Abstract
Background
Activating mutations in the NOTCH1 gene are found in over 50% of T-ALL cases. Since Notch signaling contributes to the leukemia cell survival and growth, targeting Notch signaling using γ-secretase inhibitors (GSI) has been proposed as a molecularly targeted therapy for the treatment of T-ALL. However, not all T-ALL with NOTCH1 activating mutations respond to GSI treatment.
Methods
We examined whether GSI could enhance the cytotoxic effect of anti-leukemic agents in the GSI-resistant T-ALL cells although GSI does not have anti-tumor effect as a single agent. The cytotoxic effect of the combination treatment was evaluated by measuring viable cells. To determine if Notch signaling is involved in the synergism between GSI and Vincristine (VCR), loss- and gain-of-function assays were performed. To further dissect the synergistic GSI effect in combination with VCR, cell cycle progression was analyzed and apoptosis was measured by various methods.
Results
We discovered that GSI synergized with VCR in both GSI-resistant and GSI-sensitive T-ALL in a Notch-independent manner. GSI augmented VCR-induced mitotic arrest, followed by apoptosis. GSI accelerated VCR-triggered loss of mitochondrial membrane potential and caspase-mediated apoptosis.
Conclusion
GSI promoted VCR-induced apoptosis in T-ALL. Incorporating GSI into VCR- containing therapeutic regimen may be beneficial in treating T-ALL.
Keywords: T-ALL, Vincristine, gamma secretase inhibitor, Notch, synergy, apoptosis
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is a hematologic neoplasm characterized by malignant expansion of immature T-cells [1]. Although T-ALL is an aggressive disease associated with high relapse rates and poor outcomes, the prognosis has significantly improved in recent years mainly because of advances in chemotherapy protocols [1]. However, a subset of T-ALL patients (approximately 20% of children and more than 50% of adults) fails to achieve remission or relapse after current treatment [1]. Thus, significant efforts have been made in recent years to search for molecular targets that would lead to more effective therapies [2]. The identification and molecular characterization of genetic alternations have uncovered much of the mechanisms involved in the pathogenesis of the disease [3]. NOTCH1 is one of the well characterized oncogenes associated with this disease [4,5]. Various gain-of-function mutations in the NOTCH1 are found in nearly 60% of T-ALL patients [6]. Prevalence of genetic aberrations in NOTCH1 in T-ALL highlighted the important role of Notch1 signaling in this disease. Indeed, it has been reported that overexpression of activated form of Notch1 in lymphoid progenitors caused T-ALL in mice and zebrafish [7,8]. In addition, Notch signaling was required for the continuous growth and survival of the transformed leukemic cells as well [9].
Given that NOTCH1 is the most commonly activated oncogene and Notch signaling is required for the leukemic growth, Notch signaling is a prime target for molecularly tailored therapy against T-ALL. Suppressing Notch activation could be achieved by blocking the cleavage of Notch at the membrane using pharmacologic inhibitors. Small molecule γ-secretase inhibitor (GSI), which was originally developed for the treatment of Alzheimer’s disease, has been exploited to block γ-secretase-mediated cleavage of Notch1 by Weng et al as a first proof of principle [6]. Following GSI treatment, intracellular activated Notch1 protein was cleared and NOTCH1 target genes were down-regulated in T-ALL cell lines harboring NOTCH1 mutations, resulting in G0/G1 cell cycle arrest of these T-ALL cells [6]. Subsequent studies from other groups using different GSIs repeatedly confirmed the potential of GSI for treating T-ALL [10–13]. More importantly, several GSIs showed anti-tumor effects in human T-ALL xenograft models with optimized dosing regimens [14–16]. Furthermore, several studies have proposed combination therapies of GSIs with drugs targeting other signaling pathways as well as chemotherapeutics to treat T-ALL. Combination of GSI with CDK4 inhibitor [17], mTOR inhibitor [18], and BET inhibitor [19] enhanced anti-leukemic effects compared to either GSI or these drugs alone. Notably, combined use of GSI with dexamethasone reversed glucocorticoid resistance in T-ALL [13, 20].
However, not all T-ALL with NOTCH1 activating mutations respond to GSI treatment [6,10,11,21]. There were some GSI-resistant T-ALL cells, whose growth was not inhibited by GSI treatment although GSI blocked Notch1 activation in these cell lines as effective as in GSI- sensitive cell lines [11]. These GSI-resistant T-ALL cells seem to bypass the requirement for Notch1 signaling for continuous growth and metabolism [21].
In the present study, we tested our hypothesis that suppressing Notch1 activation with GSI may enhance the cytotoxic effects of anti-leukemic agents in these GSI-resistant T-ALL cells. We found that GSI enhanced the effect of Vincristine (VCR) in GSI-resistant as well as GSI-sensitive T-ALL cells in a specific manner. Surprisingly, the GSI effect in combination with VCR was not the result of inhibition of Notch signaling. Our data suggest that GSI may offer therapeutic advantages in T-ALL when used together with VCR through inhibiting not-yet identified GS substrates.
