Abstract
A novel tetra-peptide insertion was identified in Gag-p6 ALIX-binding region which is appears in protease inhibitor (PI) failure Indian HIV-1C sequences (Odds Ratio 17.1, p<0.001) but naturally present in half of untreated Ethiopian sequences. The insertion will probably restore the ALIX mediated virus release pathway, which is lacking in HIV-1C. The clinical importance of such insertion need to be evaluated in HIV-1C dominating regions were PI-drugs are being scaled up as second line treatment options.
Keywords: Gag-p6, ALIX, HIV-1 subtype C, Protease Inhibitor failure
Subtype specific differences has been observed in Gag-p6 the motifs PTAPP and LYPxnLxxL. Among the subtypes, HIV-1 subtype C (HIV-1C) has a higher frequency of duplications in the PTAPP-motif after ART-failure [1, 2]. Also, a natural deletion of L483Y484 residues has been observed in the LYPxnLxxL motif in >95% of the sequences which abrogates the ALIX-mediated particle release in absence of PTAP/TSG101 pathway[3]. Considerable evidence also suggests that ART-induced changes in the Gag-p6 region may modulate the therapy response and the viral fitness [4]. Study have shown that selective drug pressure leads to accumulations of substitutions and insertions in Gag-p6, at sites distal from the mutations that render the virus highly resistant to PIs [5]. One of the key examples is PTAPP-duplication in TSG101binding motif in the Gag-p6 which affect the virological response to PI-drugs like amprenavir [6].
In this study we investigated the consequences of the sequence variations of Gag-p6, using HIV-1C sequences of clinical isolates from India (HIV-1CIN; n=158), Ethiopia (HIV-1CET; n=73) and Germany (HIV-1CDE; n=125) and its potential clinical impact. The patients are from several clinical cohorts from India [7, 8], Germany [9, 10] and Ethiopia [11]. Among the patients 61% (215/356) were therapy-naïve. Data were pooled with Gag-p6 sequences (n=8589) of major non-C subtypes and recombinants from the Los Alamos HIV Database.
Gag-p6, protease and partial reverse transcriptase were amplified and sequenced from the plasma viral RNA as described previously [12-15]. Primary and acquired drug resistance mutations (DRMs) were evaluated using World Health Organisation mutations list 2009 [16] and International AIDS Society list 2013[17] respectively. HIV-1 subtyping was performed as described recently using three automated bioinformatics tools [18]. Multiple-template homology models of the p6-ALIX complex were built in MODELLER 9v12 [19] using crystal structure of human ALIX/AIP1 in complex with a peptide fragment of the SIVmac239 and HIV-1 Gag-p6 proteins (PDB codes 2XS1 and 2R02) [20, 21]. The models were analysed for accuracy using the DOPE statistical potential score [22]. The efficacy of the binding was analysed by computing the electrostatics using the Adapted Poisson-Boltzmann Solver software [23] and visualized using Chimera[24]. Statistical analysis was carried out using SPSSv22.0 (IBM Corp, US). The study was approved by respective ethical review committee in India, Germany, Sweden, and Ethiopia. Written informed consent was obtained from the participants.
Cohort characteristics were presented in supplementary digital content (SDC) 1. The multiple sequence alignments of the Gag-p6 of the representative strains were shown in the SDC 2. Duplications of three to thirteen amino acids in the TSG101 binding site were observed more frequently in the HIV-1CDE (29%) and HIV-1CET (25%) sequences than in the HIV-1CIN (12%) sequences from therapy-naïve individuals (SDC 2 and Figure 1A). When therapy-naïve and therapy-failure patients were compared, the duplications occurred more frequently in the therapy-failure Indians (12% vs 25%; p<0.05) but not in the German cohort (29% vs 33%; p=0.69) (Figure 1A). Interestingly, there are subtype specific differences in the accumulation of the PTAPP-duplication after therapy-failure. The duplication occurred in greater frequency in HIV-1C (54%) compared to HIV-1B (9.3%) and HIV-1F1 (17.6%)[2]. Previous controversial findings were shown that duplications of PTAPP were associated with ART in one group of populations but not in others [2, 6, 25-28]. In our study we observed intra-HIV-1C specific preferential duplication in therapy-failure patients compared to therapy-naïve individuals.
