Abstract
Acute promyelocytic leukemia (APL) is characterized by the t(15;17)-associated PML-RARA fusion gene. We have previously found that MIR125B1 is highly expressed in patients with APL and may be associated with disease pathogenesis; however, the mechanism by which MIR125B1 exerts its oncogenic potential has not been fully elucidated. Here, we demonstrated that MIR125B1 abundance correlates with the PML-RARA status. MIR125B1 overexpression enhanced PML-RARA expression and inhibited the ATRA-induced degradation of the PML-RARA oncoprotein. RNA-seq analysis revealed a direct link between the PML-RARA degradation pathway and MIR125B1-arrested differentiation. We further demonstrated that the MIR125B1-mediated blockade of PML-RARA proteolysis was regulated via an autophagy-lysosomal pathway, contributing to the inhibition of APL differentiation. Furthermore, we identified DRAM2 (DNA-damage regulated autophagy modulator 2), a critical regulator of autophagy, as a novel target that was at least partly responsible for the function of MIR125B1 involved in autophagy. Importantly, the knockdown phenotypes for DRAM2 are similar to the effects of overexpressing MIR125B1 as impairment of PML-RARA degradation, inhibition of autophagy, and myeloid cell differentiation arrest. These effects of MIR125B1 and its target DRAM2 were further confirmed in an APL mouse model. Thus, MIR125B1 dysregulation may interfere with the effectiveness of ATRA-mediated differentiation through an autophagy-dependent pathway, representing a novel potential APL therapeutic target.
Keywords: autophagy, degradation, leukemia, microRNA, PML-RARA
Introduction
Acute promyelocytic leukemia is a unique subtype of acute myeloid leukemia (AML) that accounts for approximately 10% to 15% of all cases.1 The molecular feature of APL is an aberrant chromosomal translocation that juxtaposes the PML gene with the RARA gene,2 and the subsequent expression of the PML-RARA fusion protein blocks the differentiation of promyelocytes and leads to an accumulation of leukemic promyelocytes. All-trans retinoic acid (ATRA) has successfully been used as a leukemia therapy to target the transcriptional repression mediated by the PML-RARA fusion protein. ATRA has been found to cause disease regression specifically in patients with APL,3,4 and it induces the degradation of the chimeric protein encoded by the PML-RARA oncogene.5-7
Understanding the mechanisms involved in APL pathogenesis and its responsiveness to treatment will allow us to identify novel APL treatment targets. It has been unexpectedly found that 2 therapeutic agents, ATRA and arsenic trioxide (ATO), degrade PML-RARA. More recently, studies have demonstrated that autophagy contributes to the therapy-induced degradation of PML-RARA.7,8 Autophagy is a biological process in which cytoplasmic constituents are sequestered into double-membrane vesicles called autophagosomes and then fuse with lysosomes in which the sequestered contents are degraded.9,10 Previous studies have also revealed the proteasome system as a primary mechanism involved in the therapy-induced degradation of PML-RARA.6 However, proteasome and caspase inhibitors do not fully block PML-RARA degradation,6,11 suggesting that alternative mechanisms may be involved. Moreover, the PML-RARA oncoprotein is prone to aggregation, which makes it a preferred substrate for autophagic degradation.12
A number of studies have indicated that microRNAs (miRNAs) play crucial roles in fundamental biological processes, including the cell cycle, differentiation, development, and apoptosis.13-16 It has been demonstrated that miRNAs regulate gene expression by binding to the 3′-untranslated regions (3′UTR) of target mRNAs, which leads to translational suppression. Our group and others have reported the unique miRNA profiles associated with the main AML cytogenetic and molecular subgroups.17-19 In particular, the t(15;17) translocation leads to elevated MIR125B1 (microRNA 125b1) expression;19 however, it is not clear how MIR125B1 is correlated with PML-RARA status and in which pathways the miRNA may be involved. Although the PML-RARA oncoprotein blocks differentiation and promotes self-renewal in cell lines, this protein causes leukemia in mice only after a long latency period.20 This finding suggests that PML-RARA may require cooperating events to cause leukemia. Thus, we hypothesized that MIR125B1 upregulation may be important for the effects of PML-RARA on cell differentiation in APL or may affect the therapy-induced clearance of PML-RARA.
To test our hypothesis, we investigated correlations between MIR125B1 expression and PML-RARA fusion oncogene expression in APL in vivo and in vitro. We found that MIR125B1 could activate PML-RARA expression and repress therapy-induced PML-RARA proteolysis by an autophagy-lysosome pathway. Thus, MIR125B1 is critical for PML-RARA activities and for cell differentiation, suggesting a novel potential therapeutic target for APL.
Results
PML-RARA promotes MIR125B1 expression
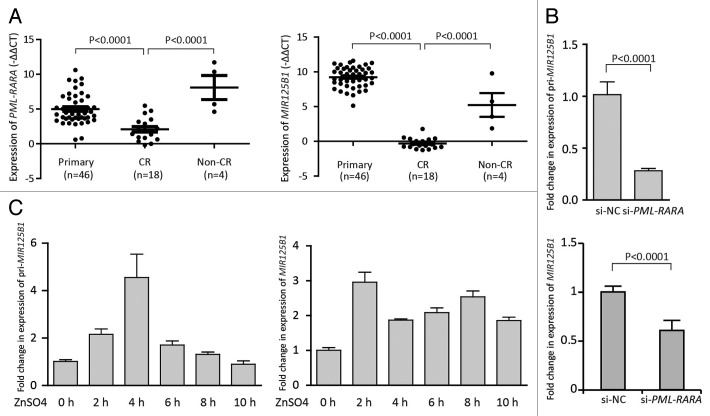
MIR125B1 overexpression has been described as an important oncogenic event, and consistent abnormalities in MIR125B1 expression were found in several cases of chromosomal translocation, including t(2;11)(p21;q23) and t(11;14)(q24;q32).14,21 Our previous study also demonstrates that MIR125B1 expression is strongly upregulated in patients with APL compared with other AML subtypes and indicated that MIR125B1 may be involved in APL pathogenesis.19 Here, we further investigated the expression of MIR125B1 and PML-RARA in the patients before and after therapy to establish the relationship between MIR125B1 and PML-RARA. We found that the MIR125B1 expression levels correlated well with the level of PML-RARA transcripts (Fig. 1A, left panel). In patients with t(15;17) primary APL, MIR125B1 expression was significantly higher than that in complete remission (CR) patients (Fig. 1A, right panel). Similarly, in non-CR patients (nonremission, including relapse samples), the MIR125B1 expression level was higher than that in CR patients. Because therapy-induced treatment of APL led to the clearance of leukemia cells and PML-RARA transcripts, we then asked whether the fusion protein is necessary for MIR125B1 expression or whether MIR125B1 promotes PML-RARA expression. To address this question, we employed a small interfering RNA (siRNA), which specifically targets the breakpoint region of PML-RARA, named as si-PML-RARA hereafter.22 Transfection of si-PML-RARA resulted in knockdown of PML-RARA expression (Fig. S1A). Knockdown of PML-RARA in NB4 cells led to the downregulation of pri-microRNA (pri-MIR125B1) (Fig. 1B, top panel) and mature miRNA (MIR125B1) (Fig. 1B, bottom panel). Next, we employed an additional cell line, U937-PR9, which carries a PML-RARA gene that can be induced with ZnSO4 treatment. We first determined the PML-RARA fusion transcript expression level upon ZnSO4 treatment and found that the PML-RARA transcript was significantly upregulated in cells after 100 μM ZnSO4 stimulation (Fig. S1B). The increased PML-RARA transcript induced both pri-MIR125B1 (Fig. 1C, left panel) and MIR125B1 (Fig. 1C, right panel) expression (Fig. 1C), indicating that MIR125B1 upregulation may be a consequence of PML-RARA accumulation.