Materials and Methods
Drugs and inhibitors
The γ-secretase inhibitors, DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-(S)-phenylglycine t-butyl ester), Compound E ((S,S)- 2-[2-(3,5-Difluorophenyl)-acetylamino]-N-(1-methyl-2-oxo-5-phenyl-2,3-dihydro-1H-benzo[e][1,4]diazepin-3-yl)-propionamide) and DBZ ((S,S)- 2-[2-(3,5-Difluorophenyl)-acetylamino]-N-(5-methyl-6-oxo-6,7-dihydro-5H-dibenzo[b,d]azepin-7-yl)-propionamide, and L-685, 458 ((5S)-(tert-Butoxycarbonylamino)-6-phenyl-(4R)-hydroxy-(2R)-benzylhexanoyl)-L-leucy-L-phenylalaninamide) were obtained from Millipore (Billerica, MA) and Tocris (Minneapolis, MN), respectively. Cytarabine (Ara C) and Methotrexate (MTX) were purchased from Sigma-Aldrich (St. Louis, MO). Vincristine (VCR) and Asparaginase (ASP) were purchased from Tocris and Raybiotech (Norcross, GA), respectively.
Cells and cell culture
Jurkat-E6 (Jurkat), CCRF-CEM (CEM), and CCRF-HSB-2 (HSB-2) cell lines were obtained from ATCC (Manassas, VA). P12-Ichikawa (P12) and KOPT-K1 (KOPT) cell lines were a gift from Dr. Aldolfo Ferrando (Columbia University, New York, USA). T-ALL cell lines except for HSB-2 were cultured in RPMI 1640 media (ATCC) supplemented with 10% fetal bovine serum, 100 U/ml penicillin G, and 100 μg/ml streptomycin at 37°C in a humidified atmosphere under 5% CO2. HSB-2 was cultured in Iscove media (ATCC) supplemented with 10% fetal bovine serum, 100 U/ml penicillin G and 100 μg/ml streptomycin.
Apoptosis assay
For detection of apoptosis, cells were treated with the drugs as indicated and harvested at 48 h and stained with Annexin V-allophycocyanin (APC) and propidium iodide (PI; BD Bioscience, San Jose, CA) according to the manufacturer’s instructions. Samples were read on flow cytometry and analyzed with Flowjo software (Tree Star, Inc. Ashland, OR). In some experiments where indicated, cells were treated with 20 μM pan-caspase inhibitor, Q-VD-OPh (Sigma-Aldrich) 1h before the drug treatment to evaluate the contribution of caspase to the observed apoptosis.
To detect active caspases in the cells treated with different drugs, sulforhodamine FLICA apoptosis detection kit (SR-VAD-FMK, Immunochemistry Technologies) was utilized. Manufacturer’s protocol was followed as provided, following which the samples were read on flow cytometry and activated caspase positive cells were quantified. For all the analyses, cell debris was excluded.
To determine the mitochondrial membrane potential, cells were incubated with tetramethylrhodamine methyl ester (200 nM; Life technologies, Grand Island, NY) for 30 min before harvest at the end of the drug treatment.
Cell cycle analysis
For cell cycle analysis, cells were harvested at the indicated times after the drug treatment. After fixing with ice-cold 70% ethanol, the cells were washed with PBS and incubated with PI staining solution (PBS containing 0.1% Triton X-100, 10 μg RNase, and 50 μg propidium iodide). The cells were read on flow cytometry for DNA contents and analyzed using ModFit LT software (Verity Software House Topsham, ME). Mitotic cells were determined by staining the cells with phospho-S10-Histone H3 antibody (Millipore). Following fixation in 70% ethanol and subsequent washing with PBS, the cells were incubated with phospho-S10-Histone H3 antibody for 1 h, followed by incubation with AlexaFluor 488-conjugated donkey anti-rabbit antibody (Life technologies). Samples were read on flow cytometry and analyzed.
RNA interference of NOTCH1 and Presenilin1 (PS1), and Stable expression of ICN1 and DN-MAML1
The lentivirus encoding shRNAs against human NOTCH1 and scramble were obtained from Santa Cruz (Dallas, Texas) and used to transduce Jurkat cells to silence Notch1. The transduced cells were selected with puromycin. To knock-down of PS1 in Jurkat cells, the pool of siRNAs against PS1 was purchased from Qiagen (Valencia, CA). PS1 siRNAs were delivered into Jurkat cells using AMAXA machine with Solution V kit and program X-001 (Lonza, Walkersville, MD). Retroviral vectors encoding the intracellular domains of human Notch1 (ICN1), dominant negative mastermind-like 1 (DN-MAML1), and control vector MigR1 were kindly provided by Dr. Jon Aster (Brigham and Women’s Hospital, Boston, MA). Retroviral supernatants were produced by transient co-transfection of the retroviral vectors along with pVSVG (Clontech, Mountain View, CA) into GP2-293 cells (Clontech) using TransIT-LT1 (Mirus Bio, Madison, WI). Supernatants were collected at 48 h after transfection and used directly for transduction of the cells after 0.45 μM filtration. The transduced cells were enriched by sorting GFP positive cells. Greater than 95% GFP positive cells were used for the assays. Knock-down efficiency was measured by real-time PCR as well as western blot and overexpression of the proteins was confirmed by western blot.
Quantitative real time PCR
Total RNA was isolated with RNAeasy kit (Qiagen) and used to synthesize complementary DNAs by ImProm II reverse transcriptase kit (Promega, Madison, WI). Real-time PCR was performed using TaqMan Universal PCR Master Mix and TaqMan probes (Applied Biosystems, Carlsbad, CA) on an automatic instrument (7900HT Fast Real-Time PCR System, Applied Biosystems). PCR probe sets for 18S, DTX1, HES1, MYC, PS1, and PS2 were obtained from Applied Biosystems (Taqman Gene Expression Assay Kits). Each sample was run in triplicates. For normalization of gene expression, 18S rRNA probe was used as an internal control.
