Abstract
Genes for cytokinin-responsive His-protein kinases (ZmHK1, ZmHK2, and ZmHK3a) were isolated from maize (Zea mays). Heterologous expression of each of the ZmHKs in Escherichia coli having the ΔrcsC and cps∷lacZ genetic background conferred cytokinin-inducibility of lacZ expression on the bacteria. In the recombinant E. coli system, ZmHK1 and ZmHK3a were more sensitive to free-base cytokinins than to the corresponding nucleosides; isopentenyladenine was most effective for ZmHK1, while ZmHK2 tended to be most sensitive to trans-zeatin and the riboside. In contrast to a known cytokinin receptor of Arabidopsis (AHK4/CRE1/WOL), all ZmHKs responded to cis-zeatin (cZ), which generally is believed to be inactive or only weakly active. In cultured maize cells, expression of ZmRR1, a cytokinin-inducible response regulator, was induced by cZ as well as by trans-zeatin. These results strongly suggest that maize cytokinin receptors differ in ligand preference, and that cZ is an active cytokinin at least in maize.
Cytokinin controls various processes in plant growth and development such as cell division, chloroplast differentiation, leaf senescence, and nutrient signaling (Mok, 1994; Forde, 2002). The expression of various genes is modulated by this hormone, and numerous cytokinin-regulated genes have been described (Schmülling et al., 1997; Rashotte et al., 2003). Phosphotransfer between His and Asp residues of signaling proteins, generally referred to as a two-component regulatory system or a His-Asp phosphorelay, is involved in cytokinin signal transduction (Inoue et al., 2001; Kieber, 2001; Schmülling, 2001; Suzuki et al., 2001; Kakimoto, 2003). In plants, phosphorelays usually consist of three protein modules, a sensory His-protein kinase (HK), a His phospho-transfer protein (HP), and a response regulator (RR; Mizuno, 1998; Aoyama and Oka, 2003).
Plant sensory HKs typically contain an input domain, an HK domain, and a receiver domain. Eleven genes encoding HKs were identified in Arabidopsis (Kieber, 2001; Aoyama and Oka, 2003). Plant HKs expressed in bacterial HK mutants can function as alternates that trigger the bacterial signal transduction cascade (Kakimoto, 1996). Exploiting this phenomenon, cytokinin-responsive HKs were identified in Arabidopsis. AHK4/CRE1/WOL (called AHK4 in the following) expressed in budding yeast (Saccharomyces cerevisiae) complemented the yeast HK mutant sln1, which is defective in osmosensing, in dependence of exogenous cytokinin (Inoue et al., 2001). Similarly, the fission yeast (Schizosaccharomyces pombe) HK mutant ΔPhk1/2/3 that shows defects in the regulation of the G2/M cell cycle progression, and the Escherichia coli HK mutant ΔrcsC that is impaired in the regulation of extracellular polysaccharide synthesis, were complemented by expressing AHK3 and AHK4 in a cytokinin-dependent manner (Suzuki et al., 2001; Yamada et al., 2001). These studies demonstrated that AHK3 and AHK4 encode cytokinin-receptors whose kinase activity is activated in response to cytokinin. In in vitro binding studies, AHK4 interacted with isopentenyladenine (iP), trans-zeatin (tZ), thidiazuron (TDZ), and benzyladenine (BA), but not with iP riboside (iPR; Yamada et al., 2001), indicating that free base cytokinins are the physiological ligands of AHK4. AHK2, a homolog of AHK3 and AHK4, also functions as a cytokinin-responsive HK (Kakimoto, 2003).
Several HPs and RRs, the downstream elements that transmit signals from HKs to target genes, have been identified. Five HPs (AHP1 to AHP5) and 22 RRs (ARR1 to ARR22) were found in Arabidopsis (Brandstatter and Kieber, 1998; Miyata et al., 1998; Suzuki et al., 1998; Imamura et al., 1999; Aoyama and Oka, 2003), whereas 3 HPs (ZmHP1 to ZmHP3) and 10 RRs (ZmRR1 to ZmRR10) were detected in maize (Zea mays; Sakakibara et al., 1998, 1999; Asakura et al., 2003). RRs can be classified into two subtypes (type A and type B), as judged from their structural designs and expression patterns (Imamura et al., 1999). Expression of most type-A RR genes is induced by cytokinin. In maize, for instance, ZmRR1, ZmRR2, and ZmRR4 to ZmRR7 are up-regulated by tZ (Sakakibara et al., 1998, 1999; Asakura et al., 2003).
The natural cytokinins are adenine-derivatives that carry either an isoprene-derived side chain or an aromatic derivative side chain at the N6-terminal. The different activities in bioassays of the members of the isoprenoid-derived group (e.g. iP, tZ, cis-zeatin [cZ], and dihydrozeatin) depend on small structural variations such as the presence and position of a hydroxyl group (Mok and Mok, 2001). For example, tZ and iP have much higher activity than cZ in Funaria hygrometrica (Spiess, 1975) and tobacco (Nicotiana tabacum; Schmitz and Skoog, 1972). In sln1 mutants of S. cerevisiae that expressed AHK4, cZ could not complement the mutation, whereas tZ and iP did (Inoue et al., 2001). Therefore, cZ had been thought to possess weak activity at best; as a result, little attention has been paid to its possible functions. However, cZ-type cytokinins are abundant in some plants such as chickpea (Cicer arietinum; Emery et al., 1998), rice (Oryza sativa; Murofushi et al., 1983), and maize (Veach et al., 2003). As recently demonstrated, the maize cytokinin O-glucosyltransferases, cisZOG1 and cisZOG2, predominantly catalyze cZ (Martin et al., 2001; Veach et al., 2003). O-glucosylation results in inactivation of cytokinins whose side chains carry a hydroxyl group. The occurrence of what appears to be a regulation of cZ by O-glucosylation suggests that cZ may be functional as a cytokinin. If cZ has physiological activity in maize, it should be recognized by cytokinin receptors. Furthermore, the small side chain variations among active cytokinins and the multiplicity of cytokinin receptor genes lead us to expect strong functional differentiation of the receptors with regard to ligand preference. However, ligand specificity of cytokinin receptors has not been characterized well so far.
