Abstract
In response to starvation, cells undergo increased levels of autophagy and cell cycle arrest but the role of autophagy in starvation-induced cell cycle arrest is not fully understood. Here we show that autophagy genes regulate cell cycle arrest in the budding yeast Saccharomyces cerevisiae during nitrogen starvation. While exponentially growing wild-type yeasts preferentially arrest in G1/G0 in response to starvation, yeasts carrying null mutations in autophagy genes show a significantly higher percentage of cells in G2/M. In these autophagy-deficient yeast strains, starvation elicits physiological properties associated with quiescence, such as Snf1 activation, glycogen and trehalose accumulation as well as heat-shock resistance. However, while nutrient-starved wild-type yeasts finish the G2/M transition and arrest in G1/G0, autophagy-deficient yeasts arrest in telophase. Our results suggest that autophagy is crucial for mitotic exit during starvation and appropriate entry into a G1/G0 quiescent state.
Keywords: autophagy, yeast, cell cycle, quiescence, starvation
Introduction
Eukaryotic cell growth and proliferation are regulated by nutrients, and the budding yeast, Saccharomyces cerevisiae, provides an excellent model for studying such regulation. In response to nutrient deprivation, cells stop proliferating, undergo cell cycle arrest, and enter a reversible state of quiescence, usually in G0 of the cell cycle.1 It is commonly accepted that the decision to proliferate is made when cells are in G1, which is called Start in budding yeast. This decision is regulated by a variety of nutrient-sensing kinases including Snf1, PKA, Tor1 and Tor2, Sch9, and Pho85-Pho80.2 Once yeast cells have passed Start, they usually traverse all phases of the cell cycle until reaching Start again. If a decision is made not to proliferate, haploid cells preferentially arrest at Start and enter quiescence with a 1N DNA content.
Yeasts can also enter quiescence—at least during carbon starvation—through other cell cycle phases besides G0/G1, although the ability to give rise to progeny is diminished.3-5 In cells that have entered quiescence upon carbon source exhaustion, quiescence exit can be triggered by glucose addition. These observations have given rise to the notion that quiescence entry and exit may be governed primarily by cellular metabolic status and are not invariably coupled to the cell cycle. Nonetheless, the cellular pathways that govern the stage of the cell cycle at which yeast enter quiescence during starvation remain poorly understood.
Autophagy is a highly conserved pathway in which cytoplasmic contents are degraded and recycled in response to stressors including starvation. Autophagy is critical for maintaining cellular energy homeostasis and regulating cell growth.6 The core yeast autophagy machinery can be divided into 4 major subgroups.7 The Atg1-Atg13-Atg17 serine/threonine kinase complex is crucial for the initiation step of autophagy. The Vps30/Atg6, Vps15, Vps34, and Atg14 lipid kinase complex is essential for the formation of the phagophore. The vesicle elongation step of autophagy includes 2 ubiquitin-like protein systems; the core members are Atg3, Atg4, Atg5, Atg7, Atg8, Atg10, Atg12, and Atg16. In addition, the Atg9 cycling system including Atg2 and Atg18 functions in autophagosome formation.
Previous studies showed that some autophagy genes participate in cell cycle regulation. For example, mice lacking Becn1, the mammalian ortholog of yeast VPS30/ATG6, have abnormal cell proliferation in mammary epithelial ducts and splenic germinal center lymphocytes;8 mice lacking Ambra1 have increased cell proliferation in fetal brain;9 and atg7−/− mouse embryonic fibroblast cells have impaired TRP53/p53-mediated cell cycle arrest.10 In the budding yeast, Saccharomyces cerevisiae, autophagy and G1/G0 cell cycle arrest are both induced after switching the cells from a nutrient-rich environment to nutrient-limited conditions.11,12 However, it is not clear whether autophagy regulates G1/G0 cell cycle arrest.
In this study, we used the budding yeast Saccharomyces cerevisiae as a model system to study the role of autophagy in cell cycle regulation. Similar to mammalian cells, which arrest in G1/G0 after serum starvation,13 the majority of wild-type yeasts arrest in G1/G0 after nitrogen starvation.11 In contrast, we found that the majority of autophagy-deficient yeasts arrest in telophase after nitrogen starvation and, interestingly, still exhibit quiescence-specific phenotypes. Based on these results, we conclude that autophagy genes are required for yeast to traverse the cell cycle and properly arrest in G1/G0 in response to starvation.