Western blotting
Cells were harvested and incubated in RIPA buffer supplemented with protease and phosphatase inhibitors. Following centrifugation, total protein concentrations in the lysates were determined using Biorad DC Assay kit (Biorad, Hercules, CA). The same amount of total protein lysates were run on an SDS-PAGE gel and transferred to PVDF membrane. After blocking with 5% milk in TBST buffer, membranes were probed with antibodies against Notch1 (Santa Cruz), phospho-S10-Histone H3 (Millipore), PARP (Santa Cruz), and β-actin (Sigma-Aldrich). Detection was done using HRP-conjugated secondary antibodies (Cell signaling, Danvers, MA) and ECL chemiluminesence (GE Healthcare, Piscataway, NJ).
Statistical Analysis
GraphPad Prism (GraphPad Software, Inc. La Jolla, CA) was used for all statistical analysis. Results were plotted on graph as mean ± SD of 3 independent experiments. Statistical p values were calculated using analysis of variance (ANOVA), as applicable (*, p < 0.05; **, p < 0.01; ***, p< 0.001).
Results
DAPT synergizes with VCR in inducing cell death of GSI-resistant T-ALL
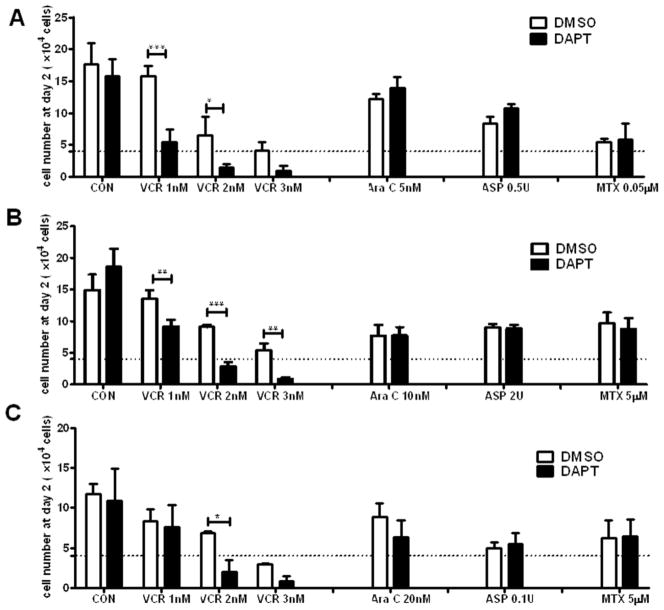
Although GSI does not have anti-tumor effect as a single agent in the GSI-resistant T-ALL, we reasoned that suppressing Notch1 activation with GSI may sensitize these cells to anti-leukemic agents. To address this question, Jurkat, CEM, and P12 cells were chosen because these cell lines have repeatedly shown to be GSI-resistant by different research groups [6,10,11,21]. We tested anti-leukemic agents that are currently used in clinics on the GSI-resistant cell lines in the presence of DAPT or DMSO (vehicle). As expected, DAPT alone did not have any effects on cell viability. However, DAPT significantly decreased viable cell numbers when combined with VCR but not with other anti-leukemic drugs (MTX, ASP, and Ara C) in all cell lines tested (Fig. 1).
Fig. 1. DAPT synergizes with VCR in killing GSI-resistant T-ALL.
GSI-resistant NOTCH1 mutant T-ALL lines (a, Jurkat; b, CEM; c P12, 4X104 cells/24well, Dotted lines) were treated with the indicated doses of chemodrugs (VCR, Vincristine; Ara C, Cytarabine; ASP, Asparaginase; MTX, Methotrexate) in the presence of DAPT (10 μM) or DMSO for 48 h. At the end of the culture, the viable cell numbers were enumerated by counting the cells with intact morphology after staining with trypan blue. All results are presented as mean ± SD of triplicate assays. The statistical significance of differences was determined by ANOVA test; *, p < 0.05; **, p < 0.01; ***, p< 0.001
DAPT enhances VCR-induced apoptosis in T-ALL
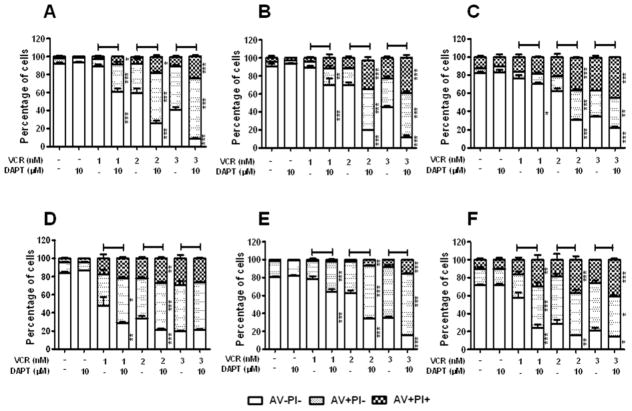
Next, we determined whether DAPT increased cell death triggered by VCR via apoptosis. Jurkat, CEM, and P12 cells were treated with increasing doses of VCR in the presence or absence of DAPT for 48 h and Annexin V and PI costaining was performed, followed by flow cytometry analysis. VCR increased early and late apoptotic populations in a dose-dependent manner (Fig. 2a-c). DAPT further increased the apoptotic Annexin V+ cell populations induced by VCR. Concomitantly, the proportion of Annexin V- live cell population significantly reduced.
Fig. 2. DAPT enhances VCR-induced apoptosis in T-ALL.
