Abstract
Background
The best method to identify women with minor cervical lesions that require diagnostic work-up remains unclear. We performed a meta-analysis to assess the accuracy of p16INK4a immunocytochemistry compared to hrHPV DNA testing with hybrid capture II (HC2) to detect cervical intraepithelial neoplasia (CIN2+ and CIN3+) in women with a cervical cytology showing atypical squamous cells of undetermined significance (ASC-US) or low-grade cervical lesions (LSIL).
Methods
A literature search was performed in three electronic databases to identify studies eligible for this meta-analysis.
Results
Seventeen studies were included in the meta-analysis. The pooled sensitivity of p16INK4a to detect CIN2+ was 83.2% (95%CI: 76.8–88.2%) and 83.8% (95%CI: 73.5–90.6%) in ASC-US and LSIL cervical cytology respectively; pooled specificities were 71.0% (95%CI: 65.0–76.4%) and 65.7% (95%CI: 54.2–75.6%). Eight studies provided both HC2 and p16INK4a triage data. p16INK4a and HC2 have a similar sensitivity and p16INK4a has significantly higher specificity in the triage of women with ASC-US (relative sensitivity: 0.95 (95%CI: 0.89–1.01); relative specificity: 1.82 (95%CI: 1.57–2.12)). In the triage of LSIL, p16INK4a has a significantly lower sensitivity but higher specificity compared to HC2 (relative sensitivity: 0.87 (95%CI: 0.81–0.94); relative specificity: 2.74 (1.99–3.76)).
Conclusion
The published literature indicates an improved accuracy of p16INK4a compared to HC2 testing in the triage of ASC-US. In LSIL triage p16INK4a is more specific but less sensitive.
Keywords: cervical cancer, cervical intraepithelial neoplasia, ASCUS, LSIL, triage, p16INK4a, cyto-immunochemistry, HPV testing, diagnostic accuracy, systematic review, meta-analysis
Introduction
Cervical cancer is the third most common cancer in women worldwide. It is estimated that approximately 530,000 women developed cervical cancer and that 275,000 died from the disease in 20081. A well-organized screening for and management of precancerous lesions could reduce the incidence of cervical cancer2. Women with high-grade cervical abnormalities should be referred immediately to colposcopy or even treatment. However, the optimal management of women with atypical squamous cells of undetermined significance (ASC-US) or low-grade squamous intraepithelial lesions (LSIL) remains elusive and continues to be the object of intensive research.
Testing for carcinogenic HPV DNA has been proposed as a triage method to identify women at increased risk of cervical cancer precursors and cervical cancer. Numerous clinical studies, most prominently ALTS3 and a meta-analysis4 indicated that the hybrid capture II assay has improved accuracy (higher sensitivity, similar specificity) than repeat Pap testing to detect CIN2+ in women with ASC-US cytology. However, for LSIL, the possible advantages of HPV triage still remain unclear5. LSIL is the morphological correlate of a productive HPV infection6. Therefore, HPV-DNA testing nearly always yields positive results and cannot provide additional risk stratification to distinguish between women with or without underlying or developing high grade lesions7.
There is a lot of research on the development of objective biomarkers that can distinguish transforming from productive HPV infections and predict disease severity. The cellular tumor suppressor protein p16INK4a has been identified as a biomarker for transforming HPV infections. It is a cyclin-dependent kinase inhibitor that decelerates the cell cycle by inactivating the cyclin-dependent kinases (CDK4/6) involved in the phosporylation of the retinoblastoma protein (pRb)8. In the presence of the HR-HPV oncogene E7, p16INK4a transcription is induced by the histone demethylase KDM6B9 and not by a pRb feedback mechanism as previously assumed10;11. As a result, p16INK4a protein accumulates in the cell and this could be considered as a surrogate of a transforming infection.
Recently, an established immunocytochemical dual-staining protocol which simultaneously detects p16INK4a and Ki-67 expression has been established. The simultaneous detection of p16INK4a over expression with the proliferation marker Ki-67 within the same cervical epithelial cell indicates deregulation of the cell cycle, and does not require morphology-based interpretations12.
A previous meta-analysis demonstrated the correlation between the frequency of p16INK4a over expression and the severity of preneoplastic cervical lesions in cellular and tissue specimen13. No hypotheses regarding clinical applications of p16INK4a immunostaining were addressed in this systematic review13. Establishing a correlation between p16INK4a expression and severity of cancer precursors is a first step in the generation of evidence for potential clinical applications in screening for cervical cancer or in management of screen-positive women14. We therefore conducted a meta-analysis to explore the performance of p16INK4a immunocytochemistry in the triage of women with minor cytological cervical lesions.
Material and methods
PICOS question
Prior to literature search, a clinical question and corresponding PICOS were defined (Population – Index test – Comparator test – Outcomes – Studies):
Can p16INK4a be used to identify women with minor cytological abnormalities who need referral to colposcopy? Is it better than repeat cytology, HPV testing (HC2, other HPV assays) or other biomarkers? In other words: is p16INK4a immunocytochemistry a good triage test to manage women with ASC-US or LSIL?
Search strategy
Three electronic databases were searched – PubMed-MedLine, Embase and CENTRAL. The following search string was used in PubMed-MedLine: (cervix OR cervical OR vaginal) AND (cancer OR carcinoma OR dysplas* OR neoplasm* OR CIN OR SIL OR “pap smear” or cytology) AND (p16* OR p16INK4a OR protein p16 OR p16 protein). No language or publication date restrictions were applied.
The references of the retrieved articles were hand-searched in order to identify other eligible studies. Eligibility of inclusion or exclusion criteria was verified independently by two investigators (JR and MR). When no consensus could be reached, a third investigator was involved (MA). Extraction of the data was done by JR and checked by MA.
Inclusion and exclusion of studies
We included all studies that assessed p16INK4a immunostaining or p16INK4a/Ki-67 dual staining with or without hybrid capture 2 (HC2) testing as comparator test on liquid based cytology (LBC) or conventional cytology (CC) specimens showing ASC-US or LSIL cytology and where the diagnosis was verified with a reference standard. Studies were excluded if the population contained less than 20 women with ASC-US or LSIL cytology. If the data were not separated according to ASC-US or LSIL cytology, separate data were requested from the authors. When the authors did not respond, the studies were excluded. When duplicate publications of the same studies were found, the most comprehensive was included.
Participants
Two groups of participants were considered: women with equivocal cervical lesions or ASC-US (triage group I) and women with low-grade cytological lesions or LSIL (triage group II).
For the first group we considered women with atypical squamous cells of undetermined significance (ASCUS) as defined in the 1988 version of The Bethesda System (TBS)15. For studies using the TBS-2001 criteria, only the data for ASC-US cases were extracted. Studies reporting data exclusively on atypical squamous cells-favor reactive (ASC-R) or atypical squamous cells-cannot exclude high-grade squamous intraepithelial lesion (ASC-H) or atypical glandular cells (AGC) were excluded. For this meta-analysis only one term “ASC-US” was used for both versions of the Bethesda System.
