Abstract
Arabidopsis aldehyde oxidase 3 (AAO3) is an enzyme involved in abscisic acid (ABA) biosynthesis in response to drought stress. Since the enzyme catalyzes the last step of the pathway, ABA production sites may be determined by the presence of AAO3. Here, AAO3 localization was investigated using AAO3 promoter:AAO3-GFP transgenic plants and by an immunohistochemical technique. AAO3-GFP protein exhibited an activity to produce ABA from abscisic aldehyde, and the transgene restored the wilty phenotype of the aao3 mutant. GFP-fluorescence was detected in the root tips, vascular bundles of roots, hypocotyls and inflorescence stems, and along the leaf veins. Intense immunofluorescence signals were localized in phloem companion cells and xylem parenchyma cells. Faint but significant GFP- and immuno-fluorescence signals were observed in the leaf guard cells. In situ hybridization with antisense AAO3 mRNA showed AAO3 mRNA expression in the guard cells of dehydrated leaves. These results indicate that the ABA synthesized in vascular systems is transported to various target tissues and cells, and also that the guard cells themselves are able to synthesize ABA.
Abscisic acid (ABA) is well known as a regulator of various stress responses in plants and for the seed maturation and dormancy (Zeevaart and Creelman, 1988). In addition, ABA is also shown to be involved in the regulation of morphogenesis of submerged plants (Anderson, 1978; Kuwabara et al., 2003) and the shoot and root growth maintenance under nonstress conditions (Sharp et al., 2000; Cheng et al., 2002; Sharp and LeNoble, 2002).
Recent studies based on the cloning of genes using ABA-deficient mutants have provided a better understanding of the ABA biosynthetic pathway in higher plants and the complex regulation of ABA biosynthesis (for review, see Cutler and Krochko, 1999; Seo and Koshiba, 2002; Schwartz et al., 2003; Xiong and Zhu, 2003). The ABA precursor, carotenoid, is converted into xanthophylls in plastids. Xanthoxin is then cleaved from xanthophyll and converted into ABA in the cytosol. The enzymes involved in the plastidial steps are zeaxanthin epoxidase (ZEP) and 9-cis-epoxycarotenoid dioxygenase (NCED). To date, it has been supposed that the carotenoid cleavage reaction is the primary regulated step in the pathway controlling ABA synthesis (Duckham et al., 1991; Rock and Zeevaart, 1991; Marin et al., 1996; Schwartz et al., 1997; Tan et al., 1997; Qin and Zeevaart, 1999). The enzymes corresponding to the cytosolic steps have been characterized by isolation of the Arabidopsis ABA2 and AAO3 genes. The ABA2 gene encodes a short-chain dehydrogenase/reductase (SDR1) which catalyzes the conversion of xanthoxin to abscisic aldehyde (Cheng et al., 2002; González-Guzmán et al., 2002). Among the four Arabidopsis aldehyde oxidase genes (AAO1-4), the AAO3 gene encodes an AO isoform, AOδ, which has an activity to produce ABA from abscisic aldehyde (Sekimoto et al., 1998; Seo et al., 2000a). Thus, the principal enzymes and genes involved in the ABA biosynthesis pathway have been identified in Arabidopsis.
The next important question to be addressed is how ABA biosynthesis is regulated in response to different stresses and whether it is regulated in a tissue-specific manner. In Arabidopsis, the presence of gene families of AtNCED and AAO might contribute to such complex regulation systems. Indeed, several studies have shown that independent regulation of these gene-expressions occurs in different tissues and/or developmental stages. For example, AtNCED3 is strongly induced by desiccation in detached leaves, while other AtNCEDs seem to be involved in ABA synthesis during seed maturation and developmental phases (Iuchi et al., 2001; Hobo et al., 2002; Tan et al., 2003). AAO3 is abundant in leaves and is necessary for drought-inducible ABA accumulation in the leaves, while other AAO(s) would be involved in ABA synthesis in the roots or seeds (Seo et al., 2000a, 2000b; Seo and Koshiba, 2002). In contrast, ABA2 is a member of a large SDR gene family, but from the sequence analysis, ABA2 (SDR1) is likely to have a unique function for the conversion of xanthoxin to abscisic aldehyde (Cheng et al., 2002). This means that ABA2 has a nonredundant function for abscisic aldehyde production, at least in Arabidopsis. Expression analysis of an ABA2 promoter-fused reporter gene (GUS) revealed that ABA2 had a highly restricted localization in vascular tissues. The spatial and temporal expression patterns of ABA2 seemed to be distinct from those of other ABA biosynthetic genes, such as ZEP and ABA3 (Cheng et al., 2002). These results led to the hypothesis that interorgan, intercellular, and interorganelle transport of ABA and/or its precursors is required for the strict ABA sorting to the physiological function site(s).
In this study, we focused on the localization of AAO3 protein in Arabidopsis, since AAO3 catalyzes the last step of ABA biosynthesis and therefore the sites of ABA production may correspond directly to AAO3 localization. The aao3 mutant exhibits a wilty phenotype and almost no increase in the endogenous ABA level after drought-treatment, and therefore AAO3 is necessary for ABA production in response to water stress (Seo et al., 2000b). Alternatively, AAO3-mediated ABA production may have an important role for ABA mobilization to the actual sites of ABA action in the water stress response. Here, we investigated AAO3 localization using two approaches. First, transgenic Arabidopsis lines expressing fusion proteins between AAO3 and GFP driven by the native AAO3 promoter were produced, and the GFP-fluorescence was observed. Second, immunohistochemical analyses using a specific mouse anti-AAO3 antibody were performed comparing the wild-type (WT) and aao3 mutant plants. The results provide new aspects for the possible sites of ABA biosynthesis in relation to the signal flow from drought stress perception to stomatal closure.