In this study, we isolated genes for cytokinin receptors from maize (ZmHKs) and determined their ligand preference. Our results strongly suggest a functional differentiation of the ZmHKs and a significant role of cZ in cytokinin signaling in maize.
RESULTS
Isolation of cDNAs Encoding Cytokinin Receptors Expressed in Maize Leaves
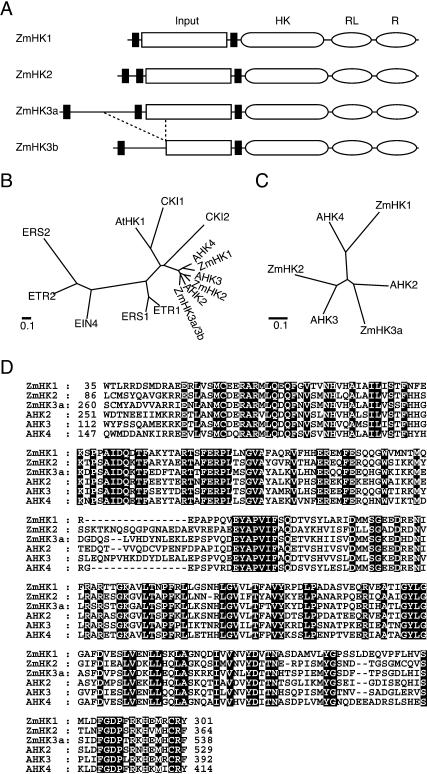
To isolate cDNAs encoding cytokinin receptors in maize, we first searched the maize expression-sequence tag database in silico using the amino acid sequence of AHK4 (Inoue et al., 2001; Suzuki et al., 2001) as a query. One partial sequence of a candidate (accession no. AI861678) was found. Reverse transcription (RT)-PCR with leaf total RNA amplified the corresponding cDNA fragment containing an HK domain of about 640 bp (data not shown). To obtain the full-length clone and possible homologs, a cDNA library was screened by nucleic acid hybridization using the above-mentioned cDNA fragment as a probe. Fifteen cDNA clones were picked up from the library, and could be classified into three groups based on their nucleotide sequence. The genes corresponding to the cDNAs were designated ZmHK1, ZmHK2, and ZmHK3. The longest cDNA clones of ZmHK1 and ZmHK2 contained reading frames of 974 and 1,007 amino acids, respectively. On the other hand, the longest cDNA of ZmHK3 lacked the amino-terminal region, and we obtained this missing region by the 5′-RACE method. Consequently, two types of cDNA with different lengths were amplified. Comparison of the two cDNA sequences with the corresponding genomic sequences (accession nos. AB121677 and AB011485) revealed that one was lacking the third exon. The longer cDNA with a reading frame of 1,201 amino acids was denoted ZmHK3a, while the shorter one (1,049 amino acids) was termed ZmHK3b. As those two types of cDNA were also obtained by additional cDNA library screening, we conclude that ZmHK3b represents a true variant of the gene.
Each of the four deduced amino acid sequences of the ZmHKs seemed to contain three parts: an input domain, an HK domain, and a receiver domain (Fig. 1A). All of the amino acid residues required for the HK and receiver function (Mizuno, 1998) were conserved (data not shown). In addition, every ZmHK also had a receiver-like domain whose functionally essential amino acid residues (Ueguchi et al., 2001) were not fully conserved. However, about one-fourth of the input domain appeared to be absent in ZmHK3b. Homology analyses revealed that ZmHK1, ZmHK2, and ZmHK3a/b were closely related to AHK4, AHK3, and AHK2, respectively, in both the HK domain (Fig. 1B) and the input domain (Fig. 1, C and D). Although the amino-terminal region of ZmHK1 was shorter than that of AHK4 by about 110 amino acids, the existence of a stop codon in the adjacent upstream region of the putative initial Met codon in ZmHK1 shows that ZmHK1 cDNA contains an entire reading frame (data not shown).
Figure 1.
Structural features of ZmHKs. A, Schematic representation of the primary structures of ZmHK1, ZmHK2, ZmHK3a, and ZmHK3b. Black rectangle, hydrophobic membrane-spanning domain; rectangle marked Input, input domain; rounded rectangle marked HK, HK domain; oval marked RL, receiver-like domain; oval marked R, receiver domain. B, Phylogenetic comparison of HK domains from maize and Arabidopsis. Amino acid alignment of the HK domains was performed using the CLUSTALW program. Bar = 0.1 amino acid substitutions per site. C, Phylogenetic relationship of the putative input domains of ZmHK1, ZmHK2, ZmHK3a, AHK2, AHK3, and AHK4. Amino acid alignment of the putative input domains was performed using the CLUSTALW program. D, Amino acid alignments of the input domains of the ZmHKs and AHKs. Gaps marked by dashes were inserted to obtain maximum homology. Amino acids that are identical in all sequences are highlighted.
