Abstract
Autophagy, a “self-eating” cellular process, has dual roles in promoting and suppressing tumor growth, depending on cellular context. PTP4A3/PRL-3, a plasma membrane and endosomal phosphatase, promotes multiple oncogenic processes including cell proliferation, invasion, and cancer metastasis. In this study, we demonstrate that PTP4A3 accumulates in autophagosomes upon inhibition of autophagic degradation. Expression of PTP4A3 enhances PIK3C3-BECN1-dependent autophagosome formation and accelerates LC3-I to LC3-II conversion in an ATG5-dependent manner. PTP4A3 overexpression also enhances the degradation of SQSTM1, a key autophagy substrate. These functions of PTP4A3 are dependent on its catalytic activity and prenylation-dependent membrane association. These results suggest that PTP4A3 functions to promote canonical autophagy flux. Unexpectedly, following autophagy activation, PTP4A3 serves as a novel autophagic substrate, thereby establishing a negative feedback-loop that may be required to fine-tune autophagy activity. Functionally, PTP4A3 utilizes the autophagy pathway to promote cell growth, concomitant with the activation of AKT. Clinically, from the largest ovarian cancer data set (GSE 9899, n = 285) available in GEO, high levels of expression of both PTP4A3 and autophagy genes significantly predict poor prognosis of ovarian cancer patients. These studies reveal a critical role of autophagy in PTP4A3-driven cancer progression, suggesting that autophagy could be a potential Achilles heel to block PTP4A3-mediated tumor progression in stratified patients with high expression of both PTP4A3 and autophagy genes.
Keywords: BECN1, cancer progression, mammalian autophagy regulation, PIK3C3, prognosis marker, PTP4A3
Introduction
Autophagy is an evolutionarily conserved process involved in selective degradation of long-lived proteins and damaged organelles.1 Initiation of autophagy begins with the formation of nascent autophagosomes, double-membrane vesicles that form around cellular components destined for destruction. Canonical autophagosome formation involves 4 steps: 1) initiation, 2) nucleation, 3) elongation, and 4) closure. Following initiation, the PIK3C3/VPS34-BECN1 complex is necessary for autophagosome nucleation. PIK3C3 is the catalytic subunit of the phosphatidylinositol (PtdIns) 3-kinase (PtdIns3K), which belongs to the class III PtdIns3K family, and phosphorylates PtdIns to generate PtdIns3P, a lipid second messenger essential for autophagosome trafficking.2,3 The interaction between BECN1 (the mammalian ortholog of yeast Vps30/Atg6) and PIK3C3 is important for the recruitment of autophagy proteins to the nucleating phagophore assembly sites (PAS).4 Subsequently, autophagosome elongation and closure occur, wherein soluble and cleaved LC3-I is conjugated to phosphatidylethanolamine (PE) to form membrane-bound LC3-II. This process, called “LC3 conversion,” requires ATG5 and constitutes a critical step for autophagosome maturation.5,6 In contrast, noncanonical autophagosome formation can occur independently of some of these autophagy proteins or even bypass some of these steps.7 Mature autophagosomes, carrying all the unwanted cellular organelles and autophagy substrate, ultimately fuse with lysosomes, where their contents are degraded and recycled to release nutrients and energy for de novo biosynthesis of new membranes and proteins.1
Generally, autophagy functions as a homeostatic cellular recycling mechanism to ensure cellular survival by minimizing accumulation of cellular damage and during starvation by maintaining cellular energy levels.8 As a consequence of this critical role, autophagy is frequently associated with mechanisms of cell death, particularly following prolonged autophagy activation that results in progressive consumption of cellular components.9 Prosurvival and/or prodeath roles for autophagy are highly dependent on cellular context, with implications in treatment of diseases where autophagy is associated with pathology. For example, modulation of autophagy using either autophagy activators or inhibitors has both been described to have anti-cancer effects.9 Reliable predictors of the consequence of a tumor response to autophagy modulation remain poorly characterized.
PTP4A3 is a prenylated protein tyrosine phosphatase we identified in 1998.10 It is best known for its role in promoting cancer metastasis.11 Upregulation of PTP4A3 in metastatic specimens from colorectal cancer was reported a decade ago.12 The oncogenic roles of PTP4A3 overexpression include increased cell migration/invasion in vitro, and enhanced metastasis in vivo through activation of the Rho GTPase family and the phosphoinositide 3-kinase-AKT pathway.13-15 PTP4A3 also promotes cell proliferation, angiogenesis, and cell-cycle progression.16-18 More importantly, PTP4A3 is upregulated in clinically malignant cancer specimens compared with their benign counterparts in multiple human cancers and is associated with poor prognosis.12,16,19,20
Recently, endosomes were shown to play an important function in autophagy, fusing with autophagosomes prior to lysosomal fusion for cargo degradation.21-23 Since PTP4A3 and its close homolog PTP4A1 both have endosomal localization,10,24 we investigated whether they were functionally linked to autophagy. Here, we report that PTP4A3, but not PTP4A1, has a novel role in canonical autophagy. Our results suggest that PTP4A3 is both an activator and a substrate of autophagy, providing a possible negative feedback mechanism for regulating autophagic flux. Notably, we provide new evidence of a critical role for autophagy during PTP4A3-mediated ovarian cancer cell growth and tumor progression, indicating that elevated PTP4A3 expression is a predictive biomarker of antiautophagy therapy for patients having high expression of autophagy genes.
Results
PTP4A3, but not PTP4A1, colocalizes with LC3 in autophagosomes and promotes LC3 puncta accumulation upon chloroquine treatment
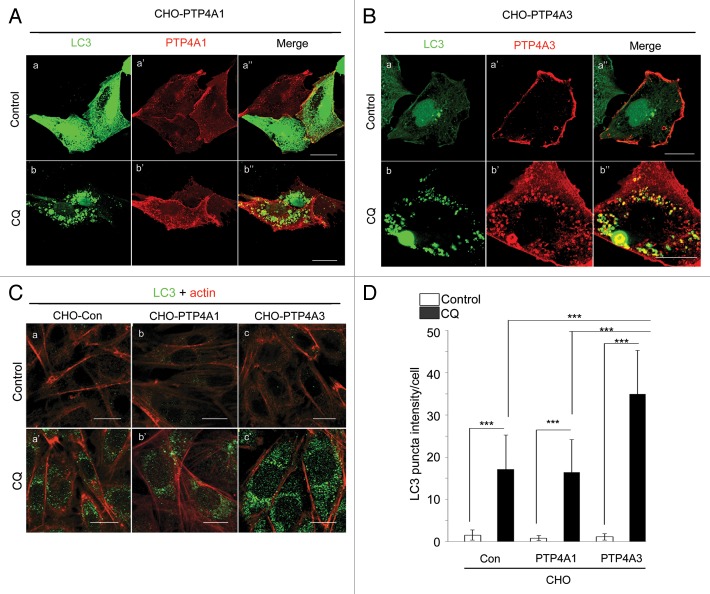
PTP4A1 and PTP4A3 are 2 closely related prenylated protein phosphatases, sharing 79% amino acid identity.10 Both are localized to the cytosolic face of the plasma membrane and endosomes.24 In light of recent reports suggesting a role for endosomes in autophagy,22 we investigated whether PTP4A1 or PTP4A3 was also localized to nascent autophagosomes. We ectopically expressed myc-tagged PTP4A1 and PTP4A3 in Chinese Hamster Ovary (CHO) cells (CHO-PTP4A1 and CHO-PTP4A3) together with EGFP-tagged LC3, and tracked autophagosome formation upon treatment with chloroquine (CQ), a lysosomal acidification inhibitor that blocks autophagic degradation. Upon CQ treatment, PTP4A1 localization remained largely unchanged and poorly colocalized with LC3 puncta (Fig. 1A, b”). In contrast, PTP4A3 accumulated and colocalized well with LC3-positive puncta-like structures (Fig. 1B, b”). Using another late stage autophagy inhibitor, bafilomycin A1 (BafA1),25 colocalization of PTP4A3 and LC3 puncta was observed as early as 1 h after BafA1 treatment (Fig. S1A; Fig. 1B, c’). Such LC3-positive puncta have been characterized as a signature of autophagosomes and amphisomes (collectively referred to as AP hereafter), 2 types of autophagic vacuoles that form prior to lysosomal fusion and cargo degradation.26,27 We next examined whether the presence of PTP4A3 in AP might implicate a role for PTP4A3 in autophagy regulation. We noted that CHO-PTP4A3 cells contained more endogenous LC3 puncta per cell compared with CHO control (CHO-Con) or CHO-PTP4A1 cells following CQ treatment (Fig. 1C). Quantification of LC3 puncta intensity confirmed a significantly higher LC3 puncta intensity per cell in CQ-treated CHO-PTP4A3 cells compared with either CHO-Con or CHO-PTP4A1 cells (Fig. 1D; P < 0.001). Collectively, our results suggest that upon CQ treatment, ectopically expressed PTP4A3 colocalizes with LC3 in AP and promotes AP accumulation.
