Abstract
Human sialic acid biology is unusual and thought to be unique among mammals. Humans lack a functional CMAH protein and cannot synthesize the sugar Neu5Gc, an innate mammalian signal of self. Losing this sugar changed how humans interact with some of our deadliest pathogens: malaria, influenza and streptococcus among others. We show that the New World monkeys, comprising a third of all primate species, have human-like sialic acid biology. They have lost Neu5Gc because of an independent CMAH inactivation ∼30 million years ago (compared to ∼3 mya in hominids). This parallel loss of Neu5Gc opens sialic acid biology to comparative phylogenetic analysis, and reveals an unexpected conservation priority. New World Monkeys risk infection by human pathogens that can recognize cells in the absence of Neu5Gc. This striking molecular convergence provides a mechanism that could explain the longstanding observation that New World monkeys are susceptible to some human diseases that cannot be transmitted to other primates.
Keywords: convergent evolution, self recognition, innate immunity, glycobiology, sialic acids, platyrrhines
Sugar molecules called glycans cover the outer surface of all cells, and their composition and chemistry influence interactions between molecules, cells, and individuals. Glycans have a tremendous diversity of natural variation (1). Their structural properties allow many different forms and modifications, and their regulation can be fine-tuned spatially and temporally. Glycans are often variable within and between species suggesting that they can evolve rapidly, perhaps in response to evolutionary conflict (2, 3). Some of the first natural genetic polymorphisms discovered are now known to be glycan variants. Blood groups are a classic example (4). The ABO blood group results from antigenicity against different glycosylation variants, and may be maintained by frequency dependent selection since pathogens target these molecules during infection (5). Natural glycan variants can thus have important effects on fitness, and glycan biology is a ubiquitous but understudied aspect of phenotypic evolution.
The first uniquely human biochemical difference discovered is also a glycan. Humans lack the sialic acid Neu5Gc. Sialylated human N-glycans, O-glycans, and gangliosides, instead terminate with the precursor form: Neu5Ac (6). The glycans of chimpanzees, gorillas, and other mammals contain both Neu5Gc and Neu5Ac, millions of each molecule on every cell. Sialic acids contribute to self-recognition by the innate immune system, and binding by Siglec proteins modulates the intensity of the innate immune response. Pathogens exploit these signals by mimicking or co-opting sialic acids to enter cells or suppress the immune response (7). Some human pathogens have evolved receptors that recognize Neu5Ac specifically, and some diseases shared by other primates cannot infect humans and vice versa. Human sialic acid biology has changed extensively to compensate for the absence of Neu5Gc (8). Sialic acid binding proteins like Siglecs often evolve by positive selection and many have uniquely human mutations. A range of human immune phenotypes – including inflammation, cancer, and even reproduction – differ from our closest relatives because of this change in cell surface chemistry (9-11).
The absence of endogenous Neu5Gc in human tissues is caused by inactivation of Cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMAH), the enzyme that adds a hydroxyl group to CMP-Neu5Ac forming CMP-Neu5Gc. The ability to synthesize Neu5Gc was eliminated in hominids ∼3 mya when an Alu element deleted exon 6 of the CMAH gene (12). Polymorphism data suggests that the CMAH(−) allele rapidly rose to high frequency in the ancestral hominid population. Selection apparently favored this loss of function allele despite its many pleiotropic effects on phenotype (10). We might therefore expect to find other organisms where CMAH function has been lost, and indeed birds and some monotremes lack Neu5Gc (13). But to date all mammals tested can synthesize Neu5Gc.