Cytokinin-Dependent Signal Transmission via ZmHK1, ZmHK2, and ZmHK3a
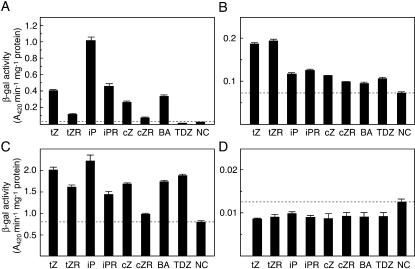
To elucidate the function of the ZmHKs, they were expressed in the ΔrcsC and cps∷lacZ mutant background of E. coli (Suzuki et al., 2001; Takeda et al., 2001). The RcsC-YojN-RcsB signaling pathway, one of the His-Asp phosphorelay systems in E. coli, is known to regulate extracellular polysaccharide synthesis by activating the cps operon (Takeda et al., 2001). AHK4 and AHK3 have been previously shown to complement the function of RcsC in a cytokinin-dependent manner (Suzuki et al., 2001; Yamada et al., 2001). We expressed the four ZmHKs in the E. coli mutants and monitored the effect of various cytokinin species on cps∷lacZ expression. Although basal levels of lacZ expression (Fig. 2; NC, negative control) varied in each strain, cytokinin-dependent expression of the reporter gene was detected in the E. coli strains expressing ZmHK1, ZmHK2, and ZmHK3a. On the other hand, ZmHK3b did not respond to any cytokinin tested (Fig. 2D). Thus, at least three ZmHKs functioned as cytokinin-responsive HKs in E. coli.
Figure 2.
Cytokinin-dependent expression of lacZ in E. coli (ΔrcsC, cps∷lacZ) expressing ZmHKs. E. coli strains harboring pIN-III-ZmHK1 (A), pIN-III-ZmHK2 (B), pIN-III-ZmHK3a (C), or pIN-III-ZmHK3b (D) were cultured in the absence (NC, negative control) or presence of the cytokinin indicated at 1 μm. tZ, trans-zeatin; tZR, trans-zeatin riboside; iP, isopentenyladenine; iPR, isopentenyladenine riboside; cZ, cis-zeatin; cZR, cis-zeatin riboside; BA, benzyladenine; TDZ, thidiazuron. tZ, iP and cZ are free-base species of isoprenoid cytokinins, and tZR, iPR, and cZR are their nucleoside forms, respectively. BA is a free-base aromatic cytokinin, and TDZ is a synthetic phenylurea cytokinin. Culture period was 12 h for ZmHK1 and ZmHK2, 4 h for ZmHK3a, and 8 h for ZmHK3b. The cells were harvested and the β-galactosidase activity was measured with o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate. The values presented are means of triplicate assays with sd. A dashed horizontal line indicates the negative control value in each case.
In this assay, different effects of the compounds on the lacZ expressions were found. The free-base cytokinins tZ, iP, cZ, and BA activated the lacZ expression via ZmHK1 significantly stronger than the corresponding nucleosides (Fig. 2A). Among them, iP was the most effective activator. A similar pattern was found in ZmHK3a-expressing E. coli. However, TDZ, a synthetic phenylurea cytokinin, was effective with ZmHK3a but not with ZmHK1 (Fig. 2A). On the other hand, in ZmHK2-expressing E. coli, tZ and tZ riboside (tZR) activated lacZ expression most strongly (Fig. 2B). These results suggested that the ZmHKs have different ligand preferences.
Responsiveness of ZmHK1 to cZ
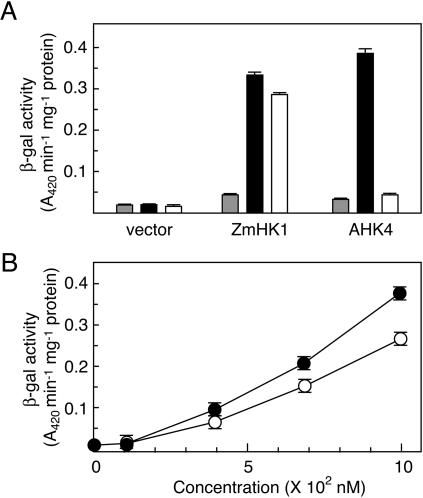
In the E. coli mutants expressing ZmHK1, ZmHK2, and ZmHK3a, cZ significantly activated the reporter gene expression (Fig. 2, A, B, and C). However, the Arabidopsis cytokinin receptor AHK4 had not been found responsive to cZ if expressed in the sln1 mutant of S. cerevisiae (Inoue et al., 2001). Therefore, the responsiveness of ZmHK1 and AHK4 to cZ was compared in the E. coli system. The response of E. coli expressing AHK4 to cZ was much less than to tZ, whereas E. coli expressing ZmHK1 showed substantial lacZ expression in response to both tZ and cZ (Fig. 3A). Analysis of the dose-response relationship showed that the sensitivity of ZmHK1 to cZ was comparable to that to tZ (Fig. 3B). These results demonstrate that ZmHK1 recognizes cZ as a ligand. It should be noted that the purity of the cZ used was checked; the contamination with tZ was below the detection limit (<0.1%, data not shown).
Figure 3.
Responses of ZmHK1 and AHK4 to cZ. A, E. coli strains (ΔrcsC, cps∷lacZ) harboring pIN-III (vector), pIN-III-ZmHK1 (ZmHK1), or pIN-III-AHK4 (AHK4) were cultured in the absence (gray bar) or presence of 1 μm tZ (black bar) or 1 μm cZ (white bar), respectively, for 8 h. The cells were harvested and the β-galactosidase activity was measured with ONPG as a substrate. B, Concentration-dependency of ZmHK1-mediated lacZ expression in response to tZ and cZ in the E. coli system. The E. coli strain harboring pIN-III-ZmHK1 was cultured with the indicated concentration of tZ (black circles) or cZ (white circles), respectively. The β-galactosidase activity of the cells was measured. The values presented are means of triplicate assays with sd.