RESULTS
Functional Expression of AAO3-GFP Fusion Protein in Transgenic Plants
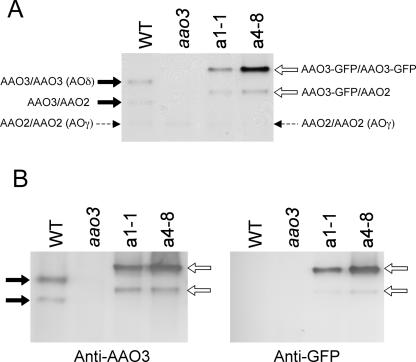
To observe the pattern of AAO3 protein localization, transgenic Arabidopsis plants expressing the AAO3-GFP fusion protein driven by the AAO3 promoter were generated in the aao3 mutant, as well as in the WT Columbia (Col) background. Eight lines for the aao3 mutant and 18 lines for the Col background were obtained, and all lines showed the same expression pattern of the fusion protein (data not shown). First, we investigated whether the AAO3-GFP protein expressed in the transgenic plants was enzymatically and physiologically functional. Crude enzyme extracts were obtained from transgenic plants (two typical lines of the aao3 background, a1-1 and a4-8) and the zymogram patterns using abscisic aldehyde as the substrate were observed. As shown in Figure 1A, two bands were detected in the WT extract. AOδ, the AAO3 homodimer, exhibited an activity band with abscisic aldehyde (upper band with a black arrow). AOγ, the AAO2 homodimer, showed almost no activity with abscisic aldehyde (Akaba et al., 1999; Seo et al., 2000a; the position is indicated by a dotted arrow); however, the heterodimer of AAO3 and AAO2 had some activity (lower band with a black arrow; Seo et al., 2000a). The aao3 mutant did not exhibit these activity bands. The transgenic plants showed different zymogram patterns from those of the WT. The upper band (white arrow in Fig. 1A), displaying a slower mobility than WT AOδ, had abscisic aldehyde oxidase activity. The intensity of the upper band varied among the transgenic lines observed: for example, in a4-8 the band showed high intensity, while in a1-1 the intensity was at the same level as the WT AOδ (Fig. 1A). This new band detected in the transgenic plants was recognized by an anti-AAO3 antibody, which had been prepared as described below (see Fig. 4), as in the WT plants (Fig. 1B, left). The new band was also recognized by an anti-GFP antibody in the transgenic plants but not in the WT plants (Fig. 1B, right). These results indicate that the upper band detected in the transgenic plants consisted of homodimers of the AAO3-GFP fusion product and had abscisic aldehyde oxidase activity. The lower band in the transgenic lines was also recognized by an anti-AAO2 antibody (data not shown), suggesting that this band is a heterodimer of AAO3-GFP and AAO2, similar to that found in the WT plants. The abscisic aldehyde oxidase activity of the AAO3-GFP fusion protein was confirmed by detecting ABA production from abscisic aldehyde incubated with crude extracts from the transgenic plants after HPLC, which was not detected with extracts from the aao3 mutant plants. Moreover, the intensity of activity bands using abscisic aldehyde showed correlation to the amount of the ABA production detected on HPLC, confirming these lines obtained to be expressing functionally the same protein as AAO3 protein intrinsically (data not shown).
Figure 1.
Analysis of the AAO3-GFP fusion protein by native PAGE. A, Zymograms of aldehyde oxidases (AOs) from Arabidopsis wild-type (WT), aao3 mutant (aao3), and AAO3 promoter:AAO3-GFP-introduced aao3 (a1-1 and a4-8) plants. Enzyme extracts were subjected to native PAGE, and activity bands were developed using abscisic aldehyde as the substrate. Each lane was loaded with 50 μg of protein. B, Immunoblotting with the same extracts as in A. After native PAGE, immunoblotting was performed using anti-AAO3 (left) and anti-GFP (right) antibodies. Black and white arrows indicate the activity bands of aldehyde oxidase isoforms in the WT and AAO3-GFP transgenic plants, respectively. Dotted arrows indicate the position of AOγ.
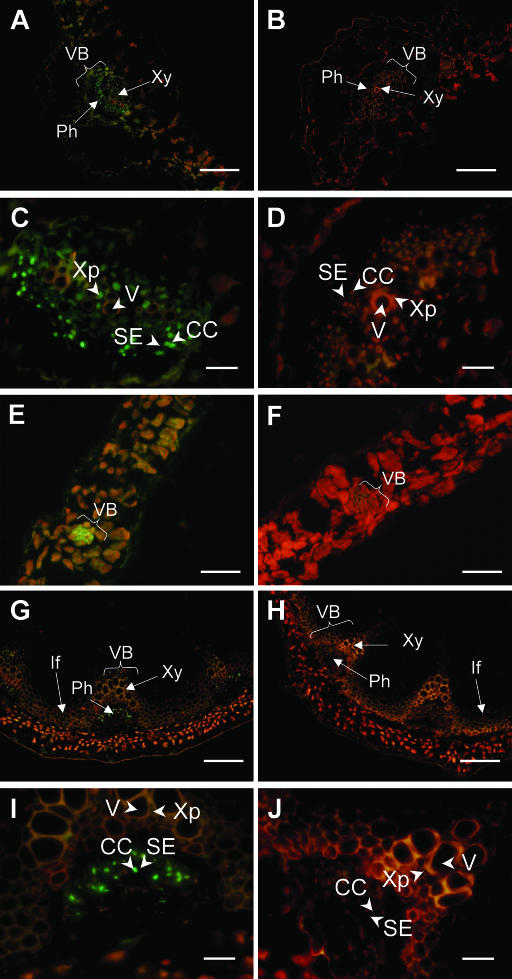
Figure 4.
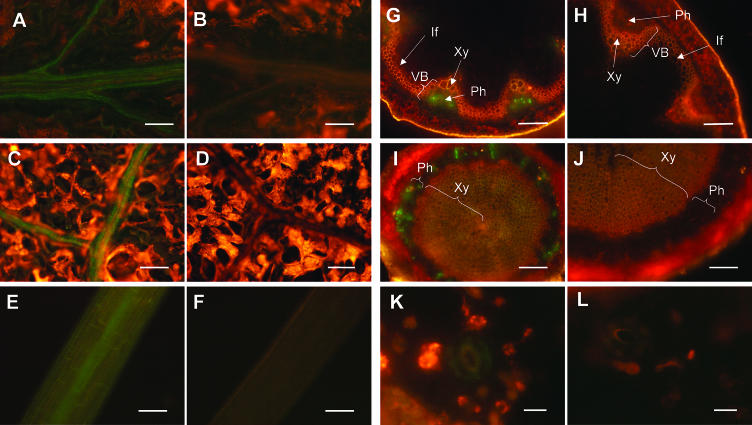
Localization of AAO3-GFP protein in AAO3-GFP transgenic aao3 plants. GFP-fluorescence images of AAO3-GFP transgenic plants (A, C, E, G, I, and K) and nontransformed plants (B, D, F, H, J, and L) are shown. The expression of AAO3-GFP protein in 1-month-old plants (A–D and G–L) and 3-d-old plants grown in the dark (E and F) were determined using epifluorescence microscopy. GFP-fluorescence images of AAO3-GFP transgenic plants are shown. A and B, GFP-fluorescence images of the main veins. C and D, GFP-fluorescence images of the lateral veins. E and F, GFP-fluorescence images of the etiolated hypocotyls. G and H, GFP-fluorescence images of the inflorescence stems. I and J, GFP-fluorescence images of the hypocotyls of a mature plant. K and L, GFP-fluorescence images of an epidermal peel. Green fluorescent signals indicate the localization of GFP-tagged AAO3 protein. Red fluorescence indicates autofluorescence of the chlorophylls. VB, vascular bundle; Ph, phloem; Xy, xylem; If, interfascicular region. Bars = 100 μm for A to J and 10 μm for K and L.