Figure 1.MIR125B1 is induced by PML-RARA. (A) The expression of PML-RARA in patients with t(15;17) APL before and after therapy. Left panel, the expression of PML-RARA mRNA in patients with t(15;17) APL before and after therapy. Right panel, the expression of MIR125B1 in patients with t(15;17) APL before and after therapy. This comparison revealed that APL therapy had broadly similar effects on PML-RARA and MIR125B1 expression. (B) qRT-PCR analysis of pri-MIR125B1 and mature MIR125B1 after knocked down the PML-RARA. (C) qRT-PCR analysis of pri-MIR125B1 and mature MIR125B1 in U937-PR9 cells treated with 100 μM ZnSO4. Experiments were performed in triplicate, and error bars represent SD in all panels. CR: samples in first complete remission; non-CR: noncomplete remission, including relapse samples.
MIR125B1 promotes PML-RARA expression and inhibits PML-RARA degradation
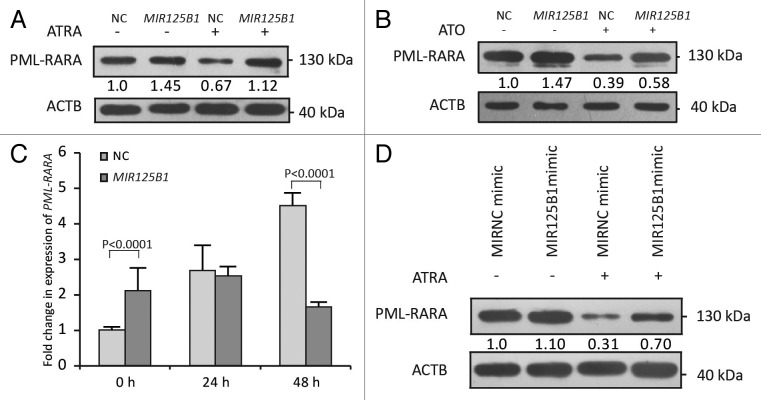
In addition to the effects of PML-RARA on MIR125B1 expression, we then tested whether MIR125B1 promotes PML-RARA expression. For this purpose, NB4 cells were infected with lentiviruses carrying the MIR125B1 gene (named as NB4-Lv-MIR125B1 cells hereafter) or a negative control (NB4-Lv-NC cells) to explore the effects of MIR125B1 on the fusion protein. ATRA treatment was also examined. The results demonstrated that the level of the PML-RARA oncoprotein was increased in cells overexpressing MIR125B1, especially when with ATRA treatment (Fig. 2A). We also confirmed these results with ATO-treated cells (Fig. 2B). Interestingly, we found that stable ectopic expression of MIR125B1 was able to promote PML-RARA expression at the transcriptional level. As shown in Figure 2C, MIR125B1 overexpression increased the expression of PML-RARA at time point 0 h. In general, PML-RARA increases in the mRNA level after being treated with ATRA,23 however, this effect was inhibited by MIR125B1 overexpression. It can be observed that PML-RARA mRNA gradually increased in NC group, while decreased in the MIR125B1 overexpression group, especially at the time point of 48h with ATRA treatment (Fig. 2C). In contrast, knockdown of MIR125B1 by antagomir-125B (a synthetic MIR125B1 antisense) decreased the expression of PML-RARA in primary APL patient bone marrow mononuclear cells, which expressed a high level of endogenous MIR125B1 (Fig. S1C). To exclude the transcriptional impact on PML-RARA expression by stable overexpression of MIR125B1, we further measured the PML-RARA oncoprotein levels via transiently transfecting cells with MIR125B1 mimics and treatment with or without ATRA. As shown in Figure 2D, when NB4 cells were treated with ATRA shortly after transiently transfecting MIR125B1, the PML-RARA level in the NC cells was not significantly changed when compared with that in MIR125B1 cells prior to treatment (lane 1 and 2 in Fig. 2D, and Fig. S1D); however, the amount of PML-RARA oncoprotein was increased in cells transfected with MIR125B1 after ATRA treatment (Fig. 2D, lane 3 and 4), suggesting that MIR125B1 could impair PML-RARA degradation. Moreover, inhibition of endogenous MIR125B1 using synthetic MIR125B1 antisense (anti-MIR125B1) facilitated PML-RARA degradation (Fig. S1E). These data indicate that MIR125B1 not only activates PML-RARA expression but also inhibits ATRA-induced PML-RARA degradation, showing a close correlation between MIR125B1 levels and PML-RARA.
Figure 2.MIR125B1 increased the PML-RARA oncoprotein level. (A) Western blot of NB4-Lv-NC and NB4-Lv-MIR125B1 cells treated with or without ATRA (1 μM) for 48 h. (B) Western blot of NB4-Lv-NC and NB4-Lv-MIR125B1 cells treated with or without ATO (1 μM) for 24 h. (C) MIR125B1 increases the PML-RARA mRNA level upon ATRA treatment. Gene expression was normalized relative to ACTB (n = 3 independently obtained biological samples). (D) Immunoblot analyses of PML-RARA after transient transfection with MIR125B1, followed by treatment with ATRA (1 μM, 48 h) or not. Cells lysates were prepared for western blotting with antibodies against RARA. ACTB expression served as a loading control.