Results
Autophagy-deficient yeasts fail to enter G1/G0 in response to starvation
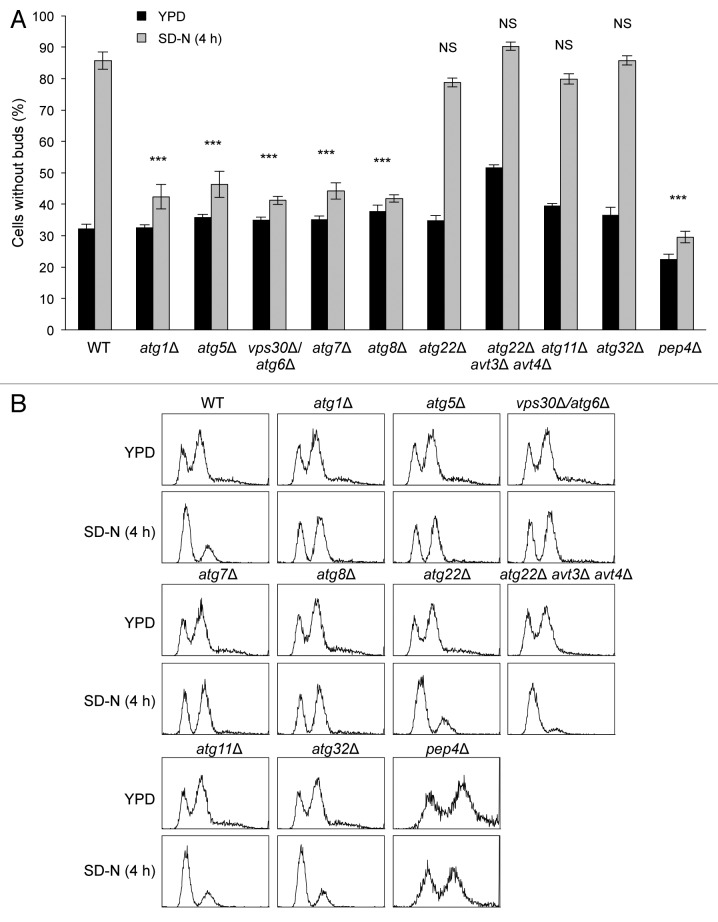
To investigate the role of autophagy in cell cycle regulation during starvation, we examined whether yeasts with null mutations in different essential autophagy genes (ATG1, ATG5, VPS30/ATG6, ATG7, and ATG8) arrest in G1/G0 after nitrogen starvation. During growth in YPD medium, about 30% of both wild-type and atg1Δ, atg5Δ, vps30Δ/atg6Δ, atg7Δ, and atg8Δ yeast cells were in G1/G0, as quantified by the percentage of cells without buds. After shifting exponentially growing yeasts to nitrogen starvation medium (SD-N) for 4 h, greater than 80% of wild-type yeasts entered G1/G0 (Fig. 1A). In contrast, the percentage of atg1Δ, atg5Δ, vps30Δ/atg6Δ, atg7Δ, and atg8Δ yeasts in G1/G0 only minimally increased in response to nitrogen starvation. The magnitude of this increase was markedly less than in wild-type yeasts indicating that autophagy-deficient yeasts have a defect in G1/G0 arrest in response to nitrogen starvation. This defect in G1/G0 arrest in autophagy-deficient yeast strains persisted at 24 h after nitrogen starvation (Fig. S1).
Figure 1. Cell-cycle analysis of wild-type and autophagy-deficient yeasts during nitrogen starvation. (A) Quantification of the percentage of wild-type and indicated atg mutant yeast strains without buds at time 0 (YPD) or after 4 h in SD-N (SD-N). Bars represent mean ± SEM of triplicate samples (at least 100 cells per sample were counted). Similar results were observed in more than 3 independent experiments. ***P < 0.001, NS, not significant; 2-way ANOVA with Bonferroni correction for comparison of magnitude of change between YPD and SD-N in the indicated atg mutant yeast strain compared with the magnitude of change between YPD and SD-N in wild-type (WT) yeasts. (B) FACS analysis of cell cycle of the indicated yeast strains at time 0 (YPD) or after 4 h in SD-N (SD-N). 10,000 cells were analyzed per strain. Similar results were observed in more than 3 independent experiments.
To gain further insight into the mechanism by which autophagy functions in permitting yeast cells to arrest in G1/G0 in response to nitrogen starvation, we examined the phenotypes of yeast strains containing null mutations in genes encoding vacuolar permeases (atg22Δ and atg22Δ avt3Δ avt4Δ),14 in selective autophagy genes (atg11Δ and atg32Δ)15,16 and in a vacuolar protease gene (pep4Δ).17 atg22Δ, atg22Δ avt3Δ avt4Δ, atg11Δ, and atg32Δ mutant yeasts did not significantly differ from wild-type yeasts with respect to the percentage of cells in G1/G0 after nitrogen starvation (Fig. 1A; Fig. S1). In contrast, pep4Δ yeasts, which are deficient in the maturation of vacuolar enzymes, showed a similar percentage of cells without buds in response to nitrogen starvation as atg1Δ, atg5Δ, vps30Δ/atg6Δ, atg7Δ, and atg8Δ yeasts (Fig. 1A; Fig. S1). Taken together, these observations suggest that the failure of atg1Δ, atg5Δ, vps30Δ/atg6Δ, atg7Δ, and atg8Δ yeasts to arrest in G1/G0 in response to nitrogen starvation is not likely due to a deficiency in general nutrient recycling or selective autophagy, but rather, may be due to defects in autophagic cargo degradation in the yeast vacuole; the pep4∆ mutant is deficient in the activity of multiple vacuolar hydrolases and accumulates autophagic bodies within the vacuole lumen.
To further confirm that autophagy-deficient yeasts fail to enter G1/G0 after nitrogen starvation, we measured DNA content using flow cytometry. Consistent with the budding assay data (Fig. 1A), the majority of wild-type, atg22∆, atg22Δ avt3Δ avt4Δ, atg11∆, and atg32∆ cells had a 1N DNA content 4 h after nitrogen starvation. In contrast, the majority of atg1Δ, atg5Δ, vps30Δ/atg6Δ, atg7Δ, atg8Δ, and pep4Δ yeasts had a 2N DNA content after starvation, indicating they were not in G1/G0 (Fig. 1B; Fig. S2). Thus, both the budding assay and DNA content measurements indicated that yeast defective in general starvation-induced autophagy or vacuolar protease activity (but not in selective autophagy or vacuolar permease-dependent nutrient recycling of large, neutral amino acids via the Atg22, Avt3 and Avt4 permeases) fail to properly arrest in G1/G0 following nitrogen starvation.