For the second group we considered women with low-grade squamous intraepithelial lesions (LSIL). Studies that used the terminology of the British Society of Clinical Cytology (BSCC)16 were translated into TBS-1988. The BSCC terms borderline and mild dyskaryosis were considered as similar to ASCUS and LSIL respectively17.
Types of outcome measures
Outcome measures were defined prior to the literature search. The primary outcome was the absolute sensitivity and specificity of p16INK4a immunocytochemistry to detect underlying disease (CIN2+ or CIN3+/AIS) in the triage of women with equivocal or low-grade cytological abnormalities. The secondary outcome was the relative sensitivity and specificity of p16INK4a immunostaining versus hrHPV testing in studies with comparator testing.
Reference standard
We considered the following categories of reference standards:
Colposcopy and LLETZ or conization on all women
Colposcopy, punch biopsies of colposcopically suspicious areas and random biopsies of colposcopic normal zones on all women
Colposcopy and more than 1 biopsy on all women (type of biopsy unknown)
Colposcopy and one or more biopsies of colposcopic suspected zone. Women are considered free of CIN2+ if colposcopy is negative
Colposcopy and/or biopsy on all women (no further information)
Retrospective collection of biopsy/histology data
Data extraction and statistical analyses
Study characteristics and covariates that could influence study outcomes were tabled: primary p16INK4a antibody used, reference standard and positivity criterion for p16INK4a. The QUADAS-checklist for evaluation of the quality of diagnostic test studies was used as a tool to evaluate the quality of the studies18. The most important quality items that were reviewed in the QUADAS-checklist are the acceptability of the reference standard, the delay between tests, blinding of results, incorporation bias and verification bias18.
A pooling of the absolute accuracy of p16INK4a immunocytochemistry and hrHPV testing was done making use of the Stata-10 procedure, metandi (Stata Corp., College Station, Texas, US). This is a two-level mixed logistic regression model, with independent binomial distributions for the true positives and true negatives conditional on the sensitivity and specificity in each study, and a bivariate normal model for the logit transforms of sensitivity and specificity between studies19;20.
The relative sensitivity and specificity of p16INK4a compared to hrHPV testing was computed using metadas, a SAS (SAS Institute Inc., Cary, NC, USA) macro for meta-analysis of diagnostic accuracy studies which allows the inclusion of “test” as a covariate making comparison of two or more tests possible21;22.
Multivariate analyses for p16INK4a immunocytochemistry were done using metadas. Different covariates were included for test-positivity criterion used for p16INK4a, primary antibody, preparation method index cytology and the reference standard used.
Results
Included studies
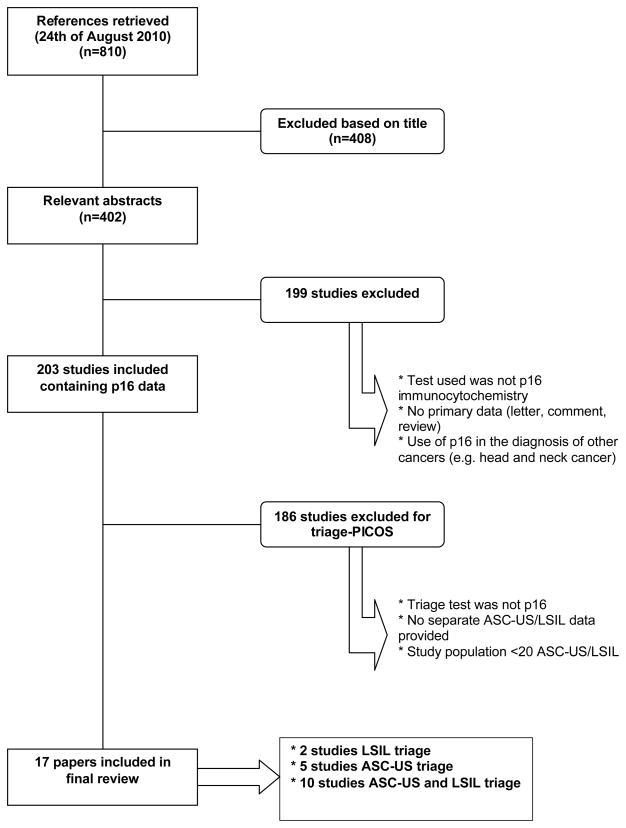
The electronic search yielded 810 articles (last search was performed on August 24, 2011). The majority of articles were found in PubMed-Medline (619). An additional 191 articles were retrieved from Embase. The CENTRAL database yielded no further results. The PRISMA flow-chart (Figure 1) shows the harvest of selected references and the reasons for exclusion. Finally, 17 reports were retained that contained data fulfilling the inclusion criteria allowing addressing the PICOS question.
Figure 1.
PRISMA-diagram: flowchart of study selection.
Two studies provided data on the accuracy of p16INK4a immunochemistry on women with LSIL cytology23;24, 5 on women with ASC-US cytology25–29 and another 10 studies on the triage of both ASC-US and LSIL cytology12;30–38. Study characteristics and technical information of included papers are shown in Table 1 and Table 2 respectively.
Table 1.
Study characteristics included studies
| Study | Country | Study size | Triage group | Triage tests | Outcomes | Gold Standard* |
|---|---|---|---|---|---|---|
| Nieh, 2005 | Taiwan | 66 | ASCUS | p16INK4a cytology | CIN2+ | 3 |
| HC2 | ||||||
| Holladay, 2006 | USA | 100 | ASC-US | p16INK4a cytology | CIN2+ | 6 |
| 100 | LSIL | HC2 | ||||
| Meyer, 2007 | USA | 28 | LSIL | p16INK4a cytology | CIN2+ | 5 |
| 15 | ASC-US | HC2 | ||||
| Monsonego, 2007 | France | 98 | ASC-US | p16INK4a cytology | CIN2+ | 3 |
| 105 | LSIL | HC2 | CIN3+ | |||
| Wentzensen, 2007 | France | 137 | ASCUS | p16INK4a cytology | CIN2+ | 3 |
| 88 | LSIL | |||||
| Schledermann, 2008 | Denmark | 43 | ASC | p16INK4a cytology | CIN2+ | 6 |
| Sweden | 36 | LSIL | ||||
| Szarewski, 2008 | UK | 104 | ASCUS | p16INK4a cytology | CIN2+ | 3 |
| 617 | LSIL | HC2 | CIN3+ | |||
| Denton, 2010 | Switzerland | 385 | ASC-US | p16INK4a cytology | CIN2+ | 6 |
| Italy | 425 | LSIL | HC2 | CIN3+ | ||
| Passamonti, 2010 | Italy | 91 | ASC-US | p16INK4a cytology | CIN2+ | 4 |
| 60 | LSIL | CIN3+ | ||||
| Samarawardana, 2010 | USA | 164 | ASC-US | p16INK4a cytology | CIN2+ | 4 |
| 42 | LSIL | |||||
| Sung, 2010 | Korea | 66 | ASC-US | p16INK4a cytology | CIN2+ | 3 |
| Tsoumpou, 2010 | Greece | 216 | LSIL | p16INK4a cytology | CIN2+ | 4 |
| Alameda, 2011 | Spain | 109 | ASCUS | p16INK4a cytology | CIN2+ | 4 |
| HC2 | ||||||
| Edgerton, 2011 | USA | 63 | ASC-US | Dual stain (p16INK4a/Ki-67) | CIN2+ | 6 |
| Guo, 2011 | USA | 65 | ASC-US | p16INK4a cytology | CIN2+ | 5 |
| CIN3+ | ||||||
| Nasioutziki, 2011 | Greece | 53 | ASCUS | p16INK4a cytology | CIN2+ | 5 |
| 277 | LSIL | HC2 | ||||
| Schmidt, 2011 | Switzerland | 361 | ASCUS | Dual stain (p16INK4a/Ki-67) | CIN2+ | 3 |
| Italy | 415 | LSIL | HC2 |
Different levels:
1= LLETZ/conization on all women
2=Punch biopsies of colposcopic abnormal zones and random biopsies of colpo-normal zones (on all women)
3= Colposcopy and (>=1) biopsy on all women included in the study (no further information given)
4= One or more biopsies of colposcopic-suspected zone; Women free of CIN lesion if colposcopy is negative
5= Colposcopy and/or biopsy, no further information given
6= Retrospective collection of biopsy/histology data (no further information given)
Table 2.