Figure 1. PTP4A3, but not PTP4A1, colocalizes with LC3-positive puncta and promotes AP accumulation upon CQ-mediated autophagic degradation inhibition. (A and B) CHO cells overexpressing myc-PTP4A1 (CHO-PTP4A1) (A) or myc-PTP4A3 (CHO-PTP4A3) (B) were transfected with EGFP-LC3 reporter construct, and allowed to grow for 24 h. The transfected cells were treated with or without CQ in full media for another 24 h before immunostaining with an anti-myc antibody (scale bar: 20 μm). (C) CHO control (CHO-Con), CHO-PTP4A1, and CHO-PTP4A3 were treated with or without CQ for 24 h prior to immunostaining for LC3 (green) and actin (red). Representative images are shown in the left panel (scale bar: 20 μm). (D) LC3 puncta intensity of at least 20 cells from 5 distinct viewing fields were quantified as described in Materials and Methods, and presented as a histogram in the right panel (mean ± S.D.). ***P < 0.001.
PTP4A3 requires catalytic activity and prenylation-dependent membrane association to promote AP accumulation via the canonical PIK3C3-BECN1 autophagy pathway
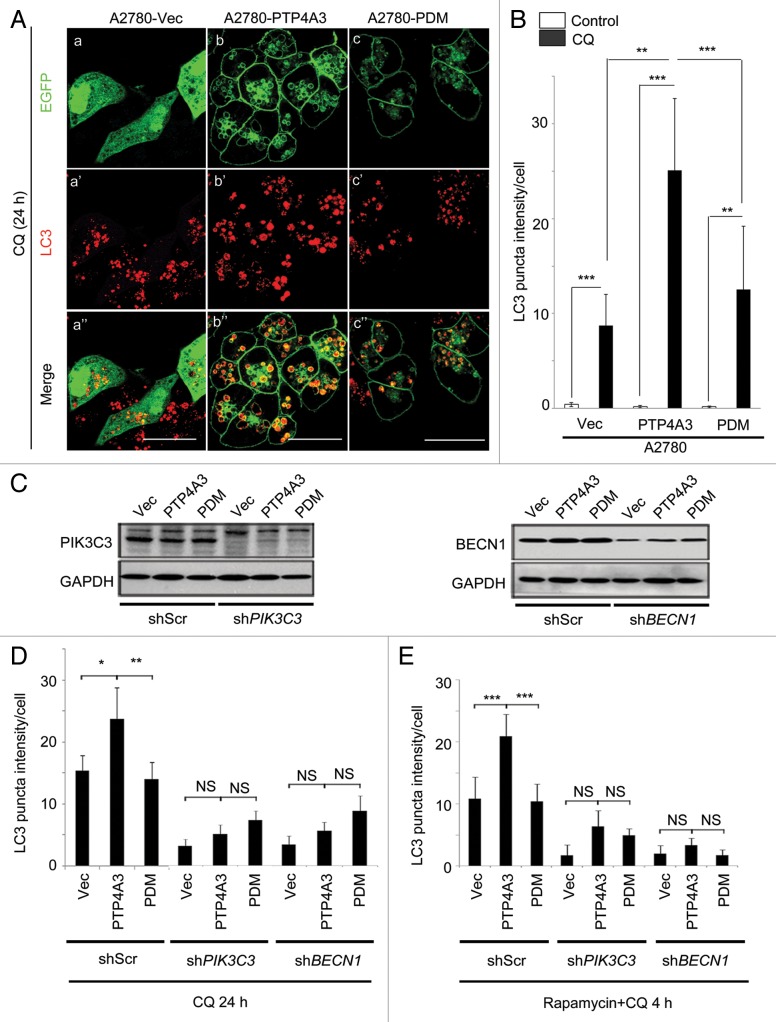
To understand the molecular factors regulating PTP4A3 recruitment into AP, we constructed 3 expression plasmids: EGFP-tagged wild-type PTP4A3 (PTP4A3), EGFP-tagged PTP4A3 catalytic-dead mutant (Cys104Ser, PDM) or EGFP-Vector control (Vec), and stably expressed the 3 constructs in A2780 human ovarian cancer cells. Upon CQ treatment, we noted that A2780-PTP4A3 cells contained more endogenous LC3 puncta (Fig. 2A, b’) per cell compared with A2780-Vec or A2780-PDM cells following CQ treatment (Fig. 2A, a’ and c’). This was despite the PDM mutant still colocalizing with AP (Fig. 2A, c”), suggesting that in addition to localization, PTP4A3 catalytic activity was also important for enhancing AP accumulation. Quantification of LC3 puncta intensity confirmed a significantly higher LC3 puncta frequency per cell in A2780-PTP4A3 cells compared with either A2780-Vec or A2780-PDM cells (Fig. 2B; P < 0.01). Prenylation was previously shown to be essential for PTP4A3 plasma membranous and endosomal localization.24 We next constructed and ectopically expressed the PTP4A3 prenylation mutant (Cys170Ser, Pre∆) in A2780 cells. As expected, prenylation-defective PTP4A3 (Pre∆) no longer colocalized with LC3 in puncta-like structures nor increased cellular LC3 puncta (Fig. S2A, c–c”). Thus, we conclude that PTP4A3 phosphatase requires both catalytic activity and prenylation-mediated membrane association to promote AP accumulation upon CQ treatment.
Figure 2. Catalytically active PTP4A3 promotes AP accumulation in a PIK3C3- and BECN1-dependent manner. (A) A2780-EGFP (Vec), A2780-EGFP-PTP4A3 (PTP4A3), and A2780-EGFP-PTP4A3 PDM (PDM) cells were treated with CQ for 24 h before immunostaining with an anti-LC3 antibody. (B) LC3 puncta intensity value of at least 20 cells from 5 distinct viewing fields were quantified as described in Materials and Methods, and presented as a histogram in the right panel (mean ± S.D.). (C) A2780-Vec, A2780-PTP4A3, and A2780-PDM cells were infected with stable shRNA knockdown constructs with either a “scrambled” sequence against non-specific targets (shScr), PIK3C3 (shPIK3C3) or BECN1 (shBECN1). The knockdown efficiency is shown by western blotting. (D) A2780-Vec, A2780-PTP4A3, and A2780-PDM cells were treated with CQ for 24 h, prior to analysis as in (B). (E) Cell lines used in (D) were treated with rapamycin in combination with CQ for 4 h, prior to analysis as in (B). *P < 0.05; **P < 0.01; ***P < 0.001; NS, P > 0.05, not significant.