Figure 1.
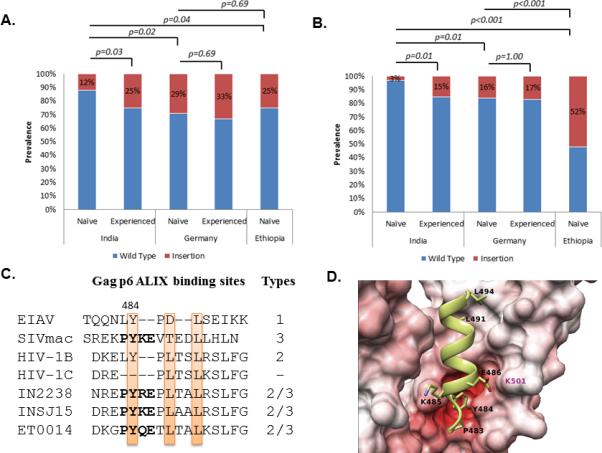
Prevalence of (A) duplications in the TSG101 binding PTAPP motif and (B) insertions in the ALIX-binding LYPxnLxxL motif. Statistically significant differences are marked. (C) Different types of ALIX-binding sites in lentiviral Gag-p6 region as describe by Zhai et al 2011 [20]. Representative clinical isolates from the cohort are presented which had PYxE insertion. All the strains had the key residues conserved (highlighted). The 3D-molecular models of the Gag-p6-ALIX complex (D) The ALIX is shown in surface representation, interacting with the Gag-p6 protein PYKE shown in ribbon and stick (side-chain only) representation. The residues that belong to the ALIX binding motif and the insertions are all labelled in black. The residues in ALIX that is crucial to mediate the interaction with Gag-p6 are labelled in magenta. The surface of the ALIX is coloured according to the electrostatic potential with blue indicating positively charged regions and red indicating negatively charged regions. The intensity of the colour reflects charge intensity. The models also establish that the Y484 residue in the insertions play a role similar to the Tyr residue in the wild type HIV-1B/SIV Gag-p6 protein by forming a specific hydrogen bond with the ALIX. Similar results were observed with PYRE and PYQE insertions (data not shown).
A novel tetra-peptide insertion [PYxE; where x represents either arginine (R), lysine (K) or glutamine (Q)] was observed in the C-terminal position of the Gag-p6 in the defective HIV-1C ALIX-binding domain. This PYxE insertion was observed in 52% of the untreated Ethiopian sequences, but significantly less often in the untreated German sequences (16%; p<0.001), and even more seldom in the untreated Indian sequences (3%; p<0.001) (Figure 1B). When analysing sequences of therapy-naïve individuals obtained from the Los Alamos HIV Database, the frequency of the PYxE insertion was much less common in therapy-naïve patients infected with non-C subtypes (0.1%; n=4263), but also less common in HIV-1C sequences from southern Africa (1%, n=2295) and eastern African (3%, n=61).
The PYxE insertion restores the key Y484 residue (SDC 2 and Figure 1C). The 3D-molecular models of the Gag-p6-ALIX complex showed a specific interaction involving the inserted Y484 residue of Gag-p6 and the ALIX (Figure 1D). Thus the insertion variant would restore the binding of Gag-p6 to ALIX that was lost due to the L483Y484 deletion in HIV-1C and probably restores the ALIX mediated virus release pathway.
Among the HIV-1CIN sequences from therapy failed patients, the PYxE insertion was found significantly more often in PI-failure patients (6/10) as compared to those failing a non-PI containing regimen (5/62) (Odds ratio; 95% CI: 17.1; 3.6 – 81.4; p<0.001). Among the other clinical and demographic parameters, the median CD4+ T-cell count was significantly lower among the individuals with the PYxE insertion in the ALIX-motif compared to those without the insertion (73 vs 160 cells/mm3; p<0.001). That Indian patients with the PYxE insertion had significantly lower CD4+ T-cell counts than those without, could possibly indicate that the virus with the insertion is more pathogenic. This suggestion was further supported by the very low CD4+ T-cell counts in the three therapy-naïve Indian individuals with the insertion before initiating ART (22, 90 and 56 cells/mm3). We further followed those three patients for two years after initiation of therapy. None of the patients gain optimal CD4+ T-cell count at two years (<350 cells/mm3) (data not shown).