Transcriptome analysis indicated that MIR125B1-arrested differentiation might partly act through the PML-RARA degradation pathway
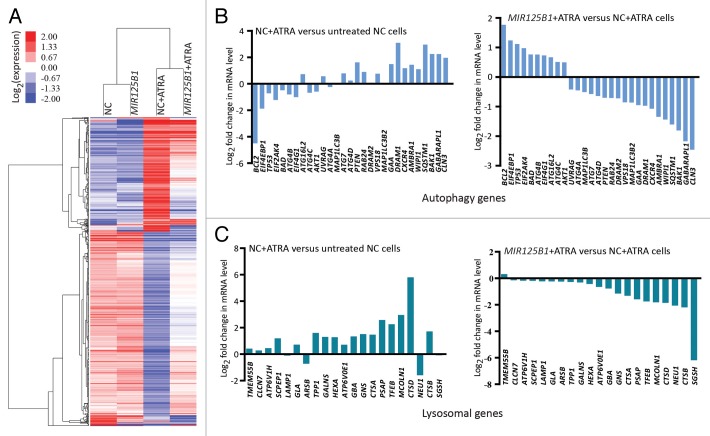
Cell differentiation is likely to be a determinant of the clinical response of APL, and MIR125B1 has been implicated in myeloid cell differentiation arrest.14 Because of the remarkable association between MIR125B1 expression and PML-RARA, we further investigated the effects of MIR125B1 on PML-RARA-regulated genes upon treatment. For this purpose, we performed high throughput RNA-seq (quantification) with RNA from NB4-Lv-MIR125B1 and NB4-Lv-NC cells treated with or without ATRA to identify genes or pathways that MIR125B1 may be mediated. We generated an average of 7279023 (range: 7140608- 7454744) reads per sample, and the gene expression profiles derived from RNA-Seq were calculated using the RPKM method.24 A hierarchical clustering analysis of the differentially expressed genes revealed that the majority of these genes are potentially regulated by MIR125B1 after treated with ATRA (Fig. 3A), including CEBPB, SPI1, IFIT3, and CEBPE, which have been reported previously.25,26 These results are consistent with the differentiation arrest effects of MIR125B1.14 The differential genes are shown in Table S1. To confirm the RNA sequencing data, we selected several genes for validation by qRT-PCR (Fig. S2A). Next, we compared our mRNA-seq results with recently published data that included potential PML-RARA targets using ChIP-on-chip experiments.27 We found that there was a significant overlap in the expression of potential PML-RARA target genes that were altered in the presence of MIR125B1 upon ATRA induction (Fig. S2B), indicating that MIR125B1 blocked PML-RARA-regulated genes which could be relieved upon ATRA treatment. This finding suggests that the effect of MIR125B1 on APL at least partially occurs via PML-RARA. In addition, using KEGG pathway analysis,28 several important signal transduction pathways, including the lysosome, MAPK, toll-like, and JAK-STAT pathways (Tables S2 and S3), were modulated by ATRA, and the effect of ATRA is impaired in the cells with MIR125B1 overexpression. Notably, previous studies have indicated an important role for lysosomes or the lysosome-dependent pathway in PML-RARA degradation and APL cell differentiation after therapeutic treatment.7,29 Collectively, these results suggest that PML-RARA degradation may be involved in MIR125B1-mediated cell differentiation arrest, and MIR125B1 expression may have a direct bearing on PML-RARA during differentiation therapy.
Figure 3.MIR125B1 attenuated the expression of a subset of PML-RARA target genes involved in myeloid differentiation. (A) Heatmap displaying the hierarchical clustering of genes in NB4-Lv-NC and NB4-Lv-MIR125B1 cells in response to ATRA treatment. The color intensity represents the degree of expression. A red-blue color scale was used to reflect standardized gene expression, with red representing a high expression and blue representing a low expression (scale shown in the upper left). A cutoff value of FDR ≤ 0.001 and a log2-fold change ratio ≥ 1 change relative to the untreated groups was selected. The identities of these genes are listed in Table S1. (B and C) Enforced MIR125B1 expression in NB4 cells repress the modification of a vast cast of lysosome and autophagy genes to ATRA stimuli, Related to (A) expression profiling of all genes whose expression is specifically altered in NB4 cells.
MIR125B1 inhibits the ATRA-induced clearance of PML-RARA by blocking the autophagy-lysosomal pathway
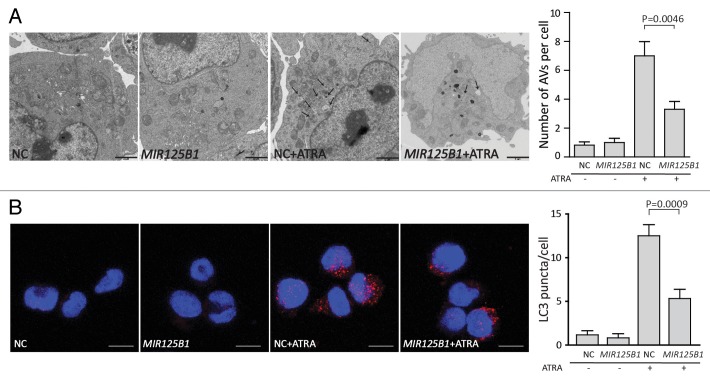
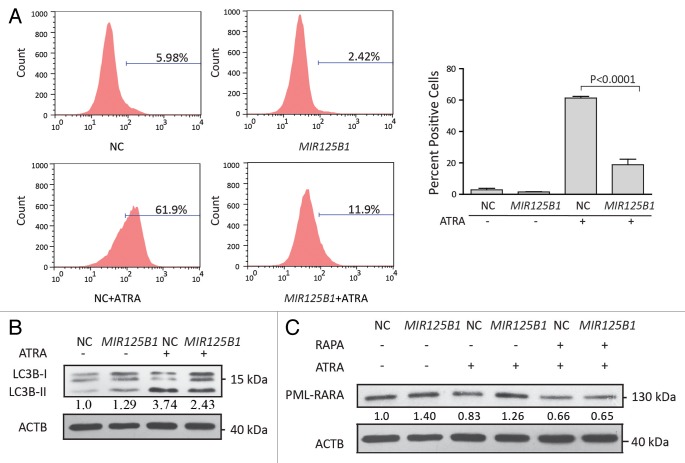
The above findings suggest that MIR125B1 may regulate the lysosomal pathway in ATRA-induced PML-RARA oncoprotein degradation and cell differentiation. Recent studies indicated a close link between the lysosomal pathway and autophagic degradation and demonstrated the importance of autophagy in APL differentiation and PML-RARA oncoprotein degradation.7,8 To better understand the role of autophagy in PML-RARA degradation, we first examined the expression of PML-RARA when degradation was blocked by 2 inhibitors chloroquine or bafilomycin A1. Both inhibitors interrupt the autophagosome-lysosome fusion step.10 Consistent with these previous studies, chloroquine and bafilomycin A1 significantly blocked ATRA-induced degradation of PML-RARA (Fig. S3A and S3B). Additionally, inhibition of autophagy by knockdown of ATG5 decreased degradation of PML-RARA (Fig. S3C). We therefore asked whether MIR125B1 could impair the degradation of PML-RARA via the autophagy-lysosomal pathway. To address this question, we first investigated the expression of a number of genes in the autophagy and lysosomal pathways in our RNA-seq data set. We found that lysosomal and autophagic genes were modulated by ATRA but the modulations were impaired in the presence of MIR125B1 (Fig. 3B and C). To further explore whether MIR125B1 inhibits the ATRA-induced clearance of PML-RARA through the autophagy-lysosomal pathway, we next examined the effects of MIR125B1 on ATRA-induced autophagy and lysosomal biogenesis. Ultrastructural analysis (Fig. 4A) and immunofluorescence (Fig. 4B) revealed that the ability of ATRA to induce autophagy was considerably reduced in the presence of MIR125B1. Accordingly, the expansion of the lysosomal compartment by ATRA was also impaired in NB4 cells that stably overexpressed MIR125B1 (Fig. 5A), indicating MIR125B1 involvement in lysosomal biogenesis. Furthermore, western blotting was used to detect LC3, whose cleaved and lipidated form (LC3-II) is widely regarded as an autophagosome marker. LC3-II levels were increased in cells treated with ATRA but repressed in cells overexpressing MIR125B1 (Fig. 5B). We further observed an increased autophagic flux in cells transfected with a synthetic MIR125B1 antisense (anti-MIR125B1) (Fig. S3D). To further validate the link between MIR125B1 and the autophagy pathway, cells were treated with an autophagy inducer (rapamycin) in the presence or absence of ATRA. We found that PML-RARA expression was markedly decreased in NB4-Lv-MIR125B1 cells after cotreatment with rapamycin and ATRA (Fig. 5C). Taken together, our findings suggest that MIR125B1 inhibits PML-RARA proteolysis via the autophagy-lysosomal pathway.