To address whether PTP4A3 promoted AP accumulation via the canonical autophagy pathway upon CQ treatment, we stably knocked down PIK3C3 or BECN1—2 critical upstream regulators of canonical autophagosome formation28—using gene-specific shRNAs in A2780-Vec, A2780-PTP4A3 and A2780-PDM cells (Fig. 2C). In agreement with our earlier observations, in the control knockdown group (shScr), significantly more LC3 puncta accumulated in A2780-PTP4A3 cells, compared with A2780-Vec or A2780-PDM cells, upon CQ treatment (Fig. 2D, first 3 columns; P < 0.01). Knockdown of either PIK3C3 (Fig. 2D, middle 3 columns) or BECN1 (Fig. 2D, last 3 columns) effectively abolished CQ-induced AP accumulation by PTP4A3 (P > 0.05, PTP4A3 vs. Vec or PDM). Our data thus suggests that PTP4A3 promotes AP formation via the canonical PIK3C3-BECN1 autophagy pathway.
Rapamycin is an inhibitor of the mechanistic target of rapamycin (MTOR), a key cellular kinase regulating autophagy in response to physiological conditions and environmental stress.29 Rapamycin treatment relieves the suppression of autophagy by MTOR.30,31 Using a combination treatment of rapamycin and CQ, we found that significantly more AP accumulated in rapamycin-treated A2780-PTP4A3 cells compared with A2780-Vec or A2780-PDM cells (P < 0.001; Fig. 2E). This suggests that while MTOR inhibition activates autophagy, PTP4A3 activity further enhances AP accumulation, potentially revealing a novel mechanism whereby PTP4A3 augments canonical autophagy signaling downstream of MTOR.
PTP4A3 promotes autophagic flux (LC3-I to LC3-II conversion) in an ATG5-dependent manner
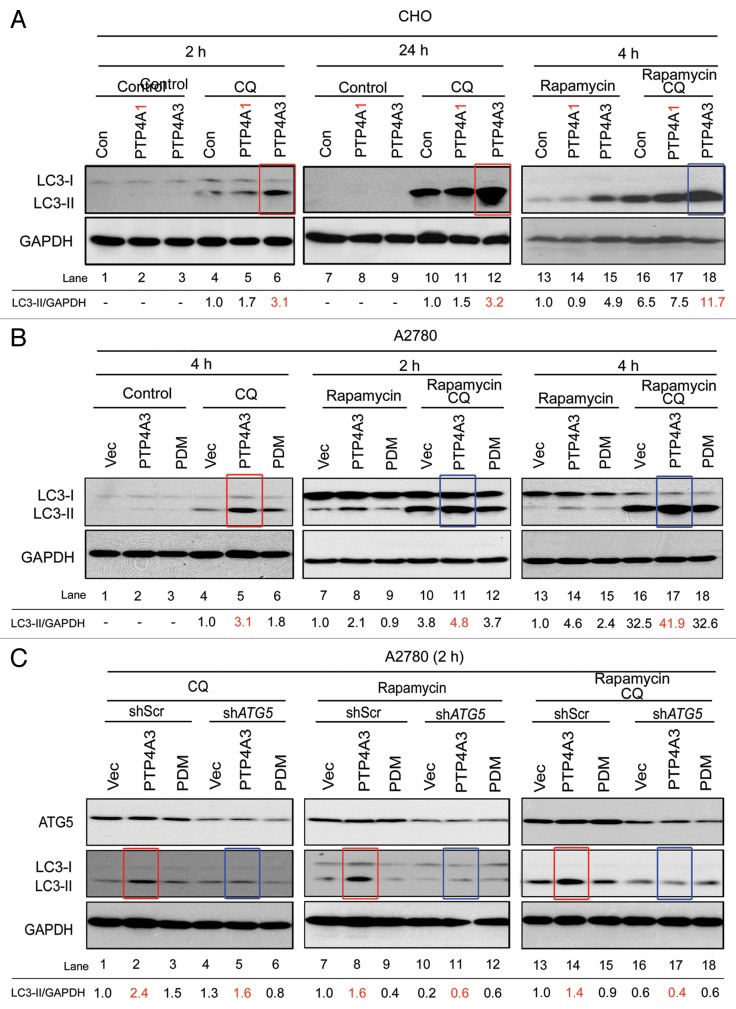
To study whether PTP4A3 could activate autophagy, immunoblotting was used to detect “autophagic flux” by comparing LC3-I to LC3-II conversion in cells either with or without CQ treatment. Lipidation of soluble LC3-I converts it into the autophagic membrane-associated LC3-II, an essential component for AP elongation and vesicle completion.31 In line with our earlier results demonstrating increased AP accumulation by PTP4A3 upon CQ treatment, we observed robust accumulation of LC3-II in CHO-PTP4A3 cells compared with CHO–Con and CHO-PTP4A1 cells at different time points after CQ treatment (Fig. 3A, red boxes) or combined treatment of CQ and rapamycin (Fig. 3A, blue boxes), suggesting that PTP4A3 (but not PTP4A1) promotes both basal and rapamycin-induced autophagy.
Figure 3. Catalytically active PTP4A3 promotes ATG5-dependent LC3 conversion. (A) CHO-Con, CHO-PTP4A1 (PTP4A1), and CHO-PTP4A3 (PTP4A3) cells were cultured in full medium with indicated treatment for the indicated durations before lysis and western blotting analysis with the anti-LC3 and anti-GAPDH antibodies. (B) A2780-Vec, A2780-PTP4A3, and A2780-PDM cells were treated as indicated, and lysed for western blotting analysis with antibodies against LC3 and GAPDH antibodies. (C) A2780-Vec, A2780-PTP4A3, and A2780-PDM cells stably expressing knockdown constructs against nonspecific targets (shScr) or ATG5 (shATG5) were treated as indicated, and lysed for western blotting analysis with the indicated antibodies.
To assess whether the catalytic activity of PTP4A3 was necessary to promote autophagic flux, the levels of LC3-I and II protein in A2780-Vec, A2780-PTP4A3, and A2780-PDM cells were analyzed. A2780-PTP4A3 cells had the highest autophagic flux compared with A2780-Vec and A2780-PDM cells under either basal (Fig. 3B, red box, lane 5) or rapamycin-induced autophagy (Fig. 3B, blue boxes, lanes 11 and 17). Prenylation-defective PTP4A3 (Pre∆) similarly failed to recapitulate the increased autophagic flux induced by wild-type PTP4A3 (Fig. S2B). Consistent with our earlier observations on promoting AP accumulation (Fig. 2C; Fig. S2A), these data reaffirm that PTP4A3 requires both catalytic activity and prenylation to drive LC3-I to LC3-II conversion and increase autophagic flux. Notably, PTP4A3 could promote autophagic flux under starvation conditions in both CHO and A2780 cell lines as well (Fig. S3A and S3B).
ATG5 has been reported as a critical autophagy regulator which plays a role in accelerating LC3 lipidation to promote autophagosome elongation.32 In cells with stable ATG5 knockdown, PTP4A3 was unable to promote LC3 conversion upon CQ, rapamycin, or combined CQ and rapamycin treatments (Fig. 3C, red boxes in lanes 2, 8, 14 vs. blue boxes in lanes 5, 11, 17). Taken together, our results show that PTP4A3 promotes autophagic flux in an ATG5-dependent manner.