GSI-resistant NOTCH1 mutant TALL lines (a, Jurkat; b, CEM; c, P12), GSI-sensitive NOTCH1 mutant T-ALL lines (d, KOPT; e, HSB-2), and a GSI-resistant wild-type NOTCH1 T-ALL line (f, Loucy) were treated with varying concentrations of VCR (1–3 nM) and/or DAPT (10 μM) as indicated for 48 h. Annexin V (AV) and Propidium Iodide (PI) binding was measured by flow cytometry. The percentage of viable cells (AV-PI-), early apoptotic cells (AV+PI-), and late apoptotic cells (AV+PI+) is graphed. All results are presented as mean ± SD of triplicate assays. The statistical significance of differences was determined by ANOVA test; *, p < 0.05; **, p < 0.01; ***, p< 0.001
We further determined if the synergistic effect of DAPT in combination with VCR was exclusive to GSI-resistant T-ALL cell lines. When GSI-sensitive cell lines, KOPT and HSB-2, were treated with DAPT in conjunction with VCR, DAPT enhanced VCR-induced apoptosis in these sensitive cell lines as well (Fig. 2d and e). Although these cell lines have been reported to be sensitive to GSI treatment [6,12], DAPT alone did not induce apoptosis at 48 h. This data is consistent with previous observations that it takes at least 4–7 days for GSI to induce apoptosis in GSI-sensitive T-ALL cell lines [6,11]. These data showed that DAPT enhanced VCR-induced apoptosis in TALL cells, irrespective of their GSI sensitivity.
The DAPT effect in combination with VCR is not off-target pharmacological effect
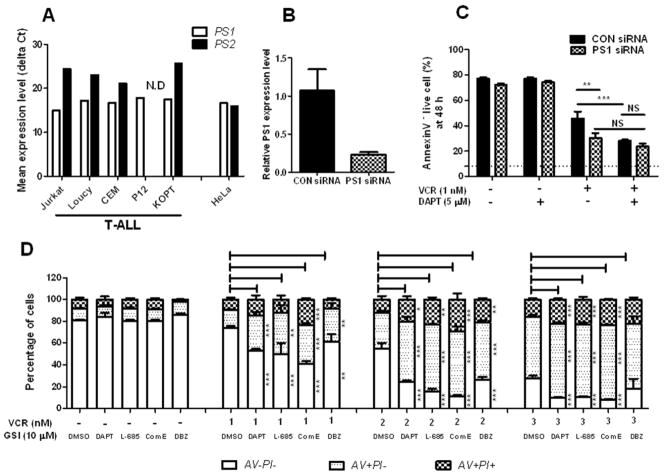
To exclude the possibility that the DAPT effect is the result of off-target pharmacological effect, PS, the catalytic component of γ-secretase complex [22,23], was knocked down in T-ALL cells. First, the expression levels of two PS isoforms (PS1 and PS2) in T-ALL cells were examined by quantitative real-time PCR. Unlike HeLa cells, which express both isoforms equally, the PS1 expression level was significantly higher than PS2 (more than 20 fold) in the T-ALL cell lines (Fig. 3a). This expression data on the transcription level was consistent with the expression on the protein level reported by Placanica et al [24]. In the report, it has been shown that HeLa expressed both PS1 and PS2 on the protein level whereas Jurkat cells expressed PS1 but not PS2 [24]. Since it appeared that PS1 is a dominant isoform in human T-ALL cell lines, PS1 was knocked down in Jurkat cells using siRNAs and the sensitivity of PS1 K/D Jurkat cells to VCR and VCR plus DAPT treatment was examined. Reducing PS1 expression sensitized Jurkat cells to VCR treatment, and decreased the DAPT effect in combination with VCR (Fig. 3c). This data indicates that the DAPT effect in combination with VCR is not off-target pharmacologic effect.
Fig. 3. The GSI effect in combination with VCR is not off-target pharmacological effect.
(a) The expression of presenilin isoforms (PS1 and PS2) in T-ALL cell lines was determined by quantitative RT-PCR. Mean expression levels (delta Ct) of each transcript are presented as cycle threshold (Ct) values normalized to the endogenous 18S rRNA. N.D; not detected. (b) PS1 was knocked down in Jurkat cells using siRNAs. Knock-down efficiency was determined by quantitative RT-PCR. PS1 expression level in CON siRNAs transfected cells was set to 1. (c) Jurkat cells transfected with PS1 siRNAs or CON siRNAs were treated with VCR alone (1 nM) or in combination with DAPT (5 μM) for 48 h and viable cells were measured with Annexin V (AV) and Propidium Iodide (PI) binding assay. Results are presented as mean ± SD of triplicate assays. The statistical significance of differences was determined by ANOVA test; *, p < 0.05; **, p < 0.01; ***, p< 0.001; NS, Not Significant. (d) Jurkat cells were treated with varying concentrations of VCR (1–3 nM) and/or 10 μM of different GSIs (DAPT, L-685,458, Compound E, and DBZ) or DMSO for 48 h. Annexin V (AV) and Propidium Iodide (PI) binding was measured by flow cytometry. The percentage of viable cells (AV-PI-), early apoptotic cells (AV+PI-), and late apoptotic cells (AV+PI+) is graphed. Results are presented as mean ± SD of triplicate assays. The statistical significance of differences was determined by ANOVA test; *, p < 0.05; **, p < 0.01; ***, p< 0.001
We further determined whether different GSIs could synergize with VCR in inducing apoptosis, similar to DAPT. When Jurkat cells were treated with other well-studied GSIs (Compound E, DBZ, and L-685,458) in conjunction with VCR, the synergistic effect was also observed with these GSIs to similar extent of DAPT (Fig. 3d).
These data suggest that GSI could synergize with VCR in inducing apoptosis of T-ALL by inhibiting γ-secretase activity.