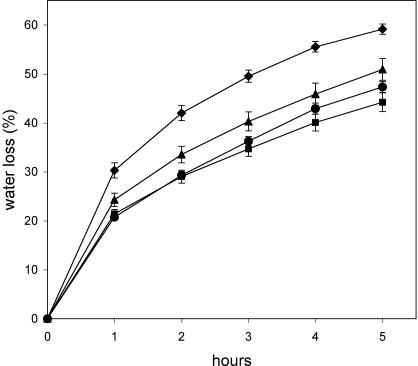
The defect in abscisic aldehyde oxidase activity in the aao3 mutant causes excess water loss and a strong wilty phenotype in the mutant plants (Seo et al., 2000b). The rate of water loss in AAO3-GFP transgenic aao3 plants was nearly the same as that in the WT plants (Fig. 2), and thus the wilty phenotype was restored. The a1-1 plants, which showed lower expression levels of AAO3-GFP protein compared to a4-8 plants, also reduced their water loss compared to the aao3 mutant. These data suggest that the AAO3-GFP fusion protein expressed in the aao3 mutant is functional and sufficient for the water stress response in vivo.
Figure 2.
Water loss in WT, aao3 mutant, and AAO3-GFP transgenic aao3 plants. The rate of water loss of detached shoots from aao3 (diamonds), Ler (squares), a1-1 (triangles), and a4-8 (circles) plants were determined by the loss of fresh weight. The figure presented is a typical result for three independent experiments. All values are means ±se.
GFP Signal in Transgenic Plants
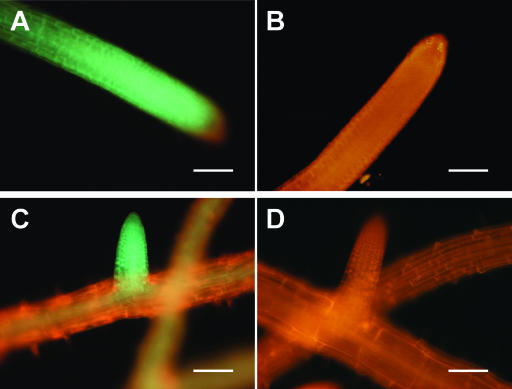
To investigate the tissue localization of the AAO3-GFP fusion protein, the GFP-fluorescence signal in the transgenic plants was observed (Figs. 3 and 4). GFP-fluorescence signals were detected throughout the whole plants, but mainly in the vasculature tissues. The vascular pattern of GFP-fluorescence was found in the roots, hypocotyls, leaves, and inflorescence stems of all stages tested (Figs. 3, A and C, and 4, A, C, E, G, and I). In the roots, in addition to the expression associated with the vasculature, such as in the root steles, AAO3-GFP was consistently highly expressed in the cell division area of both the main and lateral root tips (Fig. 3, A and C) and in the root primordium (data not shown). In the nontransgenic plants, only red signal but no GFP fluorescence was detected (Figs. 3, B and D, and 4, B, D, F, H, J, and L).
Figure 3.
Localization of AAO3-GFP protein in AAO3-GFP transgenic aao3 roots. The expressions of AAO3-GFP protein in 2-week-old plants were determined using epifluorescence microscopy as described in “Materials and Methods.” GFP-fluorescence images of AAO3-GFP transgenic plants (A and C) and nontransformed plants (B and D) are shown. A and B, GFP-fluorescence images of the main roots. C and D, GFP-fluorescence images of the lateral roots. Green fluorescent signals indicate the localization of GFP-tagged AAO3 protein. Red fluorescence indicates autofluorescence of the root tissues. Bars = 100 μm.
In the leaves, fluorescence was detected along the main and lateral leaf veins (Fig. 4, A and C). In the hypocotyls, the vasculature signal was weaker than those in the roots and leaves but was significantly detected in etiolated shoot steles (Fig. 4E). GFP-fluorescence patterns in hand-cut sections of inflorescence stems were detected in the phloem (Ph) region (Fig. 4G). Similar signals were obtained in the hypocotyls of mature plants (Fig. 4I). Since the strong red autofluorescence of the chloroplasts prevented GFP-fluorescence detection in the leaves, peeled epidermis preparations were used to investigate the guard cells. Weak signals were detected in the guard cells of transgenic plants (Fig. 4K). To confirm the presence of AAO3-GFP protein in the guard cells, an immunohistochemical analysis using an anti-GFP antibody was carried out. Faint but significant immunofluorescence was observed in the guard cells in the peeled epidermal samples (data not shown).