Technical details included studies.
| Study | p16INK4a antibody | Positivity criterion p16INK4a | Preparation method cytology | Collection device cytology |
|---|---|---|---|---|
| Nieh, 2005 | Clone E6H4 |
Nuclear/cytoplasmic staining ≥1 cytological abnormal cervical cell | Conventional cytology | Wooden spatula/cytobrush |
| Holladay, 2006 | Clone E6H4 |
Cytoplasmic/nuclear staining ≥1 cytological abnormal cervical cell | LBC (PreservCyt, ThinPrep) | ND |
| Meyer, 2007 | Clone E6H4 |
Nuclear/cytoplasmic staining ≥1 cytological abnormal cervical cell | LBC (PreservCyt, ThinPrep) | ND |
| Monsonego, 2007 | Clone E6H4 |
Nuclear/cytoplasmic staining ≥1 cytological abnormal cervical cell | LBC (PreservCyt, ThinPrep) | ND |
| Wentzensen, 2007 | Clone E6H4 |
Nuclear score* >2 | LBC (CYTO-screen system fixative fluid) | Flexible brush |
| Schledermann, 2008 | Clone E6H4 |
Nuclear staining ≥1 cytological abnormal cervical cell | LBC (ThinPrep, PreservCyt) | Plastic spatula Endocervical cytobrush |
| Szarewski, 2008 | Clone E6H4 |
Nuclear score* >2 | LBC (ThinPrep, PreservCyt) | Cervex broom |
| Denton, 2010 | Clone E6H4 |
Cytotechnologist 1 + pathologist: Presence ≥1 p16INK4a stained cervical cell Cytotechnologist 2: Nuclear score* ≥2 |
LBC | ND |
| Passamonti, 2010 | Clone JC8 | Nuclear/cytoplasmic staining ≥1 cytological abnormal cervical cell | 151 Conventional cytology 95 LBC (ThinPrep, PreservCyt) |
ND |
| Samarawardana, 2010 | 16P04 | Nuclear/cytoplasmic strong staining in ≥30 metaplastic, koilocytotic, or cytological equivocal cells | LBC (ThinPrep, PreservCyt) | Broom-like device |
| Sung, 2010 | Clone E6H4 |
Nuclear/cytoplasmic staining ≥1 cytological abnormal cervical cell | LBC | Cytobrush |
| Tsoumpou, 2010 | Clone E6H4 |
Nuclear/cytoplasmic staining ≥1 cytological abnormal cervical cell | LBC (ThinPrep, PreservCyt) | ND |
| Alameda, 2011 | Clone E6H4 |
Nuclear score* >2 | LBC (ND) | ND |
| Edgerton, 2011 | CINTec PLUS |
Simultaneous dual staining of ≥1 cervical cell | LBC (ND, SurePath) | ND |
| Guo, 2011 | Clone 6H12 |
Nuclear staining of ≥1 cytological abnormal cervical cell with/without cytoplasmic staining | LBC (SurePath) | ND |
| Nasioutziki, 2011 | Clone E6H4 |
Nuclear score* >2 | LBC (PreservCyt; ThinPrep) | Ayre’s spatula &cytobrush |
| Schmidt, 2011 | CINtec Plus Kit Clone E6H4 Clone 274-11 AC3 |
Simultaneous dual staining of ≥1 cervical cell | LBC (ThinPrep, PreservCyt) | ND |
Scoring system Bergeron, C. / Wentzensen, N 39:
Nuclear staining, four criteria:
A/ increased size
B/ granular or hyperchromatic chromatin
C/ irregular shape
D/ variable morphology from cell to cell
Positivity for any of these criteria→score 2; positivity for A + other criterion →score 3; positivity for A + >1 other criterion →4
In two studies12;26 p16INK4a/Ki-67 dual staining using CINtec Plus kit was performed, the other 15 studies23–25;27–38 applied single p16INK4a imumnocytochemistry. Twelve studies23–25;28–33;36–38 used clone E6H4 as a primary antibody for p16INK4a, other primary antibodies used were Clone 6H1227, Clone JC834 and 16P0435. Positivity criteria of p16INK4a immunostaining differed between the studies. Five studies25;30;33;37;38 made use of the nuclear scoring proposed by Wentzensen and Bergeron39. This scoring system takes into account nuclear staining and nuclear abnormalities (increased size, granular/hyperchromatic chromatin, irregular shape or variable morphology from cell to cell). When a cervical cell shows nuclear p16INK4a staining and one of the nuclear abnormalities mentioned above, a score of 2 is given. If the stained nucleus shows an increased size and 1 or more nuclear abnormality, a score of 3 and 4 is given respectively. A nuclear score of >2 or ≥2 is used as a cut-off for p16INK4a positivity. For the studies that applied p16INK4a/Ki-67 dual staining, simultaneous red nuclear- and brown cytoplasmic staining in at least one cervical cell was set as the positivity criterion12;26. The presence of staining in 1 or more or 30 or more cytological abnormal cervical cell was interpreted as a positive p16INK4a reaction in the remaining 10 studies. However, there was a difference in the localization of the immunostaining. Two studies27;36 only considered nuclear staining as a positive p16INK4a staining reaction while 8 studies23;24;28;29;31;32;34;35 considered both nuclear and/or cytoplasmic staining as a positive reaction.
Triage of atypical cells of undetermined significance
Fifteen studies contained accuracy data for p16INK4a immunostaining in the triage of women with ASC-US cytology12;25–38. A total of 1740 women were enrolled. Eight studies performed a direct comparison with HC2 triage data12;25;28;30–33;37. The study of Denton et al.30 provided p16INK4a immunocytochemistry data interpreted independently by 2 pathologists and 1 cytotechnologist. To avoid that this study should contribute too much influence each interpretation was weighted with a factor 0.33.
