Abstract
HMGB1 (high mobility group box 1) is a multifunctional, ubiquitous protein located inside and outside cells that plays a critical role in various physiological and pathological processes including cell development, differentiation, inflammation, immunity, metastasis, metabolism, and death. Increasing evidence demonstrates that HMGB1-dependent autophagy promotes chemotherapy resistance, sustains tumor metabolism requirements and T cell survival, prevents polyglutamine aggregates and excitotoxicity, and protects against endotoxemia, bacterial infection, and ischemia-reperfusion injury in vitro or in vivo. In contrast, HMGB1 may not be required for autophagy in some organs such as the liver and heart. Understanding HMGB1-dependent and -independent autophagy in more detail will provide insight into the integrated stress response and guide HMGB1-based therapeutic intervention.
Keywords: autophagy, HMGB1, knockin, knockout, phenotype
HMGB1 is an evolutionarily ancient protein that possibly originated more than 525 million years ago before the protostomes and deuterostomes split. It was first identified in 1973 by Ernest Johns and coworkers as one of a group of nonhistone, chromatin-associated proteins with 2 DNA-binding HMG-box domains (A and B box) and an acidic C-terminal tail.1 HMGB1 is normally located in the nucleus, acting as a DNA chaperone involved in the regulation of a number of DNA-associated processes such as replication, transcription, recombination, and repair. In addition to its nuclear function, HMGB1 can act as a stress sensor and translocate from the nucleus to the cytoplasm and then be released into the extracellular space during various stress conditions. Autophagy is generally a programmed cell survival process and lysosome-mediated pathway involving the degradation of cellular components (e.g., long-lived proteins and damaged organelles) and invading pathogens in a selective or nonselective manner.2-4 The dynamic process of autophagy is primarily controlled by the autophagy-related (ATG) protein family, and it shares regulators from other trafficking pathways and cell death.4,5 In the past few years, increasing evidence supports the existence of ATG pathway (e.g., ATG5, ATG7, and BECN1)-independent autophagy, making the autophagy machinery as well as autophagy monitoring extremely complicated.6-8 Indeed, HMGB1 has a context-dependent role in the regulation of autophagy and stress.9 Here, we outline the exciting new advances in our knowledge of HMGB1-dependent and -independent autophagy and discuss how these advances are driving the understanding of the integrated stress response.
HMGB1-Dependent Autophagy
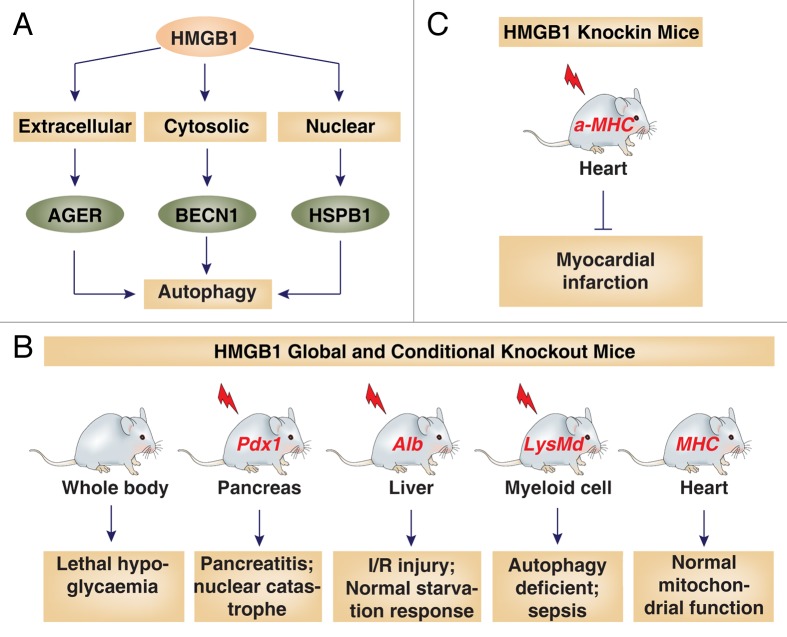
HMGB1 participates in the autophagy process at several levels (Fig. 1A). First, HMGB1 translocates to the cytoplasm following several autophagic stimuli (e.g., hydrogen peroxide, rapamycin, and starvation), which in turn promotes autophagy through direct interaction with BECN1 to dissociate it from BCL2 in immortalized mouse embryonic fibroblasts and cancer cells.10 Meanwhile, HMGB1 C23S and C45S mutants lose their ability to mediate autophagy, as they are unable to bind BECN1 and therefore cannot disrupt BCL2-BECN1 interactions.10 In addition, the HMGB1-BECN1 complex seems to be tightly controlled at the transcriptional, post-transcriptional, post-translational, and protein-protein interaction level. For example, ULK1 (unc-51 like autophagy activating kinase 1),11 MAPK (mitogen-activated protein kinase),10 and NACC1 (nucleus accumbens associated 1, BEN and BTB [POZ] domain containing)12 positively regulate HMGB1-mediated autophagy, whereas TP53,13 SNCA/α-synuclein,14 IFI30/gamma-interferon-inducible lysosomal thiol reductase,15 MIR34A,16 and MIR2217 negatively regulate HMGB1-mediated