PTP4A3 overexpression promotes autophagic degradation of SQSTM1
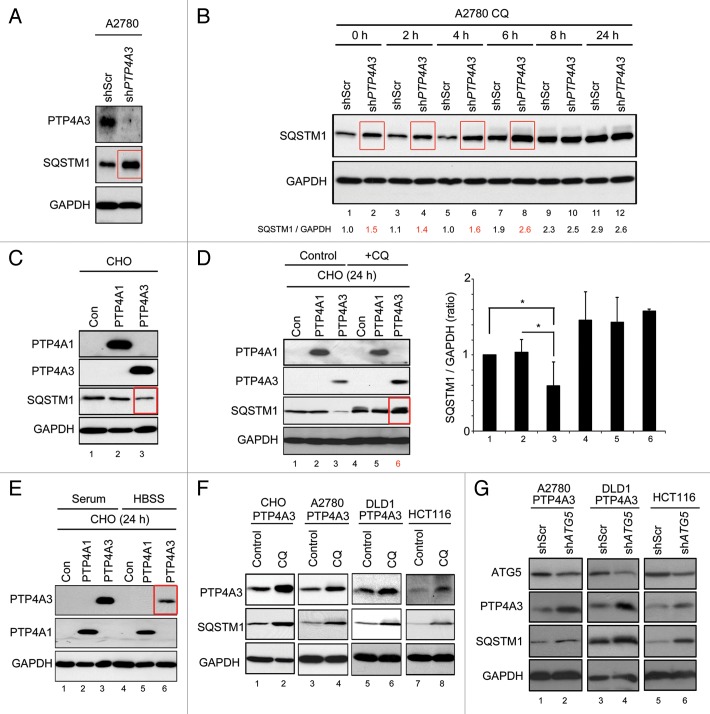
SQSTM1 is an autophagic cargo-binding protein that is degraded by autophagy.33,34 Measuring SQSTM1 protein levels has been proposed as an alternative method for determining autophagic flux, with lower SQSTM1 protein expression indicating increased autophagy activity.35 Compared with A2780 control cells, cells depleted of PTP4A3 expression had higher SQSTM1 levels (Fig. 4A, red box). Significantly, CQ treatment beyond 8 h effectively abolished this difference (Fig. 4B, lanes 10 and 12), suggesting that PTP4A3 knockdown cells had lower autophagic flux than control cells. In contrast, we found that CHO-PTP4A3 cells had lower SQSTM1 levels than CHO-Con or CHO-PTP4A1 cells (Fig. 4C, red box, lane 3), consistent with a role for PTP4A3 in promoting autophagic flux. Importantly, inhibition of lysosomal activity using CQ reversed this phenotype by preventing SQSTM1 degradation in CHO-PTP4A3 cells, with levels now comparable to CHO-Con or CHO-PTP4A1 cells (Fig. 4D, red box, lane 6; SQSTM1 quantification in right panel). Pepstatin and E64D, which are lysosomal protease inhibitors,36 could similarly rescue SQSTM1 levels (Fig. S4A, lane 3). This inverse correlation between PTP4A3 and SQSTM1 is further evidence that PTP4A3 is able to promote autophagic flux.
Figure 4. PTP4A3 promotes SQSTM1 degradation and serves as a substrate in autophagy. (A) A2780 cells were infected with stable shRNA knockdown constructs against nonspecific targets (shScr) or PTP4A3 (shPTP4A3). Exponentially growing A2780-shScr and A2780-shPTP4A3 cells were lysed and analyzed for stable-state SQSTM1 and PTP4A3 expression levels by western blotting. (B) A2780-shScr and A2780-shPTP4A3 cells were treated with CQ for indicated times before lysis and western blotting analysis of SQSTM1 expression levels. The band intensity ratio of SQSTM1/GAPDH was quantified as described in Materials and Methods, and presented as a histogram in the right panel (mean ± S.D.). (C) CHO-Con, CHO-PTP4A1, and CHO-PTP4A3 stable cells were analyzed for stable-state SQSTM1 expression levels by western blotting. (D) CHO-Con, CHO-PTP4A1, and CHO-PTP4A3 cells were cultured in full medium in the absence (control) or presence (CQ) of CQ for 24 h before lysis and western blot analysis of the indicated proteins. The ratio of SQSTM1/GAPDH was quantified as described in Materials and Methods, and presented as a histogram in the right panel (mean ± S.D.). (E) CHO-Con, CHO-PTP4A1, and CHO-PTP4A3 cells were cultured in full media (control) or in HBSS for 24 h prior to lysis and western blotting analysis of the indicated proteins. (F) CHO-PTP4A3, A2780-PTP4A3, and HCT116 cell lines were cultured in full media in the absence (control) or presence (CQ) of CQ for 24 h prior to lysis and western blotting analysis of the indicated proteins. (G) ATG5 was knocked down using shRNA in A2780-PTP4A3 and HCT116 cells. Exponentially growing cells (in full media) were lysed for western blotting analysis with the indicated antibodies. GAPDH was used as loading control.
PTP4A3 itself serves as an autophagy substrate
Unexpectedly, similar to SQSTM1, we noted that PTP4A3 (but not PTP4A1) levels decreased dramatically in cells grown in amino acid-free starvation media (Hank's balanced salt solution, HBSS) (Fig. 4E; red box, lane 6), a treatment that potently activates autophagy.1 This first indicated to us that PTP4A3 might be an autophagic substrate. To investigate this unexpected phenomenon further, we examined PTP4A3 protein levels in multiple cell lines upon autophagy inhibition by CQ. Similar to SQSTM1, CQ treatment prompted the accumulation of exogenous PTP4A3 in CHO-PTP4A3, A2780-PTP4A3 cells, and DLD1 human colorectal carcinoma cells overexpressing PTP4A3 (DLD1-PTP4A3) (Fig. 4F, lanes 2 and 4), as well as endogenous PTP4A3 in HCT116 colorectal carcinoma cells (Fig. 4F, lane 6) which express detectable levels of PTP4A3 protein. In a complimentary approach, we detected increased levels of PTP4A3 and SQSTM1 in cells with ATG5 knockdown (Fig. 4G, lanes 2, 4, and 6) or BECN1 (Fig. S4B, lanes 2, 4, and 6). These results suggest that like SQSTM1, PTP4A3 appears to be a bona fide autophagy-degraded protein.
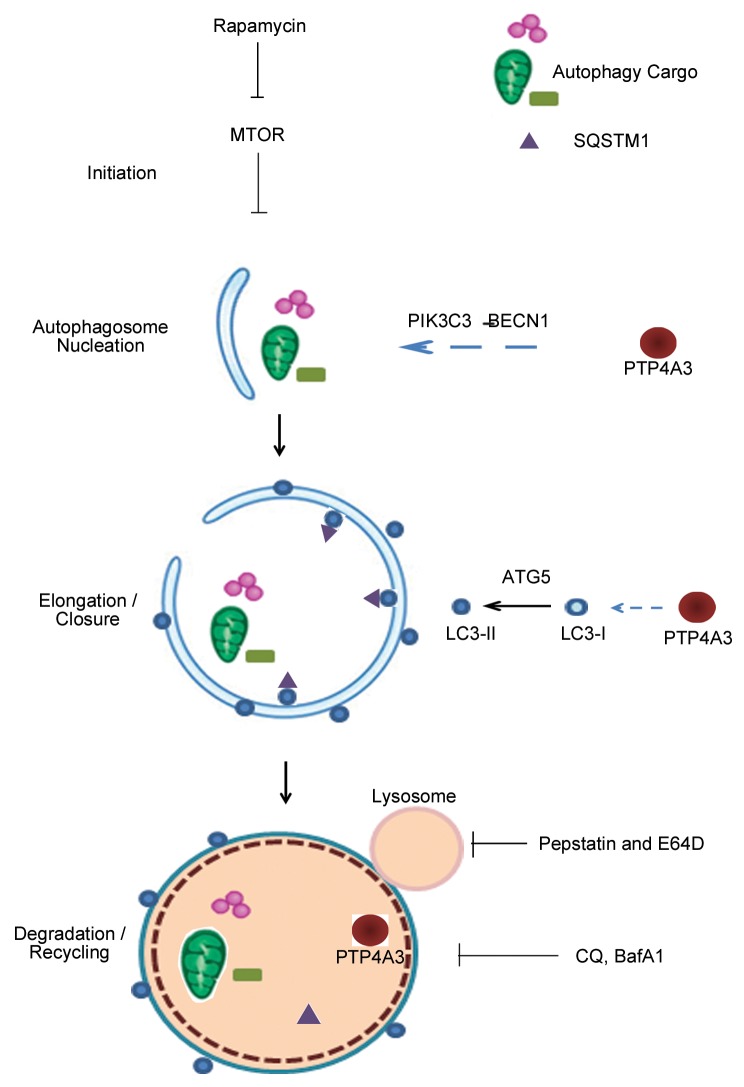
Taken together with our earlier results that PTP4A3 promotes AP formation and autophagic flux, the observation that PTP4A3 is also attenuated by autophagic degradation suggests the existence of a negative-feedback loop between PTP4A3 and autophagy activity. Herein, we propose a model on how PTP4A3 might be involved in the canonical autophagy pathway (Fig. 5): 1) PTP4A3 functions as an activator to promote autophagosome formation in a PIK3C3-BECN1-dependent manner; 2) PTP4A3 accelerates LC3-I to LC3-II conversion in an ATG5-dependent manner; 3) PTP4A3 enhances the degradation of SQSTM1, a key autophagy substrate; and 4) PTP4A3 itself serves as a novel autophagic substrate, fulfilling a negative feedback-loop to fine-tune autophagy activity.