Absolute accuracy p16INK4a-triage
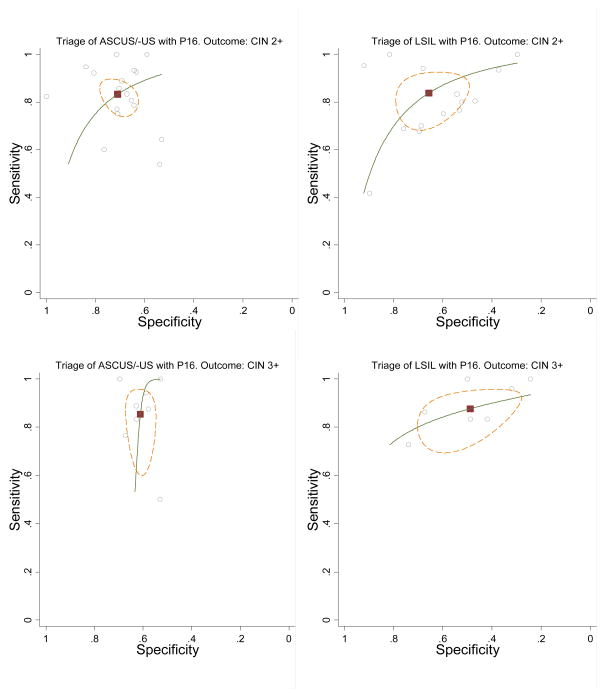
The pooled estimated absolute sensitivity and specificity values and their 95% confidence interval (CI) are shown in Table 3. The pooled sensitivity was 83.2% (95%CI: 76.8–88.2%) and 85.4% (95%CI: 71.7–93.1%) for an outcome of CIN2+ and CIN3+ respectively. To predict the absence of CIN2+ or CIN3+, the pooled absolute specificity was 71.0% (95%CI: 65.0–76.4%) and 61.1% (95%CI: 57.2–64.9%) respectively. The hierarchical summary receiver-operator curve (HSROC)-curve for p16INK4a triage for an outcome of CIN2+ is shown in Figure 2.
Table 3.
Pooled absolute accuracy estimates of p16INK4a and HC2 in the triage of women with ASCUS or LSIL cytology.
| Test | Triage group | Outcome | N° of studies | Parameter | Accuracy (%) |
|---|---|---|---|---|---|
|
| |||||
| p16INK4a | ASCUS/-US | CIN2+ | 17* | Sensitivity | 83.2 (76.8–88.2) |
| 17* | Specificity | 71.0 (65.0–76.4) | |||
| CIN3+ | 8 | Sensitivity | 85.4 (71.7–93.1) | ||
| 8 | Specificity | 61.1 (57.2–64.9) | |||
| LSIL | CIN2+ | 14 | Sensitivity | 83.8 (73.5–90.6) | |
| 14 | Specificity | 65.7 (54.2–75.6) | |||
| CIN3+ | 7 | Sensitivity | 87.7 (78.6–93.2) | ||
| 7 | Specificity | 48.9 (36.2–61.7) | |||
|
| |||||
| HC2 | ASCUS/-US | CIN2+ | 8 | Sensitivity | 91.6 (85.9–95.1) |
| 8 | Specificity | 40.5 (33.5–47.9) | |||
| CIN3+ | 3 | Sensitivity | 92.2 (85.1–99.4) § | ||
| 3 | Specificity | 41.0 (33.1–48.8) § | |||
| LSIL | CIN2+ | 7 | Sensitivity | 99.5 (82.6–100.0) | |
| 7 | Specificity | 28.9 (16.4–45.6) | |||
| CIN3+ | 3 | Sensitivity | 98.6 (95.9–101.3) § | ||
| 3 | Specificity | 22.5 (15.3–29.6) § | |||
There were only 15 papers reporting p16INK4a data but Denton, 201030 reported the results of 3 independent p16INK4a tests (2 performed by 2 different pathologists and 1 performed by a cytotechnologist).
A minimum of 4 studies is required to perform a metandi-analysis / Metandi doesn’t perform analysis. Therefore the pooled sensitivity and specificity were computed separately with a random pooling making use of the STATA metan command.
Figure 2.
Meta-analysis of the sensitivity and specificity of p16INK4a immunostaining in the triage of women with ASC-US (left) or LSIL (right) to detect CIN2+ (top) and CIN3+ (bottom). Black square: summary point, small circles: individual studies; green line: SROC curve; interrupted brown line: 95% confidence ellipse.
Relative accuracy of p16INK4a- versus HC2-triage
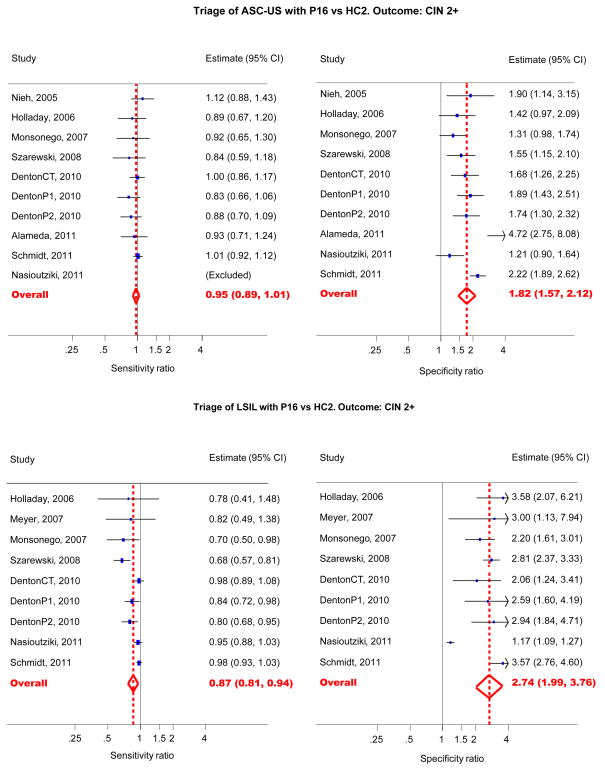
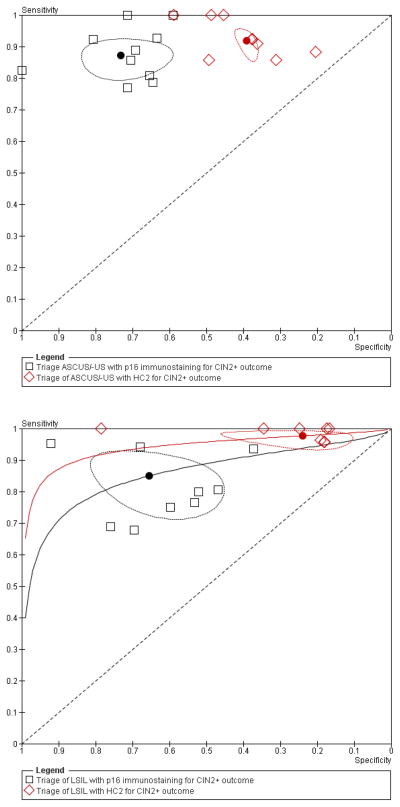
The relative accuracy measures and their CI’s are shown in Table 4. The relative sensitivity of p16INK4a versus HC2 for CIN2+ and CIN3+ was 0.95 (95%CI: 0.89–1.01) and 0.98(95% CI: 0.86–1.12) respectively. The relative specificity was 1.82 (95% CI: 1.57–2.12) and 1.64 (95% CI: 1.44–1.87) for predicting the absence of CIN2+ or CIN3+ respectively. The corresponding HSROC curve is shown in Figure 4. In the upper graph the two summary points are almost on the same height (equal sensitivity) but the summary point of p16INK4a is located more to the left (higher specificity) than that of HC2. This means that HC2 and p16INK4a have an equal sensitivity in the triage of ASC-US to detect CIN2+, however, the specificity of p16INK4a is higher than the specificity of HC2.