autophagy. Second, HMGB1 regulates the expression of HSPB1 (heat shock 27 kDa protein 1) in immortalized mouse embryonic fibroblasts and cancer cells.18 As a cytoskeleton regulator, HSPB1 is important for dynamic intracellular trafficking during autophagy and mitophagy. Thus, inhibition of the HMGB1-HSPB1 pathway impairs mitophagy and elimination of damaged mitochondria in response to mitochondrial electron-transport-chain inhibitors.18 Third, extracellular reduced HMGB1 induces autophagy and tumor growth through AGER/RAGE (advanced glycosylation end product-specific receptor), whereas oxidized HMGB1 induces apoptosis in cancer cells.19 HMGB1 released from cancer cells induces autophagy in the muscle, which sustains anaerobic energy production (namely the Warburg effect) during tumor growth in vitro and in vivo.20 These findings suggest that HMGB1 is an important mediator of systemic autophagic syndrome.
Figure 1. HMGB1 is involved in autophagy and other stress responses. (A) HMGB1 plays important nuclear, cytosolic, and extracellular roles in the regulation of autophagy. (B and C) Various phenotypes of HMGB1 knockout (B) and knockin (C) mice with or without stress (indicated by lightning bolt).
HMGB1-Independent Autophagy
HMGB1 global knockout mice die shortly after birth due to the downregulation of glucocorticoid receptor and subsequent hypoglycemia, suggesting a critical role for HMGB1 in sustaining life.21 We and others recently generated transgenic mice with conditional knockout (Fig. 1B) or knockin (Fig. 1C) of HMGB1 within the pancreas,22 liver,23,24 heart,24,25 and myeloid cells26 through a different strategy. All these mice were viable and had no significant defects such as glucose and energy metabolism defects under unstressed growth conditions. However, these mice have various, even opposite, phenotypes in response to different stressors. For example, knockout of HMGB1 in the pancreas (n = 18–25 mice per group), liver (n = 6 mice per group), and myeloid cells (n = 6–9 mice per group) make mice more sensitive to sterile inflammation (e.g., pancreatitis22 and liver ischemic reperfusion23) and infection (e.g., lipopolysaccharide and L.monocytogenes26), partly through downregulation of autophagy26 and upregulation of mitochondrial injury23 and nuclear catastrophe.22 Knockin of HMGB1 in the heart protects mice against myocardial infarction.25 In contrast, a recent study from Robert Schwabe’s lab indicates that HMGB1 is not required for mitochondrial function and autophagy in the liver. In this study, the authors crossed HMGB1 conditional liver knockout mice with GFP-LC3 mice and then starved these mice for 24 h (n = 3 mice per group). The expression patterns of GFP-LC3 puncta and GFP-LC3 cleavage were similar between these mice upon starvation, suggesting that an HMGB1-independent autophagy system exists in the liver.24 Although the exact mechanism of this phenotype is not clear, a major difference between Robert Schwabe’s engineered HMGB1 mice and other groups is the tissue-level expression of HMGB1 after knockout. Mice with hepatocyte-specific deletion of Hmgb1 from Robert Schwabe’s lab are not complete conditional knockout mice; the protein level of HMGB1 in the liver is decreased by about 70%.24 Thus, autophagy appears to correlate with HMGB1 protein level, and low HMGB1 levels may still sustain autophagy pathway activation. Moreover, the original GFP-LC3 mice study by Mizushima et al. demonstrated that the regulation of autophagy is tissue/organ-dependent and not restricted to a starvation response at 24 or 48 h.27
Conclusions