Expression Patterns of ZmHKs in Mature Maize Plants and Cultured Cells
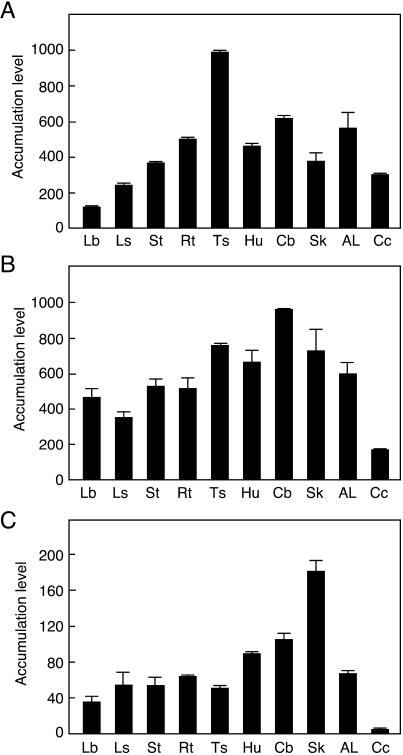
To evaluate levels of ZmHK expression in different cell types, the accumulation of ZmHK transcripts was analyzed by real-time PCR using RNA samples prepared from various tissues of mature maize plants and cultured cells. To distinguish the almost identical nucleotide sequence of the ZmHK3a and ZmHK3b mRNAs, PCR primers specific for the third exon of ZmHK3a, which is not present in ZmHK3b, were designed. The transcripts of ZmHK1, ZmHK2, and ZmHK3a were detected in all tissues tested, but the distribution patterns differed (Fig. 4). The transcript of ZmHK1 was relatively abundant in tassels, but less so in leaf blades. The accumulation of ZmHK2 transcripts was less pronounced in cultured cells than in tissues. ZmHK3a transcripts were predominantly detected in the silk. We conclude that ZmHKs are not strictly tissue-specific, and that certain ZmHKs are redundantly expressed in most tissues.
Figure 4.
Accumulation patterns of ZmHK transcripts in various maize tissues. Total RNA prepared from various maize tissues was subjected to quantitative real-time PCR. A, ZmHK1; B, ZmHK2; C, ZmHK3a. Lb, leaf blade; Ls, leaf sheath; St, stem; Rt, root; Ts, tassel; Hu, husk; Cb, cob; Sk, silk; AL, auricle and ligule; Cc, cultured cells. Accumulation levels of the transcripts are given as the copy number of ZmHK mRNA/0.5 ng total RNA. Real-time PCR was performed in triplicate, and the mean values with sd are shown.
Differential Effects of Different Cytokinins on ZmRR1 Expression in Leaves and Cultured Cells
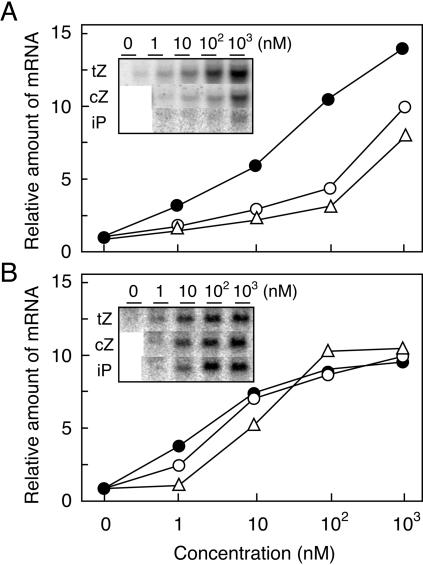
The accumulation of the ZmHK2 transcript in leaf blades was 4- and 13-fold larger than that of ZmHK1 and ZmHK3a, respectively (Fig. 4). Similarly, the transcripts of ZmHK1 and ZmHK2 were 76- and 44-fold more abundant than that of ZmHK3a, respectively, in cultured cells. To evaluate the physiological significance of the ZmHK ligand preferences and cZ observed in the transgenic E. coli system, we studied the response of ZmRR1 expression to cytokinins in detached maize leaves and cultured cells. ZmRR1 is a typical cytokinin-inducible gene encoding a response regulator (Sakakibara et al., 1998). Total RNA was prepared from the tissues that had been treated with various concentrations of iP, tZ, and cZ, and changes in the accumulation level of the ZmRR1 transcript were analyzed by northern blotting. In both leaf blades and cultured cells, all the free base cytokinins tested had positive effects on ZmRR1 induction. In detached leaves, tZ was significantly more effective than iP and cZ, which evoked similar responses (Fig. 5A). This pattern of relative effectivity resembled that observed in E. coli expressing ZmHK2 (Fig. 2B).
Figure 5.
Effects of tZ and cZ on ZmRR expression. Total RNA (20 μg), prepared from maize leaves treated with the indicated concentration of tZ, cZ, or iP for 120 min (A), or from maize cultured cells treated for 20 min (B), was subjected to electrophoresis on agarose gels and transferred to nylon membranes. The blots were treated with a 32P-labeled probe specific for ZmRR1. A probe for ubiquitin mRNA was used as an internal control (data not shown). The signal intensity of the ZmRR1 transcript relative to that of the ubiquitin transcript is given in the graphs, normalized so that the maximum signal of cZ equals 10. Black circles, tZ; white circles, cZ; white triangles, iP. Photographs of the original blots are shown in the insets.