The GSI effect in combination with VCR is not attributed to Notch inhibition
We next determined whether GSI enhances VCR-induced apoptosis by inhibiting Notch1 activation. First, we tested the combination treatment on Loucy, a T-ALL cell line that carries wild type NOTCH1 and does not have endogenous activated Notch1 [6,11]. DAPT enhanced VCR-induced apoptosis in Loucy, similar to other NOTCH1 mutant T-ALL cells (Fig. 2f). This data suggests that the GSI effect is not associated with inhibition of Notch1 signaling. To substantiate our conclusion, we performed loss-of function and gain-of-function assays. First, Notch1 was knocked down in Jurkat cells using shRNAs. The decrease in the expression of activated Notch1 (ICN1) using Notch1 shRNAs did not sensitize Jurkat cells to VCR-induced apoptosis. In addition, VCR plus DAPT induced apoptosis in Notch1 shRNAs transduced cells to the similar level as the control shRNAs transduced cells (Fig. 4a). In parallel, overexpression of activated Notch1 (ICN1) in Jurkat cells did not reverse the GSI effect on VCR-induced apoptosis (Fig. 4b). These data indicate that the GSI effect in combination with VCR is not dependent on Notch1 signaling pathway.
Fig. 4. The GSI effect in combination with VCR is not attributed to Notch inhibition.
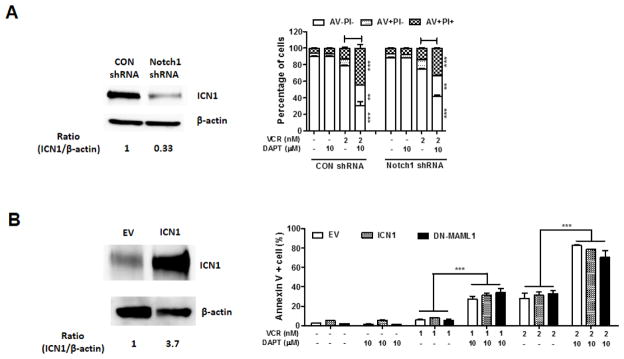
(a) Activated Notch1 protein was silenced in Jurkat cells. Sensitivity of the cells to the treatment of VCR or VCR + DAPT was evaluated by measuring apoptosis. Western blot analysis of activated Notch1 (ICN1) in Jurkat cells expressing Notch1 shRNAs or control shRNAs was performed. Beta-actin served as a loading control. (b) Jurkat cells were transduced with a construct expressing ICN1, DN-MAML1, or control empty vector (EV) and treated with VCR and/or DAPT for 48 h. Annexin V (AV) positive apoptotic cells were measured by flow cytometry to assess the sensitivity of the cells to the treatment of VCR or VCR + DAPT. Western blot analysis of ICN1 in Jurkat cells expressing ICN1 or EV was performed. Beta-actin served as a loading control. Results are presented as mean ± SD of triplicate assays. The statistical significance of differences was determined by ANOVA test; *, p < 0.05; **, p < 0.01; ***, p< 0.001
Since GSI inhibits all Notch signaling by Notch 1–4, we determined whether redundancy in Notch signaling by other Notch paralogs in T-ALL cells contributed to the GSI effect. To address that, Jurkat cells were transduced with dominant negative mastermind-like 1 (DN-MAML1), which inhibits the transcriptional activities of all Notch [9,25]. The DN-MAML1 transduced cells expressed NOTCH target genes (DTX1, HES1, and MYC) significantly less compared to the empty vector transduced cells, showing the capacity of DN-MAML1 to inhibit NOTCH transcriptional activities in Jurkat cells (Supplementary Fig. 1). However, the inhibition of Notch signaling by DN-MAML1 did not sensitize Jurkat cells to VCR treatment (Fig. 4b). Moreover, DAPT enhanced VCR-induced apoptosis in the DN-MAML1 transduced cells to the similar level as in the empty vector transduced cells (Fig. 4b). All together, these data point to the conclusion that GSI enhances VCR-induced apoptosis in a Notch signaling independent manner.
DAPT augments VCR-induced mitotic arrest
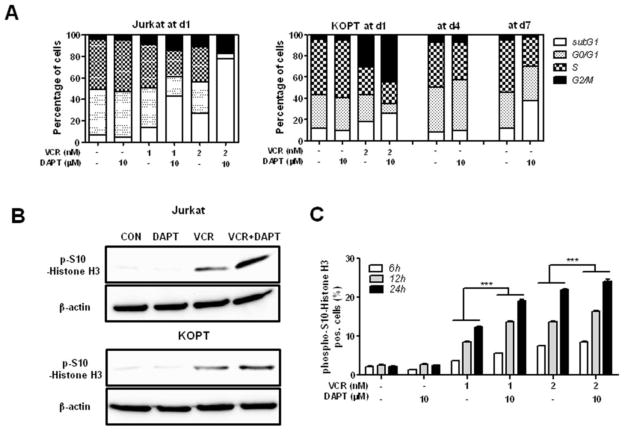
To determine the mechanism responsible for the synergism between GSI and VCR, cell cycle analysis was performed. A GSI-resistant cell line, Jurkat and a GSI-sensitive cell line, KOPT were treated with VCR and/or DAPT for 24 h and PI staining was performed to determine cell cycle progression (Fig. 5a). VCR increased the G2/M population compared to the control in both cell lines. Addition of DAPT to VCR further increased the G2/M population while DAPT treatment alone did not have any effect on cell cycle. To determine whether co-treatment of VCR and DAPT arrests cells in G2 phase or M phase, the expression of phospho-S10-histone H3 (mitosis marker) was examined by Western blot at 24 h post- treatment and compared to the single drug treatment (Fig. 5b). DAPT treatment alone showed a basal level of phospho-S10-histone H3, similar to the control. As expected, VCR treatment increased the expression of phospho-S10-histone H3, indicating an increase in the mitotic cell population. The addition of DAPT to VCR further induced phospho-S10-histone H3 expression. The kinetic experiment using KOPT cells further confirmed the western blot data. DAPT increased VCR-induced mitotic cell population as early as 6 h and gradually afterwards (Fig. 5c).