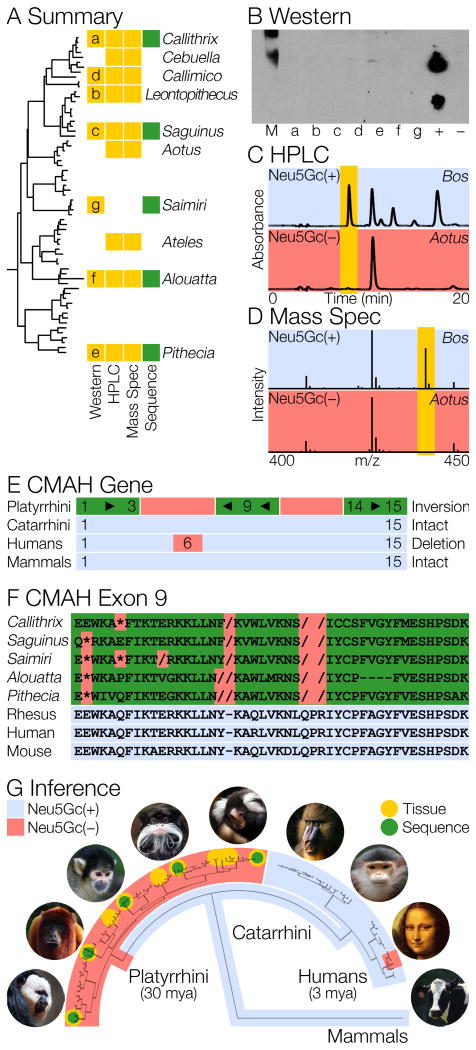
New World monkeys, the platyrrhines, are a radiation of 135 extant species which descended from a common ancestor ∼30 mya in South and Central America and have evolved an amazing diversity of forms, colors and behaviors (14-16). New World monkeys have become important disease models, in part because they can be infected by some human pathogens. For example, the malaria parasite Plasmodium falciparum binds specifically to Neu5Ac, cannot bind Neu5Gc (17, 18), and is capable of infecting humans and New World Monkeys, but does not infect other primates (19, 20). Using this as a clue, we examined the sialic acid composition of New World Monkey tissues by anti-Neu5Gc western blot, HPLC, and mass spectrometry to show that, like humans, New World monkeys do not make endogenous Neu5Gc (Figure 1). Genomic information from the marmoset (Callithrix jacchus) and squirrel monkey (Saimiri boliviensis) and direct sequence of CMAH exon 9 from several other New World monkey species reveals an inversion of exons 4 to 13 and an inactivated CMAH gene. This independent loss occurred at the base of the platyrrhine radiation: ∼30 million years ago, compared to ∼3 mya in ancestral hominids.
Fig. 1. Neu5Gc loss by CMAH inactivation in New World monkeys.
(A) Species assayed for the absence of Neu5Gc in tissues (yellow), or with CMAH sequence (green). All five platyrrhine families are represented.
(B) New World monkey sugars do not bind antiNeu5Gc antibody (a-f). M is a size standard, Neu5Gc(+) is chimpanzee milk, Neu5Gc(−) is human milk.
(C and D) HPLC and mass spectrometry do not detect Neu5Gc in New World monkey tissues. Representative results from cow and owl monkey are shown.
(E) CMAH was inactivated by an inversion that eliminated exons 4 to 13. Exon 9 is on the strand opposite the remaining exons.
(F) Frame shifts and stop codons (red) disrupt CMAH exon 9.
(G) New World monkeys lost Neu5Gc ∼30 million years ago by an independent inactivation of CMAH, the same gene that caused humans to lose Neu5Gc ∼3 mya.
The absence of Neu5Gc from New World monkeys expands their usefulness as disease models, and also raises awareness of a new conservation risk. Their sialic acid composition makes New World monkeys susceptible to some human pathogens that interact with sialic acids. This independent loss of CMAH and Neu5Gc in hominids and platyrrhines is also an interesting case of parallel evolution at the interface of pathogen interaction and self-recognition by the innate immune system. Interactions with pathogens have driven the rapid evolution of Siglecs in the human lineage (8). Siglec evolution and it's influence on self recognition can now be subject to comparative analysis, to study the range of responses that follow Neu5Gc loss and the repeatability of evolution following the parallel loss of a self-signal. Because New World monkeys have had 10 times as long to compensate, their sialic acid biology may suggest new treatments for human diseases that result from our unusual sialic acid composition.