Table 4.
Pooled relative accuracy of p16INK4a vs HC2 in the triage of women with ASCUS or LSIL cytology.
| Triage group | Outcome | Parameter | Ratio (p16INK4a vs HC2) | p-value |
|---|---|---|---|---|
|
| ||||
| ASCUS/-US | CIN2+ | Sensitivity | 0.95 (0.89–1.01) | 0.1287 |
| Specificity | 1.82 (1.57–2.12) | <0.0001 | ||
| CIN3+ | Sensitivity* | 0.98 (0.86–1.12) | 0.780 | |
| Specificity* | 1.64 (1.44–1.87) | 0.000 | ||
|
| ||||
| LSIL | CIN2+ | Sensitivity | 0.87 (0.81–0.94) | 0.0002 |
| Specificity | 2.74 (1.99–3.76) | <0.0001 | ||
| CIN3+ | Sensitivity | 0.88 (0.81–0.95) | 0.0013 | |
| Specificity | 2.81 (2.38–3.33) | <0.0001 | ||
The metadas SAS macro failed to converge. Therefore the pooled relative sensitivity and specificity were computed separately as ratios of 2 proportions using a random effect model.
Figure 4.
Forest plot sensitivity (left) and specificity (right) ratios of p16INK4a triage versus HC2 in women with ASC-US (top) or LSIL (bottom) to detect CIN2+.
Triage of low grade squamous intraepithelial lesions
Two thousand nineteen women were enrolled in 12 studies12;23;24;30–38 reporting p16INK4a triage accuracy data for LSIL. Seven studies allowed comparison of p16INK4a with HC2 triage12;23;30–33;37.
Absolute accuracy p16 INK4a -triage
The pooled absolute sensitivity was similar to that in the triage of ASC-US, with 83.8% (95%CI: 73.5–90.6%) and 87.7% (95%CI: 78.6–93.2%) to predict respectively CIN2+ or CIN3+ lesions. The absolute specificity to predict the absence of CIN2+ or CIN3+ lesions was a bit lower than in ASC-US triage, pooled estimates were respectively 65.7% (95% CI: 54.2–75.6%) and 48.9% (95%CI: 36.2–61.7%). (Table 3, Figure 2)
Relative accuracy p16 INK4a - versus HC2-triage
In contrast with ASC-US triage, p16INK4a showed a lower sensitivity than HC2 to predict CIN2+ or CIN3+ lesions. The relative sensitivity for CIN2+ and CIN3+ lesions was 0.87 (95%CI: 0.81–0.94) and 0.88 (95%CI: 0.81–0.95) respectively. In concordance to ASC-US triage, p16INK4a showed a statistically significantly higher specificity than HC2 with pooled values of 2.74 (95%CI: 1.99–3.76) and 2.81 (95% CI: 2.38–3.33) for CIN2+ and CIN3+ outcome respectively. The corresponding HSROC curve is shown in Figure 4, lower graph. The summary point of p16INK4a is located lower (lower sensitivity) and more to the left (higher specificity) than that of HC2 testing, which means that there is a difference in sensitivity and specificity between p16INK4a and HC2 to triage LSIL. p16INK4a-triage has a higher specificity but a lower sensitivity than HC2 to detect CIN2+ lesions in women with LSIL cytology. (Table 4, Figure 4 and Figure 6)
Influence of study characteristics
The multivariate analysis showed a higher sensitivity and specificity for studies that used the nuclear scoring system to interpret p16INK4a results and studies that applied dual staining for p16INK4a and Ki-67 compared to studies that only looked at simple p16INK4a expression in cytologically abnormal cells (Table 6). However, these differences were not statistically significant (p>0.05).
The studies that applied both p16INK4a and HC2 triage tests showed no significant differences in sensitivity and equal specificity compared to studies that only assessed p16INK4a immunocytochemistry. The type of p16INK4a antibody used also did not significantly influence the accuracy measures.
Discussion
Our meta-analysis showed better accuracy of p16INK4a triage of ASC-US than HC2 (similar sensitivity but better specificity) considering both outcomes CIN2+ and CIN3+. In LSIL triage, p16INK4a-staining was more specific than HC2 but less sensitive.
Triage of ASC-US
It has been shown in large randomized trials and meta-analyses that HC2 performs better than repeat cytology to triage women with ASC-US4;5;40;41. Nevertheless, the triage specificity of HC2 still is not optimal (often in the 40–60% range), resulting in colposcopy referral of many women without disease. With a pooled specificity of 71% (1.82 times higher than HC2), p16INK4a immunostaining appears to be a test that meets the demand for a more specific triage test without loosing sensitivity. The specificity of HC2 in ASC-US triage including 8 studies (40.5%, 95%CI: 33.5–47.9%) seems lower in our meta-analysis compared to previous meta-analysis including 20 studies (62.5%, 95% CI: 57.8–67.3%)5, but not so different from the specificity reported in the ALTS study (48%)3, which could be explainable by differences in age composition of study populations. Age could not be controlled for throughout previous meta-analyses since age-stratified data were not sufficiently reported in the included studies. However, within each of the 8 studies included in our meta-analysis, age could not cause bias since the two compared tests were done on the same women.
Triage of LSIL
HC2 does not perform well in many studies because of its very low specificity7;40;42. However, these findings are not universal and depend on quality of cytological interpretation and the HPV test used. In our meta-analysis, the pooled specificity values for HC2 were very similar to previous meta-analyses (in the range 22% to 28% for CIN3+ and CIN2+ outcomes5. There is clearly a need for more specific assays universally usable in triage of LSIL, which are as sensitive as and more specific than HC2. Our meta-analysis shows that p16INK4a is indeed more specific, but in contrast to triage of ASC-US, it is less sensitive.