On the other hand, iP, tZ, and cZ had almost identical effects on ZmRR1 expression in cultured cells (Fig. 5B). Since cZ and tZ can be reversibly interconverted by zeatin cis-trans isomerase (Bassil et al., 1993), it would appear possible that the apparent cZ effect on ZmRR1 expression in the cultured cells was caused by tZ that had been enzymatically produced from cZ. To evaluate the extent of metabolic conversion during the experiment, cytokinin contents of cultured cells that had been treated identically as those used for the northern-blot analysis were measured (Table I). Levels of cZ, cZR, and cZR 5′-monophosphate generally increased following application of cZ, suggesting that cZ was converted to the corresponding nucleosides and nucleotides. However, isomeric conversion of cZ to tZ and its derivatives was low; tZ contents in the cultured cells did not rise after treatment with 1 nm or 10 nm cZ, and increased only 2-fold after treatment with 100 nm cZ. These results strongly suggest that cZ is the physiologically active cytokinin for the induction of ZmRR1 expression in maize. The cytokinin responses vary between the detached leaves and the cultured cells and this could be potentially explained by having a different complement of the ZmHKs.
Table I.
| CKsc
|
cZ
|
||||
|---|---|---|---|---|---|
| 0 nm | 1 nm | 10 nm | 100 nm | 1,000 nm | |
| pmol g−1 fresh wt | |||||
| tZ | 0.64 ± 0.02 | 0.60 ± 0.01 | 0.54 ± 0.03 | 1.18 ± 0.02 | 5.96 ± 0.04 |
| tZR | 2.00 ± 0.04 | 2.15 ± 0.10 | 2.05 ± 0.05 | 2.67 ± 0.05 | 7.40 ± 0.17 |
| tZRMP | 3.20 ± 0.26 | 3.29 ± 0.38 | 3.79 ± 0.59 | 4.87 ± 0.27 | 27.57 ± 2.14 |
| cZ | 0.23 ± 0.03 | 0.49 ± 0.00 | 2.66 ± 0.04 | 26.84 ± 0.99 | 45.70 ± 0.93 |
| cZR | 2.33 ± 0.02 | 2.80 ± 0.12 | 6.33 ± 0.24 | 43.53 ± 1.91 | 181.32 ± 8.97 |
| cZRMP | 4.70 ± 0.31 | 4.63 ± 0.44 | 14.84 ± 1.55 | 119.45 ± 18.65 | >QLd |
Mean value from two replicate samples with the average deviation.
After treatment of maize cultured cells with cZ for 20 min, cells were harvested and washed twice with water. Then, cytokinin content was analyzed as described in “Materials and Methods.”
CKs, cytokinins; tZ, trans-zeatin; tZR, tZ riboside; tZRMP, tZR 5′-monophosphate; cZ, cis-zeatin; cZR, cZ riboside; cZRMP, cZR 5′-monophosphate.
>QL, content larger than the quantifiable limit (193 pmol g−1 fresh wt).
DISCUSSION
In this study, we identified cytokinin-responsive HKs in maize and characterized their ligand preference. Although cytokinin receptors had been described before in Arabidopsis, our analysis of the maize orthologs provides new insights into cytokinin signaling.
Sequence analysis of cytokinin receptors in Arabidopsis and maize suggested one-to-one correspondence between ZmHKs and AHKs (Fig. 1). Such close correspondence is not found in the downstream modules, ZmHPs and AHPs, and ZmRRs and ARRs, respectively (Asakura et al., 2003). The conservation of HKs implies that these cytokinin receptors had diverged from the ancestral gene into the three isogenes before the divergence of monocots and dicots, and that they play physiologically similar roles in cytokinin signaling in all angiosperms.
We found two mRNA species corresponding to ZmHK3, and only ZmHK3a had a cytokinin-responsive HK activity. At present, we can only speculate about the physiological function of ZmHK3b. ZmHK3b lacks a part of the input domain that is well conserved among all cytokinin receptors. In AHK4, two alternatively spliced products have been described, but both contained the complete input domain (Inoue et al., 2001). Thus, the physiological significance of the multiplicity would appear to be different in AHK4 and ZmHK3. Levels of ZmHK3a and ZmHK3b transcripts appeared similar as judged from quantitative real-time PCR (data not shown).
It is generally assumed that cZ is an inactive or weakly active cytokinin, and that its isomerization to tZ catalyzed by zeatin cis-trans isomerase is necessary for the induction of biological activity. However, our findings strongly suggest that cZ itself functions as an active cytokinin in maize. Supporting the idea, recent analyses revealed that maize contains substantial amounts of cZ-type cytokinins in various tissues (Veach et al., 2003). cZ may play specific roles in certain plant species or at certain stages of development; future studies will have to evaluate these possibilities. Presumably, cZ originates from the degradation of tRNA (Murai, 1994). Therefore, in addition to the conjugation of isoprenoid side chains to free adenosine phosphates catalyzed by adenosine phosphate-isopentenyltransferase (Kakimoto, 2001; Takei et al., 2001a; Sakakibara and Takei, 2002), the metabolism of tRNAs also may be important for cytokinin homeostasis.
ZmHK1-mediated signaling is more sensitive to iP, whereas the ZmHK2-mediated pathway is more responsive to tZ. The different affinities of the receptors to ligands that differ by minute variations in their side chains suggests that these variations carry physiological meaning. In Pisum sativum, tZ was much more effective than iP in releasing buds from apical dominance (King and Van Staden, 1988). Distinct distributions of iPR and tZR in phloem and xylem sap, respectively, were found in Sinapis alba (Lejeune et al., 1994). However, differences in the biological roles of iP and tZ are not well understood at present, which calls for further study.