In conclusion, for the first time we have identified a PYxE tetra-peptide insertion in the ALIX-binding motif, which appeared in PI-therapy-failure cases in HIV-1CIN sequences, but occurred naturally in more than half of the therapy-naïve HIV-1CET sequences examined. We therefore hypothesised that the genetic background might have influenced the preferential selection of these insertions. The insertion probably restores the ALIX-mediated virus release pathway, which is lacking in HIV-1C and the virus with this insertion might be more pathogenic. To better elucidate the clinical importance of such insertion, in countries with a high prevalence of HIV-1C, further investigations are needed when the PI-drugs are used in the ART regimen.
Supplementary Material
Acknowledgements
The study was partially funded by European Union FP7, Swedish International Developing Agency, CHAIN FP7 EU, the Swedish Civil Contingencies Agency [SWE-2009-151], and the Swedish Research Council [521-2012-3476 and 2007-7092]. MLE acknowledge the funds received from R01MH067513 from the National Institute of Mental Health (NIMH) (Bethesda, MD, USA) for the study Examining ART Adherence Issues in Bangalore India. V.R.P. and V.R.R. wish to acknowledge support from the National Institute of Health, USA (NIH) grant 1R01MH083579 (to V.R.P.).
Footnotes
Declaration of interests
The author(s) declare that they have no competing interests
References
- 1.Marlowe N, Flys T, Hackett J, Jr., Schumaker M, Jackson JB, Eshleman SH. Analysis of insertions and deletions in the gag p6 region of diverse HIV type 1 strains. AIDS Res Hum Retroviruses. 2004;20:1119–1125. doi: 10.1089/aid.2004.20.1119. [DOI] [PubMed] [Google Scholar]
- 2.Martins AN, Arruda MB, Pires AF, Tanuri A, Brindeiro RM. Accumulation of P(T/S)AP late domain duplications in HIV type 1 subtypes B, C, and F derived from individuals failing ARV therapy and ARV drug-naive patients. AIDS Res Hum Retroviruses. 2011;27:687–692. doi: 10.1089/aid.2010.0282. [DOI] [PubMed] [Google Scholar]
- 3.Patil A, Bhattacharya J. Natural deletion of L35Y36 in p6 gag eliminate LYPXnL/ALIX auxiliary virus release pathway in HIV-1 subtype C. Virus Res. 2012;170:154–158. doi: 10.1016/j.virusres.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 4.Dam E, Quercia R, Glass B, Descamps D, Launay O, Duval X, et al. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 2009;5:e1000345. doi: 10.1371/journal.ppat.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fun A, Wensing AM, Verheyen J, Nijhuis M. Human Immunodeficiency Virus Gag and protease: partners in resistance. Retrovirology. 2012;9:63. doi: 10.1186/1742-4690-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lastere S, Dalban C, Collin G, Descamps D, Girard PM, Clavel F, et al. Impact of insertions in the HIV-1 p6 PTAPP region on the virological response to amprenavir. Antivir Ther. 2004;9:221–227. [PubMed] [Google Scholar]
- 7.Shet A, Antony J, Arumugam K, Kumar Dodderi S, Rodrigues R, DeCosta A. Influence of adverse drug reactions on treatment success: prospective cohort analysis of HIV-infected individuals initiating first-line antiretroviral therapy in India. PLoS One. 2014;9:e91028. doi: 10.1371/journal.pone.0091028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekstrand ML, Chandy S, Heylen E, Steward W, Singh G. Developing useful highly active antiretroviral therapy adherence measures for India: the Prerana study. J Acquir Immune Defic Syndr. 2010;53:415–416. doi: 10.1097/QAI.0b013e3181ba3e4e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balduin M, Oette M, Daumer MP, Hoffmann D, Pfister HJ, Kaiser R. Prevalence of minor variants of HIV strains at reverse transcriptase position 103 in therapy-naive patients and their impact on the virological failure. J Clin Virol. 2009;45:34–38. doi: 10.1016/j.jcv.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Oette M, Reuter S, Kaiser R, Lengauer T, Fatkenheuer G, Knechten H, et al. Epidemiology of transmitted drug resistance in chronically HIV-infected patients in Germany: the RESINA study 2001-2009. Intervirology. 2012;55:154–159. doi: 10.1159/000332015. [DOI] [PubMed] [Google Scholar]