Figure 5. A model illustrates the involvement of PTP4A3 in multiple steps of canonical autophagy pathway. 1) PTP4A3 acts as an activator to promote autophagosome formation in a PIK3C3-BECN1-dependent manner; 2) PTP4A3 accelerates LC3-I to LC3-II conversion in an ATG5-dependent manner; 3) PTP4A3 enhances the degradation of SQSTM1, a key autophagy substrate; 4) PTP4A3 itself is a novel autophagic substrate. Sites of action of drug treatments (rapamycin, pepstatin and E64D, CQ, Baf A1) are indicated at where appropriate.
PTP4A3 exploits autophagy to promote ovarian cancer cell proliferation
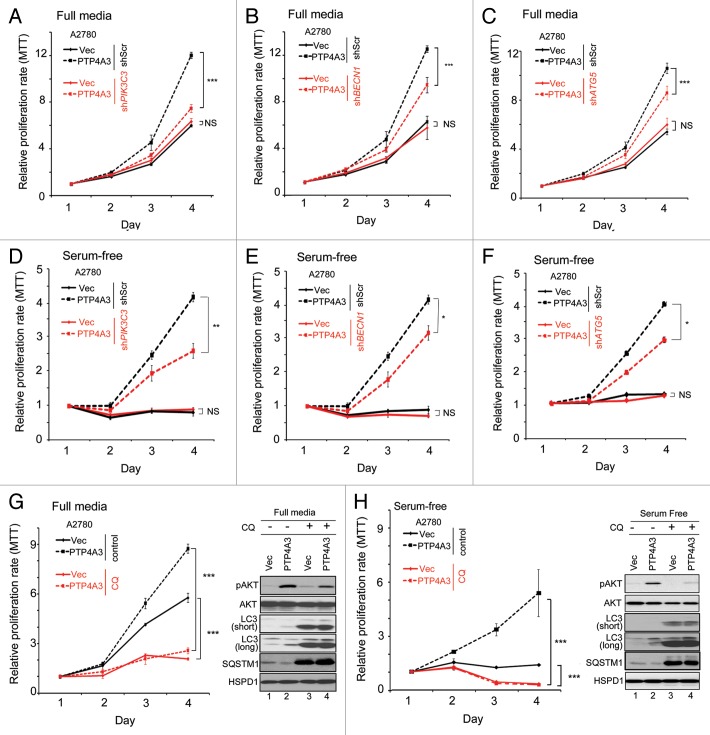
PTP4A3 has been shown to promote cancer cell growth, migration, invasion, angiogenesis, epithelial-mesenchymal transition, and ultimately metastasis.11 In light of our data suggesting that PTP4A3 could promote autophagy, which in turn has established associations with tumor biology,9 we further investigated the role of autophagy in PTP4A3-mediated cancer progression. First, we compared the growth of A2780-Vec and A2780-PTP4A3 cells following depletion of PIK3C3, BECN1, or ATG5 expression. Under serum-replete conditions, PTP4A3 promoted an increase in cell growth in control knockdown cells (shScr), as measured by MTT assays (Fig. 6A–C, black dashed lines). Importantly, knockdown of PIK3C3, BECN1, or ATG5 resulted in a significant decrease in the growth of A2780-PTP4A3 cells (Fig. 6A–C, red vs. black dashed lines; P < 0.001). In contrast, knockdown of these autophagy regulators had no significant effect on A2780-Vec cell growth (Fig. 6A–C, red vs. black solid lines; P > 0.05), consistent with a previous report demonstrating that A2780 cells are insensitive to autophagy inhibition under serum-replete conditions.37 We next repeated and measured the growth of these cells under serum starvation, a common autophagy inducer in cultured cells.1 Interestingly, under serum starvation, A2780-PTP4A3 cells could survive and grow for up to 4 d, with knockdown of PIK3C3, BECN1, or ATG5 resulting in reduced growth (Fig. 6D–F, red vs. black dashed lines; P < 0.001). In contrast, A2780-Vec cells were unable to grow under serum-starved conditions (Fig. 6D–F, red vs. black solid lines; P > 0.05). Collectively, these results suggest that PTP4A3 promotes ovarian cancer cell growth and survival via an autophagy-dependent mechanism, a particularly important manifestation under serum starvation stress.
Figure 6. Overexpression of PTP4A3 promotes cell proliferation in A2780 cells in an autophagy-dependent manner. (A–C) A2780-vec and A2780-PTP4A3 cells infected with shScr or shPIK3C3 (A), shBECN1 (B), or shATG5 (C) were cultured in full media. Relative proliferation rates were measured using an MTT assay. (D and E) Cell lines used in (A–C) were cultured in serum-free RPMI media. Relative proliferation rates were measured using an MTT assay. (G and H) A2780-Vec and A2780-PTP4A3 cells were cultured in full media (G) or serum-free (H) in the absence (control) or presence of CQ. A representative relative proliferation rate curve is shown. Similarly treated cells were analyzed (48 h treatment) in parallel by western blotting for pAKT, total AKT, LC3, SQSTM1, and HSPD1 levels (right panel).
Next, we inhibited lysosomal degradation with CQ. We observed a significant decrease in growth of both A2780-Vec (Fig. 6G, red vs. black solid lines; P < 0.001) and A2780-PTP4A3 cells (Fig. 6G, red vs. black dashed lines; P < 0.001) upon CQ treatment under serum-replete conditions. Critically, the enhanced cell growth induced by PTP4A3 overexpression was effectively abolished by CQ treatment alone, suggesting that cells harboring increased PTP4A3 expression are more sensitive to autophagy inhibitors.
PTP4A3 overexpression promotes AKT activity by phosphorylation (pAKT) on Ser473 of the kinase.11,38 In A2780-PTP4A3 cells grown in serum-replete conditions, we detected increased pAKT compared with A2780 control cells (Fig. 6G, right panel, lanes 1–2). Moreover, treatment with CQ caused pAKT levels to decrease in A2780-PTP4A3 cells, but not in A2780-Vec cells (Fig. 6G, right panel, lanes 3 and 4). Under serum-depleted conditions, A2780-PTP4A3 cells were also able to survive and continue growth despite the absence of serum and glutamine supplementation, whereas A2780 control cells could not (Fig. 6H, black lines; P < 0.001). Importantly, CQ treatment abolished the difference in growth rate between A2780-Vec and A2780-PTP4A3 cells (Fig. 6H, red lines; P > 0.05). Since PTP4A3 overexpression has been shown to promote pAKT levels under growth-factor deprived conditions,39 we tested whether AKT activity was similarly involved in the ability of PTP4A3 to sustain cell growth under serum-starved conditions. Indeed, under serum-free conditions, relative to control cells, A2780-PTP4A3 cells were able to sustain pAKT levels to a greater extent than A2780-Vec cells (Fig. 6H, western blot lanes 1 and 2). Importantly, CQ treatment reduced the difference in pAKT levels between these 2 cell lines (Fig. 6H, western blot lanes 3 and 4), thereby uncoupling PTP4A3-dependent pAKT signaling. Taken together, our data suggests that PTP4A3 overexpression increased autophagy activity and thus endowed cells with a growth advantage regardless of growth factor availability—an advantage that can be effectively suppressed by autophagy inhibition.