Fig. 5. GSI augments VCR-induced mitotic arrest.
Jurkat and KOPT cells were treated with DAPT (10 μM) and/or VCR (1, 2 nM) for different time periods. (a) Cell cycle progression was analyzed after PI staining. The percentage of cells in each cell cycle phase is graphed, and one representative data is presented. (b) Cell lysates at 24 h were analyzed for phospho-S10-Histone H3 by Western blot. Beta-actin served as a loading control. (c) Cell population in mitotic phase was measured by staining with phospho-S10-Histone H3. The percentage of phospho-S10-Histone H3 positive cells is presented. All results are presented as mean ± SD of triplicate assays. The statistical significance of differences was determined by ANOVA test; *, p < 0.05; **, p < 0.01; ***, p< 0.001
Since it has been reported that GSI-sensitive T-ALL cell lines such as KOPT arrest at G0/G1 phase of cell cycle following GSI treatment as a result of Notch inhibition, we also monitored cell cycle progression in KOPT cells for long term (7 days) (Fig. 5a). Although DAPT treatment for a short term (1–2 days) did not show any effect on cell cycle progression, DAPT notably reduced S phase cell population and induced apoptotic subG1 population at d7, consistent with the previous reports [6,11,26].
Taken together, these results indicate that GSI induces a significant accumulation of mitotic cells when combined with VCR.
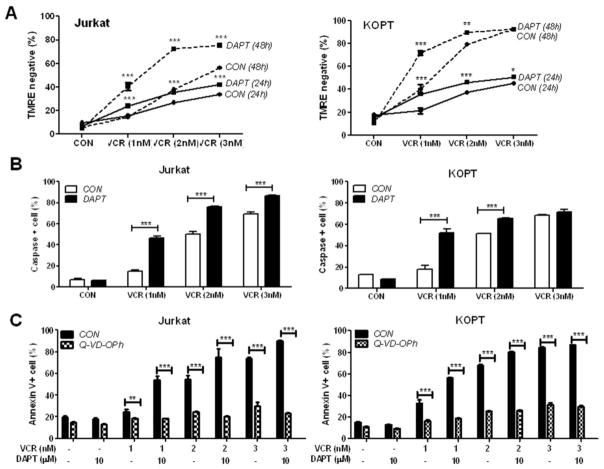
DAPT accelerates VCR-triggered loss of mitochondrial membrane integrity and caspase-mediated apoptosis
The addition of DAPT to VCR led to an increase in apoptosis following mitotic arrest evidenced by an increase in the subG1 population (Fig. 5a) as well as Annexin V+ population (Fig. 2). Hence, the apoptotic pathway induced by VCR plus DAPT was further investigated. Since it has been reported that anti-leukemic drugs including VCR activate the intrinsic apoptotic pathway in ALL samples [27], we first examined the integrity of mitochondrial membrane in Jurkat and KOPT, following treatment with VCR or VCR plus DAPT using TMRE staining (Fig. 6a). Although loss of mitochondrial membrane potential (MMP) is an early event in the intrinsic apoptotic pathway, mitochondrial depolarization was more profound at 48 h than 24 h post-treatment in both cell lines. In addition, DAPT significantly increased TMRE-negative, MMP-compromised population when combined with VCR. DAPT alone did not have any effect on the mitochondrial membrane permeability.
Fig. 6. GSI accelerates VCR-triggered loss of mitochondrial membrane integrity and caspase-mediated apoptosis.
Jurkat and KOPT were treated with varying concentrations of VCR and/or 10 μM DAPT for 48 h unless indicated. (a) Loss of mitochondrial membrane potential was assessed by TMRE staining. TMRE negative cells were quantified and are presented as percentage. (b) Poly-caspase positive cells were quantified after staining with polycaspase FLICA SR-VAD-FMK reagent and are presented as percentage. (c) Cells were treated as indicated with or without preincubation with 20 μM pan- caspase inhibitor, Q-VD-OPh. Apoptosis was assessed after 48 h using Annexin V/PI staining. Apoptotic Annexin V-positive cells are shown as percentage. All results are presented as mean ± SD of triplicate assays. The statistical significance of differences was determined by ANOVA test; *, p < 0.05; **, p < 0.01; ***, p< 0.001
We further examined the involvement of caspases that are activated upon cytochrome c release following loss of MMP (Fig. 6b). To detect active caspases, fluorochrome inhibitor of poly-caspases (SR-VAD-FMK) reagent which binds to active caspases was added to the cells at 48 h after the drug treatment and active caspase-positive cells were quantified using FACS analysis. Adding DAPT to VCR led to a significant increase in the caspase-positive population, consistent with TMRE staining data shown in Fig. 6a. The effector caspase-3 activation was further confirmed by the cleavage of PARP (Supplementary Fig. 2). The cleaved form of PARP induced by VCR was increased by addition of DAPT in both cell lines. To evaluate whether caspase activation is required for the observed apoptosis, the cells were treated with VCR and/or DAPT in the presence or absence of pan-caspase inhibitor, Q-VD-OPh (Fig. 6c). Pretreatment with Q-VD-OPh almost completely abrogated apoptosis induced by VCR as well as VCR plus DAPT in both Jurkat and KOPT cells.