AAO3-Specific Antibody Preparation
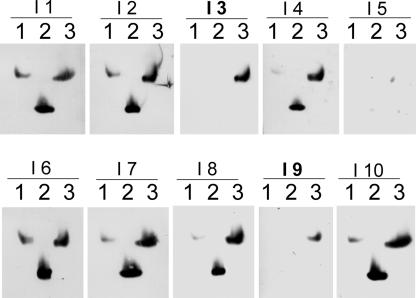
To directly elucidate the precise localization patterns of AAO3 protein, the production of AAO3-specific antibodies had been attempted, because of the high homology among the AAO gene family. Polyclonal rabbit antibodies raised against a partial peptide of AAO3 could not discriminate among AAO1, AAO2, and AAO3 proteins by immunoblotting (Akaba et al., 1999; Seo et al., 2000a, 2000b). Using the purified recombinant AAO3 protein as an antigen, 10 mice were immunized independently and the obtained immune sera (named as I1–I10) were checked for their specificity to AAO1, AAO2, and AAO3 recombinant protein by immunoblotting (Fig. 5). Unfortunately, AAO4 protein has not been obtained at either AO activity or protein level, so we could not test for crossreactivity for AAO4 protein. The result showed that among these I3 and I9 antisera specifically recognized AAO3 protein (Fig. 5). The I3 antiserum showed a higher titer than I9; thus, the I3 antiserum was used for further analyses. Protein extracts from WT ecotype Landsberg erecta of Arabidopsis (Ler) and aao3 mutant plants defective AAO3 proteins showed that the mutant plant extracts showed no immunoreactive band with the I3 antisera (see Fig. 1B).
Figure 5.
Preparation of AAO3-specific antibodies. Immunoblot analysis with various mouse antisera against AAO3 protein. Recombinant AAO1, AAO2, and AAO3 proteins were subjected to native PAGE followed by immunoblot analysis using the I1 to I10 immune serum obtained from 10 individually AAO3-injected mice. Lanes 1, 2, and 3 contain recombinant purified AAO1, AAO2, and AAO3 protein, respectively. Each lane contains 100 ng of purified protein.
Immunolocalization of AAO3 Protein
Immunohistochemical detection of AAO3 localization using the I3 antibody (Fig. 5) was performed on WT and aao3 mutant plants. In this experiment, ecotype Col was used as the WT because of the facility for sectioning tissues. The results revealed that AAO3 protein was expressed most abundantly in the vascular systems of the main and lateral veins of leaves (Fig. 6, A and E). These expression patterns were essentially the same as the GFP-fluorescence patterns in the AAO3-GFP transgenic plants (see Fig. 4, A and C). AAO3 protein was also observed in the mesophyll cells (Fig. 6, A and E), although the autofluorescence of the chloroplasts again prevented clear resolution. Signals were not detected in aao3 mutant plants (Fig. 6, B and F). Higher magnification of the main vascular bundles of leaves clearly showed that AAO3 protein was predominantly localized in the phloem companion cells (CC) next to the phloem sieve element (SE; Fig. 6, C and D). Furthermore, the xylem parenchyma cells (Xp) adjacent to the xylem vessel (V) were also labeled significantly with the anti-AAO3 antibody (Fig. 6C). In inflorescence stems, signal was detected in the phloem (Ph; Fig. 6G) which was essentially the same as the GFP-fluorescence in the transgenic plants (Fig. 4G), and higher magnification showed AAO3 localization in the companion cells (CC) and no significant signal in the xylem (Fig. 6I). The phloem cells labeled by the anti-AAO3 antibody were identified as companion cells based on their size, arrangement, and cytoplasmic density as described in DeWitt and Sussman (1995). In the hypocotyls of seedlings, a clear signal was also detected in the phloem tissue (data not shown).
Figure 6.
Immunolocalization of AAO3 protein in WT and aao3 mutant plants. Immunofluorescence images of WT (A, C, E, G, and I) and aao3 mutant (B, D, F, H, and J) plants. A and B, Transverse sections of main veins of rosette leaves. C and D, Magnified images of A and B, respectively. E and F, Transverse sections of minor veins of rosette leaves. G and H, Transverse sections of inflorescence stems. I and J, Magnified images of G and H, respectively. VB, vascular bundle; Ph, phloem; Xy, xylem; If, interfascicular region; Xp, xylem parenchyma cell; V, vessel; SE, sieve element; CC, companion cell. Bars = 100 μm for A, B, G, and H; 20 μm for C, D, I, and J; 50 μm for E and F.
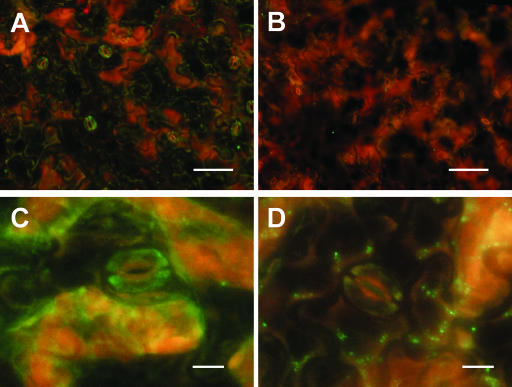
The presence of AAO3 protein in the guard cells was much weaker than that in the vascular tissues, but it was clearly observed (Fig. 7A), similar to the cases of GFP-fluorescence (Fig. 4K) and anti-GFP antibody staining in the AAO3-GFP transgenic plants mentioned above. Higher magnification showed AAO3 localization inside the guard cells (Fig. 7C). These data strongly suggest that AAO3 protein, which catalyzes the last step of ABA biosynthesis, is expressed in guard cells and acts in ABA synthesis inside the guard cells. The signals shown in Figures 6, A, C, E, G, and I, and 7, A and C, were not detected in the aao3 mutant, which has impaired AAO3 protein (Figs. 6, B, D, F, H, and J, and 7, B and D). The same patterns as those with ecotype Col were also observed with ecotype Ler that is in the background of the aao3 mutant (data not shown).
Figure 7.
Immunolocalization of AAO3 protein in WT and aao3 mutant epidermal peels. A, Immunofluorescence image of an epidermal peel from a WT plant. B, Immunofluorescence image of an epidermal peel from an aao3 mutant plant. C, Magnified image of the epidermal peel from a WT plant. D, Magnified image of the epidermal peel from an aao3 mutant plant. Bars = 50 μm for A and B, and 10 μm for C and D.