Figure 4.MIR125B1 impaired the autophagy-lysosomal-dependent degradation of PML-RARA. (A) The ultrastructure of NB4-Lv-NC and NB4-Lv-MIR125B1 cells was observed by electron microscopy after ATRA treatment. The number of autophagosomes observed by transmission electron microscopy was calculated (right panel, n = 10 cells). NB4-Lv-NC and NB4-Lv-MIR125B1 cells treated with ATRA (1 μM, 48 h) or not. (B) Confocal microscopy after staining with an antibody directed against LC3. NB4-Lv-NC and NB4-Lv-MIR125B1 cells treated with ATRA (1 μM, 48 h) or not. A representative image of 3 independent experiments is shown. Scale bar: 10 μm.
Figure 5. Effect of MIR125B1 overexpression on autophagy. (A) Fluorescence-activated cell sorting (FACS) analysis after staining with the lysosome-specific dye LysoTracker Red. The analysis was performed in triplicate, and 30,000 cells were analyzed. The bars indicate the proportion of cells with fluorescence intensity greater than the indicated threshold. (B) Western blot analyses of LC3 in NB4-Lv-NC and NB4-Lv-MIR125B1 cells treated with ATRA. ACTB was used as a loading control. LC3-II/ACTB densitometric ratios were recorded. (C) Western blot of NB4-Lv-NC and NB4-Lv-MIR125B1 cells treated with 1 μM ATRA or in combination with rapamycin (100 nM) for 24 h. ACTB was used as a loading control. PML-RARA/ACTB densitometric ratios were recorded.
MIR125B1 inhibits degradation of PML-RARA fusion protein by targeting DRAM2, a critical regulator of autophagy
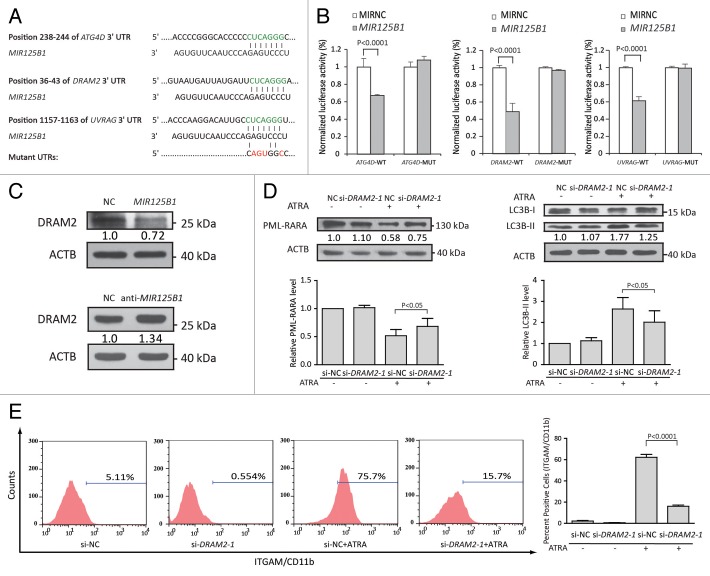
We then explored the molecular mechanism of MIR125B1 in autophagy. TargetScan was used to generate a list of MIR125B1 target candidates. Several genes (DRAM2, ATG4D, UVRAG) associated with autophagy were found to have MIR125B1-binding sites in their 3′UTRs (Fig. 6A). Dual-luciferase reporter analysis revealed that MIR125B1 directly suppressed the luciferase activity of DRAM2, ATG4D, and UVRAG (Fig. 6B). Notably, both MIR125B1 and the binding site in the DRAM2 are highly conserved across species, and DRAM2 was recently found to be involved in autophagy.30 To further confirm that the DRAM2 protein is suppressed by MIR125B1, we performed both MIR125B1 overexpression and knockdown experiments in NB4 cells and examined the expression of DRAM2. As shown in Figure 6C, MIR125B1 overexpression caused an apparent decrease in the amount of DRAM2 in NB4 cells (upper panel), while inhibition of endogenous MIR125B1 using a synthetic MIR125B1 inhibitor increased DRAM2 expression (bottom panel). To gain further insight into whether MIR125B1 suppresses autophagy by regulating DRAM2, we examined the function of DRAM2 in ATRA induction. We employed an siRNA (si-DRAM2) to knock down DRAM2 and then used immunoblotting assay to confirm that DRAM2 expression was effectively silenced (Fig. S3E). We next investigated the function of DRAM2. When treated with ATRA, DRAM2 knockdown resulted in increasing amounts of PML-RARA oncoprotein (Fig. 6D, left panel), inhibition of autophagy (Fig. 6D, right panel and Fig. S4A), and myeloid cell differentiation arrest (Fig. 6E). We have also employed a second siRNA sequence against DRAM2 to confirm the functions of DRAM2 in APL cell differentiation and autophagy (Fig. S4B–S4D). These results showed that knockdown of DRAM2 phenocopied the effects of overexpressing MIR125B1, further validating that DRAM2 is a functional target of MIR125B1. All together, this suggested that MIR125B1 may suppress the autophagy-lysosomal pathway, at least in part, by repressing DRAM2 and subsequently impairment of PML-RARA proteolysis and myeloid cell differentiation.
Figure 6.MIR125B1 directly targeted autophagy genes involved in autophagy, PML-RARA degradation, and differentiation. (A) Schematic representation of the interaction of miRNA with the 3′UTR of its corresponding targets. The miRNA response elements (MRE) in the 3′UTR of human were predicted by TargetScan and in green. Each predicted MRE 3′UTR was inserted into a psiCHECK-2 vector immediately downstream from the Renilla luciferase gene. (B) Luciferase reporter assays analyzing the putative targets of MIR125B1. NC or MIR125B1 duplexes were cotransfected with psiCHECK-2 plasmids with the 3′UTR of target genes-wt or the target genes-mut. The firefly luciferase activity of each sample was normalized to the Renilla luciferase activity (n = 3 independent experiments performed in triplicate). (C) Western blot analysis of DRAM2 in NB4 cells transduced with lentiviral vectors expressing MIR125B1 (top panel) or shRNAs targeting MIR125B1 (bottom panel). (D) Western blot analyses of PML-RARA and LC3 in DRAM2-knockdown NB4 cells, followed by treatment with ATRA. LC3-II/ACTB densitometric ratios were recorded. Quantification of data from 3 independent experiments (bottom panel). (E) Flow cytometry analysis of ITGAM/CD11b cell-surface expression in NB4 cells transfected with si-DRAM2 or si-NC, followed by treatment with or without ATRA. Values are derived from n = 3 independent experiments data are reported as mean ± SD.