Influence of study characteristics
The use of p16INK4a immunocytochemistry in clinical applications remains controversial due to the variation in procedures used. The most important difference between the different studies is the interpretation of p16INK4a expression31. Since a purely color-based approach to identify abnormal cells in cervical smears using p16INK4a is hampered by the fact that few normal endocervical, squamous metaplastic, or atrophic cells also may display some p16INK4a expression, Wentzensen et al.38 defined morphologic criteria that would enable scoring of p16INK4a –positive squamous cells. A major concern of using morphology-based biomarkers is achieving adequate reproducibility. While the nuclear scoring showed high reproducibility in the initial reports38–39, it was not consistently applied in subsequent studies, and reproducibility was not evaluated on a larger scale. The recent p16INK4a/Ki-67 dual staining could eliminate the need for a standardized methodology because it allows identifying cells with deregulated cell cycle in cervical cytology specimens independent of morphology-based parameters. We presumed that the studies applying the nuclear scoring system or p16INK4a/Ki-67 dual staining would have a greater accuracy (higher sensitivity and specificity) to identify women with CIN2+ compared to studies that only looked at simple p16INK4a expression in cytological abnormal cells without scoring. Multivariate analyses showed higher sensitivity and specificity of the ASC-US studies applying nuclear scoring or dual staining compared to those applying simple p16INK4a immunostaining but, in general, these differences were not statistically significant. Only the specificity of p16INK4a immunostaining with nuclear scoring in women with ASC-US was significantly higher compared to the other studies (p=0.04). p16INK4a/Ki-67 dual staining was used in only 2 ASC-US-triage studies12;26. One study12 reported excellent sensitivity (92%) and specificity (81%) for CIN2+ using dual staining, where the sensitivity was similar to that of HC2 (ratio 1.01, 95% CI: 0.92–1.16) but with increased specificity (ratio 2.22, 95% CI: 1.89–2.62). Another study26 using dual staining showed substantially lower sensitivity (64%) and specificity (53%) for the same outcome without comparison with HC2. This could be due to the fact that this study did not follow the manufacturer’s instructions for CINtec PLUS dual staining. In LSIL triage only one study used dual staining with similar findings as for ASC-US-triage: high sensitivity (94%) and rather good specificity (68%) for CIN2+ which was similar to sensitivity of HC2 (ratio 0.98, 95% CI: 0.93–1.03) but with higher specificity than HC2 (ratio 3.57, 95% CI: 2.76–4.60)12.
The gold standard used can influence accuracy estimates of the triage test. In this meta-analysis we considered colposcopy and histology as the gold standard and distinguished 6 types of verification. However, none of these methods of verification influenced significantly accuracy estimates of triage. In addition, staining of biopsies also can impact on the outcome assessment. Two studies12;30 used p16 immunohistochemistry in addition to the normal haematoxylin & eosin (HE) staining for the histological interpretation of biopsies. Previous studies have shown that this improved gold standard increases the sensitivity of the histological interpretation43;44. Multivariate analysis showed no significant difference in absolute sensitivity of triage using p16INK4a immunocytochemistry between studies that used HE staining compared to p16INK4a staining of biopsies (p=0.17 and p=0.22 for ASC-US and LSIL respectively). Furthermore, outcome adjudication using p16 will bias results in favor of p16 cytology because of autocorrelation.
Future research on triage of ASC-US and LSIL
The meta-analysis presented in this paper is part of an international effort including a series of ongoing meta-analyses addressing the accuracy of triage of minor cytological abnormalities using other methods, such as other hrHPV DNA tests than HC2, assays detecting viral RNA, picking up a restricted number HPV types (in particular HPV types 16 and 18), as well as other protein markers such as ProExC BD Diagnostics—TriPath, Burlington, NC, USA). All these meta-analyses will address questions of follow-up of screen-positive women participating in cytology-based screening. Investigators and authors should be recommended to follow STARD guidelines for good diagnostic research involving application of one or more markers followed by verification with colposcopy and colposcopy-targeted biopsies with or without additional random punch biopsies for all patients with ASC-US and LSIL14;45. This gold standard verification should preferentially be blinded to the results of the markers and take place in a short delay (<10 weeks) to avoid development of disease after the triage tests. Future research should also target longitudinal outcomes, in particular the risk of developing CIN3 in women triage+ and triage- results over 3 to 5 years (longitudinal PPV and 1-NPV).
Conclusion
Based on the currently published data, we can conclude that p16INK4a immunocytochemistry could be recommended for use in the triage of women with ASC-US due to the higher specificity without loss of sensitivity compared to HC2 testing. In LSIL triage, p16INK4a is less sensitive but more specific than HC2. It can therefore be used as a first step triage justifying further diagnostic work-up of p16INK4a-positive women. However, women with LSIL testing p16INK4a negative cannot be referred back to normal screening. Those women should be re-invited for a repeat testing. Dual staining in LSIL triage could be as sensitive as HC2 but this was observed in only one observational study, which is insufficient to justify clinical recommendations. More studies using the dual stain are currently ongoing and may have an influence on the current conclusions.
Figure 3.
HSROC plot of the relative sensitivity and specificity of p16INK4a immunostaining versus HC2 in the triage of women with ASC-US (top) or LSIL (bottom) to detect CIN2+ lesions.
Table 5.
Multivariate meta-analysis of the absolute sensitivity and specificity of p16INK4a triage of ASC-US and LSIL for an outcome of CIN2+ according to covariates
| Triage group | Covariate | Covariate level | N° studies | Sensitivity (%) | p-value | Specificity (%) | p-value |
|---|---|---|---|---|---|---|---|
|
| |||||||
| ASC-US | Test cutoff criterion | p16INK4a expression in >1 cell | 10 | 81.6 (70.2–89.3) | REF | 66.8 (62.3–70.9) | REF |
| NS>2 | 5 | 85.9 (75.5–92.3) | 0.504* | 83.1 (60.8–94.0) | 0.036* | ||
| Dual staining >1 cell | 2 | 84.6 (69.1–93.1) | 0.696* | 70.2 (56.7–80.9) | 0.595* | ||
|
| |||||||
| Nb of triage tets evaluated | Both triage tests§ | 10 | 87.0 (81.1–91.3) | 0.119* | 73.3 (62.9–81.7) | 0.452* | |
| Only p16INK4a testing** | 7 | 77.3 (65.1–86.1) | REF | 68.7 (60.9–75.6) | REF | ||
|
| |||||||
| LSIL | Test cutoff criterion | p16INK4a expression in >1 cell | 9 | 79.7 (65.8–88.9) | REF | 58.9 (46.1–70.7) | REF |
|
| |||||||
| NS>2 | 4 | 82.1 (65.9–91.6) | 0.778* | 77.1 (56.4–89.7) | 0.086* | ||
|
| |||||||
| Dual staining >1 cell | 1 | 94.4 (55.0–99.6) | 0.106* | 68.0 (45.5–84.4) | 0.445* | ||
|
| |||||||
| Nb of triage tets evaluated | Both triage tests | 9 | 85.2 (74.4–91.1) | 0.644* | 64.1 (49.3–76.6) | 0.681* | |
|
| |||||||
| Only p16INK4a testing | 5 | 80.2 (54.9–93.1) | REF | 68.7 (49.9–82.8) | REF | ||
Compared to reference covariate level (ref)
Studies that assessed both p16INK4a immunostaining and HC2
Studies that only assessed p16INK4a immunostaining
Acknowledgments
Financial support was received from: (1) the European Commission through the PREHDICT Network, coordinated by the Free University of Amsterdam (the Netherlands), funded by the 7th Framework program of DG Research (Brussels, Belgium), and through the ECCG (European Cooperation on development and implementation of Cancer screening and prevention Guidelines, via IARC, Lyon, France), funded by Directorate of SANCO (Luxembourg, Grand-Duchy of Luxembourg); (2) The Belgian Foundation Against Cancer, Brussels, Belgium; (3) the Gynaecological Cancer Cochrane Review Collaboration (Bath, United Kingdom).