The absence of Neu5Gc can no longer be considered a unique feature of human glycans. New World monkeys also lack this key signal of self. This parallel evolutionary change will broaden our understanding of the many roles of glycans in health and disease, allow new tests of selection and constraint in primate evolution, and refine our ability to protect the fantastic diversity of New World monkeys.
Materials and Methods
Neu5Gc Detection
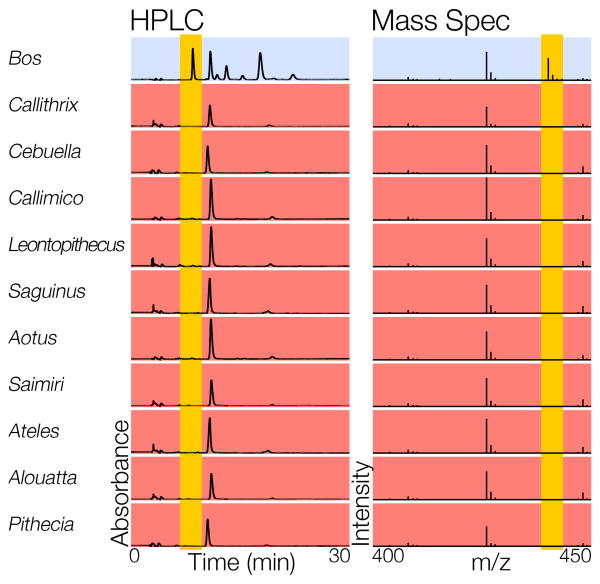
New World monkey tissues were obtained from John Kaas (Vanderbilt University), The San Diego Zoological Society, and Alison Muotri (UC San Diego). We analyzed the Sia content of New World monkey tissues by anti-Neu5Gc western blot (Figure 1B), high-pressure liquid chromatography (HPLC), and mass spectrometry (Figures 1C and 1D respectively, and Figure 2). Neu5Gc could not be detected in New World monkey tissues by any of these three methods.
Fig. 2.
Neu5Gc is absent from New World monkey tissues. HPLC and Mass Spectrometry analysis of New World monkey tissue samples. Neu5Gc(+) positive control (blue) is from cow (Bos). HPLC. Absorbance range: 0 to 1×105 units on all plots. All New World monkey samples (red) are missing the peak that indicates the presence of Neu5Gc (elution time: ∼8.5 mins). Mass Spectrometry. Intensity range: 0 to 8×106 on all plots. All New World monkey samples are missing fragments associated with Neu5Gc (m/z ratio: ∼440).
Anti-Neu5Gc antibody Western blots
We used 250 ug of spleen or liver tissue which are typically Neu5Gc rich in mammals. Samples were placed in 1 mL lysis buffer (PBS with 0.2% Triton-X100) and homogenized on a Polytron (three 20 second bursts). Tissue lysates were ultracentrifuged (100 000 g, 1 hour). Protein concentrations in the resulting supernatant were determined by BCA (Pierce). Samples were run on a 10% denaturing SDS-Page gel (10 ug of protein extract per well). Consistent loading across samples was verified by Ponceau staining. Separated proteins were transferred to a nitrocellulose membrane and blocked overnight in TBS-T. Western blots were incubated with affinity purified anti Neu5Gc chicken IgY (20°C, 2 hours) (21). Membranes were washed in TBS-T (5 washes, 5 minutes each), and bound with HRP conjugated secondary goat anti-chicken antibody (20°C, 2 hours). We ran two controls on binding: unspecific chicken IgY, and secondary antibody only, and two controls on sialic acid composition: Neu5Gc(+) chimpanzee milk, and Neu5Gc(−) human milk.