It has become clear that HMGB1-dependent autophagy promotes chemotherapy resistance,11,12,28-35 sustains the tumor metabolism requirement19,20 and T cell survival,36 prevents polyglutamine aggregates37 and excitotoxicity,38 and protects against endotoxemia, bacterial infection, and ischemia-reperfusion injury.26,39-41 However, many questions remain unanswered regarding HMGB1-independent autophagy in the liver, including its tissue-specific role. HMGB1 dysfunction has been implicated in various forms of liver disease ranging from liver damage to fibrosis, as well as tumorigenesis.42 Extensive research is needed to determine the relationship between HMGB1, autophagy, and liver diseases. Of note, primary cells and cell lines have different baseline levels of autophagy as well as HMGB1 because transformed cell lines display different gene expression profiles.43 Understanding HMGB1-dependent and -independent autophagy in more detail will provide insight into the integrated stress response and guide HMGB1-based therapeutic intervention in cancer and other diseases.44
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Christine Heiner (Department of Surgery, University of Pittsburgh) for her critical reading of the manuscript. This work was supported by the National Institutes of Health (R01CA160417 to DT), 2013 Pancreatic Cancer Action Network-AACR Career Development Award (Grant Number 13-20-25-TANG), The National Natural Science Foundation-Guangdong Joint Fund (U1132005 to XS), and Science and Information Technology of Guangzhou Key Project (2011Y1-00038 to XS). Work done in support of findings reviewed in this manuscript was aided by the Core Support of the University of Pittsburgh Cancer Institute (P30CA047904).
Glossary
Abbreviations:
- ATG
autophagy-related
- AGER/RAGE
advanced glycosylation end product-specific receptor
- HMGB1
high mobility group box 1
- HSPB1
heat shock 27 kDa protein 1
- MAPK
mitogen-activated protein kinase
- NACC1
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
- ULK1
unc-51 like autophagy activating kinase 1
References
- 1.Goodwin GH, Sanders C, Johns EW. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem. 1973;38:14–9. doi: 10.1111/j.1432-1033.1973.tb03026.x. [DOI] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 7.Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013;14:143–51. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, Kang R, Zeh HJ, 3rd, Lotze MT, Tang D. DAMPs and autophagy: cellular adaptation to injury and unscheduled cell death. Autophagy. 2013;9:451–8. doi: 10.4161/auto.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, 3rd, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–92. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R, Vernon P, Cao L, Tang D. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72:230–8. doi: 10.1158/0008-5472.CAN-11-2001. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Cheng Y, Ren X, Zhang L, Yap KL, Wu H, Patel R, Liu D, Qin ZH, Shih IM, et al. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene. 2012;31:1055–64. doi: 10.1038/onc.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livesey KM, Kang R, Vernon P, Buchser W, Loughran P, Watkins SC, Zhang L, Manfredi JJ, Zeh HJ, 3rd, Li L, et al. p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Res. 2012;72:1996–2005. doi: 10.1158/0008-5472.CAN-11-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song JX, Lu JH, Liu LF, Chen LL, Durairajan SS, Yue Z, Zhang HQ, Li M. HMGB1 is involved in autophagy inhibition caused by SNCA/α-synuclein overexpression: a process modulated by the natural autophagy inducer corynoxine B. Autophagy. 2014;10:144–54. doi: 10.4161/auto.26751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang HS, Maric M. Lysosomal thiol reductase negatively regulates autophagy by altering glutathione synthesis and oxidation. Free Radic Biol Med. 2011;51:688–99. doi: 10.1016/j.freeradbiomed.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Liu K, Huang J, Xie M, Yu Y, Zhu S, Kang R, Cao L, Tang D, Duan X. MIR34A regulates autophagy and apoptosis by targeting HMGB1 in the retinoblastoma cell. Autophagy. 2014;10:442–52. doi: 10.4161/auto.27418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Wang S, Chen Y, Liu G, Yang X. miR-22 targets the 3′ UTR of HMGB1 and inhibits the HMGB1-associated autophagy in osteosarcoma cells during chemotherapy. Tumour Biol. 2014;35:6021–8. doi: 10.1007/s13277-014-1797-0. [DOI] [PubMed] [Google Scholar]