Previously, in vitro binding analyses had suggested that only free base cytokinins were recognized by AHK4 (Yamada et al., 2001). We observed a similar specificity of ZmHK1 in the E. coli system, but also detected a weak response of ZmHK1 to iPR. This is probably due to the recognition of iP derived from iPR degradation by internal nucleosidase activity, because shortening the period of treatment (2 and 8 h) reduced the iPR response (data not shown). In ZmHK2, there were no significant differences in the responses to free base cytokinins and nucleosides (Fig. 2B). This tendency was also observed when the period of treatment was shortened (8 h; data not shown). We cannot totally exclude the possibility of free base formation by nucleoside degradation during the experiments. However, the incubation periods in tests with E. coli strains expressing ZmHK1 and ZmHK2, respectively, were identical, arguing against the idea that nucleoside degradation could be responsible for the differences observed. We therefore suggest that the observed differences in the responses of the two ZmHKs to nucleoside cytokinins are probably due to a genuine sensitivity of ZmHK2 to nucleosides.
The extent of ZmHK-dependent cytokinin-induced lacZ expression in E. coli differed between the ZmHKs (Fig. 2). This had also been observed in the Arabidopsis counterparts, AHK4 and AHK3 (Yamada et al., 2001). The differences may simply be the result of different expression levels for the ZmHKs in the bacterial system. It must be cautioned that these results may reflect various levels of expression and compatibility of plant HKs in the E. coli system, rather than differences in the cytokinin responsiveness of the HKs as such. In the E. coli ΔrcsC genetic background (Suzuki et al., 2001), differences in the affinities between the plant HK and YojN, a bacterial HP, may result in apparent differences in sensitivity of the HKs to cytokinins.
In conclusion, we have demonstrated here functional differentiation of ZmHKs in terms of ligand preference. Although the physiological significance of structural varieties of the cytokinin isoprenoid side chains and, in particular, the function of cZ are not well understood, it appears clear that structural variations play a regulatory role as the variants are discriminated by the receptors. These findings raise further questions to be solved. First, how do plants modulate the levels of tZ- and cZ-type cytokinins? Previous studies demonstrated that cZ-type cytokinins are derived from tRNA degradation, and that the isoprenoid side chains originate from the mevalonate pathway (Murai, 1994). On the other hand, the metabolic origin of tZ-type side chains is not well characterized yet. The elucidation of regulatory mechanism of their biosyntheses and that of physiological contribution of the cis-trans isomerization will provide clues to the physiological roles of tZ and cZ. Second, how do ZmHKs transmit signals differentially to downstream factors? At present, 3 ZmHPs and 10 ZmRRs are known in maize (Sakakibara et al., 1998; Asakura et al., 2003). Signaling cascades via His-Asp phosphorelays must be tightly regulated to transmit specific signals to the various target genes. Future work will have to focus on these issues to fully unravel the intricate network of cytokinin signaling in plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Maize (Zea mays L. cv Golden Cross Bantam T51) was grown in a growth chamber (Koitotron KG-201SHL-D, Koito Industries, Tokyo) for 16 d after germination in vermiculite with Hoagland nutrient solution (Arnon and Hoagland, 1940) with reduced nitrogen (1.6 mm NO3−) at 600 μE m−2 s−1 and a photoperiod of 14 h light (28°C)/10 h dark (20°C). Treatment of detached leaves with cytokinins was carried out as described previously (Sakakibara et al., 1997). For the analysis of mature plants, maize was grown in a greenhouse under natural light conditions with Hoagland nutrients (16 mm NO3−) for 2 months. For tests on maize protoplasts, the suspension-cultured cell line Z86 (Kawaguchi et al., 1991) was grown in Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 3% Suc, 1 mg L−1 thiamine, 20 mg L−1 KH2PO4, 100 mg L−1 myoinositol, and 1 mg L−1 2,4-dichlorophenoxyacetic acid, pH 5.6, at 25°C with shaking (120 rpm) in the dark.
RT-PCR
cDNA of maize leaf poly(A)+ RNA was synthesized with SuperScript II RT (Invitrogen, Carlsbad, CA) using an oligo(dT) primer. Then, 30 PCR cycles were carried out (94°C for 0.5 min, 55°C for 0.5 min, and 72°C for 1 min per cycle) with the primers 5′-GGATGATCTTGAGGGCGAAAAC-3′ and 5′-GTCGGCTTCAGAGGTGACC-3′.
Isolation of Full-Length cDNA Clones and Sequence Analysis
The full-length cDNA clones for ZmHK1 and its homologs (ZmHK2 and ZmHK3) were isolated from a cDNA library that had been prepared from cytokinin-treated maize leaves (Asakura et al., 2003) by nucleic hybridization screening (Sambrook et al., 1989) using the RT-PCR-amplified ZmHK1 cDNA as a probe. To obtain the 5′-missing region of ZmHK3 cDNA, 5′-RACE was performed using the 5′-Full RACE Core set (TaKaRa, Kyoto). The sequences of the cDNA insets were determined with an automated DNA sequencer (3100 Genetic Analyzer, Applied Biosystems, Foster City, CA). A phylogenetic tree was generated using the CLUSTALW program at the DNA Data Bank of Japan (www.ddbj.nig.ac.jp).
Heterologous Expression of ZmHKs in E. coli
The reading frames of ZmHK1, ZmHK2, ZmHK3a, and ZmHK3b were amplified by PCR and ligated into pIN-III vectors (Masui et al., 1983). The resulting plasmids were designated pIN-III-ZmHK1, pIN-III-ZmHK2, pIN-III-ZmHK3a, and pIN-III-ZmHK3b, respectively. The plasmids were transformed into an E. coli strain with the ΔrcsC and cps∷lacZ genetic background (Suzuki et al., 2001). Transformants were grown in Luria-Bertani medium supplemented with 50 mm potassium phosphate, pH 7.0, 40 mm Glc, and 50 μg mL−1 ampicillin. The overnight culture (1 mL) was inoculated in 100 mL of the fresh culture medium supplemented with various concentrations of cytokinins, and was further grown for appropriate periods at 25°C with vigorous shaking. The cells were harvested and suspended in an appropriate volume of Z buffer (100 mm sodium phosphate, 10 mm KCl, 1 mm MgSO4, pH 7.0). After disruption of the cells by sonication, the supernatant was recovered by centrifugation and was assayed for β-galactosidase activity with o-nitrophenyl β-d-galactopyranoside as a substrate.