The authors acknowledge M. Nasioutziki, M. Guo 2010 for the provision of additional data.
References
- 1.Arbyn M, Castellsagué X, de Sanjosé S, et al. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–86. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 2.Arbyn M, Rebolj M, de Kok IM, et al. The challenges for organising cervical screening programmes in the 15 old member states of the European Union. Eur J Cancer. 2009;45:2671–8. doi: 10.1016/j.ejca.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 3.Solomon D, Schiffman MA, Tarone B. Comparison of three management strategies for patients with atypical squamous cells of undetermined significance (ASCUS): baseline results from a randomized trial. J Natl Cancer Inst. 2001;93:293–9. doi: 10.1093/jnci/93.4.293. [DOI] [PubMed] [Google Scholar]
- 4.Arbyn M, Buntinx F, Van Ranst M, et al. Virologic versus cytologic triage of women with equivocal Pap smears: a meta-analysis of the accuracy to detect high-grade intraepithelial neoplasia. J Natl Cancer Inst. 2004;96:280–93. doi: 10.1093/jnci/djh037. [DOI] [PubMed] [Google Scholar]
- 5.Arbyn M, Sasieni P, Meijer CJ, et al. Chapter 9: Clinical applications of HPV testing: a summary of meta-analyses. Vaccine. 2006;24(SUPPL 3):S78–S89. doi: 10.1016/j.vaccine.2006.05.117. [DOI] [PubMed] [Google Scholar]
- 6.Zuna RE, Wang SS, Rosenthal DL, et al. Determinants of human papillomavirus-negative, low-grade squamous intraepithelial lesions in the atypical squamous cells of undetermined significance/low-grade squamous intraepithelial lesions triage study (ALTS) Cancer. 2005;105:253–62. doi: 10.1002/cncr.21232. [DOI] [PubMed] [Google Scholar]
- 7.Arbyn M, Martin-Hirsch P, Buntinx F, et al. Triage of women with equivocal or low-grade cervical cytology results. A meta-analysis of the HPV test positivity rate. J Cell Mol Med. 2009;13:648–59. doi: 10.1111/j.1582-4934.2008.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benevolo M, Vocaturo A, Mottolese M, et al. Clinical role of p16INK4a expression in liquid-based cervical cytology: correlation with HPV testing and histologic diagnosis. Am J Clin Pathol. 2008;129:606–12. doi: 10.1309/BEPQXTCQD61RGFMJ. [DOI] [PubMed] [Google Scholar]
- 9.McLaughlin-Drubin ME, Crum CP, Munger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci U S A. 2011;108:2130–5. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klaes R, Friedrich T, Spitkovsky D, et al. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer. 2001;92:276–84. doi: 10.1002/ijc.1174. [DOI] [PubMed] [Google Scholar]
- 11.Wentzensen N, von Knebel DM. Biomarkers in cervical cancer screening. Dis Markers. 2007;23:315–30. doi: 10.1155/2007/678793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt D, Bergeron C, Denton KJ, Ridder R. p16/ki-67 dual-Stain cytology in the triage of ASCUS and LSIL papanicolaou cytology: Results from the european equivocal or mildly abnormal papanicolaou cytology study. Cancer Cytopathol. 2011;119:158–66. doi: 10.1002/cncy.20140. [DOI] [PubMed] [Google Scholar]
- 13.Tsoumpou I, Arbyn M, Kyrgiou M, et al. p16INK4a immunostaining in cytological and histological specimens from the uterine cervix: a systematic review and meta-analysis. Cancer Treat Rev. 2009;35:210–20. doi: 10.1016/j.ctrv.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arbyn M, Ronco G, Cuzick J, Wentzensen N, Castle PE. How to evaluate emerging technologies in cervical cancer screening? Int J Cancer. 2009;125:2489–96. doi: 10.1002/ijc.24774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundberg GD National Cancer Institute. The 1988 Bethesda System for Reporting Cervical/Vaginal Cytologic Diagnoses. JAMA. 1989;262:931–4. [PubMed] [Google Scholar]
- 16.Evans DM, Hudson EA, Brown CL, et al. Terminology in gynaecological cytopathology: report of the Working Party of the British Society for Clinical Cytology. J Clin Pathol. 1986;39:933–44. doi: 10.1136/jcp.39.9.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudding N, Sutton J. BSCC Terminology Conference, Koilocytosis and Mild Dyskaryosis. Cytopathology. 2002;13:379–81. doi: 10.1046/j.1365-2303.2002.00448_1.x. [DOI] [PubMed] [Google Scholar]
- 18.Whiting P, Rutjes AWS, Reitsma JB, Bossuyt PM, Kleijnen J. The development of QUADAS : a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol. 2003;3:1–13. doi: 10.1186/1471-2288-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harbord RM, Deeks JJ, Egger M, Whiting P, Sterne JA. A unification of models for meta-analysis of diagnostic accuracy studies. Biostatistics. 2007;8:239–51. doi: 10.1093/biostatistics/kxl004. [DOI] [PubMed] [Google Scholar]
- 20.Harbord RM, Whiting P. metandi: Meta-analysis of diagnostic accuracy using hierarchical logistic regression. The Stata Journal. 2009;9:211–29. [Google Scholar]
- 21.Takwoingi Y Diagnostic Test Accuracy Working Group. METADAS: A SAS macro for meta-analysis of diagnostic accuracy studies. 2009 [Accessible via: http://srdta.cochrane.org/Files/Website/METADAS_Readme_v1.0_beta.pdf]
- 22.Diagnostic Test Accuracy Working Group. Handbook for Diagnostic Test Accuracy Reviews. 2011 Available from: http://srdta.cochrane.org/handbook-dta-reviews.