AAO3 mRNA Expression in Epidermal Cells
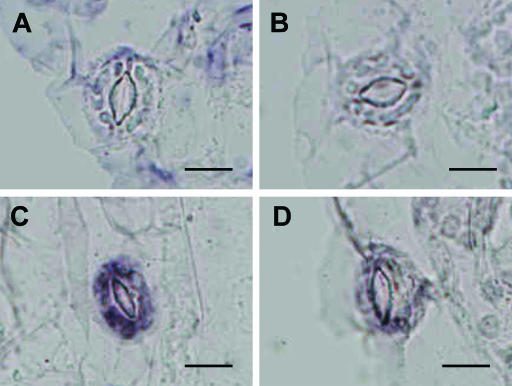
It has been demonstrated by RNA gel blotting analysis that AAO3 mRNA is rapidly induced in leaves by drought stress (Seo et al., 2000a), and it is considered that such a rapid response will be correlated with a rapid stomatal response to dehydration stress. In order to examine the cellular expression patterns of AAO3, especially in guard cells, AAO3 mRNA localization was investigated by in situ RNA hybridization using digoxigenin-labeled riboprobes. When AAO3 antisense RNA was used as the probe in sections parallel to the surface of the leaf epidermis, no detectable hybridization signal was observed in the guard cells of nonstressed rosette or wet-control leaves (Fig. 8, A and B). In contrast, intense signals were observed within the guard cells of water-stressed rosette leaves (Fig. 8C). Only background levels were detected when the sense RNA probe was used for the hybridization (Fig. 8D). Vascular tissues displayed a high background, even with the sense probe, and thus the expressions of AAO3 transcripts in vascular tissues such as phloem companion cells or xylem parenchyma cells have yet to be verified.
Figure 8.
Detection of AAO3 mRNA in guard cells by in situ hybridization. Sections of epidermis from water-stressed or nonstressed leaves were hybridized with AAO3 sense and antisense RNA probes. AAO3 mRNA-positive signals are dark blue. The AAO3 RNA sense probe was used as a negative control. A, Hybridization of the AAO3 antisense probe to a 0-h stressed leaf. B, Hybridization of the AAO3 antisense probe to a well-watered leaf. C, Hybridization of the AAO3 antisense probe with a 3-h stressed leaf. D, Hybridization of the AAO3 sense probe with a 3-h stressed leaf. Bars = 10 μm.
DISCUSSION
Recently, the possibility of dynamic mobilization of the ABA precursor and/or ABA in Arabidopsis has been proposed, since ABA2 expression was shown to be highly restricted to specific vascular tissues which are not thought to be target sites of ABA actions (Cheng et al., 2002). In the present study, we demonstrated that in GFP-AAO3 transgenic Arabidopsis plants, the fusion protein was enzymatically functional and the gene could complement the wilty phenotype of aao3 mutants. Observation of the fusion protein showed that AAO3 protein was abundantly expressed in the vascular systems of roots, hypocotyls, and inflorescence stems and in leaf veins (Figs. 3 and 4). More precise localization of AAO3 protein determined by immunohistochemical analysis clearly indicated the presence of AAO3 in the phloem companion cells and xylem parenchyma cells in the vascular tissues (Fig. 6). Furthermore, our preliminary histochemical observations with an anti-ABA2 antibody also showed expression of ABA2 protein in the vascular systems of leaves and inflorescence stems (A. Endo, H. Koiwai, T. Koshiba, unpublished data). These data suggest that at least the last two steps in the ABA biosynthesis pathway are taking place in the vascular bundle regions.
The present results show that AAO3 protein is expressed in vascular tissues, especially in phloem companion cells and xylem parenchyma cells next to the phloem sieve cells and xylem vessels, respectively, in the leaf veins and inflorescence stems (Fig. 6). Recently, much attention has been paid to the sieve element and companion cell complex (SE-CC). Due to its central roles in trafficking solutes and macromolecules from companion cells and mesophyll cells throughout the plant, and in monitoring the nature of substances passing into and out of the phloem, the complex has been proposed to be like a “busy international airport” (Oparka and Turgeon, 1999; van Bel, 2003). For example, SOS1, a putative plasma membrane Na+/H+ antiporter in Arabidopsis, is expressed preferentially in xylem parenchyma cells and root tips (Shi et al., 2002). This expression pattern indicates a role for this transporter in controlling the Na+ load of the vascular system. In addition to SOS1, the promoter activity of a hypothetical osmosensor, ATHK1, seemed to be higher in vascular bundles, especially in the roots (Urao et al., 1999). Some stress-inducible and tolerance genes/proteins, such as late-embryogenesis abundant (LEA) proteins and Ca2+-dependent protein kinase, are also expressed very highly in the vascular area (Pearce et al., 1998; Saijo et al., 2001). Furthermore, one of the ABA-inducible genes, rab16A, is detected in vascular bundles and is induced in the leaves and roots by ABA, NaCl, and desiccation treatments (Ono et al., 1996). Considering such an active function of the vascular systems, the vascular localization of ABA biosynthetic enzymes leads us to propose the following hypothesis: osmotic- and salt-stresses are monitored in the vascular tissues, and the rate of ABA biosynthesis and amount of ABA loading are regulated in the companion cells and/or xylem parenchyma cells. Then, the exuded ABA is transported to the target tissues or organs via the sieve element and/or xylem vessels. The actual movement of ABA should be determined by introducing a microtracer and using a microinjection technique, or by molecular genetic approaches.
It is well known that guard cells are the targets of ABA action for stomatal closure. Despite enormous efforts to obtain experimental evidence for the movement of ABA from the roots, leaf veins, or leaf mesophyll cells to the guard cells and to isolate ABA receptors from guard cell membranes, no clear answers have been obtained for either the source of the ABA or the ABA receptors for stomatal closure. It has been supposed that the redistribution of preexisting ABA in leaves is important for stomatal closure under stress conditions (Hartung et al., 1998), whereas the guard cells themselves have been suggested to have the capacity to produce stress-induced ABA from experiments using isolated guard cells (Weiler et al., 1982; Cornish and Zeevaart, 1986) or immunological techniques (Harris et al., 1988). In this study, it was revealed that both AAO3 protein and mRNA were localized inside in guard cells (Figs. 4K, 7, A and C, and 8C). To our knowledge, this is the first experimental evidence to show the presence of an ABA biosynthetic enzyme inside guard cells. A result reported for Commelina communis, where stomatal closure was induced by abscisic aldehyde supplied directly on epidermal strips (Raschke et al., 1975), also suggests the occurrence of the conversion activity from abscisic aldehyde to ABA in guard cells. AAO3 transcripts showed a drastic increase in response to drought stress within the guard cells (Fig. 8C) as well as at the bulk leaf level, while the bulk leaf AAO3 protein was not increased rapidly after dehydration (Seo et al., 2000a). It is still possible that, in restricted regions of a leaf such as the guard cells, AAO3 protein is increased concomitantly with the mRNA increase. However, in the present study, at least the rapid and strong induction of the AAO3 protein level in guard cells could not been detected. Precise detection of AAO3 protein in the guard cells before and after desiccation treatment using immunocytochemical analysis is now being conducted. Furthermore, the presence of posttranslational modifications of AAO3 protein such as those by ABA3 or Moco sulfurase (Bittner et al., 2001; Xiong et al., 2001) and the regulation of the protein turnover rate after water stress also need to be considered.