Autophagy-deficient yeasts fail to progress through G2/M in response to starvation
Next we examined whether autophagy-deficient yeasts continued to actively divide after nitrogen starvation. We measured the growth curve of wild-type and vps30Δ/atg6Δ yeast strains in both nutrient-rich YPD medium and in SD-N starvation medium (Fig. 2A and B). In YPD medium, wild-type and vps30Δ/atg6Δ yeasts grew at a similar rate (Fig. 2A). In nitrogen-starvation conditions, the growth of wild-type yeasts did not cease until after 4 h whereas, in contrast, autophagy-deficient yeasts stopped dividing almost immediately (Fig. 2B). To exclude the possibility that the decreased numbers of vps30Δ/atg6Δ yeasts reflected increased cell death, we measured colony formation in wild-type and vps30Δ/atg6Δ yeasts after 4 h nitrogen starvation and found no difference in cell survival (Fig. 2C).
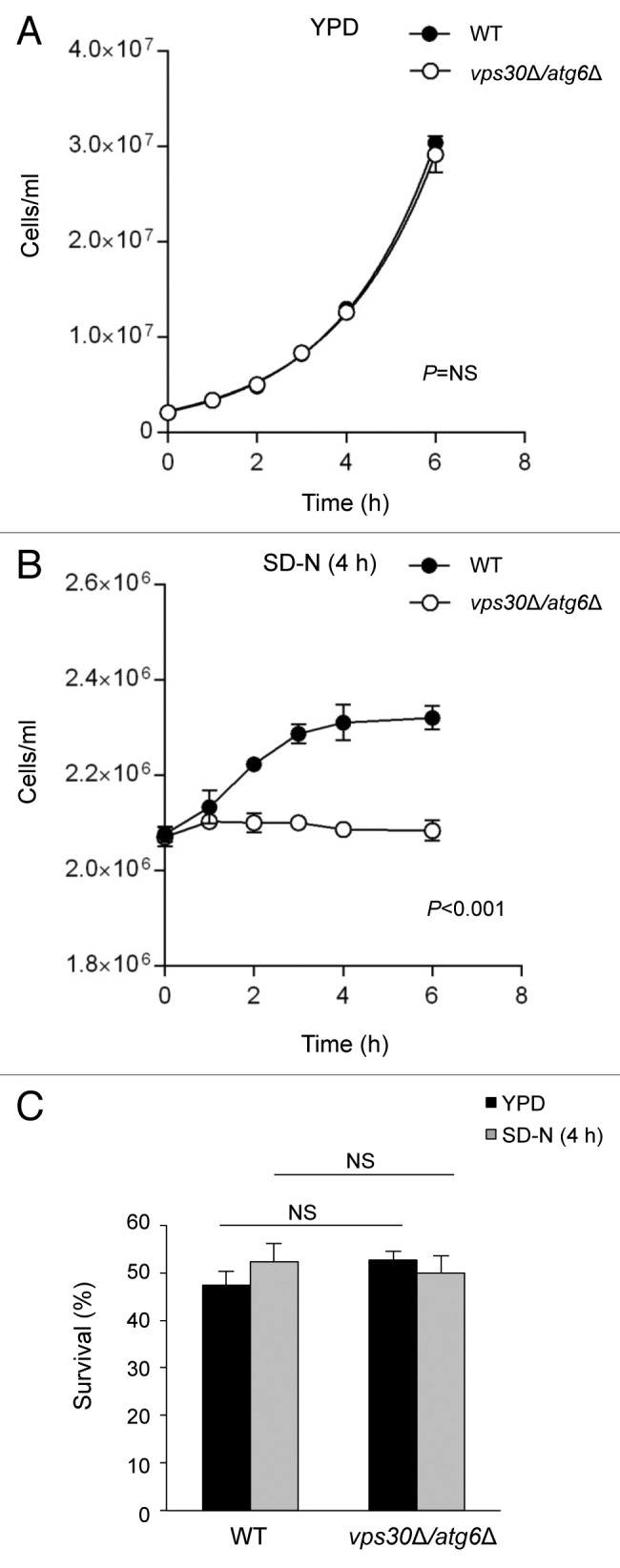
Figure 2. Cell growth and cell survival of wild-type and autophagy-deficient yeast during nitrogen starvation. (A and B) The number of yeast cells grown in YPD (A) or SD-N (B) was quantified at serial time points. Values represent mean ± SEM of triplicate samples. Similar results were observed in 2 independent experiments. In (A) cell growth curves were fitted to an exponential growth model to determine their doubling time. The difference between growth curves in wild-type (WT) vs. vps30Δ/atg6Δ yeast strains was analyzed using an ANOVA model; statistical results are indicated in each graph. NS, not significant. (C) Wild-type (WT) and vps30Δ/atg6Δ strains were grown in YPD or SD-N for 4 h and tested for survival by plating 100 cells on YPD plates and counting colonies after 48 h at 30 °C. Cell survival was calculated as the percentage of colonies over the total number of plated cells. Bars represent mean ± SEM of triplicate samples. Similar results were observed in 2 independent experiments. NS, not significant; one-way ANOVA for indicated comparison.