MIR125B1 overexpression inhibits PML-RARA proteolysis and leukemic cell differentiation in a mouse model
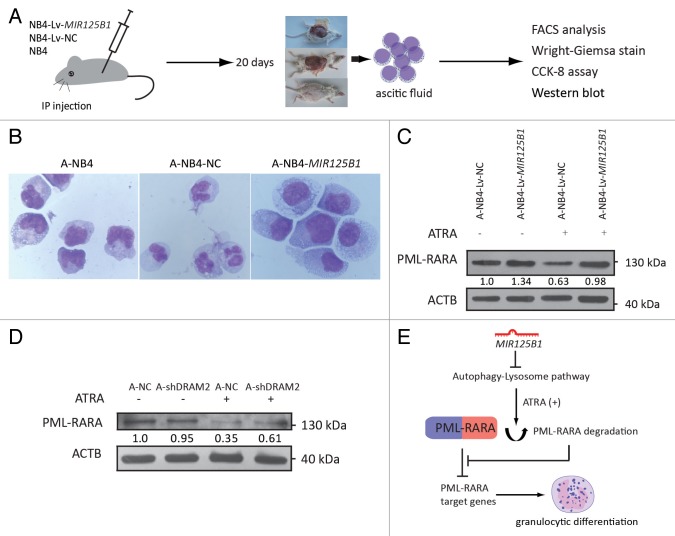
The observations above indicate that MIR125B1 is associated with PML-RARA expression and may be involved in the development and treatment of APL. To further understand the function of MIR125B1 in APL pathogenesis and cell differentiation therapy, a human APL-ascites SCID mouse model was employed to study the response of MIR125B1 to therapy. A diagram illustrating the experimental design used for the APL-ascites NOD-SCID mouse model is shown in Figure 7A. When the NOD-SCID mice developed an ascites burden, the ascitic fluid was withdrawn, and the ascitic cells carrying the MIR125B1, a miRNA negative control, or NB4 cells alone (called A-NB4-MIR125B1, A-NB4-NC and A-NB4, respectively) were cultured in RPMI 1640 medium and treated with 1 μM ATRA. Cell differentiation was detected by FACS analysis (Fig. S5A) and Wright-Giemsa staining (Fig. 7B). A-NB4-MIR125B1 cells demonstrated less differentiation than the other cell types. In addition, we measured cell proliferation at 24, 48, and 72 h using a CCK-8 assay and found that the proliferation of A-NB4-MIR125B1 cells was stimulated by MIR125B1 overexpression (Fig. S5B). Overexpression of MIR125B1 resulted in inhibition of autophagy and PML-RARA proteolysis (Fig. 7C; Fig. S5C). We also generated a transplantation model with DRAM2 knockdown cells. As shown in Figure 7C, silencing DRAM2 expression by infecting with lentivirus expressing shRNAs targeting DRAM2 impaired PML-RARA degradation in vivo. These results indicate that MIR125B1 overexpression impairs degradation of PML-RARA via the autophagy-lysosomal pathway in SCID-APL cells undergoing differentiation therapy (Fig. 7E).
Figure 7. Effects of MIR125B1 overexpression in a human APL-ascites SCID mouse model. (A) Diagram illustrating the experimental design of the mice xenograft experiment. IP, intraperitoneal transplantation. (B) Wright-Giemsa staining was performed to evaluate cell differentiation upon 72 h of ATRA treatment. (C) Western blot analyses of PML-RARA in A-NB4-Lv-NC and A-NB4-Lv-MIR125B1 cells treated with ATRA. A-NB4: ascitic cells developed from the injection of NB4 cells; A-NB4-NC: ascitic cells developed from the injection of NB4-Lv-NC cells; A-NB4-MIR125B1: ascitic cells developed from the injection of NB4-Lv-MIR125B1 cells. (D) Western blot analyses of PML-RARA in A-shDRAM2-NB4 and A-shNC-NB4 cells. A-shNC-NB4, ascitic cells developed from the injection of shNC-NB4 cells; A-shDRAM2-NB4, ascitic cells developed from the injection of shDRAM2-NB4 cells. ACTB was used as a loading control. (E) A proposed model for MIR125B1 and PML-RARA in APL. See Discussion for details.
Discussion
In this study, we found that MIR125B1 abundance correlated with PML-RARA status in patients with APL and activated PML-RARA expression. Furthermore, we provided the in vitro and in vivo evidence that the degradation of PML-RARA via the autophagy-lysosomal pathway is impaired by MIR125B1 and that this impairment subsequently arrests cell differentiation.
PML-RARA is the initiating factor for APL development and blocks promyelocyte differentiation. However, leukemia development in transgenic mice expressing PML-RARA occurs after a long latency period,31 which strongly suggests that additional genetic events collaborate with PML-RARA to block differentiation and stimulate a number of APL features. Previous studies have demonstrated that MIR125B1 is able to confer a competitive advantage on engrafting hematopoietic cells,32 and MIR125B1 overexpression is a consistent abnormality in several patients with chromosomal translocations.14,21 In particular, MIR125B1 is highly expressed in patients with APL, suggesting that it could be a primary oncogenic event in APL leukemogenesis. However, the underlying molecular mechanisms of MIR125B1 in APL remain largely unknown. The observations in this study indicated that loss of PML-RARA decreased MIR125B1 expression. Interestingly, MIR125B1 also increased PML-RARA expression, suggesting that PML-RARA and MIR125B1 might form a positive feedback loop. In line with the increased PML-RARA expression by MIR125B1, we found that a stable ectopic expression of MIR125B1 affected some of the mRNA transcript levels of hypothetical target genes involved in PML-RARA regulation (Fig. S5D). Thus, it is possible that MIR125B1 regulates the expression of PML-RARA through altered interaction of the PML-RARA promoter with transcription factors affected by target genes of MIR125B1 (Fig. S6). Given that a high level of PML-RARA appears to be required during the initiation stages of cancer,5,33 the strong MIR125B1 upregulation may promote the initiation of oncogenesis by PML-RARA. This finding is the first evidence that an miRNA cooperates with a fusion protein and has a critical role in leukemia development and response to treatment. However, whether this cooperation is essential for APL initiation requires further investigation.