- 18.Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, Van Houten B, Zeh HJ, 3rd, Lotze MT. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab. 2011;13:701–11. doi: 10.1016/j.cmet.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, Benschop R, Sparvero LJ, Amoscato AA, Tracey KJ, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29:5299–310. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y, Yoneda J, Ohmori H, Sasaki T, Shimbo K, Eto S, Kato Y, Miyano H, Kobayashi T, Sasahira T, et al. Cancer usurps skeletal muscle as an energy repository. Cancer Res. 2014;74:330–40. doi: 10.1158/0008-5472.CAN-13-1052. [DOI] [PubMed] [Google Scholar]
- 21.Calogero S, Grassi F, Aguzzi A, Voigtländer T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22:276–80. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 22.Kang R, Zhang Q, Hou W, Yan Z, Chen R, Bonaroti J, Bansal P, Billiar TR, Tsung A, Wang Q, et al. Intracellular Hmgb1 inhibits inflammatory nucleosome release and limits acute pancreatitis in mice. Gastroenterology. 2014;146:1097–107. doi: 10.1053/j.gastro.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H, Nace GW, McDonald KA, Tai S, Klune JR, Rosborough BR, Ding Q, Loughran P, Zhu X, Beer-Stolz D, et al. Hepatocyte-specific high-mobility group box 1 deletion worsens the injury in liver ischemia/reperfusion: a role for intracellular high-mobility group box 1 in cellular protection. Hepatology. 2014;59:1984–97. doi: 10.1002/hep.26976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huebener P, Gwak GY, Pradere JP, Quinzii CM, Friedman R, Lin CS, Trent CM, Mederacke I, Zhao E, Dapito DH, et al. High-mobility group box 1 is dispensable for autophagy, mitochondrial quality control, and organ function in vivo. Cell Metab. 2014;19:539–47. doi: 10.1016/j.cmet.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitahara T, Takeishi Y, Harada M, Niizeki T, Suzuki S, Sasaki T, Ishino M, Bilim O, Nakajima O, Kubota I. High-mobility group box 1 restores cardiac function after myocardial infarction in transgenic mice. Cardiovasc Res. 2008;80:40–6. doi: 10.1093/cvr/cvn163. [DOI] [PubMed] [Google Scholar]
- 26.Yanai H, Matsuda A, An J, Koshiba R, Nishio J, Negishi H, Ikushima H, Onoe T, Ohdan H, Yoshida N, et al. Conditional ablation of HMGB1 in mice reveals its protective function against endotoxemia and bacterial infection. Proc Natl Acad Sci U S A. 2013;110:20699–704. doi: 10.1073/pnas.1320808110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livesey KM, Kang R, Vernon P, Buchser W, Loughran P, Watkins SC, Zhang L, Manfredi JJ, Zeh HJ, 3rd, Li L, et al. p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Res. 2012;72:1996–2005. doi: 10.1158/0008-5472.CAN-11-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y, Xie M, Yin X, Livesey KM, Lotze MT, et al. HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia: official journal of the Leukemia Society of America. Leukemia Research Fund, UK. 2011;25:23–31. doi: 10.1038/leu.2010.225. [DOI] [PubMed] [Google Scholar]
- 30.Zhao M, Yang M, Yang L, Yu Y, Xie M, Zhu S, Kang R, Tang D, Jiang Z, Yuan W, et al. HMGB1 regulates autophagy through increasing transcriptional activities of JNK and ERK in human myeloid leukemia cells. BMB Rep. 2011;44:601–6. doi: 10.5483/BMBRep.2011.44.9.601. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Yu Y, Kang R, Yang M, Xie M, Wang Z, Tang D, Zhao M, Liu L, Zhang H, et al. Up-regulated autophagy by endogenous high mobility group box-1 promotes chemoresistance in leukemia cells. Leuk Lymphoma. 2012;53:315–22. doi: 10.3109/10428194.2011.616962. [DOI] [PubMed] [Google Scholar]