High expression levels of PTP4A3 predict poorer survival of ovarian cancer patients coexpressing high levels of autophagy genes
To investigate the clinical relevance of our in vitro findings obtained from CHO and A2780 ovarian cell lines, we extracted microarray and clinical-pathological data from an ovarian cancer patient cohort (GSE 9899), the largest ovarian cancer data set (n = 285) currently available in the GEO database. In agreement with previous studies showing that PTP4A3 overexpression promotes ovarian cancer progression,40 we found that mRNA expression of PTP4A3 was significantly increased in malignant ovarian tumors compared with those with low malignant potential tumors (Fig. S5A; P < 0.001). Increased PTP4A3 mRNA expression was associated with both higher histological grade 2 and 3 (Fig. S5B; P = 0.001) and pathological stage IV (Fig. S5C; P = 0.006). Moreover, a higher level of PTP4A3 mRNA was significantly associated with a shorter survival time in this ovarian cancer patient cohort (Fig. S5D; P = 0.007, Kaplan-Meier analysis, log-rank test). By Cox-regression analysis, increasing expression of PTP4A3 mRNA was significantly associated with higher risk of recurrence-associated death (P = 0.009; HR = 1.223, 95% CI = 1.051–1.424). Thus, ovarian cancer patients having low PTP4A3 expression did profoundly better than those having high levels of PTP4A3, supporting the oncogenic character of PTP4A3.
To investigate whether autophagy contributed to the prognostic significance of PTP4A3 in this ovarian cancer patient cohort, we then stratified patients into “high” or “low” groups using the median mRNA expression of PIK3C3 and BECN1 as cut-off points. Neither PIK3C3 nor BECN1 individually had significant prognostic value on patient survival using this cut-off criterion (Fig. S6A and S6B). Nonetheless, in patients with high PIK3C3 mRNA expression, PTP4A3 mRNA expression was significantly associated with pathological stage III and IV (Fig. 7A; P = 0.002) and patient recurrence-free survival (Fig. 7B; P = 0.003). In contrast, in patients with low PIK3C3 mRNA expression, PTP4A3 mRNA level was not significantly associated with any of these clinical-pathological parameters (Fig. S7A and S7B). Similar results were obtained when BECN1 mRNA expression was used to stratify the patients. In patients with high BECN1 expression, PTP4A3 mRNA expression was significantly associated with pathological stage IV (Fig. 7C; P = 0.003) and shorter recurrence-free survival (Fig. 7D; P = 0.001). Intriguingly, in patients with low BECN1 expression, PTP4A3 expression was again not significantly associated with any of these clinical pathological parameters (Fig. S7C and S7D). Significantly, Cox-regression analysis revealed that increasing mRNA expression of PTP4A3 was significantly associated with a higher risk of recurrence-associated death only in patients with a high level of PIK3C3 (Table S1) or BECN1 (Table S2) expression.
Figure 7. High PTP4A3 expression levels predict poorer survival of ovarian cancer patients coexpressing high levels of autophagy genes in the GSE9899 patient cohort. (A) In ovarian cancer patients with high levels of PIK3C3, PTP4A3 expression levels were significantly correlated with higher pathological stage. (B) Patients with high expression levels of PIK3C3 and PTP4A3 have significantly reduced recurrence-free survival. (C and D) Similar results as in (A and B) were obtained when the tumors were stratified by BECN1 instead of PIK3C3.
Discussion
In this study, we demonstrated that metastasis-associated PTP4A3 phosphatase functions as an activator of canonical autophagy and also serves as a substrate for canonical autophagy. Autophagy inhibition effectively curtailed PTP4A3-driven cell growth, as well as its ability to enhance AKT phosphorylation. We showed that autophagy plays an important role in PTP4A3-mediated ovarian cancer progression and, more importantly, characterized how the prognostic significance of PTP4A3 correlates with high expression levels of various autophagy-related genes in clinical patient samples. Taken together, our results indicate that autophagy inhibition represents an attractive therapeutic approach against ovarian cancers having high expression of both PTP4A3 and autophagy genes.
Our results provide evidence that PTP4A3, but not PTP4A1, localizes to autophagosomes and promotes autophagy. This is surprising, given that these 2 phosphatases share 78% amino acid sequence identity and are both endosomally localized.9 In agreement with autophagosomal localization, we report here that overexpressed PTP4A3, but not PTP4A1, could be degraded via autophagy. Importantly, this applies for endogenous PTP4A3 as well, suggesting that autophagic degradation of PTP4A3 was not simply due to nonspecific aggregation of overexpressed protein.41 Such specificity potentially sets the stage for a highly tuned feedback mechanism by PTP4A3 on canonical autophagic activity—as PTP4A3 levels accumulate, autophagy flux is enhanced and consequently becomes degraded by autophagy, thereby eventually returning autophagy activity to a homeostatic functional state. Indeed, PTP4A3 is not the only autophagy-regulated protein with such a feedback role on autophagy. The essential autophagy core component and regulator ULK1/ATG1 has recently been reported as an autophagy substrate, similarly constituting a negative feedback loop believed to play a role in the maintenance of balanced autophagic activity.42 Such negative feedback loops might serve to prevent autophagic cell death as a consequence of unrestricted autophagic activity which would result in progressive consumption of cellular components and subsequent autophagic cell death.43 Our findings here suggest a role for PTP4A3 in balancing autophagy activity in nutrient-demanding cancer cells under starvation conditions, where nutrient recycling via autophagy might be critical to tolerate such stress and protect cells from apoptosis.44
Herein, we show that PTP4A3 promotes PIK3C3-, BECN1-, and ATG5-dependent canonical autophagy. Suppression of these critical autophagy regulators or treatment with CQ inhibited autophagy flux in ovarian cancer cells and effectively crippled the ability of PTP4A3 to drive cell proliferation. The physiological importance of this is that PTP4A3 requires functional autophagy to promote ovarian cancer proliferation. Clinical data supports this—we show that the prognostic effect of PTP4A3 in human ovarian cancers is significant in cases where there is a genetic background with elevated autophagy-related gene (BECN1, PIK3C3, and ATG5) expression. Recently, activated KRAS was shown to increase cancer cell dependence on autophagy for survival and tumorigenesis.43 Interestingly, there are several parallels between PTP4A3 and KRAS: 1) both PTP4A3 expression and KRAS activity increases in more aggressive cancers; 2) autophagy activation by PTP4A3 or KRAS is critical to allow cancer cell proliferation under starvation conditions; and, 3) both potentiate receptor tyrosine kinase signaling.44-46 Intriguingly, RAS and PTP4A3 also share similar membrane-targeting modules—a polybasic motif and prenylation—in their carboxy termini domains, presumably localizing them in close proximity on cellular membranes.