It has been reported that the treatment of cultured APL cells with ATRA triggers the differentiation of malignant promyelocytes via transcriptional regulation.34 Similarly, in our study, numerous differentially expressed genes after ATRA treatment were initially repressed by PML-RARA; this repression can be relieved by ATRA and restored by MIR125B1. The involvement of PML-RARA target genes, together with enrichment for genes belonging to the PML-RARA degradation pathway, led to the assumption that there is a direct link between MIR125B1-mediated differentiation arrest and PML-RARA degradation. These findings are of particular interest because MIR125B1 and PML-RARA have been closely linked with APL.
It has been demonstrated that ATRA exerts its therapeutic effect by promoting the degradation of the PML-RARA oncogenic protein.6-8 Therapy-induced PML-RARA degradation allows the transcriptional derepression of PML-RARA target genes, and PML-RARA knockdown results in spontaneous differentiation in APL models.22 Moreover, the inhibition of PML-RARA degradation leads to a differentiation block.7,8 We conclude that PML-RARA destruction is not merely a consequence of differentiation, but it plays an important role in the induction of differentiation. Indeed, other studies have found that PML-RARA remains a potent repressor even in the presence of ATRA.23,35 In line with these observations, the MIR125B1-mediated inhibition of transcriptional derepression occur simultaneously with high PML-RARA oncoprotein levels and a differentiation block, and the degradation impairment contributes to differentiation arrest. Although our data suggest a role for PML-RARA degradation in the effects of MIR125B1 on cell differentiation, we cannot exclude that MIR125B1 upregulates PML-RARA at the transcriptional level and leads to an increase in the amount of fusion protein, which may also contribute to the function of MIR125B1 during differentiation.
The degradation of PML-RARA has been shown to be involved in the activity of the ubiquitin-proteasome and autophagy-lysosomal pathways.6-8 However, the PML-RARA oncoprotein is prone to aggregation, indicating that it may represent a preferable substrate for autophagic degradation. In this study, we provided evidence that MIR125B1 inhibited the autophagy-lysosomal pathway induced by ATRA. Importantly, autophagy appears to rely on the cooperation of autophagosomes and lysosomes. Indeed, lysosomal and autophagic genes were regulated by MIR125B1. Bioinformatic analysis was also used to address the important role of the lysosome in ATRA-mediated differentiation of APL. The repression of the autophagy-lysosome system imposed by MIR125B1 was demonstrated by the decreased number of autophagosomes and this was accompanied by inhibition of lysosomal and autophagic genes (Fig. 3B and C). Thus, it is suggested that impairment of PML-RARA degradation partially occurs through abrogation of the autophagic-lysosomal system. Furthermore, we identified DRAM2 as a novel target that was at least partly responsible for the function of MIR125B1 in autophagy. Importantly, the knockdown phenotype for DRAM2 is similar to the phenotype of overexpressing MIR125B1, including impairment of PML-RARA degradation, inhibition of autophagy, and myeloid cell differentiation arrest. Although there are conflicting reports as to the role of DRAM2 in autophagy,30,36 our results support the finding that DRAM2 is involved in autophagy induction. The discrepancy in these studies may be caused by differences in cell type, stimuli, and autophagy monitoring systems.
In summary, we provided evidence for a direct link between MIR125B1 and PML-RARA in t(15;17) APL. MIR125B1 overexpression can significantly increase the expression of the APL fusion oncogene PML-RARA and repress the therapy-induced proteolysis of PML-RARA by an autophagy-lysosomal pathway. MIR125B1 therefore inhibits the ATRA-induced transcription of PML-RARA target genes that were initially repressed by PML-RARA, resulting in cell differentiation arrest. The oncogenic activity of MIR125B1 in the treatment of t(15;17) APL was also demonstrated in vivo. Moreover, the data presented in this study suggested that the expression levels of MIR125B1 are associated with the therapeutic status of the disease.
Materials and Methods
Patients and samples
A total of 80 bone marrow samples including 46 APL samples at diagnosis, 22 APL samples after therapy, and 12 normal donors, from the First and Second Affiliated Hospital of Sun Yat-sen University were enrolled in this study. Patients’ characteristics are summarized in Table 1. Written informed consent was obtained from the parents and guardians. This study was approved by the ethics committee of the affiliated hospitals of Sun Yat-sen University.
Table 1. APL patients’ characteristicsa.
| APL Primary (n = 46) | Characteristics | Median (range) | No. (%) |
|---|---|---|---|
| Cytogenetics | |||
| t(15;17) | |||
| Age at diagnosis, y | 8.11 (0–14) | 46 (100) | |
| Sex | |||
| Male | 19 (41.3) | ||
| Female | 27 (56.7) | ||
| WBC count, × 109/L | 15.67 | ||
| Less than 10 | 30 (65.2) | ||
| 10–50 | 9 (19.6) | ||
| 50 or higher | 5 (10.9) | ||
| N/A | 2 (4.3) | ||
| APL after therapy (n = 22) | CR | 18 (81.8) | |
| NR | 2 (9.1) | ||
| Relapse | 2 (9.1) |
a WBC, white blood cell; CR, samples in first complete remission; relapse, samples at relapse. NR, Non-remission; N/A, not available. CR and Relapse samples are from patients of 46 primary patients with 3 y of clinical follow-up.
Cell lines and cell cultures
NB4 cells were cultured in RPMI 1640 (HyClone, SH30027) containing 10% fetal bovine serum (HyClone, SV30160). The cells were cultured in a humidified atmosphere containing 5% CO2 at 37 °C. All drugs were purchased from Sigma-Aldrich and used at the following final concentrations: 1 μM ATRA (Sigma-Aldrich, R2625, stock 10 mM in EtOH) and 100 nM rapamycin (Sigma-Aldrich, R8781, stock 100 mM in dimethyl sulfoxide). Cell differentiation was assessed by measuring the surface ITGAM/CD11b (eBioscience, 11-0113) and FUT4/CD15 (eBioscience, 46-0159) antigen expression by flow cytometry analysis or cell morphology changes using Wright-Giemsa staining.14
Lysosomal staining and FACS analysis
Cells were maintained in 50 nM of the acidotropic dye LysoTracker Red DND-99 (Molecular Probes, L7528) for 60 min. The red lysosomal fluorescence of 30,000 cells per sample was determined by flow cytometry using a BD FACSAria cytometer (BD Biosciences, San Jose, CA, USA).
RNA isolation and RNA-SEQ (quantification)
The total RNA was extracted from samples with TRIzol (Invitrogen, 15596) according to the manufacturer’s instructions. cDNA libraries were prepared as previously described.37 The library products were then ready for sequencing analysis via an IlluminaHiSeq™ 2000 (San Diego, CA, USA). Results from reads that could be uniquely mapped to a gene were used to calculate the expression level. The numbers of gene reads were further measured by assessing the number of uniquely mapped reads per kilobase of exon region per million mappable reads (RPKM) using Chen’s method,38 which is based on the Audic method39 for analyzing differential expression. We identified differentially expressed genes between samples using the following criteria: FDR ≤ 0.001 and a log2-fold change ratio ≥ 1.
