis of podosome fractions from macrophages reveals similarities to spreading initiation centres. Eur J Cell Biol. 2012;91:908–22. doi: 10.1016/j.ejcb.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Meddens MB, Rieger B, Figdor CG, Cambi A, van den Dries K. Automated podosome identification and characterization in fluorescence microscopy images. Microsc Microanal. 2013;19:180–9. doi: 10.1017/S1431927612014018. [DOI] [PubMed] [Google Scholar]
- 31.van den Dries K, Meddens MB, de Keijzer S, Shekhar S, Subramaniam V, Figdor CG, Cambi A. Interplay between myosin IIA-mediated contractility and actin network integrity orchestrates podosome composition and oscillations. Nat Commun. 2013;4:1412. doi: 10.1038/ncomms2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cervero P, Panzer L, Linder S. Podosome reformation in macrophages: assays and analysis. Methods Mol Biol. 2013;1046:97–121. doi: 10.1007/978-1-62703-538-5_6. [DOI] [PubMed] [Google Scholar]
- 33.Burgstaller G, Gimona M. Actin cytoskeleton remodelling via local inhibition of contractility at discrete microdomains. J Cell Sci. 2004;117:223–31. doi: 10.1242/jcs.00839. [DOI] [PubMed] [Google Scholar]
- 34.Clark K, Langeslag M, van Leeuwen B, Ran L, Ryazanov AG, Figdor CG, Moolenaar WH, Jalink K, van Leeuwen FN. TRPM7, a novel regulator of actomyosin contractility and cell adhesion. EMBO J. 2006;25:290–301. doi: 10.1038/sj.emboj.7600931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133:1403–15. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, Ballestrem C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol. 2007;179:1043–57. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelham RJ, Jr., Wang Yl. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–72. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 39.van den Dries K, van Helden SF, te Riet J, Diez-Ahedo R, Manzo C, Oud MM, van Leeuwen FN, Brock R, Garcia-Parajo MF, Cambi A, et al. Geometry sensing by dendritic cells dictates spatial organization and PGE(2)-induced dissolution of podosomes. Cell Mol Life Sci. 2012;69:1889–901. doi: 10.1007/s00018-011-0908-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu CH, Rafiq NB, Krishnasamy A, Hartman KL, Jones GE, Bershadsky AD, Sheetz MP. Integrin-Matrix Clusters Form Podosome-like Adhesions in the Absence of Traction Forces. Cell Rep 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–4. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 43.Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677–95. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 44.Ross TD, Coon BG, Yun S, Baeyens N, Tanaka K, Ouyang M, Schwartz MA. Integrins in mechanotransduction. Curr Opin Cell Biol. 2013;25:613–8. doi: 10.1016/j.ceb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–27. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–52. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geblinger D, Addadi L, Geiger B. Nano-topography sensing by osteoclasts. J Cell Sci. 2010;123:1503–10. doi: 10.1242/jcs.060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chellaiah MA. Regulation of podosomes by integrin alphavbeta3 and Rho GTPase-facilitated phosphoinositide signaling. Eur J Cell Biol. 2006;85:311–7. doi: 10.1016/j.ejcb.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Tsuboi S, Takada H, Hara T, Mochizuki N, Funyu T, Saitoh H, Terayama Y, Yamaya K, Ohyama C, Nonoyama S, et al. FBP17 Mediates a Common Molecular Step in the Formation of Podosomes and Phagocytic Cups in Macrophages. J Biol Chem. 2009;284:8548–56. doi: 10.1074/jbc.M805638200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collin O, Tracqui P, Stephanou A, Usson Y, Clément-Lacroix J, Planus E. Spatiotemporal dynamics of actin-rich adhesion microdomains: influence of substrate flexibility. J Cell Sci. 2006;119:1914–25. doi: 10.1242/jcs.02838. [DOI] [PubMed] [Google Scholar]
- 51.Geblinger D, Zink C, Spencer ND, Addadi L, Geiger B. Effects of surface microtopography on the assembly of the osteoclast resorption apparatus. J R Soc Interface. 2012;9:1599–608. doi: 10.1098/rsif.2011.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayakawa K, Tatsumi H, Sokabe M. Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J Cell Biol. 2011;195:721–7. doi: 10.1083/jcb.201102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Courtemanche N, Lee JY, Pollard TD, Greene EC. Tension modulates actin filament polymerization mediated by formin and profilin. Proc Natl Acad Sci U S A. 2013;110:9752–7. doi: 10.1073/pnas.1308257110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Labernadie A, Thibault C, Vieu C, Maridonneau-Parini I, Charrière GM. Dynamics of podosome stiffness revealed by atomic force microscopy. Proc Natl Acad Sci U S A. 2010;107:21016–21. doi: 10.1073/pnas.1007835107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cougoule C, Van Goethem E, Le Cabec V, Lafouresse F, Dupré L, Mehraj V, Mège JL, Lastrucci C, Maridonneau-Parini I. Blood leukocytes and macrophages of various phenotypes have distinct abilities to form podosomes and to migrate in 3D environments. Eur J Cell Biol. 2012;91:938–49. doi: 10.1016/j.ejcb.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 56.Van Goethem E, Guiet R, Balor S, Charrière GM, Poincloux R, Labrousse A, Maridonneau-Parini I, Le Cabec V. Macrophage podosomes go 3D. Eur J Cell Biol. 2011;90:224–36. doi: 10.1016/j.ejcb.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 57.Van Goethem E, Poincloux R, Gauffre F, Maridonneau-Parini I, Le Cabec V. Matrix architecture dictates three-dimensional migration modes of human macrophages: differential involvement of proteases and podosome-like structures. J Immunol. 2010;184:1049–61. doi: 10.4049/jimmunol.0902223. [DOI] [PubMed] [Google Scholar]
- 58.Lidke DS, Lidke KA. Advances in high-resolution imaging--techniques for three-dimensional imaging of cellular structures. J Cell Sci. 2012;125:2571–80. doi: 10.1242/jcs.090027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–45. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]