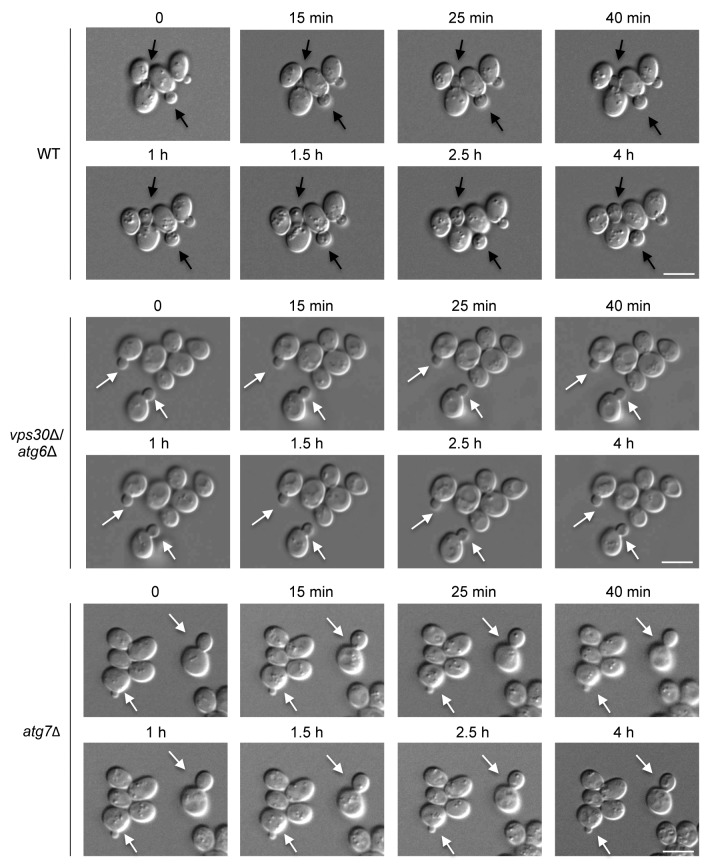
The accumulation of autophagy-deficient yeasts in G2/M (rather than G1/G0) coupled with their failure to divide after nitrogen starvation raised the possibility that they have a defect in cell cycle progression through G2/M. To examine this, we performed live cell imaging of wild-type and autophagy-deficient yeasts. Following nitrogen starvation, we found that the buds of wild-type yeast grew larger and eventually finished cytokinesis before the yeast cells became dormant. In contrast, the buds of autophagy-deficient vps30Δ/atg6Δ and atg7∆ yeasts stayed attached to the mother cells and failed to grow larger (Fig. 3). Thus, autophagy may be necessary for proper transit through G2/M after nitrogen starvation.
Figure 3. Autophagy-deficient yeasts fail to complete cell division after nitrogen starvation. Yeasts were grown on an SD-N agarose pad and images were taken at serial time points using a Zeiss Axioplan 2 microscope. The time above each image indicates the time in nitrogen starvation (SD-N). Black arrows denote buds that separate to become daughter cells during the imaging period. White arrows denote buds that fail to separate to become daughter cells during the imaging period. Scale bar: 5 μm. At least 15 cells were examined per time-lapse photography experiment and similar results were observed in 5 different experiments.
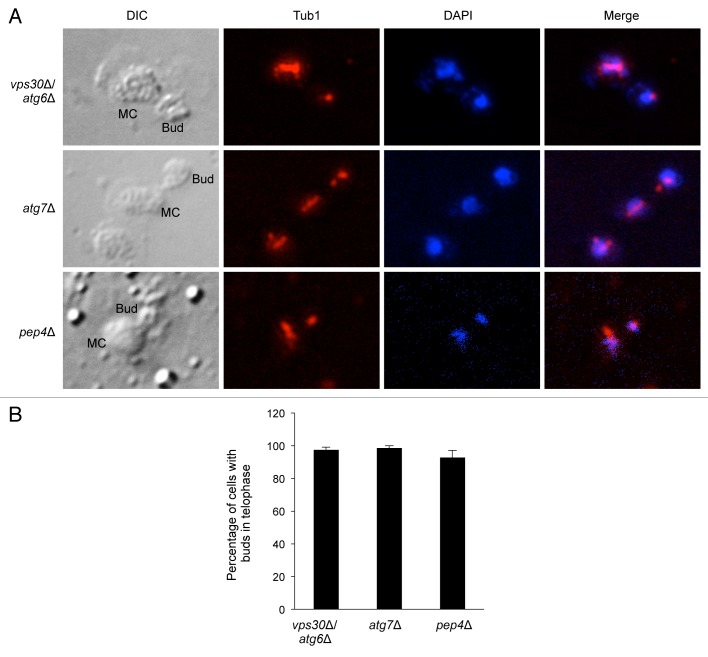
To further clarify the stage of G2/M arrest in autophagy-deficient yeasts in response to nitrogen starvation, we stained DNA with DAPI and examined the morphology of the yeast spindle microtubules by immunofluorescence staining with an antibody recognizing the yeast tubulin, Tub1. We examined cells at 4 h after nitrogen starvation—a time point when the majority of wild-type yeasts lacked buds and were in G1/G0 and the majority of atg6Δ, atg7∆, and pep4∆ yeasts were in G2/M (Fig. 1). For the mutant cells, we quantified the percentage of cells among those arrested with buds in different stages of G2/M; this was not possible for the wild-type yeasts as the vast majority did not have buds. In atg6Δ, atg7∆, and pep4∆ yeasts, nearly 100% of the cells arrested with buds had complete spindle disassembly and separation of the mother and daughter chromosomes, signifying an arrest in telophase with a failure to undergo cytokinesis (Fig. 4A and B). Thus, genes essential for autophagy and for vacuolar protease activity are required for mitotic exit during nitrogen starvation.