These data indicate that the intrinsic apoptosis pathway including loss of MMP and caspase activation significantly contributes to apoptosis induced by the combination of VCR and GSI.
Discussion
Blocking Notch signaling by inhibiting γ-secretase activity with GSI has been proposed as a new therapeutic strategy to treat T-ALL, in which more than 50% of patients have aberrant Notch signaling due to activation mutations in NOTCH1. Now, several GSIs are in the clinical trials to test their efficacy for T-ALL (http://clinicaltrials.gov). Although GSI showed some promise in the preclinical settings, there were some hindrances to be overcome before being translated into clinic. One of them is the lack of a correlation between the presence of NOTCH1 activating mutations and the response to GSI treatment. In some T-ALL cells harboring activating mutations in NOTCH1, GSI does not show anti-leukemic effects although GSI depletes the intracellular activated Notch1 protein in these GSI-resistant cells [11]. In the present study, we tested the hypothesis that GSI may enhance the cytotoxic effects of current standard anti-leukemic agents in these GSI-resistant T-ALL cells. In the process, we unexpectedly discovered that GSI synergizes with VCR (anti-mitotic drug) in a specific manner. GSI augments VCR-induced mitotic arrest, which eventually leads to apoptotic cell death. The synergism between VCR and GSI was not limited to GSI-resistant T-ALL cells but also observed in GSI-sensitive TALL cells.
The synergism between GSI and VCR in GSI-sensitive T-ALL cells seems to be at odds with the previous reports that GSI induces cell cycle block at G0/G1 phase in GSI-sensitive T-ALL cells because VCR induces cell cycle arrest at mitotic phase and a concurrent reduction in the population of G0/G1 phase. One might think that these two drugs antagonize each other. However, we found that GSI treatment alone for a short time such as 1–2 days did not have any effect on either cell cycle progression or cell viability in GSI-sensitive T-ALL cells, similar to GSI-resistant T-ALL cells. Thus, GSI appeared not to interfere with the action of VCR. Worth mentioning is that a previous report showed mixed results for combination of VCR and GSI (i.e., Compound E) in GSI-sensitive T-ALL cells [12]. In that report, Compound E synergized with VCR in some T-ALL cells while antagonized in others [12]. The discrepancy between our data and theirs may be explained by the difference in the treatment schedule. In our study, the cells were treated with GSI and VCR simultaneously and cell viability was assessed at day 2 whereas cells were pre-treated with Compound E for 5–9 days and then treated with the combination of VCR and Compound E for another 2–3 days in the study reported by De Keersmaecker et al.
Furthermore, we presented evidence that the synergistic GSI effect in combination with VCR is not due to inhibition of Notch signaling. First, depletion of endogenously activated Notch signal by either knocking down Notch1 or overexpressing DN-MAML1 did not sensitize the cells to VCR-induced apoptosis. Second, overexpression of activated Notch1 did not reverse the GSI effect on VCR-induced apoptosis. These data indicate that the mode of action of GSI in conjunction with VCR is different from the disclosed GSI effect as Notch inhibitors. This novel GSI effect appears not to be limited to T-ALL cell lines. Previously, Katano group reported that GSI synergized with Paclitaxel but not with other chemotherapeutics (i.e., 5-FU, Gemcitabine, Cisplatin, and Camptothecin) in colon and pancreatic cancer cells in a Notch independent manner [28,29]. Taken together, it is likely that GSI has a functional role in the mitotic phase of cell cycle.
We hypothesize that there may be one or more of γ-secretase substrates that play a role in mitosis-related events in the γ-secretase processed form, and blocking the cleavage of the substrates using GSI may affect the fidelity of mitosis. A couple of reports seem to support our hypothesis. First, PS1, an essential component for γ-secretase activity, turned out to be one of the signature genes in mitotic phase of HeLa cells when a global gene expression profiling study was performed [30]. Second, overexpression of PS1 in an immortalized epithelial cell line (i.e., hTERT-HME1) resulted in frequent abnormalities in mitotic spindle structures [31].
Although the target γ-secretase substrates that are involved in the observed GSI effect remain to be identified, our data have implications in treating T-ALL. VCR is used universally as a first-line agent in the treatment of leukemia. Despite the fact that VCR is only one of multiple drugs used in combination chemotherapy regimens to treat leukemia, a patient’s initial response to VCR is considered one of the strongest predictors of outcome. Consistently, VCR resistance in mouse xenografts have shown to be associated with strong prognosis of relapse [32,33]. Hence, incorporating GSI into the conventional regimen containing VCR may offer therapeutic advantage by potentiating VCR treatment in leukemia patients. However, we acknowledge that our study has several limitations. First, it still remains to be determined whether the observed synergism with the cell lines is reproducible with primary patient samples. Second, in vivo studies using representative and well-characterized xenograft and genetic animal models of TALL will be necessary to evaluate the benefit of this combination therapy. Third, selective toxicity of the combination treatment has to be assessed.
Overall, we demonstrated here that GSI could potentiate VCR treatment in T-ALL cells. This raises the possibility that GSI may be used as adjuvant in combination with VCR in T-ALL patients.
Supplementary Material
Acknowledgments
We thank the Louisiana Cancer Research Consortium FACS core for flow cytometry analysis (P20GM103518). This work was supported by NIH grants P20GM103501 to SOY and R01CA121039 to YSC.