- 11.Abdurahman S, Barqasho B, Nowak P, Cuong do D, Amogne W, Larsson M, et al. Pattern of microbial translocation in patients living with HIV-1 from Vietnam, Ethiopia and Sweden. J Int AIDS Soc. 2014;17:18841. doi: 10.7448/IAS.17.1.18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bontell I, Cuong do D, Agneskog E, Diwan V, Larsson M, Sonnerborg A. Transmitted drug resistance and phylogenetic analysis of HIV CRF01_AE in Northern Vietnam. Infect Genet Evol. 2012;12:448–452. doi: 10.1016/j.meegid.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 13.Neogi U, Prarthana BS, Gupta S, D'Souza G, De Costa A, Kuttiatt VS, et al. Naturally occurring polymorphisms and primary drug resistance profile among antiretroviral-naive individuals in Bangalore, India. AIDS Res Hum Retroviruses. 2010;26:1097–1101. doi: 10.1089/aid.2010.0092. [DOI] [PubMed] [Google Scholar]
- 14.Neogi U, Sahoo PN, Kumar R, De Costa A, Shet A. Characterization of HIV type 1 subtype C protease gene: selection of L63P mutation in protease inhibitor-naive Indian patients. AIDS Res Hum Retroviruses. 2011;27:1249–1253. doi: 10.1089/AID.2011.0078. [DOI] [PubMed] [Google Scholar]
- 15.Verheyen J, Litau E, Sing T, Daumer M, Balduin M, Oette M, et al. Compensatory mutations at the HIV cleavage sites p7/p1 and p1/p6-gag in therapy-naive and therapy-experienced patients. Antivir Ther. 2006;11:879–887. [PubMed] [Google Scholar]
- 16.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 2009. 4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer RW, et al. Update of the drug resistance mutations in HIV-1: March 2013. Top Antivir Med 2013. 21:6–14. [PMC free article] [PubMed] [Google Scholar]
- 18.Neogi U, Haggblom A, Santacatterina M, Bratt G, Gisslen M, Albert J, et al. Temporal Trends in the Swedish HIV-1 Epidemic: Increase in Non-B Subtypes and Recombinant Forms over Three Decades. PLoS One. 2014;9:e99390. doi: 10.1371/journal.pone.0099390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 20.Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J Virol. 2011;85:632–637. doi: 10.1128/JVI.01683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 22.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 25.Brumme ZL, Chan KJ, Dong WW, Wynhoven B, Mo T, Hogg RS, et al. Prevalence and clinical implications of insertions in the HIV-1 p6Gag N-terminal region in drug-naive individuals initiating antiretroviral therapy. Antivir Ther. 2003;8:91–96. [PubMed] [Google Scholar]
- 26.Gallego O, de Mendoza C, Corral A, Soriano V. Changes in the human immunodeficiency virus p7-p1-p6 gag gene in drug-naive and pretreated patients. J Clin Microbiol. 2003;41:1245–1247. doi: 10.1128/JCM.41.3.1245-1247.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters S, Munoz M, Yerly S, Sanchez-Merino V, Lopez-Galindez C, Perrin L, et al. Resistance to nucleoside analog reverse transcriptase inhibitors mediated by human immunodeficiency virus type 1 p6 protein. J Virol. 2001;75:9644–9653. doi: 10.1128/JVI.75.20.9644-9653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibe S, Shibata N, Utsumi M, Kaneda T. Selection of human immunodeficiency virus type 1 variants with an insertion mutation in the p6(gag) and p6(pol) genes under highly active antiretroviral therapy. Microbiol Immunol. 2003;47:71–79. doi: 10.1111/j.1348-0421.2003.tb02788.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