Northern Analysis
Total RNA was prepared from various maize tissues by the guanidine thiocyanate procedure (McGookin, 1984). The RNA was subjected to electrophoresis on a 1.2% (w/v) agarose gel (Sambrook et al., 1989), and blotted onto nylon membranes (Hybond-N+; Amersham Biosciences, Buckinghamshire, UK). The blots were probed with subfragments of ZmRR1 cDNA (Sakakibara et al., 1999) that had been labeled by the random-primer method in the presence of [α-32P]dCTP (Feinberg and Vogelstein, 1984). Hybridization and washing of the filters were performed as described previously (Sakakibara et al., 1991). Signal intensity was determined with a Bio-imaging analyzer (Typhoon 8600, Amersham Biosciences).
Quantitative Real-Time PCR
cDNA was synthesized using SuperScript II RT (Invitrogen) with oligo(dT) primers. Accumulation levels of the ZmHK transcripts were analyzed by a real-time PCR method, with ABIPRISM 7000 Sequence Detection System (Applied Biosystems) monitoring the amplification with the SYBR-Green I dye (Applied Biosystems). The primers for PCR were designed using Primer Express software (Applied Biosystems) and checked for the specific product formation by polyacrylamide gel analysis. Sequences of the primers used were: 5′-AAGTAGGAGCTTGACAGAGGCACTA-3′ and 5′-CGTACAGGTCTCCCATCTACCAA-3′ for ZmHK1, 5′-AGCATTGGGTGGGATAGATAAACT-3′ and 5′-GAAGGCTGCCCAGTGTTGAA-3′ for ZmHK2, and 5′-TGGAATCAGTGGCGTGAATG-3′ and 5′-GCTCCCCAAAAAAGCAGATAGA-3′ for ZmHK3a. In each case, plasmid DNA containing the corresponding ZmHK was used as a template to generate calibration curve.
Cytokinin Analysis
Extraction and fractionation of cytokinins were performed as described previously (Dobrev and Kaminek, 2002). Fractions for cytokinin nucleotides and other cytokinin species were separately purified further by immuno-affinity columns (Takei et al., 2001b). After desalting, the samples were dissolved in 0.005% acetic acid and analyzed with a LC-MS system (model 2695/ZQ2000MS, Waters, Milford, MA). Cytokinins were separated using an ODS column (Symmetry Shield RP18, 3.5 μm, 2.1 mm × 150 mm, Waters) at a flow rate of 0.25 mL/min, with the gradients of solvent A (water), B (methanol), and C (0.1% acetic acid) set according to the following profile: 0 min, 95% A + 5% C; 1 min, 95% A + 5% C; 16 min, 45% A + 50% B + 5% C; 22 min, 25% A + 70% B + 5% C; 30 min, 25% A + 70% B + 5% C. The column temperature was 40°C. Quantification was performed in the selected ion recording mode. Cone voltage was 35 V, source temperature 110°C, and capillary voltage 4.0 kV. Data were analyzed using the MassLynx version 3.5 software (Waters).
Chemicals
iP, iPR, tZ, tZR, cZR, BA, and TDZ were purchased from Sigma (St. Louis). cZ was purchased from ICN Biomedicals (Irvine, CA).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AB042270 for ZmHK1, AB102956 for ZmHK2, AB102957 for ZmHK3a, and AB121445 for ZmHK3b.
Acknowledgments
We thank Dr. T. Mizuno, Nagoya University, for providing us with E. coli strains with the ΔrcsC and cps∷lacZ genetic background and pIN-III-AHK4. We are grateful to Dr. K. Syono, Nihon Women's University, for supplying cultured cell lines of maize.
This study was partly supported by Grants-in-Aids for Scientific Research on Priority Areas (grant no. 12142202 to H.S.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.037176.