At present, the localization of ABA biosynthetic enzymes in guard cells has only been studied for AAO3. However, due to the presence of plastids in these cells, it is possible that whole enzyme sets for ABA biosynthesis are present inside guard cells. Additional studies on the guard cell localization of other ABA biosynthetic enzymes, such as ABA2, NCED, ZEP, and ABA3, are required, although intercellular transport of some precursors is, of course, possible (Raschke et al., 1975; Cheng et al., 2002). The introduction of microtechniques, such as precursor injection, transient expression of biosynthetic genes, and ABA determination within a guard cell, is necessary to resolve the detailed picture of ABA biosynthesis in guard cells. Furthermore, if it is confirmed that ABA synthesis occurs inside in guard cells, we also need to identify cytosolic receptors in the cells.
Root tips have been regarded as the main sites of ABA biosynthesis for a long time, and the synthesized ABA is transported to the target tissues. Intense AAO3-GFP expression was detected in the root tips (Fig. 3, A and C) and lateral root primordium (data not shown). AAO3 protein expression in roots was also confirmed by western blotting using the I3 antibody, and relatively strong signals were detected on the root sample (data not shown). These results indicate that the roots are one of the ABA synthetic sites in Arabidopsis.
The gene expression pattern of an AtNCED family in Arabidopsis has been reported recently using a quantitative real-time PCR and promoter:GUS analysis (Tan et al., 2003). It was demonstrated that the promoter activity of AtNCED3, the major stress-induced NCED in Arabidopsis, was detected in the roots, pericycle cells, flowers, developing anthers, and developing seeds, and that AtNCED3 mRNA was the dominant NCED transcript in leaves and inflorescence stems. The GUS signals were also detected in the stomatal guard cells of cotyledon leaves and petioles, and stem regions. Unfortunately, there was no investigation of the histochemical localization of NCEDs at the protein level, and it is thus currently difficult to discuss the spatial relationships between AtNCED3 and AAO3 localizations. However, it is most likely that the vascular system has some important roles in ABA production for ABA mobilization to functional targets in Arabidopsis, and that the guard cells themselves have an ABA synthetic ability. To clarify these important aspects, we are presently conducting simultaneous determinations of ABA2, AtNCED3, and AAO3 proteins in the same plants using immunohistochemical and immunocytochemical methods, in order to investigate whether or not these ABA biosynthetic protein expressions overlap in the same tissues and/or at the cellular level. Moreover, in situ hybridization experiments with their transcripts will also be valuable for elucidating the gene expression regulation in response to drought stress.
Transgenic lines of aao3 in which the tissue- or cell-specific promoter:AAO3 gene is introduced, and enhancer-trap lines of aao3 in which the AAO3 gene is expressed ectopically, will be useful in elucidating the role of the restricted localization of AAO3 in ABA biosynthesis in relation to environmental stress responses.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of Arabidopsis WT ecotype Landsberg erecta (Ler), Columbia (Col), aao3 mutant (Seo et al., 2000b), and transgenic lines were sown in pots containing vermiculite and watered with nutrient solution under constant light at 22°C. After approximately one month the plants were used for enzyme extraction, water loss determination, immunohistochemical analysis, and in situ hybridization analysis. For the detection of GFP-fluorescence, transgenic plants were grown on agar plates containing 0.5× Murashige and Skoog medium (Murashige and Skoog, 1962) without antibiotics for 2 weeks. For the observation of etiolated seedlings, seeds were sown on Murashige and Skoog agar plates and grown in the dark for 3 d.
Vector Construction and Plant Transformation
To produce transgenic plants in which GFP-tagged AAO3 protein was expressed under the control of the AAO3 promoter, the construct for the aao3 complementation test, described previously (Seo et al., 2000b), was modified. The sGFP (S65T) gene (Chiu et al., 1996) and the nopaline synthase (Nos) 3′-transcription terminator were inserted in-frame in front of the stop codon of the AAO3 gene. Transformation to the aao3 mutant was performed according to the procedure described by Bechtold et al. (1993). Transformants were selected on Murashige and Skoog agar with 50 μg mL−1 kanamycin. Plants with a single copy of the transgene insertion were selected based on segregation of the selection marker gene in T2 progeny, and homozygous transgenic lines were generated.
Enzyme Assay, Native PAGE, and Immunoblotting
For enzymatic characterization, rosette leaves were cut from aao3 mutant, WT (Ler), and transgenic plants, and stored at −80°C until use. Enzyme extraction, activity staining after native PAGE, and immunoblotting were carried out according to the methods described by Seo et al. (2000a, 2000b). An anti-GFP antibody (Santa Cruz Biotechnology) was commercially purchased.
Measurement of Water Loss in Rosettes
Water loss was determined by weighing well-watered plants that had just started to blot as described previously (Seo et al., 2000b). Aerial parts were cut from aao3 mutant, WT (Ler), and transgenic plants, placed on filter paper on a bench at ambient temperature, and weighed every hour.
Determination of AAO3-GFP Protein Localization
For the detection of GFP-fluorescence, Arabidopsis foliage leaves and roots detached from 2-week-old seedlings and etiolated hypocotyls were mounted on slides. For observation of leaf veins, fully expanded leaves were detached from mature plants and fixed onto cellotape with the adaxial side stuck down. The epidermal cells and spongy parenchyma cells were subsequently peeled off by using another strip of cellotape, and peels including leaf veins left stuck to the cellotape were used for observation (Desikan et al., 2002). Sections of inflorescence stems and hypocotyls of mature plants were cut by hand using carbon steel razor blades (0.1 mm thickness, FEATHER, Osaka). The GFP-fluorescence in epidermal peels from 1-month-old rosettes sown in pots was observed as described above. For the control, nontransgenic Arabidopsis, Ler, were used. GFP-fluorescence was observed using an epifluorescence microscope (model BX51, Olympus, Tokyo) equipped with a digital camera system (model DP50, Olympus). GFP was excited using a mercury lamp and filter unit (FI/TR, Olympus). Images were captured with the digital camera and photographs were processed for optimal presentation using Photoshop 7.0 (Adobe Systems Japan, Tokyo) and MS Office X (Microsoft Japan, Tokyo) software packages.