To translate our in vitro findings into a more clinically relevant context, we investigated the survival outcomes of ovarian cancer patients with high or low PTP4A3 and autophagy-related genes mRNA expression levels. We found that in the presence of high autophagy gene expression, patients coexpressing high levels of PTP4A3 mRNA had significantly shorter survival than patients coexpressing low levels of PTP4A3 mRNA. In light of our in vitro data suggesting that PTP4A3 is dependent on the autophagy pathway for cancer progression, we propose that inhibition of autophagy might effectively block the oncogenic functions of PTP4A3 in such patient tumors. Currently, autophagy inhibition is at the forefront of cancer therapy, with approximately 20 ongoing clinical trials in multiple types of cancers.47 In view of our current understanding of the molecular factors that determine the opposing roles of autophagy during cancer progression, the identification of PTP4A3 as a candidate biomarker to predict the favorable response to autophagy inhibition is envisioned to have significant clinical utility. This is especially pertinent, given that drugs that result in either autophagy activation or inhibition have been demonstrated to provide therapeutic value in combating cancers, highlighting the essential need for proper patient diagnostics and stratification prior to treatment. Recent research has shown that some cancer cells—such as those driven by active KRAS—depend upon and get addicted on elevated levels of autophagy for survival even in the absence of external stressors.47 In view of this, we propose that the targeted inhibition of autophagy in PTP4A3-positive tumors might be a significant advancement in developing a novel therapeutic approach for treating similarly “autophagy-addicted” ovarian tumors due to overexpression of both PTP4A3 and autophagy genes.
Materials and Methods
Cell culture conditions
CHO-K1 cells (ATCC), human ovarian carcinoma cell line A2780 (ATCC), and human colorectal carcinoma cell line HCT116 (ATCC) were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (HyClone, SV30160.03), 2 mM L-glutamine (Life Technologies, 25030-081), and 1% antibiotic/anti-mycotic (Life Technologies, 15140-122) (“full media”). Where indicated, cells were starved in the absence of serum (“serum free”) or HBSS. The viral packaging cell line Phoenix Ampho (a gift from the Nolan Laboratory, Stanford University) and 293T cells (ATCC) were maintained in DMEM 4.5 g/L high-glucose medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, and 1% antibiotic/antimycotic. All cells were maintained in a 5% CO2 atmosphere at 37 °C.
Plasmids and transfection
CHO-PTP4A1, CHO-PTP4A3, and CHO-EGFP-PTP4A3 cells were generated previously.13 A2780-Vec, A2780-PTP4A3, and A2780-PTP4A3-PDM cells were generated from A2780 parental cells (ATCC), and the plasmids used including pEGFPC1 vector, pEGFP-PTP4A3, and pEGFP-PTP4A3-PDM were generated previously.14 Lentiviral-mediated knockdown of PIK3C3, BECN1, ATG5, and PTP4A3, (Mission shRNA, Sigma-Aldrich) was performed as previously described.48 Transfection was done using JetPRIME according to the manufacturer’s instructions (Polyplus-transfection, 114-75).
Reagents
Unless otherwise specified, chemicals were used at the following final concentrations; chloroquine diphosphate salt (50 μM, C6628, Sigma-Aldrich); rapamycin (2 μM, R8781, Sigma-Aldrich); E64D (10 μg/ml, E8640, Sigma-Aldrich); pepstatin A (10 μg/ml, P5318, Sigma-Aldrich); bafilomycin A1 (100 nM, B1793, Sigma-Aldrich). HBSS (phenol red-free; 14025-126) was purchased from Life Technologies.
Immunofluorescence staining
Immunofluorescence staining was done as described previously.48 Briefly, cells were fixed in 4% formaldehyde for 15 min, washed with PBS, and then permeabilized and blocked with 0.3% Triton X-100 (BDH Chemicals, 9002-93-1) and 5% BSA (Sigma-Aldrich, 05470), respectively, for 1 h. Cells were then incubated with primary antibodies overnight at 4 °C. Anti-LC3 antibody (3868, Cell Signaling Technology) was used at a dilution of 1:200 and PTP4A3 antibody49 was used at a dilution of 1:400. Cells were then washed thrice with PBS and then incubated with secondary antibody (1:400 dilution; Life Technologies, A10040, A10036) conjugated with appropriate fluorophore for 1 h at room temperature. Slides were mounted with Prolong Gold Anti-fade Reagent and staining was analyzed immediately using an LSM700 confocal microscope (Carl Zeiss AG). The average LC3 channel pixel intensity value within the cytoplasmic area per cell was determined using Photoshop (Adobe Systems) to represent the overall amount of LC3 puncta accumulation in each cell. The colocalzation percentage of LC3 puncta with PTP4A3 puncta was counted using visual inspection. The results were presented as mean ± SD.
Western blotting
Western immunoblotting was performed as previously described.48 BECN1 (3495), LC3 (3868), ATG5 (8540), SQSTM1 (5114), phospho-AKT-Ser473 (4060), and AKT (4691) were purchased from Cell Signaling Technology. GAPDH (MAD372) was purchased from Millipore. HSPD1 (611562) antibody was purchased from BD Biosciences. PTP4A1 and PTP4A3 antibodies were generated in-house.49 Anti-rabbit HRP-labeled secondary antibody (7074) was purchased from Cell Signaling Technology, and anti-mouse HRP-labeled secondary antibody (115-035-003) was purchased from Jackson ImmunoResearch Laboratories. Immunoreactive bands were visualized using chemiluminescent substrate (Thermo Scientific, 34080) and detected on radiography film. Quantification of band intensity was done using ImageJ software.
MTT cell proliferation assay
A2780 cells were seeded at a concentration of 5,000 cells per well in 96-well plates in triplicate. Cells were allowed to grow for 24 h and the medium was replaced with desired media with or without drugs, as indicated. MTT reagent (M5655, Sigma-Aldrich) was added at a final concentration of 0.2 mg/mL in each well. Cells were incubated with MTT reagent at 37 °C for 3 h. Medium was completely removed and the retained formazan crystals were then dissolved in DMSO and the absorbance at 595 nm was measured.
Analysis of ovarian cancer patient microarray data
The GEO-accessible GSE9899 data set consisting of 285 ovarian cancer patients with corresponding microarray, clinic-pathological, and survival data was used in this study. This is the largest ovarian cancer cohort data set available in the GEO database. Microarray and patient data were extracted as previously described.48 Patients were divided into 4 subgroups, including weak, moderate, strong, and very strong, based on their PTP4A3 mRNA expression levels using quartiles as the cut-off point. The highest quartile of PTP4A3 was used as a cut-off point to separate patients into high and low subgroup for Kaplan-Meier survival analysis. The association between PTP4A3 expression and clinical-pathological parameters was tested by Chi-Square, while the association between PTP4A3 and patient survival was tested by Kaplan-Meier analysis together with log-rank test. Patients were further stratified based on their PIK3C3 and BECN1 mRNA expression levels using the median value as a cut-off point to distinguish a more autophagy-competent subgroup (“high”) and a less autophagy-competent subgroup (“low”).
Statistical analysis
All the statistical analyses were performed using SPSS19.0 software (IBM Corporation). One-way ANOVA was performed for comparison of means between groups. Repeated measures ANOVA was performed for MTT assay where data was obtained at multiple time points. The P value for comparison of 2 groups in the same experimental setting was generated by ANOVA post-hoc test, either Tukey HSD or Games-Howell, depending on the results from the homogeneity test by Levene statistics, where applicable. A P value of < 0.05 was considered as significant in all tests.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by research grant from the Agency of Science, Technology and Research (A*STAR), Singapore. We are grateful to Drs Eng Chon Boon and Rajeev Singh from the NUH-NUS Tissue Repository for their help in providing human cancer samples, and to Dr Low Kee Chung for assistance with lentiviral transduction of cells.