Quantitative real-time PCR analysis
qRT-PCR was performed to detect mature miRNAs expression. Briefly, RNA was reverse-transcribed to cDNA using M-MLV reverse transcriptase (Promega, M1705) and amplified with specific miRNA RT primers and PCR amplification primers. U6 served as internal control. For PML-RARA, primers were designed as previously reported.40 PML breakpoints were rapidly assigned by qRT-PCR using a common PML-RARA reverse primer in conjunction with PML intron 3 (bcr3) breakpoint-specific forward primers. ACTB was used as internal controls. The oligonucleotide sequences are available in Table S4.
Transfection
Cells were transfected using the Neon® Transfection System (Invitrogen) with 100 pmol of oligonucleotides in 10 μl reactions. The hsa-MIR125B1 miRNA mimics (Ambion, AM17100) and a mimics negative control (Ambion, AM17111) were purchased from Ambion. The MIR125B1 inhibitor (RiboBio, anti-MIR125B1, miR10004669-1-2) and inhibitor negative control (RiboBio, miR02201-1-5) were purchased from RiboBio. The antagomir (chemically engineered oligonucleotides anti-miRNAs that could be easily transfected into cells and inhibited endogenous miRNA) against human MIR125B1 (RiboBio, miR30000423-1-10) and antagomir negative control (RiboBio, miR03201-1-10) were obtained from RiboBio. The sequences of small interfering RNA (siRNA) that specifically targets the breakpoint region of PML-RARA were designed as previously described.22 The nucleotide sequence for DRAM2 siRNA was 5′ GUGCUUCACA UGAUCACUAC UG 3′ (si-DRAM2-1) and 5′ GAACACGGCU ACUUUCCAG 3′ (si-DRAM2-2). The following siRNA sequences targeting the DRAM2 were designed and expressed as shRNA: si-DRAM2-1 5′ GUGCUUCACA UGAUCACUAC UG 3′. The nucleotide sequence for ATG5 siRNA was 5′CAUCUGAGCU ACCCGGAUA3′.
Western blot
Protein extracts were boiled in RIPA buffer (Beyotime, P0013) and separated in a sodium dodecyl sulfate polyacrylamide electrophoresis (SDS-PAGE) gel. The proteins were then transferred to a polyvinylidene fluoride membrane (Millipore, HVPPEAC12) and probed with anti-RARA (C-20; Santa Cruz Biotechnology, sc-551), anti-MTOR (Cell Signaling Technology, 2983), anti-LC3B (Sigma-Aldrich, L7543), anti-DRAM2 (Sigma-Aldrich, HPA018036) and anti-ACTB (Sigma-Aldrich, A2103) antibodies.
Animal models
NB4 cells were transfected with Lv-MIR125B1 (lentivirus vector expressing MIR125B1) or Lv-NC (lentivirus vector expressing miRNA negative control) according to the recommended protocol (GeneChem). The transfected cells (NB4-Lv-MIR125B1 and NB4-Lv-NC) were sorted by flow cytometry (FACStar+ instrument, BD Biosciences, San Jose, CA, USA), and the GFP-positive cells were used for in vivo experiments.
Five-wk-old NOD-SCID mice were maintained under specific pathogen-free conditions in the Laboratory Animal Center of Sun Yat-sen University, and all experiments were performed in accordance with our Institutional Animal Guidelines. The human APL-ascites SCID mouse model was used as previously described with modifications.41 The tumor xenografts were established by single intraperitoneal transplantation in NOD-SCID mice with 1 × 106 NB4 cells in 0.1 ml RPMI-1640 medium.
Immunofluorescence
Cells were fixed in 4% formaldehyde for 30 min at room temperature prior to cell permeabilization with 0.1% Triton X-100 (4 °C, 10 min). The cells were blocked for 2 h with 2.5% BSA in PBS (Beyotime, ST023 and C0221A). After blocking, they cells were then incubated for 20 h with an anti-LC3 antibody, washed 3 times for 5 min with PBS, and then incubated for 1 h with Alexafluor 594-conjugated (Invitrogen, A21207) secondary antibodies. Fluorescence signals were analyzed using a Leica TCS SP5 II confocal microscope (Leica Corp., Wetzlar, Hesse, Germany).
Transmission electron microscopy analysis
Cells were washed with PBS and fixed in 3% glutaraldehyde dissolved in 0.1 mol/l phosphate buffer (pH 7.4) for 30 min at room temperature. The cells were then postfixed for 1 h in 1% OsO4 (Sigma-Aldrich, O5500). After dehydration, the cells were embedded in Epon 812 (SPI Supplies, 02635-AB) and polymerized at 60 °C for 24 h. EM images were acquired from thin sections using a JEM1400 electron microscope (JEOL, Akishima, Tokyo, Japan). Images were digitally acquired from a randomly selected pool under each condition. At least 10 cells per group were analyzed.
Statistical analysis
Data were expressed as the mean ± SD of 3 independent experiments. The significance of the differences between groups was determined by a 2-tailed Student t test. A P value < 0.05 was considered significant.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the funds from National Science and Technology Department (973, 2011CB8113015 and 2011CBA0110) and National Science Foundation of China (No. 81270629).