- 32.Kang R, Tang D, Schapiro NE, Livesey KM, Farkas A, Loughran P, Bierhaus A, Lotze MT, Zeh HJ. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell Death Differ. 2010;17:666–76. doi: 10.1038/cdd.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan Z, Li Q, Wu P, Ye Y, Tseng HY, Zhang L, Zhang XD. Autophagy-mediated HMGB1 release antagonizes apoptosis of gastric cancer cells induced by vincristine via transcriptional regulation of Mcl-1. Autophagy. 2012;8:109–21. doi: 10.4161/auto.8.1.18319. [DOI] [PubMed] [Google Scholar]
- 34.Liu K, Huang J, Xie M, Yu Y, Zhu S, Kang R, Cao L, Tang D, Duan X. MIR34A regulates autophagy and apoptosis by targeting HMGB1 in the retinoblastoma cell. Autophagy. 2014;10:442–52. doi: 10.4161/auto.27418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ni Z, Dai X, Wang B, Ding W, Cheng P, Xu L, Lian J, He F. Natural Bcl-2 inhibitor (-)- gossypol induces protective autophagy via reactive oxygen species-high mobility group box 1 pathway in Burkitt lymphoma. Leuk Lymphoma. 2013;54:2263–8. doi: 10.3109/10428194.2013.775437. [DOI] [PubMed] [Google Scholar]
- 36.Zong M, Jorholt J, Winter J, Lindroos E, Harris HE, Lundberg IE. A8.24 autophagy may contribute to glucocorticoid resistance in myositis patients by maitaining muscle T cells homeostasis. Ann Rheum Dis. 2014;73(Suppl 1):A85–6. doi: 10.1136/annrheumdis-2013-205124.198. [DOI] [Google Scholar]
- 37.Min HJ, Ko EA, Wu J, Kim ES, Kwon MK, Kwak MS, Choi JE, Lee JE, Shin JS. Chaperone-like activity of high-mobility group box 1 protein and its role in reducing the formation of polyglutamine aggregates. J Immunol. 2013;190:1797–806. doi: 10.4049/jimmunol.1202472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pérez-Carrión MD, Ceña V. Knocking down HMGB1 using dendrimer-delivered siRNA unveils its key role in NMDA-induced autophagy in rat cortical neurons. Pharm Res. 2013;30:2584–95. doi: 10.1007/s11095-013-1049-9. [DOI] [PubMed] [Google Scholar]
- 39.Hagiwara S, Iwasaka H, Hasegawa A, Kudo K, Kusaka J, Oyama Y, Noguchi T. Infusion of a glucose solution reduces autophagy in the liver after LPS-induced systemic inflammation. Inflammation. 2012;35:249–58. doi: 10.1007/s10753-011-9311-y. [DOI] [PubMed] [Google Scholar]
- 40.Shen M, Lu J, Dai W, Wang F, Xu L, Chen K, He L, Cheng P, Zhang Y, Wang C, et al. Ethyl pyruvate ameliorates hepatic ischemia-reperfusion injury by inhibiting intrinsic pathway of apoptosis and autophagy. Mediators Inflamm. 2013;2013:461536. doi: 10.1155/2013/461536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang H, Liu A, Dahmen U, Dirsch O. Dual role of chloroquine in liver ischemia reperfusion injury: reduction of liver damage in early phase, but aggravation in late phase. Cell Death Dis. 2013;4:e694. doi: 10.1038/cddis.2013.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen R, Hou W, Zhang Q, Kang R, Fan XG, Tang D. Emerging role of high-mobility group box 1 (HMGB1) in liver diseases. Mol Med. 2013;19:357–66. doi: 10.2119/molmed.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordon K, Clouaire T, Bao XX, Kemp SE, Xenophontos M, de Las Heras JI, Stancheva I. Immortality, but not oncogenic transformation, of primary human cells leads to epigenetic reprogramming of DNA methylation and gene expression. Nucleic Acids Res. 2014;42:3529–41. doi: 10.1093/nar/gkt1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang R, Chen R, Zhang Q, Hou W, Wu S, Cao L, Huang J, Yu Y, Fan XG, Yan Z, et al. HMGB1 in health and disease. Mol Aspects Med. 2014 doi: 10.1016/j.mam.2014.05.001. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]