Glossary
Abbreviations:
- ATG5
autophagy-related 5
- BafA1
bafilomycin A1
- BECN1
Beclin 1, autophagy related
- CQ
chloroquine
- EGFP
enhanced green fluorescent protein
- MAP1LC3/LC3
microtubule-associated protein 1 light chain 3
- GEO
Gene Expression Omnibus
- HBSS
Hank's balanced salt solution
- HSPD1
heat shock 60 kDa protein 1
- KRAS
Kirsten rat sarcoma viral oncogene homolog
- MTOR
mechanistic target of rapamycin
- PAS
phagophore assembly site
- PDM
phosphatase-defective mutant
- PE
phosphatidylethanolamine
- PIK3C3
phosphatidylinositol 3-kinase, catalytic subunit type 3
- PtdIns3P
phosphatidylinositol 3-phosphate
- PTP4A3/PRL-3
protein tyrosine phosphatase type IVA, member 3
- SQSTM1
sequestosome 1
- ULK1
unc-51 like autophagy activating kinase 1
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Takegawa K, DeWald DB, Emr SD. Schizosaccharomyces pombe Vps34p, a phosphatidylinositol-specific PI 3-kinase essential for normal cell growth and vacuole morphology. J Cell Sci. 1995;108:3745–56. doi: 10.1242/jcs.108.12.3745. [DOI] [PubMed] [Google Scholar]
- 3.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–8. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 4.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–9. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20:444–54. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 7.Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2012;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 8.Shanware NP, Bray K, Abraham RT. The PI3K, metabolic, and autophagy networks: interactive partners in cellular health and disease. Annu Rev Pharmacol Toxicol. 2013;53:89–106. doi: 10.1146/annurev-pharmtox-010611-134717. [DOI] [PubMed] [Google Scholar]
- 9.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nature reviews. Clin Oncol. 2011;8:528–39. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 10.Zeng Q, Hong W, Tan YH. Mouse PRL-2 and PRL-3, two potentially prenylated protein tyrosine phosphatases homologous to PRL-1. Biochem Biophys Res Commun. 1998;244:421–7. doi: 10.1006/bbrc.1998.8291. [DOI] [PubMed] [Google Scholar]
- 11.Al-Aidaroos AQO, Zeng Q. PRL-3 phosphatase and cancer metastasis. J Cell Biochem. 2010;111:1087–98. doi: 10.1002/jcb.22913. [DOI] [PubMed] [Google Scholar]
- 12.Saha S, Bardelli A, Buckhaults P, Velculescu VE, Rago C, St Croix B, Romans KE, Choti MA, Lengauer C, Kinzler KW, et al. A phosphatase associated with metastasis of colorectal cancer. Science. 2001;294:1343–6. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 13.Zeng Q, Dong JM, Guo K, Li J, Tan HX, Koh V, Pallen CJ, Manser E, Hong W. PRL-3 and PRL-1 promote cell migration, invasion, and metastasis. Cancer Res. 2003;63:2716–22. [PubMed] [Google Scholar]
- 14.Wang H, Quah SY, Dong JM, Manser E, Tang JP, Zeng Q. PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Res. 2007;67:2922–6. doi: 10.1158/0008-5472.CAN-06-3598. [DOI] [PubMed] [Google Scholar]
- 15.Fiordalisi JJ, Keller PJ, Cox AD. PRL tyrosine phosphatases regulate rho family GTPases to promote invasion and motility. Cancer Res. 2006;66:3153–61. doi: 10.1158/0008-5472.CAN-05-3116. [DOI] [PubMed] [Google Scholar]
- 16.Polato F, Codegoni A, Fruscio R, Perego P, Mangioni C, Saha S, Bardelli A, Broggini M. PRL-3 phosphatase is implicated in ovarian cancer growth. Clin Cancer Res. 2005;11:6835–9. doi: 10.1158/1078-0432.CCR-04-2357. [DOI] [PubMed] [Google Scholar]
- 17.Guo K, Li J, Wang H, Osato M, Tang JP, Quah SY, Gan BQ, Zeng Q. PRL-3 initiates tumor angiogenesis by recruiting endothelial cells in vitro and in vivo. Cancer Res. 2006;66:9625–35. doi: 10.1158/0008-5472.CAN-06-0726. [DOI] [PubMed] [Google Scholar]
- 18.Basak S, Jacobs SBR, Krieg AJ, Pathak N, Zeng Q, Kaldis P, Giaccia AJ, Attardi LD. The metastasis-associated gene Prl-3 is a p53 target involved in cell-cycle regulation. Mol Cell. 2008;30:303–14. doi: 10.1016/j.molcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radke I, Götte M, Kersting C, Mattsson B, Kiesel L, Wülfing P. Expression and prognostic impact of the protein tyrosine phosphatases PRL-1, PRL-2, and PRL-3 in breast cancer. Br J Cancer. 2006;95:347–54. doi: 10.1038/sj.bjc.6603261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, Wang S, Lu J, Li J, Ding Y. Over-expression of phosphatase of regenerating liver-3 correlates with tumor progression and poor prognosis in nasopharyngeal carcinoma. Int J Cancer. 2009;124:1879–86. doi: 10.1002/ijc.24096. [DOI] [PubMed] [Google Scholar]
- 21.Razi M, Chan EYW, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–21. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen TE, Johansen T. Following autophagy step by step. BMC Biol. 2011;9:39. doi: 10.1186/1741-7007-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai Q, Lu L, Tian JH, Zhu YB, Qiao H, Sheng ZH. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 2010;68:73–86. doi: 10.1016/j.neuron.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng Q, Si X, Horstmann H, Xu Y, Hong W, Pallen CJ. Prenylation-dependent association of protein-tyrosine phosphatases PRL-1, -2, and -3 with the plasma membrane and the early endosome. J Biol Chem. 2000;275:21444–52. doi: 10.1074/jbc.M000453200. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 26.Berg TO, Fengsrud M, Strømhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883–92. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 27.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–50. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–95. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–62. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol. 2004;36:2445–62. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 35.Bjørkøy G, Lamark T, Pankiv S, Øvervatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–97. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 36.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–66. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Cheng Y, Ren X, Zhang L, Yap KL, Wu H, Patel R, Liu D, Qin ZH, Shih IM, et al. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene. 2012;31:1055–64. doi: 10.1038/onc.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Vardy LA, Tan CP, Loo JM, Guo K, Li J, Lim SG, Zhou J, Chng WJ, Ng SB, et al. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell. 2010;18:52–62. doi: 10.1016/j.ccr.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 39.Jiang Y, Liu XQ, Rajput A, Geng L, Ongchin M, Zeng Q, Taylor GS, Wang J. Phosphatase PRL-3 is a direct regulatory target of TGFbeta in colon cancer metastasis. Cancer Res. 2011;71:234–44. doi: 10.1158/0008-5472.CAN-10-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS, Semplicio G, Stoffel I, Brezovich A, Verma M, Hansmann I, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–30. doi: 10.1016/S0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 42.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–41. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JMS, Karantza V, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walls CD, Iliuk A, Bai Y, Wang M, Tao WA, Zhang ZY. Phosphatase of regenerating liver 3 (PRL3) provokes a tyrosine phosphoproteome to drive prometastatic signal transduction. Mol Cell Proteomics. 2013;12:3759–77. doi: 10.1074/mcp.M113.028886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Aidaroos AQO, Yuen HF, Guo K, Zhang SD, Chung TH, Chng WJ, Zeng Q. Metastasis-associated PRL-3 induces EGFR activation and addiction in cancer cells. J Clin Invest. 2013;123:3459–71. doi: 10.1172/JCI66824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mancias JD, Kimmelman AC. Targeting autophagy addiction in cancer. Oncotarget. 2011;2:1302–6. doi: 10.18632/oncotarget.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuen HF, Chan YK, Grills C, McCrudden CM, Gunasekharan V, Shi Z, Wong ASY, Lappin TR, Chan KW, Fennell DA, et al. Polyomavirus enhancer activator 3 protein promotes breast cancer metastatic progression through Snail-induced epithelial-mesenchymal transition. J Pathol. 2011;224:78–89. doi: 10.1002/path.2859. [DOI] [PubMed] [Google Scholar]
- 49.Li J, Guo K, Koh VWC, Tang JP, Gan BQ, Shi H, Li HX, Zeng Q. Generation of PRL-3- and PRL-1-specific monoclonal antibodies as potential diagnostic markers for cancer metastases. Clin Cancer Res. 2005;11:2195–204. doi: 10.1158/1078-0432.CCR-04-1984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.