Figure 4. Autophagy-deficient yeasts and pep4∆ yeasts arrest in telophase after nitrogen starvation. (A) Anti-Tub1 staining and DNA staining with DAPI of indicated yeast strains starved in SD-N for 4 h. MC, mother cell. (B) Quantification of percentage of cells with buds that are in telophase. Values represent mean ± SEM of triplicate samples (> 50 cells analyzed per sample). Similar results were observed in 3 independent experiments.
Autophagy-deficient yeasts have quiescence-specific phenotypes after nitrogen starvation
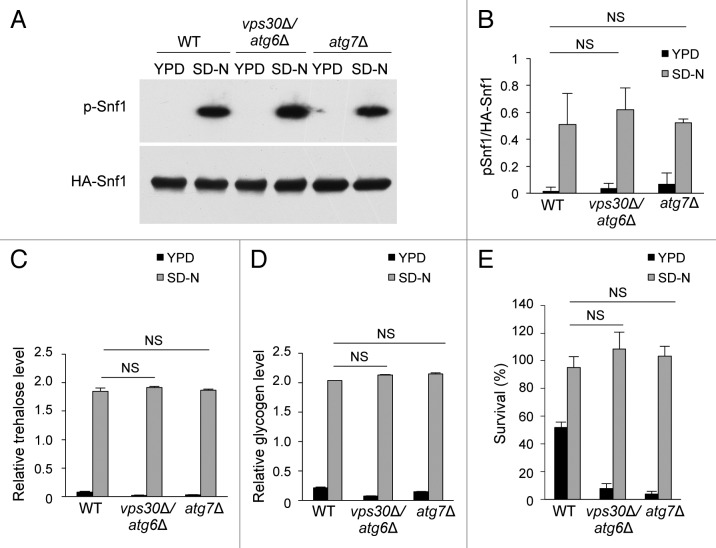
Previous studies have shown that cell cycle mutants arrested in G2/M can acquire characteristics of quiescence.3 We investigated whether autophagy-deficient yeasts, although they fail to properly enter G1/G0 in response to nitrogen starvation, also display features of quiescence. First, we asked whether the yeast ortholog of mammalian AMP-activated protein kinase (AMPK), Snf1, a crucial regulator of quiescence entry11 is activated normally during nitrogen starvation in autophagy-deficient yeast strains. We found that Snf1 was phosphorylated within 30 min after nitrogen starvation in wild-type, vps30Δ/atg6Δ, and atg7∆ yeasts (Fig. 5A and B). Thus, Snf1 activation can occur in the absence of G1/G0 entry, and defects in Snf1 activation are unlikely to explain the failure of autophagy-deficient yeast to properly arrest in G1/G0 in response to nitrogen starvation.
Figure 5. Analysis of quiescence-specific phenotypes in wild-type and autophagy-deficient yeasts after nitrogen starvation. (A) Western blot detection of p-Snf1 during growth in YPD at time 0 (YPD) or 30 min after transfer to SD-N (SD-N) in indicated yeast strains transformed with a plasmid expressing HA-Snf1. (B) Densitometry quantification of the ratio of p-Snf1/total Snf1 in indicated yeast strains transformed with a plasmid expressing HA-Snf1 and cultured as described in (A). Bars represent mean ± SEM values from 3 independent experiments, including the representative gels shown in (A). (C and D) Measurement of trehalose (C) and glycogen (D) levels in yeasts grown in YPD or SD-N for 4 h. Bars represent mean ± SEM of triplicate samples. Similar results were observed in 3 independent experiments. (E) Assessment of heat-shock resistance in indicated yeast strains during growth in YPD or SD-N for 4 h. Bars represent mean ± SEM of triplicate samples. Similar results were observed in 3 independent experiments. NS, not significant; one-way ANOVA with Tukey test for multiple comparisons.
Some of the phenotypic properties that characterize quiescence in yeast are metabolic changes, such as increased trehalose and glycogen accumulation and resistance to heat shock.2,11 In response to nitrogen starvation, wild-type, vps30Δ/atg6Δ, and atg7∆ yeasts accumulated trehalose and glycogen (Fig. 5C and D). In addition, all 3 strains showed starvation-induced heat resistance (Fig. 5E). During exponential growth in YPD medium, the viability of wild-type, vps30Δ/atg6Δ, and atg7∆ strains was reduced after exposure to 50 °C for 20 min. Consistent with the known prosurvival function of autophagy in response to environmental stress, vps30Δ/atg6Δ and atg7∆ yeasts were more sensitive than wild-type yeasts to heat shock when grown in YPD. However, after 4 h of nitrogen starvation, vps30Δ/atg6Δ and atg7∆ yeasts were as heat-resistant as wild-type yeasts (Fig. 5E). The same results were also observed for other autophagy-deficient yeast strains, including atg1∆, atg5∆, and atg8∆ (Fig. S3). Together, these results suggest that autophagy-deficient yeasts arrest in telophase and enter quiescence in response to nitrogen starvation.