- 23.Meyer JL, Hanlon DW, Andersen BT, Rasmussen OF, Bisgaard K. Evaluation of p16(INK4a) expression in ThinPrep cervical specimens with the CINtec p16(INK4a) assay: correlation with biopsy follow-up results. Cancer. 2007;111:83–92. doi: 10.1002/cncr.22580. [DOI] [PubMed] [Google Scholar]
- 24.Tsoumpou I, Valasoulis G, Founta C, et al. High-risk human papillomavirus DNA test and p16(INK4a) in the triage of LSIL: A prospective diagnostic study. Gynecol Oncol. 2010;121:49–53. doi: 10.1016/j.ygyno.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Alameda F, Alameda F, Piujan L, et al. The value of p16 in ASCUS cases: A retrospective study using frozen cytologic material. Diagn Cytopathol. 2011;39:110–4. doi: 10.1002/dc.21349. [DOI] [PubMed] [Google Scholar]
- 26.Edgerton N, Cohen C, Siddiqui MT. Evaluation of CINtec PLUS(R) testing as an adjunctive test in ASC-US diagnosed SurePath(R) preparations. Diagn Cytopathol. 2011 doi: 10.1002/dc.21757. in-press. [DOI] [PubMed] [Google Scholar]
- 27.Guo M, Warriage I, Mutyala B, et al. Evaluation of p16 immunostaining to predict high-grade cervical intraepithelial neoplasia in women with Pap results of atypical squamous cells of undetermined significance 68. Diagn Cytopathol. 2011;39:482–8. doi: 10.1002/dc.21415. [DOI] [PubMed] [Google Scholar]
- 28.Nieh S, Chen SF, Chu TY, et al. Is p16(INK4A) expression more useful than human papillomavirus test to determine the outcome of atypical squamous cells of undetermined significance-categorized Pap smear? A comparative analysis using abnormal cervical smears with follow-up biopsies. Gynecol Oncol. 2005;97:35–40. doi: 10.1016/j.ygyno.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 29.Sung CO, Kim SR, Oh YL, Song SY. The use of p16(INK4A) immunocytochemistry in “Atypical squamous cells which cannot exclude HSIL” compared with “Atypical squamous cells of undetermined significance” in liquid-based cervical smears. Diagn Cytopathol. 2010;38:168–71. doi: 10.1002/dc.21164. [DOI] [PubMed] [Google Scholar]
- 30.Denton KJ, Bergeron C, Klement P, et al. The Sensitivity and Specificity of p16INK4a Cytology vs HPV Testing for Detecting High-Grade Cervical Disease in the Triage of ASC-US and LSIL Pap Cytology Results. Am J Clin Pathol. 2010;134:12–21. doi: 10.1309/AJCP3CD9YKYFJDQL. [DOI] [PubMed] [Google Scholar]
- 31.Holladay EB, Logan S, Arnold J, Knesel B, Smith GD. A comparison of the clinical utility of p16(INK4a) immunolocalization with the presence of human papillomavirus by hybrid capture 2 for the detection of cervical dysplasia/neoplasia. Cancer. 2006;108:451–61. doi: 10.1002/cncr.22284. [DOI] [PubMed] [Google Scholar]
- 32.Monsonego J, Pollini G, Evrard MJ, et al. P16(INK4a) immunocytochemistry in liquid-based cytology samples in equivocal Pap smears: added value in management of women with equivocal Pap smear. Acta Cytol. 2007;51:755–66. doi: 10.1159/000325839. [DOI] [PubMed] [Google Scholar]
- 33.Nasioutziki M, Daniilidis A, Dinas K, et al. The evaluation of p16INK4a immunoexpression/immunostaining and human papillomavirus DNA test in cervical liquid-based cytological samples. Int J Gynecol Cancer. 2011;21:79–85. doi: 10.1097/IGC.0b013e3182009eea. [DOI] [PubMed] [Google Scholar]
- 34.Passamonti B, Gustinuci D, Rechia P, et al. Expression of p16 in abnormal pap-tests as an indicator of CIN2+ lesions: a possible role in the low grade ASC/US and L/Sil (lg) cytologic lesions for screening prevention of uterine cervical tumours. Pathologica. 2010;102:6–11. [PubMed] [Google Scholar]
- 35.Samarawardana P, Dehn DL, Singh M, et al. p16(INK4a) is superior to high-risk human papillomavirus testing in cervical cytology for the prediction of underlying high-grade dysplasia. Cancer Cytopathol. 2010;118:146–56. doi: 10.1002/cncy.20078. [DOI] [PubMed] [Google Scholar]
- 36.Schledermann D, Andersen BT, Bisgaard K, et al. Are adjunctive markers useful in routine cervical cancer screening? Application of p16(INK4a) and HPV-PCR on ThinPrep samples with histological follow-up. Diagn Cytopathol. 2008;36:453–9. doi: 10.1002/dc.20822. [DOI] [PubMed] [Google Scholar]
- 37.Szarewski A, Ambroisine L, Cadman L, et al. Comparison of Predictors for High-Grade Cervical Intraepithelial Neoplasia in Women with Abnormal Smears. Cancer Epidemiol Biomarkers Prev. 2008;17:3033–43. doi: 10.1158/1055-9965.EPI-08-0508. [DOI] [PubMed] [Google Scholar]
- 38.Wentzensen N, Bergeron C, Cas F, Vinokurova S, von Knebel DM. Triage of women with ASCUS and LSIL cytology: use of qualitative assessment of p16INK4a positive cells to identify patients with high-grade cervical intraepithelial neoplasia. Cancer. 2007;111:58–66. doi: 10.1002/cncr.22420. [DOI] [PubMed] [Google Scholar]
- 39.Wentzensen N, Bergeron C, Cas F, et al. Evaluation of a nuclear score for p16(INK4a)-stained cervical squamous cells in liquid-based cytology samples. Cancer. 2005:461–7. doi: 10.1002/cncr.21378. [DOI] [PubMed] [Google Scholar]
- 40.ALTS group Anonymous. Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. J Natl Cancer Inst. 2000;92:397–402. doi: 10.1093/jnci/92.5.397. [DOI] [PubMed] [Google Scholar]
- 41.ASCUS-LSIL Triage Study Group. Results of a randomized trial on the management of cytology interpretations of atypical squamous cells of undetermined significance. Am J Obstet Gynecol. 2003;188:1383–92. doi: 10.1067/mob.2003.457. [DOI] [PubMed] [Google Scholar]
- 42.Arbyn M, Roelens J, Martin-Hirsch P, Leeson S, Wentzensen N. Use of HC2 to triage women with borderline and mild dyskaryosis in the UK. Br J Cancer. 2011;105:877–80. doi: 10.1038/bjc.2011.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q, Kuhn L, Denny LA, et al. Impact of utilizing p16(INK4A) immunohistochemistry on estimated performance of three cervical cancer screening tests. Int J Cancer. 2006;120:351–6. doi: 10.1002/ijc.22172. [DOI] [PubMed] [Google Scholar]
- 44.Bergeron C, Ordi J, Schmidt D, et al. Conjunctive p16INK4a testing significantly increases accuracy in diagnosing high-grade cervical intraepithelial neoplasia. Am J Clin Pathol. 2010;133:395–406. doi: 10.1309/AJCPXSVCDZ3D5MZM. [DOI] [PubMed] [Google Scholar]
- 45.Bossuyt PM, Reitsma JB, Bruns DE, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. BMJ. 2003;326:41–4. doi: 10.1136/bmj.326.7379.41. [DOI] [PMC free article] [PubMed] [Google Scholar]