Preparation of a Specific Antibody for AAO3 Protein
For AAO3 expression in Pichia pastoris, AAO3 cDNA was cloned into the pPICZA vector (Invitrogen, Carlsbad, CA) to produce a fusion protein with the MYC epitope and polyhistidine tags as described previously (Cheng et al., 2002). Transformation, expression, extraction, and partial purification of recombinant AAO3 protein were carried out as described previously (Koiwai et al., 2000). AAO3 protein was purified in a native form by using the Xpress Protein Purification System (Invitrogen) and subsequently by a DEAE-column chromatography. The purified recombinant AAO3 protein was used for the antibody production. Ten mice were injected with the purified AAO3 protein in order to obtain specific antibodies. The antibody preparation was performed as described previously (Koshiba et al., 1996). The immune sera from the mice were named I1 to I10 and checked for their specificity for AAO3 protein by immunoblotting. For the immunoblotting, recombinant AAO1, AAO2, and AAO3 proteins were expressed using the Pichia expression system and purified as described above.
Immunohistochemical Analysis of AAO3
Detection of AAO3 protein by indirect immunofluorescence staining was performed essentially according to methods described previously (Koshiba, 1993). Arabidopsis rosette leaves and inflorescence stems of WT (Col) and aao3 mutant plants were fixed for 3 h in 10% formalin, 0.1% Nonidet P-40, and 10% dimethylsulfoxide in 20 mm sodium phosphate buffer, pH 7.5, containing 0.15 m NaCl (PBS). The fixed tissues were incubated successively in 10% (w/v) and 20% (w/v) Suc in PBS for 1 h each, and then incubated overnight in PBS supplemented with 30% (w/v) Suc. The samples were then embedded in Tissue-Tek II (Sakura Finetechnical, Tokyo) and frozen at −80°C. Frozen samples were transversely sectioned at a thickness of 16 to 20 μm using a cryostat (Cryocut 1800; Reichert-Jung, Heidelberg, Germany), and sections were air-dried on 3-aminopropyltriethoxysilane (APS)-coated glass slides (Matsunami, Osaka). Samples on slides were treated with cold acetone for 30 min at −20°C and then with 0.3% Triton X-100 for 30 min at 30°C. The pretreated samples were blocked with 5% bovine serum albumin (BSA) in PBS, and then incubated with an anti-AAO3 antibody (mouse antiserum at a 500- to 1,000-fold dilution) for 1 h at 37°C. After rinsing, slides were incubated at room temperature with Bodipy-labeled goat anti-mouse IgG (Molecular Probes, Eugene, OR; 5 μg mL−1 PBS) for 1 h. After washing, the samples were sealed with 50% glycerol-PBS containing 2 mg mL−1 of 1,4-diazabicyclo-[2.2.2] octane. The prepared samples were observed with an epifluorescence microscope as described above.
Immunohistochemical analysis of epidermal tissues was performed by the method of Sang et al. (2001) with some modifications. Epidermal peels were collected from the abaxial side of Arabidopsis rosette leaves immediately following detachment, and fixed as described above. The fixed peels were washed in PBS for 30 min with three changes of solution, then spread onto APS-coated glass slides and air-dried. Peels adhering to slides were treated with cold acetone for 30 min at −20°C, incubated with 0.1% pectolyase (Pectolyase Y-23, Seishin Pharmaceutical, Tokyo) in PBS for 30 min at 30°C, and then treated with 0.3% Triton X-100 in PBS for 1 h at 30°C. Immunostaining of epidermal peels was performed as described above.
Detection of AAO3 mRNA by in Situ Hybridization
Fully expanded leaves were cut and subjected to dehydration treatment for 0 (as a nonstressed control) and 3 h as described above. For the turgid control, leaves were kept on wet filter paper, and the lids of the petri dishes were sealed with plastic film for 3 h. These tissues were fixed in 50 mm sodium phosphate buffer (pH 7.2) containing 4% paraformaldehyde and 0.25% glutaraldehyde. The tissues were dehydrated with a graded series of ethanol solutions. Ethanol was replaced by t-butyl alcohol and then by liquid Paraplast+ (Sigma, St. Louis). Tissues were embedded in Paraplast+ blocks, cut into 8-μm sections, and mounted on microscope slides. The Paraplast+ was removed using xylene. The sections were hydrated with a graded series of ethanol and then treated with proteinase K (5 μg mL−1) at 37°C for 15 min. Tissues were hybridized with digoxigenin-labeled antisense or sense riboprobes prepared from a 500 bp cDNA of AAO3 including the 3′ untranslated region. Signals were imaged using alkaline phosphatase-conjugated anti-digoxigenin antibodies (Roche Diagnostics, Mannheim, Germany).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial purposes.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AB016622.
Acknowledgments
We thank Dr. T. Asami (RIKEN, Saitama, Japan) for kindly providing the abscisic aldehyde, and Dr. Y. Niwa (University of Shizuoka, Japan) for providing the GFP expression vector [pblue-sGFP (S65T) nos SK]. We also thank Drs. T. Kaneta (Ehime University), I. Yazaki (Tokyo Metropolitan University), A. Kadota (Tokyo Metropolitan University), and S. Sawa (University of Tokyo) for their advice regarding the histochemical analyses.
This work was supported in part by the Ministry of Education, Science, Sports and Culture of Japan (Grant-in-Aid for Scientific Research (B) no. 13490024 to T.K.) and by the Japan Society for the Promotion of Science for Young Scientists (Research Fellowship to H.K.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036970.