References
- Aoyama T, Oka A (2003) Cytokinin signal transduction in plant cells. J Plant Res 116: 221–231 [DOI] [PubMed] [Google Scholar]
- Arnon DI, Hoagland DR (1940) Crop production in artificial solutions and soils with special reference to factors influencing yield and absorption of inorganic nutrients. Soil Sci 50: 463–471 [Google Scholar]
- Asakura Y, Hagino T, Ohta Y, Aoki K, Yonekura-Sakakibara K, Deji A, Yamaya T, Sugiyama T, Sakakibara H (2003) Molecular characterization of His-Asp phosphorelay signalling factors in maize leaves: implications of the signal divergence by cytokinin-inducible response regulators in the cytosol and the nuclei. Plant Mol Biol 52: 331–341 [DOI] [PubMed] [Google Scholar]
- Bassil NV, Mok D, Mok MC (1993) Partial purification of a cis-trans-isomerase of zeatin from immature seed of Phaseolus vulgaris L. Plant Physiol 102: 867–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstatter I, Kieber JJ (1998) Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrev PI, Kaminek M (2002) Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J Chromatogr A 950: 21–29 [DOI] [PubMed] [Google Scholar]
- Emery RJN, Leport L, Barton JE, Turner NC, Atkins A (1998) cis-Isomers of cytokinins predominate in chickpea seeds throughout their development. Plant Physiol 117: 1515–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1984) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 139: 266–267 [DOI] [PubMed] [Google Scholar]
- Forde BG (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol 53: 203–224 [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Kiba T, Ueguchi C, Sugiyama T, Mizuno T (1999) Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol 40: 733–742 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (1996) CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274: 982–985 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (2001) Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol 42: 677–685 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (2003) Perception and signal transduction of cytokinins. Annu Rev Plant Biol 54: 605–627 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Kobayashi M, Sakurai A, Syono K (1991) The presence of an enzyme that converts indole-3-acetamide into IAA in wild and cultivated rice. Plant Cell Physiol 32: 143–149 [Google Scholar]
- Kieber JJ (2001) Cytokinins. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0063 http://www.aspb.org/publications/arabidopsis/
- King RA, Van Staden J (1988) Differential responses of buds along the shoot of Pisum sativum to isopentenyladenine and zeatin application. Plant Physiol Biochem 26: 253–259 [Google Scholar]
- Lejeune P, Bernier G, Requier M-C, Kinet J-M (1994) Cytokinins in phloem and xylem saps of Sinapis alba during floral induction. Physiol Plant 90: 522–528 [Google Scholar]
- Martin RC, Mok MC, Habben JE, Mok DW (2001) A maize cytokinin gene encoding an O-glucosyltransferase specific to cis-zeatin. Proc Natl Acad Sci USA 98: 5922–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui Y, Coleman J, Inoue M (1983). In M Inoue, ed, Experimental Manipulation of Gene Expression. Academic Press, New York, pp 15–32
- McGookin R (1984) RNA extraction by the guanidine thiocyanate procedure. In JM Walker, ed, Methods in Molecular Biology, Vol 2. Humana Press, Clifton, NJ, pp 113–116 [DOI] [PubMed]
- Miyata S, Urao T, Yamaguchi-Shinozaki K, Shinozaki K (1998) Characterization of genes for two-component phosphorelay mediators with a single HPt domain in Arabidopsis thaliana. FEBS Lett 437: 11–14 [DOI] [PubMed] [Google Scholar]
- Mizuno T (1998) His-Asp phosphotransfer signal transduction. J Biochem (Tokyo) 123: 555–563 [DOI] [PubMed] [Google Scholar]
- Mok DW, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52: 89–118 [DOI] [PubMed] [Google Scholar]
- Mok MC (1994) Cytokinins and plant development: an overview. In DWS Mok, MC Mok, eds, Cytokinins: Chemistry, Activity, and Function. CRC Press, Boca Raton, FL, pp 155–166
- Murai N (1994) Cytokinin biosynthesis in tRNA and cytokinin incorporation into plant RNA. In DWS Mok, MC Mok, eds, Cytokinins: Chemistry, Activity, and Function. CRC Press, Boca Raton, FL, pp 87–99
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Murofushi N, Inoue A, Watanabe N, Yasua O, Takahashi N (1983) Identification of cytokinins in root exudate of the rice plant. Plant Cell Physiol 24: 87–92 [Google Scholar]
- Rashotte AM, Carson SD, To JP, Kieber JJ (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132: 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Hayakawa A, Deji A, Gawronski S, Sugiyama T (1999) His-Asp phosphotransfer possibly involved in a nitrogen signal transduction mediated by cytokinin in maize: molecular cloning of cDNAs for two-component regulatory factors and demonstration of phosphotransfer activity in vitro. Plant Mol Biol 41: 563–573 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kobayashi K, Deji A, Sugiyama T (1997) Partial characterization of signalling pathway of nitrate-dependent expression of the genes for nitrogen-assimilatory enzymes using detached maize leaves. Plant Cell Physiol 38: 837–843 [Google Scholar]
- Sakakibara H, Suzuki M, Takei K, Deji A, Taniguchi M, Sugiyama T (1998) A response-regulator homolog possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J 14: 337–344 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takei K (2002) Identification of cytokinin biosynthesis genes in Arabidopsis: a breakthrough for understanding the metabolic pathway and the regulation in higher plants. J Plant Growth Regul 21: 17–23 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Watanabe M, Hase T, Sugiyama T (1991) Molecular cloning and characterization of complementary DNA encoding for ferredoxin-dependent glutamate synthase in maize leaf. J Biol Chem 266: 2028–2035 [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schmitz RY, Skoog F (1972) Cytokinins: synthesis and biological activity of geometric and position isomers of zeatin. Plant Physiol 50: 702–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmülling T (2001) CREam of cytokinin signalling: receptor identified. Trends Plant Sci 6: 281–284 [DOI] [PubMed] [Google Scholar]
- Schmülling T, Schafer S, Romanov G (1997) Cytokinins as regulators of gene expression. Physiol Plant 100: 505–519 [Google Scholar]
- Spiess LD (1975) Comparative activity of isomers of zeatin and ribosyl-zeatin on Funaria hygrometrica. Plant Physiol 55: 583–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Imamura A, Ueguchi C, Mizuno T (1998) Histidine-containing phosphotransfer (HPt) signal transducers implicated in His-to-Asp phosphorelay in Arabidopsis. Plant Cell Physiol 39: 1258–1268 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T (2001) The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol 42: 107–113 [DOI] [PubMed] [Google Scholar]
- Takeda S, Fujisawa Y, Matsubara M, Aiba H, Mizuno T (2001) A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC--> YojN--> RcsB signalling pathway implicated in capsular synthesis and swarming behavior. Mol Microbiol 40: 440–450 [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Sugiyama T (2001. a) Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J Biol Chem 276: 26405–26410 [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T (2001. b) Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol 42: 85–93 [DOI] [PubMed] [Google Scholar]
- Ueguchi C, Koizumi H, Suzuki T, Mizuno T (2001) Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol 42: 231–235 [DOI] [PubMed] [Google Scholar]
- Veach YK, Martin RC, Mok DW, Malbeck J, Vankova R, Mok MC (2003) O-glucosylation of cis-zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol 131: 1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42: 1017–1023 [DOI] [PubMed] [Google Scholar]