Discussion
Our results indicate that, during nitrogen starvation in S. cerevisiae, autophagy gene products are essential for cell cycle progression and G1/G0 arrest. However, these proteins are dispensable for acquisition of a quiescent phenotype, including acquisition of thermo-resistance and accumulation of storage carbohydrates. This requirement for autophagy genes is consistent with a previous report demonstrating that atg1∆ and atg2∆ yeasts are deficient in G2/M progression during starvation.18 Our observation of a similar phenotype in yeast with null mutations in multiple other autophagy genes, including ATG5, VPS30/ATG6, ATG7, and ATG8, provides additional support for the finding that the autophagy pathway is essential in cell cycle progression following nitrogen starvation. Moreover, our results also demonstrate a somewhat unexpected finding—namely, that core autophagy genes are NOT essential for key aspects of starvation-induced cellular quiescence, at least in the budding yeast, S. cerevisiae. In addition, our results confirm and extend the importance of autophagy genes and vacuolar degradation in mitotic exit during nitrogen starvation; based on analysis of spindle morphology, we found that vps30Δ/atg6Δ, atg7∆, and pep4∆ yeasts arrest in telophase. Consistent with our findings, Matsui et al. also reported a role for Atg2 in completion of cytokinesis.18
The mechanism(s) by which autophagy genes are required for mitotic exit during nitrogen starvation are not yet fully understood. In the study by Matsui et al., it was postulated that starved cells rely on autophagy to generate catabolites necessary for synthesizing proteins essential for cell cycle progression; indeed, the defect in G2/M progression in atg2∆ yeasts with auxotrophy for tryptophan is rescued when tryptophan is added to the nitrogen-depleted medium.18 However, in our study, it seems unlikely that the lack of cytoplasmic availability of amino acids, nucleic acids, and/or other catabolites is responsible for the defect in starvation-induced G1/G0 arrest as yeasts lacking the vacuolar permeases Atg22, Avt3, and Avt4 were similar to wild-type yeasts with respect to G1/G0 arrest in response to nitrogen starvation. It is possible that the role of nutrient recycling in G2/M progression is different in a synchronous population following α-factor release (as studied by Matsui et al.) compared with the asynchronous population of yeasts examined in the present study. We also cannot definitively rule out the possibility that recycling of certain amino acids or another nutrient factor is important in mediating mitotic exit in nitrogen starvation conditions, but just not through the vacuolar permeases Atg22, Avt3, and Avt4; these permeases export large, neutral amino acids and are not the only amino acid permeases in the vacuole membrane.
In contrast to vacuolar permeases, we found that Pep4, a vacuolar protease required for function of multiple hydrolases and, hence, autophagic cargo degradation,17 was required for proper starvation-induced G1/G0 arrest. Since the formation of autophagosomes is normal in pep4 mutant yeasts (and presumably the sequestration of cell cycle regulators within the vacuole should be unimpaired), the mitotic arrest phenotype observed in our study is unlikely due to a specific requirement for autophagy-dependent proteolysis of a cell regulator(s) during nitrogen starvation conditions. Rather, our findings are more consistent with a role for vacuolar nutrient sensing, which depends on both autophagosome formation (hence the phenotype of atg1Δ, atg5Δ, vps30/atg6Δ, atg7Δ, and atg8Δ mutant yeasts) and the generation of free amino acids generated by vacuolar proteases within the vacuole (hence the phenotype of pep4Δ mutant yeasts), in mediating mitotic exit. In mammalian cells, there is increasing evidence that amino acids inside the lysosome are sensed by the v-ATPase-Ragulator complex, which signals to the mechanistic target of rapamycin complex 1 (MTORC1), a critical regulator of the cell cycle including telophase and cytokinesis.19-24 Although Matsui et al. conclude that yeast TORC1 may be dispensable for cell cycle progression during nitrogen starvation (based on the lack of an observed effect of rapamycin in wild-type yeasts),18 it is notable that rapamycin does not suppress mammalian MTORC1 serine 2481 phosphorylation which enriches MTORC1 near the contractile ring during telophase and cytokinesis.25 Thus, our findings highlight the need for further studies to investigate the role of TORC1-dependent and/or other potential yeast vacuolar nutrient sensing pathways in mediating mitotic exit during nitrogen starvation.
The entry of autophagy-deficient yeasts into quiescence with 2N DNA content may have important implications for understanding broader aspects of yeast biology. First, since autophagy-deficient yeasts have impaired survival during prolonged nitrogen starvation,26 our findings suggest that the quiescence program of autophagy-deficient yeasts is qualitatively different than the G1/G0 quiescence program in terms of permitting cells to adapt to nutrient deprivation. Second, it has been previously shown that yeasts that arrest in G2/M have diminished ability to give rise to progeny,3-5 but the mechanistic basis for this has been unclear. Matsui et al. found that atg2∆ yeasts with a defect in G2/M progression during nitrogen starvation have increased aneuploidy following replenishment with a nitrogen source and cell division.18 Therefore, our findings, taken together with these previous observations, suggest a model in which entry into quiescence in G2/M (such as that which occurs in autophagy-deficient yeasts) may increase genome instability, and thereby diminish production of progeny. Clearly, despite having similar markers of quiescence that signify similar adaptation to environmental stress (such as the acquisition of thermotolerance), there are fundamental as-of-yet undefined differences between G1/G0 and quiescence entry during other stages of the cell cycle that may explain the beneficial effects of G1/G0 quiescence on eukaryotic cell survival and reproduction.
Materials and Methods
Strains and culture conditions
S. cerevisiae strains used in this study are listed in Table 1. Unless noted otherwise, yeasts were grown in YPD (20 g/L peptone, 10 g/L yeast extract and 20 g/L glucose). For nitrogen starvation, yeasts were grown in SD-N (1.7 g/L yeast nitrogen base without amino acids and ammonium sulfate and 20 g/L glucose). Solid medium contained 20 g/L agar.