References
- Akaba S, Seo M, Dohmae N, Takio K, Sekimoto H, Kamiya Y, Furuya N, Komano T, Koshiba T (1999) Production of homo- and hetero-dimeric isozymes from two aldehyde oxidase genes of Arabidopsis thaliana. J Biochem (Tokyo) 126: 395–401 [DOI] [PubMed] [Google Scholar]
- Anderson LWJ (1978) Abscisic acid induces formation of floating leaves in the heterophyllous aquatic angiosperm Potamogeton nodosus. Science 201: 1135–1138 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In-planta agrobacterium-mediated gene-transfer by infiltration of adult Arabidopsis-thaliana plants. C R Acad Sci Ser III Life Sci 316: 1194–1199 [Google Scholar]
- Bittner F, Oreb M, Mendel RR (2001) ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J Biol Chem 276: 40381–40384 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, Koshiba T, Sheen J (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Cornish K, Zeevaart JAD (1986) Abscisic acid accumulation by in situ and isolated guard cells of Pisum sativum L. and Vicia faba L. in relation to water stress. Plant Physiol 81: 1017–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler AJ, Krochko JE (1999) Formation and breakdown of ABA. Trends Plant Sci 4: 472–478 [DOI] [PubMed] [Google Scholar]
- Desikan R, Griffiths R, Hancock J, Neill S (2002) A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 16314–16318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt ND, Sussman MR (1995) Immunocytological localization of an epitope-tagged plasma membrane proton pump (H+-ATPase) in phloem companion cells. Plant Cell 7: 2053–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckham SC, Linforth RST, Taylor IB (1991) Abscisic-acid-deficient mutants at the aba gene locus of Arabidopsis are impaired in the epoxidation of zeaxanthin. Plant Cell Environ 14: 601–606 [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodriguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MJ, Outlaw WH Jr, Mertens R, Weiler EW (1988) Water-stress-induced changes in the abscisic acid content of guard cells and other cells of Vicia faba L. leaves as determined by enzyme-amplified immunoassay. Proc Natl Acad Sci USA 85: 2584–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung W, Wilkinson S, Davies WJ (1998) Factors that regulate abscisic acid concentrations at the primary site of action at the guard cell. J Exp Bot 49: 361–367 [Google Scholar]
- Hobo T, Kobayashi M, Shinozaki K, Shinozaki K (2002) Characterization of the AtNCED2 gene, encoding 9-cis-epoxycarotenoid dioxygenase, a key enzyme in ABA biosynthesis in Arabidopsis seed maturation. Plant Cell Physiol 43: s93 [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Koiwai H, Akaba S, Seo M, Komano T, Koshiba T (2000) Functional expression of two Arabidopsis aldehyde oxidases in the yeast Pichia pastoris. J Biochem (Tokyo) 127: 659–664 [DOI] [PubMed] [Google Scholar]
- Koshiba T (1993) Cytosolic ascorbate peroxidase in seedlings and leaves of maize (Zea mays). Plant Cell Physiol 34: 713–721 [Google Scholar]
- Koshiba T, Saito E, Ono N, Yamamoto N, Satô M (1996) Purification and properties of flavin- and molybdenum-containing aldehyde oxidase from coleoptiles of maize. Plant Physiol 110: 781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara A, Ikegami K, Koshiba T, Nagata T (2003) Effects of ethylene and abscisic acid upon heterophylly in Ludwigia arcuata (Onagraceae). Planta 217: 880–887 [DOI] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J 15: 2331–2342 [PMC free article] [PubMed] [Google Scholar]
- Murashige TR, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Ono A, Izawa T, Chua NH, Shimamoto K (1996) The rab16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiol 112: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka KJ, Turgeon R (1999) Sieve elements and companion cells-traffic control centers of the phloem. Plant Cell 11: 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RS, Houlston CE, Atherton KM, Rixon JE, Harrison P, Hughes MA, Alison Dunn M (1998) Localization of expression of three cold-induced genes, blt101, blt4.9, and blt14, in different tissues of the crown and developing leaves of cold-acclimated cultivated barley. Plant Physiol 117: 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Zeevaart JAD (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA 96: 15354–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke K, Firn RD, Pierce M (1975) Stomatal closure in response to xanthoxin and abscisic acid. Planta 125: 149–160 [DOI] [PubMed] [Google Scholar]
- Rock CD, Zeevaart JAD (1991) The aba mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc Natl Acad Sci USA 88: 7496–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Kinoshita N, Ishiyama K, Hata S, Kyozuka J, Hayakawa T, Nakamura T, Shimamoto K, Yamaya T, Izui K (2001) A Ca2+-dependent protein kinase that endows rice plants with cold- and salt-stress tolerance functions in vascular bundles. Plant Cell Physiol 42: 1228–1233 [DOI] [PubMed] [Google Scholar]
- Sang Y, Zheng S, Li W, Huang B, Wang X (2001) Regulation of plant water loss by manipulating the expression of phospholipase Dα. Plant J 28: 135–144 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JAD (2003) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol 131: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JAD, McCarty DR (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Sekimoto H, Seo M, Kawakami N, Komano T, Desloire S, Liotenberg S, Marion-Poll A, Caboche M, Kamiya Y, Koshiba T (1998) Molecular cloning and characterization of aldehyde oxidases in Arabidopsis thaliana. Plant Cell Physiol 39: 433–442 [DOI] [PubMed] [Google Scholar]
- Seo M, Koiwai H, Akaba S, Komano T, Oritani T, Kamiya Y, Koshiba T (2000. a) Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J 23: 481–488 [DOI] [PubMed] [Google Scholar]
- Seo M, Koshiba T (2002) Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJM, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JAD, Koornneef M, Kamiya Y, Koshiba T (2000. b) The Arabidopsis aldehyde oxidase 3 (AA03) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA 97: 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp RE, LeNoble ME (2002) ABA, ethylene and the control of shoot and root growth under water stress. J Exp Bot 53: 33–37 [PubMed] [Google Scholar]
- Sharp RE, LeNoble ME, Else MA, Thorne ET, Gherardi F (2000) Endogenous ABA maintains shoot growth in tomato independently of effects on plant water balance: evidence for an interaction with ethylene. J Exp Bot 51: 1575–1584 [DOI] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JAD, McCarty DR (1997) Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA 94: 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel AJE (2003) Transport phloem: low profile, high impact. Plant Physiol 131: 1509–1510 [PMC free article] [PubMed] [Google Scholar]
- Weiler EW, Schnabl H, Hornberg C (1982) Stress-related levels of abscisic acid in guard cell protoplasts of Vicia faba L. Planta 154: 24–28 [DOI] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK (2001) The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK (2003) Regulation of abscisic acid biosynthesis. Plant Physiol 133: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol 39: 439–473 [Google Scholar]