Abbreviations
- T-ALL
T-cell acute lymphoblastic leukemia
- GSI
γ-secretase inhibitor
- PS
Presenilin
- ICN1
intracellular domains of human Notch1
- DN-MAML1
dominant negative mastermind-like 1
- VCR
Vincristine
- Ara C
Cytarabine
- MTX
Methotrexate
- ASP
Asparaginase
- Jurkat
Jurkat-E6
- CEM
CCRF-CEM
- P12
P12-Ichikawa
- KOPT
KOPT-K1
- HSB-2
CCRF-HSB-2
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- 1.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 2.Kraszewska MD, Dawidowska M, Szczepanski T, Witt M. T-cell acute lymphoblastic leukaemia: recent molecular biology findings. Br J Haematol. 2012;156:303–315. doi: 10.1111/j.1365-2141.2011.08957.x. [DOI] [PubMed] [Google Scholar]
- 3.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122:3398–3406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 5.Koch U, Radtke F. Notch in T-ALL: new players in a complex disease. Trends Immunol. 2011;32:434–442. doi: 10.1016/j.it.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 7.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Jette C, Kanki JP, Aster JC, Look AT, et al. NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia. 2007;21:462–471. doi: 10.1038/sj.leu.2404546. [DOI] [PubMed] [Google Scholar]
- 9.Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110:278–286. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Keersmaecker K, Lahortiga I, Mentens N, Folens C, Van Neste L, et al. In vitro validation of gamma-secretase inhibitors alone or in combination with other anti-cancer drugs for the treatment of T-cell acute lymphoblastic leukemia. Haematologica. 2008;93:533–542. doi: 10.3324/haematol.11894. [DOI] [PubMed] [Google Scholar]
- 13.Samon JB, Castillo-Martin M, Hadler M, Ambesi-Impiobato A, Paietta E, et al. Preclinical analysis of the gamma-secretase inhibitor PF-03084014 in combination with glucocorticoids in T-cell acute lymphoblastic leukemia. Mol Cancer Ther. 2012;11:1565–1575. doi: 10.1158/1535-7163.MCT-11-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda S, Kumano K, Suzuki T, Tomita T, Iwatsubo T, et al. Dual antitumor mechanisms of Notch signaling inhibitor in a T-cell acute lymphoblastic leukemia xenograft model. Cancer Sci. 2009;100:2444–2450. doi: 10.1111/j.1349-7006.2009.01328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tammam J, Ware C, Efferson C, O'Neil J, Rao S, et al. Down-regulation of the Notch pathway mediated by a gamma-secretase inhibitor induces anti-tumour effects in mouse models of T-cell leukaemia. Br J Pharmacol. 2009;158:1183–1195. doi: 10.1111/j.1476-5381.2009.00389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei P, Walls M, Qiu M, Ding R, Denlinger RH, et al. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010;9:1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- 17.Rao S, O’Neil J, Liberator C, Hardwick J, Dai X, et al. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res. 2009;69:3060–3068. doi: 10.1158/0008-5472.CAN-08-4295. [DOI] [PubMed] [Google Scholar]
- 18.Cullion K, Draheim K, Hermance N, Tammam J, Sharma V, et al. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009;113:6172–6181. doi: 10.1182/blood-2008-02-136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46:364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, et al. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 23.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, et al. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 24.Placanica L, Chien JW, Li YM. Characterization of an atypical gamma-secretase complex from hematopoietic origin. Biochemistry. 2010;49:2796–2804. doi: 10.1021/bi901388t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maillard I, Weng AP, Carpenter AC, Rodriguez CG, Sai H, et al. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 26.Lewis HD, Leveridge M, Strack PR, Haldon CD, O'Neil J, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14:209–219. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 27.Holleman A, den Boer ML, Kazemier KM, Janka-Schaub GE, Pieters R. Resistance to different classes of drugs is associated with impaired apoptosis in childhood acute lymphoblastic leukemia. Blood. 2003;102:4541–4546. doi: 10.1182/blood-2002-11-3612. [DOI] [PubMed] [Google Scholar]
- 28.Akiyoshi T, Nakamura M, Yanai K, Nagai S, Wada J, et al. Gamma-secretase inhibitors enhance taxane-induced mitotic arrest and apoptosis in colon cancer cells. Gastroenterology. 2008;134:131–144. doi: 10.1053/j.gastro.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Tasaka T, Akiyoshi T, Yamaguchi K, Tanaka M, Onishi H, et al. Gamma–secretase complexes regulate the responses of human pancreatic ductal adenocarcinoma cells to taxanes. Anticancer Res. 2010;30:4999–5010. [PubMed] [Google Scholar]
- 30.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boeras DI, Granic A, Padmanabhan J, Crespo NC, Rojiani AM, et al. Alzheimer's presenilin 1 causes chromosome missegregation and aneuploidy. Neurobiol Aging. 2008;29:319–328. doi: 10.1016/j.neurobiolaging.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lock RB, Liem N, Farnsworth ML, Milross CG, Xue C, et al. The nonobese diabetic/severe combined immunodeficient (NOD/SCID) mouse model of childhood acute lymphoblastic leukemia reveals intrinsic differences in biologic characteristics at diagnosis and relapse. Blood. 2002;99:4100–4108. doi: 10.1182/blood.v99.11.4100. [DOI] [PubMed] [Google Scholar]
- 33.Liem NL, Papa RA, Milross CG, Schmid MA, Tajbakhsh M, et al. Characterization of childhood acute lymphoblastic leukemia xenograft models for the preclinical evaluation of new therapies. Blood. 2004;103:3905–3914. doi: 10.1182/blood-2003-08-2911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.