HPLC
Total Sia extracts were obtained by 2M acetic acid hydrolysis. Released Sias were filtered through microcon 10 spin columns (Millipore) by centrifugation (13,000 g, 20 minutes) and derivatized in 1,2-diamino-4,5-methylenedoxybenzene (DMB) reagent for 2.5 hours at 50°C in the dark. Derivatized sias were separated by HPLC using a C18 reverse-phase column (Varian) and eluted (0.9 mL/minute, 50 minutes) in an isocratic solvent (85% water, 7% methanol, 8% acetonitrile)(22). Sia standards were isolated from bovine submaxillary mucin using the same protocol.
Mass spectrometry
DMB derivatives of sialic acids were run on a Finnigan MAT HPLC with online mass spectrometry on an LCQ-Mass Spectrometer System. The HPLC eluent from above was simultaneously monitored by absorbance at 373nm and by mass spectrometry (capillary temperature 210°C, capillary voltage 31 V, lens offset voltage 0 V). Mass spectra were acquired by scanning from 150-2000 m/z in the positive ion mode. MS/MS was acquired by selecting the parent mass and using a 20% normalized collision energy. Data analysis was performed with the manufacturer's Xcalibur data analysis program.
Genomic Information
Draft genome assemblies of marmoset (Callithrix jacchus: WUGSC 3.2, Mar 2009) and squirrel monkey (Saimiri boliviensis: saiBol1, Oct 2011) show that the CMAH gene has been disrupted in both species. Local synteny around CMAH is conserved despite a large inversion on the chromosome: CMAH is flanked by CANT1 and FAM65b as it is in other mammals, though there are other inversions along the chromosome. Chain-Net alignments accessed with the UCSC genome browser identify CMAH exons 1-3, 9, 14, and 15 in marmoset and squirrel monkey. Several of the recognizable exons are truncated or contain premature stop codons. CMAH exons 4 to 8 and 10 to 13 are not identifiable in either species. CMAH exon 9 is recognizable, but exists on the strand opposite the other CMAH exons. Thus, the center of the CMAH gene appears to have undergone an inversion. The cause of this event is not clear, but the inverted region is roughly 20 thousand base pairs longer than the corresponding region in catarrhines, and contains stretches of unrecognizable DNA in addition to regions with similarity to CMAH introns. BLAT searches do not find other regions with similarity to CMAH in either New World monkey genome.
PCR and Sequencing
We isolated DNA (Qiagen: DNeasy Blood and Tissue Kit), from various tissues of species representing 4 of the 5 platyrrhine families (all but Aotidae, which is a sub-clade of the Cebidae). PCR primers that bracket exon 9 were designed from Multiz alignments of several primate species (howlers: F 5′ ttcatgctctctgttcttcacc 3′, R 5′ tgcaacattttcctagcaacc 3′; other platyrrhines: F 5′ tttccttctcatgtcacattgc 3′, R 5′ tggaactttgctctatttctgc 3′). Diploid PCR products were amplified using standard reagents (Invitrogen: Taq, buffer and dNTPs) and Sanger sequenced using standard protocols (Applied Biosystems: Big Dye v3.1). Exon sequences are shown in Fig 1, and deposited in Genbank (accession numbers: X to X). Sequences of CMAH exon 9 contain stop codons and other disruptions in every platyrrhine species sequenced.
Acknowledgments
We thank Jim Mullikin for checking the disruption of CMAH in the marmoset genome prior to its final assembly, and members of the UCSD Gagneux and Varki labs for stimulating discussions. This work was supported by the G. Harold and Leila Y. Mathers Charitable Foundation, Grant CSD081 (to P.G.) and by National Institutes of Health Grant R01 1065732 (to P.G.). S.D. was supported by a National Institutes of Health Grant R01 GM32373 (to Ajit Varki).