Glossary
Abbreviations:
- 3′UTR
3′-untranslated region
- AML
acute myeloid leukemia
- APL
acute promyelocytic leukemia
- ATRA
all-trans retinoic acid
- ATO
arsenic trioxide
References
- 1.Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–215. [PubMed] [Google Scholar]
- 2.de Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–84. doi: 10.1016/0092-8674(91)90113-D. [DOI] [PubMed] [Google Scholar]
- 3.Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L, Gu LJ, Wang ZY. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72:567–72. [PubMed] [Google Scholar]
- 4.Breitman TR, Collins SJ, Keene BR. Terminal differentiation of human promyelocytic leukemic cells in primary culture in response to retinoic acid. Blood. 1981;57:1000–4. [PubMed] [Google Scholar]
- 5.Nasr R, Guillemin MC, Ferhi O, Soilihi H, Peres L, Berthier C, Rousselot P, Robledo-Sarmiento M, Lallemand-Breitenbach V, Gourmel B, et al. Eradication of acute promyelocytic leukemia-initiating cells through PML-RARA degradation. Nat Med. 2008;14:1333–42. doi: 10.1038/nm.1891. [DOI] [PubMed] [Google Scholar]
- 6.Zhu J, Gianni M, Kopf E, Honoré N, Chelbi-Alix M, Koken M, Quignon F, Rochette-Egly C, de Thé H. Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor alpha (RARalpha) and oncogenic RARalpha fusion proteins. Proc Natl Acad Sci U S A. 1999;96:14807–12. doi: 10.1073/pnas.96.26.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isakson P, Bjørås M, Bøe SO, Simonsen A. Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein. Blood. 2010;116:2324–31. doi: 10.1182/blood-2010-01-261040. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Cao L, Kang R, Yang M, Liu L, Zhao Y, Yu Y, Xie M, Yin X, Livesey KM, et al. Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARα oncoprotein. Autophagy. 2011;7:401–11. doi: 10.4161/auto.7.4.14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, Liang WX, Song AX, Lallemand-Breitenbach V, Jeanne M, et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–3. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]
- 12.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–5. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 13.Jiang F, Liu T, He Y, Yan Q, Chen X, Wang H, Wan X. MiR-125b promotes proliferation and migration of type II endometrial carcinoma cells through targeting TP53INP1 tumor suppressor in vitro and in vivo. BMC Cancer. 2011;11:425. doi: 10.1186/1471-2407-11-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bousquet M, Quelen C, Rosati R, Mansat-De Mas V, La Starza R, Bastard C, Lippert E, Talmant P, Lafage-Pochitaloff M, Leroux D, et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–76. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng CW, Zhang XJ, Lin KY, Ye H, Feng SY, Zhang H, Chen YQ. Camptothecin induces apoptosis in cancer cells via microRNA-125b-mediated mitochondrial pathways. Mol Pharmacol. 2012;81:578–86. doi: 10.1124/mol.111.076794. [DOI] [PubMed] [Google Scholar]
- 17.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Löwenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–85. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 18.Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG, Schnittger S, Haferlach T, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105:3945–50. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS, Wu J, Zhou H, Qu LH, Xu L, Chen YQ. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One. 2009;4:e7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter MJ, Park JS, Ries RE, Lau SK, McLellan M, Jaeger S, Wilson RK, Mardis ER, Ley TJ. Reduced PU.1 expression causes myeloid progenitor expansion and increased leukemia penetrance in mice expressing PML-RARalpha. Proc Natl Acad Sci U S A. 2005;102:12513–8. doi: 10.1073/pnas.0504247102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapiro E, Russell LJ, Struski S, Cavé H, Radford-Weiss I, Valle VD, Lachenaud J, Brousset P, Bernard OA, Harrison CJ, et al. A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukemia. Leukemia. 2010;24:1362–4. doi: 10.1038/leu.2010.93. [DOI] [PubMed] [Google Scholar]
- 22.Ward SV, Sternsdorf T, Woods NB. Targeting expression of the leukemogenic PML-RARα fusion protein by lentiviral vector-mediated small interfering RNA results in leukemic cell differentiation and apoptosis. Hum Gene Ther. 2011;22:1593–8. doi: 10.1089/hum.2011.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamashev D, Vitoux D, De Thé H. PML-RARA-RXR oligomers mediate retinoid and rexinoid/cAMP cross-talk in acute promyelocytic leukemia cell differentiation. J Exp Med. 2004;199:1163–74. doi: 10.1084/jem.20032226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 25.Yu L, Zhang YD, Zhou J, Yao DM, Li X. Identification of target genes of transcription factor CEBPB in acute promyelocytic leukemia cells induced by all-trans retinoic acid. Asian Pac J Trop Med. 2013;6:473–80. doi: 10.1016/S1995-7645(13)60077-2. [DOI] [PubMed] [Google Scholar]
- 26.Yu M, Tong JH, Mao M, Kan LX, Liu MM, Sun YW, Fu G, Jing YK, Yu L, Lepaslier D, et al. Cloning of a gene (RIG-G) associated with retinoic acid-induced differentiation of acute promyelocytic leukemia cells and representing a new member of a family of interferon-stimulated genes. Proc Natl Acad Sci U S A. 1997;94:7406–11. doi: 10.1073/pnas.94.14.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Wang P, Shi J, Zhu X, He M, Jia X, Yang X, Qiu F, Jin W, Qian M, et al. PML/RARalpha targets promoter regions containing PU.1 consensus and RARE half sites in acute promyelocytic leukemia. Cancer Cell. 2010;17:186–97. doi: 10.1016/j.ccr.2009.12.045. [DOI] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitareewan S, Roebuck BD, Demidenko E, Sloboda RD, Dmitrovsky E. Lysosomes and trivalent arsenic treatment in acute promyelocytic leukemia. J Natl Cancer Inst. 2007;99:41–52. doi: 10.1093/jnci/djk004. [DOI] [PubMed] [Google Scholar]
- 30.Yoon JH, Her S, Kim M, Jang IS, Park J. The expression of damage-regulated autophagy modulator 2 (DRAM2) contributes to autophagy induction. Mol Biol Rep. 2012;39:1087–93. doi: 10.1007/s11033-011-0835-x. [DOI] [PubMed] [Google Scholar]
- 31.Walter MJ, Park JS, Lau SK, Li X, Lane AA, Nagarajan R, Shannon WD, Ley TJ. Expression profiling of murine acute promyelocytic leukemia cells reveals multiple model-dependent progression signatures. Mol Cell Biol. 2004;24:10882–93. doi: 10.1128/MCB.24.24.10882-10893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A. 2010;107:14235–40. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grignani F, Valtieri M, Gabbianelli M, Gelmetti V, Botta R, Luchetti L, Masella B, Morsilli O, Pelosi E, Samoggia P, et al. PML/RAR alpha fusion protein expression in normal human hematopoietic progenitors dictates myeloid commitment and the promyelocytic phenotype. Blood. 2000;96:1531–7. [PubMed] [Google Scholar]
- 34.Liu TX, Zhang JW, Tao J, Zhang RB, Zhang QH, Zhao CJ, Tong JH, Lanotte M, Waxman S, Chen SJ, et al. Gene expression networks underlying retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Blood. 2000;96:1496–504. [PubMed] [Google Scholar]
- 35.Lin RJ, Evans RM. Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Mol Cell. 2000;5:821–30. doi: 10.1016/S1097-2765(00)80322-6. [DOI] [PubMed] [Google Scholar]
- 36.O’Prey J, Skommer J, Wilkinson S, Ryan KM. Analysis of DRAM-related proteins reveals evolutionarily conserved and divergent roles in the control of autophagy. Cell Cycle. 2009;8:2260–5. doi: 10.4161/cc.8.14.9050. [DOI] [PubMed] [Google Scholar]
- 37.Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X, Cui Z, Zhang J, Yi K, Xu W, et al. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012;22:806–21. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S, Yang P, Jiang F, Wei Y, Ma Z, Kang L. De novo analysis of transcriptome dynamics in the migratory locust during the development of phase traits. PLoS One. 2010;5:e15633. doi: 10.1371/journal.pone.0015633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–95. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 40.Grimwade D, Jovanovic JV, Hills RK, Nugent EA, Patel Y, Flora R, Diverio D, Jones K, Aslett H, Batson E, et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J Clin Oncol. 2009;27:3650–8. doi: 10.1200/JCO.2008.20.1533. [DOI] [PubMed] [Google Scholar]
- 41.Zhang SY, Zhu J, Chen GQ, Du XX, Lu LJ, Zhang Z, Zhong HJ, Chen HR, Wang ZY, Berger R, et al. Establishment of a human acute promyelocytic leukemia-ascites model in SCID mice. Blood. 1996;87:3404–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.