Table 1. Yeast strains used in this study.
| Strain | Genotype | Refs. |
|---|---|---|
| AHY001 | SEY6210 atg11Δ::HIS5 | 15 |
| JCY3000 | SEY6210 atg6∆::HIS3 | 27 |
| MGY101 | SEY6210 atg5∆::LEU2 | 28 |
| SEY6210 | MATα leu2-3, 112 ura3-52 his∆200 trp1-∆901 lys2-801 suc2-∆9 | 29 |
| TKYM139 | SEY6210 atg32∆::LEU2 | 16 |
| TVY1 | SEY6210 pep4∆::LEU2 | 17 |
| VDY101 | SEY6210 atg7∆::LEU2 | 30 |
| WHY001 | SEY6210 atg1∆::HIS5 | 31 |
| WPHYD7 | SEY6210 atg8∆::LEU2 | 32 |
| ZFY6 | SEY6210 atg22∆::TRP1 | 14 |
| ZFY22 | SEY6210 avt3Δ::HIS3 avt4Δ::URA3 atg22Δ::TRP1 | 14 |
Budding assay
Five × 105 cells/ml were grown in 5 ml fresh YPD for 4 h to early midlog phase (OD600 0.2–0.3). Samples were washed 3 times with SD-N and 1/3 of the culture (labeled “YPD”) was fixed in 70% ethanol for 2 h and washed with 50 mM sodium citrate, pH 7.4. After brief sonication, cells with or without buds were quantified using differential interference contrast microscopy. The remainder of the culture was starved in SD-N for 4 h or 24 h. Samples were then processed and analyzed as described for the YPD samples.
Quantification of yeast cell numbers
Yeast cells were washed 3 times in autoclaved H2O prior to resuspension in either YPD or SD-N. Cell numbers were determined by using a hemocytometer (Hausser Scientific, 3100) according to the manufacturer’s instructions.
Yeast transformation
Yeasts were transformed with a plasmid encoding HA-Snf133 using the Frozen-EZ Yeast Transformation II Kit (Zymo Research, T2001) according to the manufacturer’s instructions.
Flow cytometry
Five × 105 cells/ml were grown in 5 ml fresh YPD for 4 h to early midlog phase (OD600 0.2–0.3). Samples were washed 3 times with SD-N and 1 ml of the culture was removed and fixed with 70% ethanol at 4 °C overnight. The remainder of the culture was starved in SD-N for 4 h before fixation with 70% ethanol. After overnight fixation, cells were washed twice with 50 mM sodium citrate, pH 7.4, incubated for 2 h at 37 °C with 0.25 mg/ml RNase A (Sigma, R4875), washed twice with 50 mM sodium citrate, pH 7.4, and stained with 5 μg/ml propidium iodide (Roche, 11348639001) for 30 min in the dark. Cell cycle analysis was performed on a FACSCalibur (BD Biosciences) in the UT Southwestern Flow Cytometry Core Facility and the data were analyzed using FlowJo 8.8.6.
Protein extraction, immunoprecipitation, and western blot analysis
Protein was extracted from yeast cells grown in YPD or SD-N. Two × 108 cells were harvested from each sample. Yeasts were lysed in 500 μl RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% SDS, 1% NP40 substitute IGEPAL CA-630 (Sigma, I8896), 0.5% sodium deoxycholate (Sigma, D6750) containing the complete protease inhibitor cocktail (Roche, 11836170001) and the halt phosphatase inhibitor cocktail (Thermo Scientific, 78427). HA-Snf1 was immunoprecipitated using anti-HA agarose conjugate (Santa Cruz, SC7392AC). Proteins were resolved using 4–15% TGX gels (Bio-Rad, 456-1086) and transferred to PVDF membranes. HA-Snf1 was detected with an HA-specific HRP-conjugated mouse monoclonal antibody (Santa Cruz, SC7392HRP). Phospo-Snf1 was detected with a phospho-AMPK α (Thr172) rabbit monoclonal antibody (Cell Signaling Technology, 2535).
Glycogen and trehalose assays
Glycogen and trehalose were hydrolyzed to glucose as previously described.34 2 × 108 cells were harvested. Glucose levels were determined using the Glucose (GO) Assay Kit (Sigma, GAGO20). OD540 was measured using a POLARstar Omega multi-mode microplate reader (BMG Labtech).
Immunofluorescence
Yeast cells were grown to early midlog phase (OD600 0.2–0.3), washed 3 times with SD-N and starved in SD-N for 4 h. Cells were then fixed with 4% paraformaldehyde for 45 min at 30 °C, washed once with 100 mM potassium phosphate, pH 6.5, and washed twice with P solution (100 mM potassium phosphate, pH 6.5, 1.2 M sorbitol). Fifteen microliters zymolase (100T; Zymo Research, E1005) and 2.5 μL β-mercaptoethanol were added to 500 μL cells in P solution. After 1 h incubation at 30 °C, cells were washed twice with P solution and placed on poly-d-lysine-coated chamber slides (Electron Microscopy Sciences, 63410-01) for 10 min. Cells were washed with phosphate-buffered saline (Sigma, D8537) containing 5 mg/ml BSA (Sigma, A9647) (PBS-BSA). Anti-Tub1 antibody (Novus Biologicals, NB100-690) was added at a dilution of 1:100 in PBS-BSA, and slides were incubated overnight at 4 °C. The slides were washed 3 times with PBS-BSA and secondary antibody (Alexa 594; Invitrogen, A21203) was added at a dilution of 1:2,000 in PBS-BSA and incubated in the dark for 2 h. Cells were washed 3 times with PBS-BSA, mounted with DAPI-mounting medium (Vector Laboratories, H-1200) and sealed with nail polish. Images were captured using a Zeiss Axioplan 2 fluorescence microscope.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Martin Schmidt for the HA-Snf1 construct and advice on Snf1 western blotting; Shuyi Huang and Sunil Laxman for advice and technical help with yeast live cell imaging; and Haley Harrington for assistance with manuscript preparation. This work was supported by CPRIT grant RP120718-P1 (BL) and NIH grants R01CA84254 (BL), R01CA109618 (BL) R01CA172211 (RZ, GX), R01CA108947 (RF), and R01GM053396 (DJK).