References
- 1.Varki AP, et al. Evolution of Glycan Diversity. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]
- 2.Gagneux P, Varki AP. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9:747–755. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- 3.Springer S, Gagneux P. Glycan Evolution in Response to Collaboration, Conflict, and Constraint. J Biol Chem. 2013 doi: 10.1074/jbc.R112.424523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haldane JBS. The Blood-Group Frequencies of European Peoples and Racial Origins. Human Biology. 1940;12:457–480. [PubMed] [Google Scholar]
- 5.Ségurel L, et al. The ABO blood group is a trans-species polymorphism in primates. Proc Natl Acad Sci USA. 2012;109:18493–18498. doi: 10.1073/pnas.1210603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou HH, et al. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci USA. 1998;95:11751–11756. doi: 10.1073/pnas.95.20.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varki AP. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology. 2011;21:1121–1124. doi: 10.1093/glycob/cwr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varki AP. Colloquium Paper: Uniquely human evolution of sialic acid genetics and biology. Proc Natl Acad Sci USA. 2010;107:8939–8946. doi: 10.1073/pnas.0914634107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearce OMT, et al. Inverse hormesis of cancer growth mediated by narrow ranges of tumor-directed antibodies. Proceedings of the National Academy of Sciences. 2014;111:5998–6003. doi: 10.1073/pnas.1209067111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghaderi D, et al. Sexual selection by female immunity against paternal antigens can fix loss of function alleles. Proc Natl Acad Sci USA. 2011;108:17743–17748. doi: 10.1073/pnas.1102302108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pham T, et al. Evidence for a novel human-specific xeno-auto-antibody response against vascular endothelium. Blood. 2009;114:5225–5235. doi: 10.1182/blood-2009-05-220400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayakawa T, Aki I, Varki AP, Satta Y, Takahata N. Fixation of the human-specific CMP-N-acetylneuraminic acid hydroxylase pseudogene and implications of haplotype diversity for human evolution. Genetics. 2006;172:1139–1146. doi: 10.1534/genetics.105.046995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schauer R, Srinivasan GV, Coddeville B, Zanetta JP, Guérardel Y. Low incidence of N-glycolylneuraminic acid in birds and reptiles and its absence in the platypus. Carbohydr Res. 2009;344:1494–1500. doi: 10.1016/j.carres.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Perez SI, Tejedor MF, Novo NM, Aristide L. Divergence Times and the Evolutionary Radiation of New World Monkeys (Platyrrhini, Primates): An Analysis of Fossil and Molecular Data. PLoS ONE. 2013 doi: 10.1371/journal.pone.0068029.s009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santana SE, Alfaro JL, Alfaro ME. Adaptive evolution of facial colour patterns in Neotropical primates. Proc Biol Sci. 2012;279:2204–2211. doi: 10.1098/rspb.2011.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perelman P, et al. A Molecular Phylogeny of Living Primates. In: Brosius J, editor. PLoS Genet. Vol. 7. 2011. p. e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin MJ, Rayner JC, Gagneux P, Barnwell JW, Varki AP. Evolution of human-chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. Proc Natl Acad Sci USA. 2005;102:12819–12824. doi: 10.1073/pnas.0503819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varki AP, Gagneux P. Human-specific evolution of sialic acid targets: explaining the malignant malaria mystery. Proc Natl Acad Sci USA. 2009;106:14739–14740. doi: 10.1073/pnas.0908196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward JM, Vallender EJ. The resurgence and genetic implications of New World primates in biomedical research. Trends Genet. 2012;28:586–591. doi: 10.1016/j.tig.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galland GG. Role of the squirrel monkey in parasitic disease research. ILAR J. 2000;41:37–43. doi: 10.1093/ilar.41.1.37. [DOI] [PubMed] [Google Scholar]
- 21.Diaz SL. Sensitive and Specific Detection of the Non-Human Sialic Acid N-Glycolylneuraminic Acid In Human Tissues and Biotherapeutic Products. In: Batzer MA, editor. PLoS ONE. Vol. 4. 2009. p. e4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manzi AE, Diaz S, Varki AP. High-pressure liquid chromatography of sialic acids on a pellicular resin anion-exchange column with pulsed amperometric detection: a comparison with six other systems. Anal Biochem. 1990;188:20–32. doi: 10.1016/0003-2697(90)90523-c. [DOI] [PubMed] [Google Scholar]