Glossary
Abbreviations:
- Atg
autophagy-related
- MTORC1
mechanistic target of rapamycin complex 1
- SD-N
nitrogen starvation medium
References
- 1.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–27. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 2.Smets B, Ghillebert R, De Snijder P, Binda M, Swinnen E, De Virgilio C, Winderickx J. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr Genet. 2010;56:1–32. doi: 10.1007/s00294-009-0287-1. [DOI] [PubMed] [Google Scholar]
- 3.Daignan-Fornier B, Sagot I. Proliferation/Quiescence: When to start? Where to stop? What to stock? Cell Div. 2011;6:20. doi: 10.1186/1747-1028-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laporte D, Lebaudy A, Sahin A, Pinson B, Ceschin J, Daignan-Fornier B, Sagot I. Metabolic status rather than cell cycle signals control quiescence entry and exit. J Cell Biol. 2011;192:949–57. doi: 10.1083/jcb.201009028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei W, Nurse P, Broek D. Yeast cells can enter a quiescent state through G1, S, G2, or M phase of the cell cycle. Cancer Res. 1993;53:1867–70. [PubMed] [Google Scholar]
- 6.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 10.Lee IH, Kawai Y, Fergusson MM, Rovira II, Bishop AJ, Motoyama N, Cao L, Finkel T. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336:225–8. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abeliovich H, Klionsky DJ. Autophagy in yeast: mechanistic insights and physiological function. Microbiol Mol Biol Rev. 2001;65:463–79. doi: 10.1128/MMBR.65.3.463-479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zetterberg A, Larsson O. Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc Natl Acad Sci U S A. 1985;82:5365–9. doi: 10.1073/pnas.82.16.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17:5094–104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr., Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–96. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerhardt B, Kordas TJ, Thompson CM, Patel P, Vida T. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J Biol Chem. 1998;273:15818–29. doi: 10.1074/jbc.273.25.15818. [DOI] [PubMed] [Google Scholar]
- 18.Matsui A, Kamada Y, Matsuura A. The role of autophagy in genome stability through suppression of abnormal mitosis under starvation. PLoS Genet. 2013;9:e1003245. doi: 10.1371/journal.pgen.1003245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–83. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med. 2012;18:524–33. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekim B, Magnuson B, Acosta-Jaquez HA, Keller JA, Feener EP, Fingar DC. mTOR kinase domain phosphorylation promotes mTORC1 signaling, cell growth, and cell cycle progression. Mol Cell Biol. 2011;31:2787–801. doi: 10.1128/MCB.05437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vazquez-Martin A, Sauri-Nadal T, Menendez OJ, Oliveras-Ferraros C, Cufí S, Corominas-Faja B, López-Bonet E, Menendez JA. Ser2481-autophosphorylated mTOR colocalizes with chromosomal passenger proteins during mammalian cell cytokinesis. Cell Cycle. 2012;11:4211–21. doi: 10.4161/cc.22551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaba A, Bianchi V, Borini A, Johnson J. A putative mitotic checkpoint dependent on mTOR function controls cell proliferation and survival in ovarian granulosa cells. Reprod Sci. 2008;15:128–38. doi: 10.1177/1933719107312037. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Bonet E, Vazquez-Martin A, Pérez-Martínez MC, Oliveras-Ferraros C, Pérez-Bueno F, Bernadó L, Menendez JA. Serine 2481-autophosphorylation of mammalian target of rapamycin (mTOR) couples with chromosome condensation and segregation during mitosis: confocal microscopy characterization and immunohistochemical validation of PP-mTOR(Ser2481) as a novel high-contrast mitosis marker in breast cancer core biopsies. Int J Oncol. 2010;36:107–15. [PubMed] [Google Scholar]
- 25.Hornberger TA, Chien S. Mechanical stimuli and nutrients regulate rapamycin-sensitive signaling through distinct mechanisms in skeletal muscle. J Cell Biochem. 2006;97:1207–16. doi: 10.1002/jcb.20671. [DOI] [PubMed] [Google Scholar]
- 26.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 27.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–48. doi: 10.1128/MCB.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–37. doi: 10.1016/S1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol Biol Cell. 2000;11:969–82. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seaman MN, Marcusson EG, Cereghino JL, Emr SD. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell. 1999;10:1337–51. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Huang WP, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCartney RR, Schmidt MC. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem. 2001;276:36460–6. doi: 10.1074/jbc.M104418200. [DOI] [PubMed] [Google Scholar]
- 34.Parrou JL, François J. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem. 1997;248:186–8. doi: 10.1006/abio.1997.2138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.